The myelodysplastic syndromes (MDS) comprise a heterogeneous group of clonal haematopoietic neoplasms with a variable propensity to progress to acute myeloid leukaemia (AML). This emphasises the need to differentiate MDS patients as low or high risk for progression, so that therapy is tailored according to the risk group. Several prognostic models combining traditional morphological and clinical criteria have been developed as default staging systems to risk-stratify patients with MDS, including the International Prognostic Scoring System (IPSS) (Greenberg et al, 1997), the World Health Organization (WHO) classification-based Prognostic Scoring System (WPSS) (Malcovati et al, 2007), the MD Anderson Prognostic Scoring System (Kantarjian et al, 2008) and the revised IPSS (IPSS-R) (Greenberg et al, 2012). The IPSS-R, based on large international databases of more than 7000 patients and integrated detailed disease-related and patient-related factors, has been proved as an excellent predictor of MDS prognosis and is most commonly used.
Over the past decade, a MDS-associated gene mutation profile has been confirmed by several large studies (Papaemmanuil et al, 2013; Haferlach et al, 2014; Makishima et al, 2017), ascertaining that some somatic mutations in certain genes could predict patient outcomes (Papaemmanuil et al, 2013; Haferlach et al, 2014; Makishima et al, 2017). For example, TP53, EZH2, ETV6, RUNX1, ASXL1 and SRSF2 mutations predict poor outcomes, whereas SF3B1 mutations are associated with better clinical outcomes. However, because of the various combinations of different gene mutations in different patients and overlap between some clinical parameters and gene mutations, there is no consensus about how to use these genetic data in the prognostic scoring systems (Haferlach et al, 2014; Gerstung et al, 2015).
Recently, Makishima et al (2017) developed a prognostic model that was based only on genetic data (type-1 mutation: FLT3, PTPN11, WT1, IDH1, NPM1, IDH2 and NRAS; type-2 mutation: GATA2, KRAS, TP53, RUNX1, STAG2, ASXL1, ZRSR2 and TET2; SF3B1) according to the clonal evolution in MDS. Four risk groups were identified: low (Group III, who had SF3B1 mutation with no type-1 or type-2 mutations), intermediate-1 (Group IV, with no type-1, type-2 or SF3B1 mutations), intermediate-2 (Group II, who had type-2 mutations but lacked type-1 mutations) and high (Group I, with type-1 mutations) with significantly different overall survival (OS). Nazha et al (2016) developed a novel dynamic prognostic model that combined mutation of EZH2, SF3B1 and TP53, IPSS-R score and age to predict OS in patients with MDS. By using this model, four risk groups were identified: low, intermediate-1, intermediate-2 and high with a median OS of 37·4, 23·2, 19·9 and 12·2 months, respectively. In this study, we applied these two novel prognostic models to an independent group of 457 patients with MDS in order to validate the models and compared the OS predictive values of these two models and IPSS-R.
One hundred and twelve genes were detected by targeted sequencing in 457 successive MDS patients who had evaluable karyotypes and had been reclassified according to the 2016 revised WHO criteria (Arber et al, 2016). Details about targeted gene sequencing are described in the Data S1. All patients provided informed consent in compliance with the Declaration of Helsinki. The cohort comprised 277 (61%) males and 180 (39%) females, with a median age of 52 years (range, 14–83 years) (Table I). Two hundred and twenty-six patients (49%) received immune suppressive drugs including ciclosporin and thalidomide. Fifteen patients (3%) received anti-cancer therapy(ies) including aclacinomycin or homo-harringtonine combined with cytarabine and granulocyte-colony stimulating factor (G-CSF; termed CAG or HAG), idarubicin or daunorubicin combined with cytarabine (IA or DA) or melphalan. Seventy-eight patients (17%) received erythropoietin with or without G-CSF, red blood cell and/or platelet transfusions and/or iron chelation with desferrioxamine. Fifty-eight patients (13%) received decitabine, 34 (7%) an allotransplant and 46 (10%) traditional Chinese medicines. Follow-up data was available for 429 (94%) patients. Median follow-up for survivors was 20 months (range, 1–144). Survival distributions were estimated by the Kaplan–Meier method. Cox regression model and the likelihood ratio test were used to evaluate the predictive power. P < 0·05 were considered statistically significant.
Table I.
Clinical characteristics of 457 patients with myelodysplastic syndromes.
| Patients (n) | % | |
|---|---|---|
| Age, years, Median (range) | 52 (14–83) | |
| Male | 277 | 61 |
| WHO subtype | ||
| MDS-SLD | 15 | 3 |
| MDS-RS-SLD | 12 | 3 |
| MDS-MLD | 251 | 55 |
| MDS-RS-MLD | 3 | 0·7 |
| MDS-EB-1 | 70 | 15 |
| MDS-EB-2 | 92 | 20 |
| MDS with isolated 5q- | 6 | 1·3 |
| MDS-U | 8 | 2 |
| Hb, g/l, median (range) | 77 (31–153) | |
| ANC, × 109/l, median (range) | 1·1 8 (0·04–11·19) | |
| PLT, × 109/l, median (range) | 63 (2–1561) | |
| IPSS-R karyotype (Greenberg et al, 2012) | ||
| Very good | 5 | 1 |
| Good | 270 | 59 |
| Median | 113 | 25 |
| Poor | 26 | 6 |
| Very poor | 43 | 9 |
| IPSS-R risk group (Greenberg et al, 2012) | ||
| Very low | 12 | 3 |
| Low | 115 | 25 |
| Intermedia | 152 | 33 |
| High | 92 | 20 |
| Very high | 86 | 19 |
| Nazha model (Nazha et al, 2016) | ||
| Low | 131 | 29 |
| Intermediate-1 | 165 | 36 |
| Intermediate-2 | 117 | 26 |
| High | 44 | 10 |
| Makishima model (Makishima et al, 2017) | ||
| Low | 27 | 6 |
| Intermediate-1 | 263 | 58 |
| Intermediate-2 | 121 | 27 |
| High | 46 | 10 |
ANC, absolute neutrophil count; Hb, haemoglobin; IPSS-R, Revised International Prognostic Scoring System; MDS, myelodysplastic syndrome; MDS-EB-1/2, MDS with excess blasts type 1/2; MDS-MLD, MDS with multilineage dysplasia; MDS-RS-MLD, MDS with ring sideroblasts (MDS-RS) with multilineage dysplasia; MDS-RS-SLD, MDS-RS with single lineage dysplasia; MDS-SLD, MDS with single lineage dysplasia; MDS-U, MDS unclassifiable; PLT, platelet count; WHO, World Health Organization.
According to the IPSS-R, the risk for 12 (3%) patients was very low, 115 (25%) patients were low, 152 (33%) were intermediate, 92 (20%) were high and 86 patients (19%) were very high risk, with a 5-year OS of 90·9%, 88·1%, 74·5%, 39·8% and 20·1%, respectively (P < 0·001, Fig 1A). Based on the Makishima model (Makishima et al, 2017), 27 patients (6%) were low risk, 263 (58%) were intermediate-1, 121 (27%) were intermediate-2 and 46 (10%) patients were high risk, with a 5-year OS of 80·2%, 71·8%, 55·5% and 35%, respectively (P = 0·003, Fig 1B). The Nazha model (Nazha et al, 2016) classified 131 patients (29%) as low risk, 165 (36%) as intermediate-1, 117 (26%) as intermediate-2 and 44 (10%) as high risk, with a 5-year OS of 88·1%, 68%, 47% and 16·5%, respectively (P < 0·001, Fig 1C). When comparing the prognostic value of these three prognostic scoring systems using the Cox regression model and the likelihood ratio test, a significantly higher predictive power for OS became evident for the Nazha scoring system, compared with the IPSS-R and Makishima model (2 log-likelihood ratios of Nazha model: 989; IPSS-R: 1012; Makishima model: 1258; Nazha model vs. IPSS-R: P = 0·044; Nazha model vs. Makishima model: P < 0·001).
Fig 1.
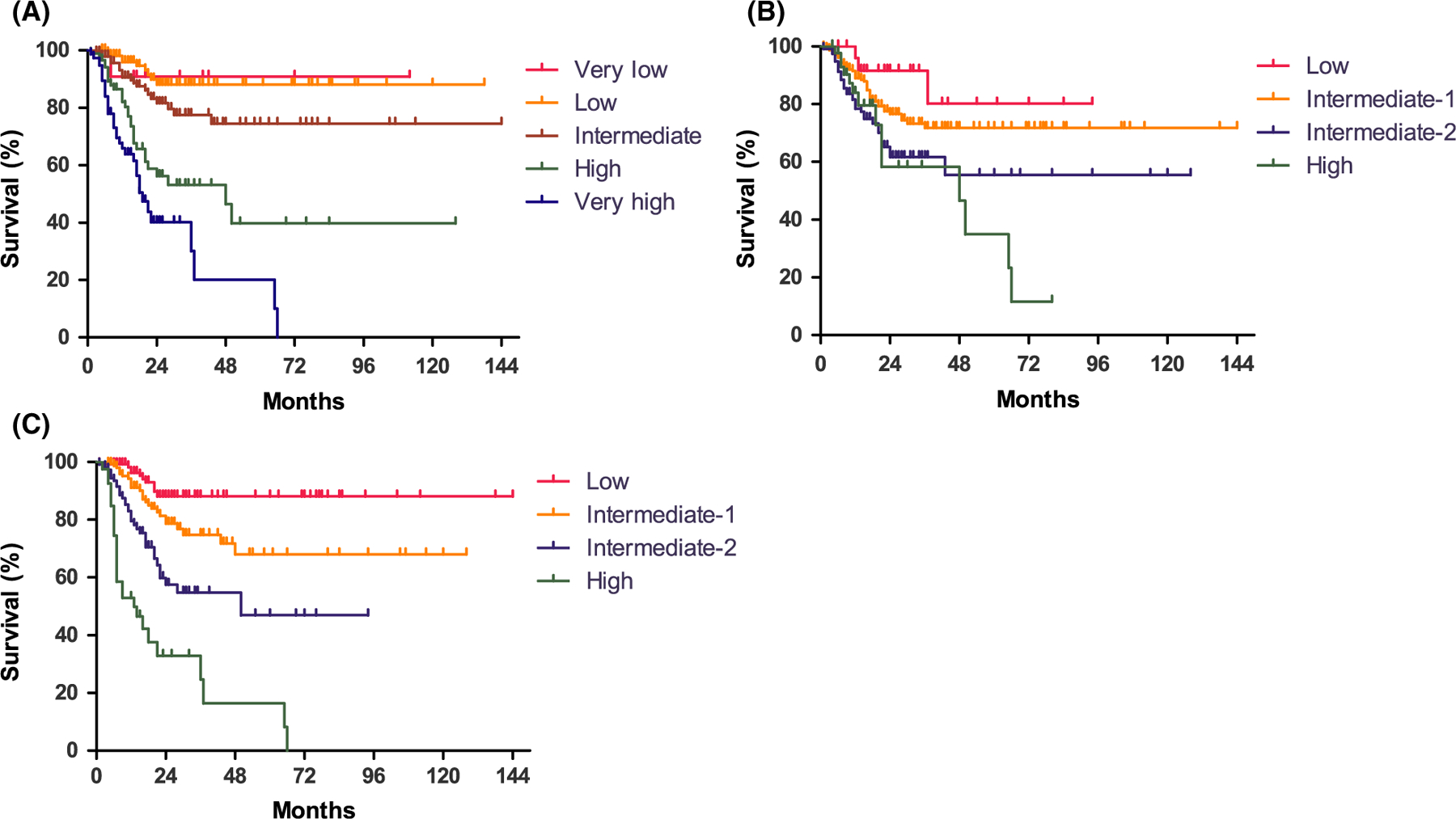
The overall survival was significantly different in myelodysplastic syndromes (MDS) patients classified according to the (A) Revised International Prognostic Scoring System (IPSS-R; Greenberg et al, 2012), (B) Makishima model (Makishima et al, 2017) and (C) Nazha model (Nazha et al, 2016) as shown by the Kaplan–Meier method.
Comparing the IPSS-R with the Nazha model, 12% of patients in the IPSS-R lower risk group (low/very low) were shifted into the higher risk intermediate-2 category in the Nazha model) and 19% of patients in the IPSS-R intermediate risk groupwere shifted into higher risk (intermediate-2). Meanwhile, 7% of the IPSS-R higher-risk (high/very high) patients were down staged into low-risk categories by the Nazha model. Detailed information is shown in Figure S1.
In our study, the utility of the Makishima and Nazha models was confirmed in a cohort of 457 patients with clinical and molecular data available. Moreover, the Nazha model showed higher predictive value than both the IPSS-R and Makishima model. Furthermore, our data indicated that over 10% of patients in the IPSS-R lower risk (very low/low) were reclassified as intermediate-2 risk using the Nazha model. Our data demonstrated that the Nazha model could help clinicians to more precisely define the probability of poor outcome in the lower MDS risk groups, which represent the majority of MDS patients, in whom new approaches, including allogeneic stem-cell transplantation, should be addressed.
In conclusion, a prognostic model that includes clinical and common mutational data can more precisely predict survival in MDS, and the confirmation and revision of novel molecular-clinical scoring systems should continue under international collaboration.
Supplementary Material
Acknowledgements
Supported in part by National Natural Science Funds (No. 81530008, No. 81370611, No. 81600098, No. 81270585, No. 81470295), Program for Peking Union Scholars and Innovative Research Team, PUMC Youth Fund & Fundamental Research Funds for the Central Universities (No. 3332016089) and Science and technology project of Tianjin (No. 15ZXLCSY00010).
Footnotes
Conflict-of-Interest disclosure
The authors declare no competing financial interests.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Data S1. Supplemental methods.
Figure S1. Distribution (%) of MDS patients who previously had been categorized by IPSS-R now categorized by Nazha et al model.
References
- Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, Bloomfield CD, Cazzola M & Vardiman JW (2016) The 2016 revision to the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia. Blood, 127, 2391–2405. [DOI] [PubMed] [Google Scholar]
- Gerstung M, Pellagatti A, Malcovati L, Giagounidis A, Porta MG, Jädersten M, Dolatshad H, Verma A, Cross NC, Vyas P, Killick S, Hellstrom-Lindberg E, Cazzola M, Papaemmanuil E, Campbell PJ & Boultwood J (2015) Combining gene mutation with gene expression data improves outcome prediction in myelodysplastic syndromes. Nature communications, 6, 5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg P, Cox C, LeBeau MM, Fenaux P, Morel P, Sanz G, Sanz M, Vallespi T, Hamblin T, Oscier D, Ohyashiki K, Toyama K, Aul C, Mufti G & Bennett J (1997) International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood, 89, 2079–2088. [PubMed] [Google Scholar]
- Greenberg PL, Tuechler H, Schanz J, Sanz G, Garcia-Manero G, Solé F, Bennett JM, Bowen D, Fenaux P, Dreyfus F, Kantarjian H, Kuendgen A, Levis A, Malcovati L, Cazzola M, Cermak J, Fonatsch C, Le Beau MM, Slovak ML, Krieger O, Luebbert M, Maciejewski J, Magalhaes SM, Miyazaki Y, Pfeilstocker M, Sekeres M, Sperr WR, Stauder R, Tauro S, Valent P, Vallespi T, van de Loosdrecht AA, Germing U & Haase D (2012) Revised international prognostic scoring system for myelodysplastic syndromes. Blood, 120, 2454–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G, Schnittger S, Sanada M, Kon A, Alpermann T, Yoshida K, Roller A, Nadarajah N, Shiraishi Y, Shiozawa Y, Chiba K, Tanaka H, Koeffler HP, Klein HU, Dugas M, Aburatani H, Kohlmann A, Miyano S, Haferlach C, Kern W & Ogawa S (2014) Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia, 28, 241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantarjian H, O’brien S, Ravandi F, Cortes J, Shan J, Bennett JM, List A, Fenaux P, Sanz G, Issa JP, Freireich EJ & Garcia-Manero G (2008) Proposal for a new risk model in myelodysplastic syndrome that accounts for events not considered in the original International Prognostic Scoring System. Cancer, 113, 1351–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makishima H, Yoshizato T, Yoshida K, Sekeres MA, Radivoyevitch T, Suzuki H, Przychodzen B, Nagata Y, Meggendorfer M, Sanada M, Okuno Y, Hirsch C, Kuzmanovic T, Sato Y, Sato-Otsubo A, LaFramboise T, Hosono N, Shiraishi Y, Chiba K, Haferlach C, Kern W, Tanaka H, Shiozawa Y, Gomez-Sequi I, Husseinzadeh HD, Thota S, Guinta KM, Dienes B, Nakamaki T, Miyawaki S, Saunthararajah Y, Chiba S, Miyano S, Shih LY, Haferlach T, Ogawa S & Maciejewski JP (2017) Dynamics of clonal evolution in myelodysplastic syndromes. Nature genetics, 49, 204–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcovati L, Germing U, Kuendgen A, Della Porta MG, Pascutto C, Invernizzi R, Giagounidis A, Hilderbrandt B, Bernasconi P, Knipp S, Strupp C, Strupp C, Lazzarino M, Aul C & Cazzola M (2007) Time-dependent prognostic scoring system for predicting survival and leukemic evolution in myelodys-plastic syndromes. Journal of Clinical Oncology, 25, 3503–3510. [DOI] [PubMed] [Google Scholar]
- Nazha A, Narkhede M, Radivoyevitch T, Seastone DJ, Patel BJ, Gerds AT, Mukherjee S, Kalaycio M, Advani A, Przychodzen B, Carraway HE, Maciejewski JP & Sekeres MA (2016) Incorporation of molecular data into the Revised International Prognostic Scoring System in treated patients with myelodysplastic syndromes. Leukemia, 30, 2214–2220. [DOI] [PubMed] [Google Scholar]
- Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, Yoon CJ, Ellis P, Wedge DC, Pellagatti A, Shlien A, Groves MJ, Forbes SA, Raine K, Hinton J, Mudie LJ, McLaren S, Hardy C, Latimer C, Della Porta MG, O’Meara S, Ambaglio I, Galli A, Butler AP, Walldin G, Teague JW, Quek L, Sternberg A, Gambacorti-Passerini C, Cross NC, Green AR, Boultwood J, Vyas P, Hellstrom-Lindberg E, Bowen D, Cazzola M, Stratton MR & Campbell PJ; Chronic Myeloid Disorders Working Group of the International Cancer Genome Consortium. (2013). Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood, 122, 3616–3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


