Abstract
The secondary intracellular symbiotic bacterium (S-symbiont) of the pea aphid Acyrthosiphon pisum was investigated to determine its prevalence among strains, its phylogenetic position, its localization in the host insect, its ultrastructure, and the cytology of the endosymbiotic system. A total of 14 aphid strains were examined, and the S-symbiont was detected in 4 Japanese strains by diagnostic PCR. Two types of eubacterial 16S ribosomal DNA sequences were identified in disymbiotic strains; one of these types was obtained from the primary symbiont Buchnera sp., and the other was obtained from the S-symbiont. In situ hybridization and electron microscopy revealed that the S-symbiont was localized not only in the sheath cells but also in a novel type of cells, the secondary mycetocytes (S-mycetocytes), which have not been found previously in A. pisum. The size and shape of the S-symbiont cells were different when we compared the symbionts in the sheath cells and the symbionts in the S-mycetocytes, indicating that the S-symbiont is pleomorphic under different endosymbiotic conditions. Light microscopy, electron microscopy, and diagnostic PCR revealed unequivocally that the hemocoel is also a normal location for the S-symbiont. Occasional disordered localization of S-symbionts was also observed in adult aphids, suggesting that there has been imperfect host-symbiont coadaptation over the short history of coevolution of these organisms.
To date, about 4,400 species of aphids (Homoptera, Aphididae) have been described (5). Almost all of them have an intracellular symbiotic bacterium, Buchnera sp. (4, 7, 41, 44): the exceptions are some cerataphidine aphids in which the bacterial symbiont has been replaced by an ascomycetous yeastlike endosymbiotic fungus (6, 17, 21, 25, 38). In the aphid body, Buchnera cells are harbored in the cytoplasm of mycetocytes (or bacteriocytes), which are hypertrophied cells in the abdomen that are specialized for endosymbiosis. The aphids and their Buchnera symbionts are considered intimately mutualistic; the symbionts cannot live when they are removed from the host cells (3), and the aphids become sterile or die when they are deprived of their symbionts (35, 36, 46). Aphids are supposed to provide their Buchnera symbionts with a stable niche and nutrients, and it has been demonstrated that Buchnera cells synthesize essential amino acids and other nutrients for the host (4, 10, 11). Since Buchnera cells are passed from one generation to the next by ovarial transmission and have no free-living state (7), they are considered a maternally inherited genetic element. The evolutionary origin of Buchnera symbionts is believed to be quite ancient. Morphological, histological, biochemical, and molecular phylogenetic lines of evidence have consistently suggested that the Buchnera symbionts of various distantly related aphid species had a single origin; these bacteria descended from a bacterium that was acquired by the common ancestor of extant aphids (7, 20, 41, 43, 45). Because of their predominance and importance in aphids, Buchnera spp. and the mycetocytes harboring them are often referred to as the primary symbionts (P-symbionts) and the primary mycetocytes (P-mycetocytes), respectively. Phylogenetically, the Buchnera P-symbiont belongs to the γ subdivision of the division Proteobacteria (51). Buchnera represents one of the most extensively investigated endosymbiotic microbes of insects.
In addition to Buchnera P-symbionts, a number of aphids are known to contain a second type of intracellular symbiotic bacteria (7, 20, 22, 26). These additional bacteria are harbored separately in a different type of mycetocytes, which constitute a mycetome (or bacteriome) with the P-mycetocytes, and are vertically transmitted to the aphid offspring (7, 18). They are found in many but not all lineages of aphids, and differ remarkably in morphology and localization in different lineages. It is thought that they have polyphyletic evolutionary origins (7, 20, 22, 26). These symbionts are collectively called secondary symbionts (S-symbionts). In contrast to the studies of P-symbionts, only a few modern studies of the S-symbionts of aphids have been performed (8, 20, 21, 26, 51); some early exceptions were histological studies in which conventional light microscopy was used. The only previous molecular phylogenetic characterization of S-symbionts was a study performed with the pea aphid Acyrthosiphon pisum, whose P- and S-symbionts belong to distinct lineages in the γ subdivision of the Proteobacteria (51).
The previous findings for the S-symbiont of A. pisum are, however, rather fragmentary and somewhat confusing. Griffiths and Beck (30) and McLean and Houk (40) first described the S-symbiont of A. pisum by using electron microscopy. In these studies, the S-symbionts were found in syncytial sheath cells that were located at the periphery of the mycetome and were closely associated with the P-mycetocytes. In the cytoplasm of the sheath cells, small rod-shaped bacteria occurred together with well-developed endoplasmic reticulum, the Golgi apparatus, and mitochondria. Using light microscopy, Douglas and Dixon (12) observed that rod-shaped S-symbionts were present in the sheath cells of young larvae but were only loosely associated with the mycetocytes in older insects. In contrast, Fukatsu and Ishikawa (19, 20) did not detect any intracellular bacteria other than the P-symbionts in immunohistochemical studies. Grenier et al. (29) also found no rod-shaped intracellular bacteria but discovered that one strain of A. pisum contained tubular extracellular microorganisms in its hemocoel. Although Unterman et al. (51) identified the 16S ribosomal DNA (rDNA) sequence of the S-symbiont, they did not confirm that the sequence was derived from the rod-shaped bacteria in the sheath cells. Using a specific PCR technique, Chen and Purcell (8) detected the S-symbiont sequence in more than 80% of the California clones of A. pisum which they examined. When hemolymph of S-symbiont-positive insects was microinjected into S-symbiont-negative insects, the S-symbiont sequence was successfully transferred to the recipients and, notably, inherited by their offspring. Thus, it was suggested that the carrier of the S-symbiont sequence occurs freely in the hemocoel, although bacterial cells were not found in the hemolymph when microscopy was used. Based on these results, it is difficult to find consensus concerning the morphology, localization, and microbial nature of the S-symbiont in A. pisum. Furthermore, the presence of other types of facultative bacterial associates makes the situation more complicated. For instance, Chen et al. (9) identified a rod-shaped, maternally inherited bacterium which was a member of the genus Rickettsia in the hemocoel of many strains of A. pisum, and Harada and Ishikawa (31), Grenier et al. (29), and Harada et al. (32, 33) have isolated many types of gut bacteria from A. pisum.
In the present study, we investigated the prevalence, phylogenetic position, localization, cytology, and ultrastructure of the S-symbiont of A. pisum by using diagnostic PCR, molecular phylogeny, histology, in situ hybridization, and electron microscopy. Notably, we discovered a novel type of cells harboring the S-symbionts in addition to the sheath cells and characterized the facultative and pleomorphic nature of the S-symbiont in A. pisum; based on our findings previous reports could be reinterpreted and synthesized.
MATERIALS AND METHODS
Materials.
The strains of A. pisum used in this study are listed in Table 1. All of these strains are laboratory-maintained strains. Because they originated from a single parthenogenetic female or a few parthenogenetic females and were maintained through numerous parthenogenetic generations, the lines are clonal isofemale lines. Japanese strains were reared on seedlings of the broad bean, Vicia faba, at 20°C by using a long-day regimen (16 h of light and 8 h of darkness). Newly molted unwinged adults were preserved in acetone until molecular and histological analyses were conducted (16). Unwinged adults of European and American strains maintained at INRA-INSA, Lyon, France, were shipped to us by Y. Rahbe in acetone.
TABLE 1.
Strains of A. pisum (Harris) used in this study
| Strain | Original locality | Collection date | Original host plant | Lab host plant | Provider | P-symbiont | S-symbiont |
|---|---|---|---|---|---|---|---|
| AIST99 | Tsukuba, Ibaraki, Japan | 7 April 1999 | Vicia sativa | Vicia faba | T. Fukatsu | +a | − |
| EF99 | Suginami-ku, Tokyo, Japan | 22 April 1999 | Vicia sativa | Vicia faba | T. Fukatsu | + | − |
| HG99 | Bunkyo-ku, Tokyo, Japan | 22 April 1999 | Vicia sativa | Vicia faba | T. Fukatsu | + | + |
| IS | NDb | ND | ND | Vicia faba | H. Ishikawa | + (AB033776)c | + (AB033778) |
| MR88 | Morioka, Iwate, Japan | 15 August 1988 | Pisum sativum | Vicia faba | K. Honda | + (AB033775) | + (AB033779) |
| SM | ND | ND | ND | Vicia faba | Y. Narai | + (AB033774) | − |
| TKC93 | Tokachi, Hokkaido, Japan | 23 June 1993 | Glycine max | Vicia faba | K. Honda | + (AB033772) | + (AB033777) |
| LL01 | Lusignan, France | Before 1988 | Medicago sativa | Vicia faba | Y. Rahbe | + (AB033773) | − |
| LL02 | Lusignan, France | Before 1988 | Medicago sativa | Vicia faba | Y. Rahbe | + | − |
| LL04 | Lusignan, France | 29 March 1995 | Medicago sativa | Vicia faba | Y. Rahbe | + | − |
| LL05 | Lusignan, France | 29 March 1995 | Medicago sativa | Vicia faba | Y. Rahbe | + | − |
| LL06 | Cornell University, Ithaca, N.Y. | 19 July 1996 | Trifolium sp. | Vicia faba | Y. Rahbe | + | − |
| LF08 | Fleurieu-sur-Saone, France | 21 July 1996 | Trifolium pratense | Vicia faba | Y. Rahbe | + | − |
| LC09 | Cornell University, Ithaca, N.Y. | 19 July 1996 | Medicago sativa | Medicago sativa | Y. Rahbe | + | − |
+, present, as determined in this study; −, absent, as determined in this study.
ND, no data.
The numbers in parentheses are accession numbers of the 16S rDNA sequences.
PCR, cloning, and sequencing of 16S rDNA.
The DNA of an individual insect kept in acetone was extracted by using a QIAamp tissue kit (Qiagen). Using the whole-insect DNA, we amplified almost all of the bacterial 16S rDNA (length, about 1.5 kb) by PCR by using primers 16SA1 (5′-AGAGTTTGATCMTGGCTCAG-3′) and 16SB1 (5′-TACGGYTACCTTGTTACGACTT-3′) and the following temperature profile: 94°C for 2 min, followed by 30 cycles consisting of 94°C for 1 min, 50°C for 1 min, and 70°C for 2 min. Cloning of the PCR products, typing of the clones by restriction fragment length polymorphism (RFLP) analysis, and DNA sequencing were conducted as previously described (23).
Diagnostic PCR.
Using specific reverse primers PASScmp (5′-GCAATGTCTTATTAACACAT-3′) and ApisS (5′-GCCATCAGGCAGTTTC-3′) for the S-symbiont, primer ApisP (5′-TCTTTTGGGTAGATCC-3′) for the P-symbiont, and primer Rick16SR (5′-CATCCATCAGCGATAAATCTTTC-3′) for Rickettsia spp. in combination with universal forward primer 16SA1, we performed a diagnostic PCR detection analysis of the 16S rDNA of the endosymbiotic bacteria by using the following temperature profile: 94°C for 2 min, followed by 30 cycles consisting of 94°C for 1 min, 55°C for 1 min, and 70°C for 2 min.
Molecular phylogenetic analysis.
A multiple alignment of 16S rDNA sequences was prepared by using the methods of Feng and Doolittle (15) and Gotoh (28). The final alignment was inspected and corrected manually. Ambiguously aligned regions were excluded from the phylogenetic analysis. Nucleotide sites that included an alignment gap(s) were also omitted from the aligned data set. Neighbor-joining trees (47) were constructed with Kimura's two-parameter distance (37) by using the CLUSTAL W program package (50). Maximum-likelihood trees (13) were constructed by using the MORPHY 2.3 program package (1). Maximum-parsimony trees were constructed by using the PAUP 4.0b2 program package (49). A bootstrap test (14) was conducted with 1,000 resamplings.
Histology.
Histological preparation, in situ hybridization, and enzymatic probe detection were performed essentially as previously described (26). Insects preserved in acetone were transferred to alcoholic formalin (ethanol-formalin, 3:1), and their heads and thoraxes were removed with forceps to facilitate infiltration of reagents. After the insects were kept in the fixative overnight, they were dehydrated and cleared with an ethanol-xylene series and embedded in paraffin. Serial tissue sections (thickness, 5 μm) were cut with a rotary microtome and were mounted on silane-coated glass slides. The sections were dewaxed with a xylene-ethanol series and air dried prior to in situ hybridization.
In situ hybridization.
BIO-PASScmp (5′-<biotin>GCAATGTCTTATTAACACAT-3′) targeting the S-symbiont was complementary to primer PASS-5′ (8). BIO-ApisS (5′-<biotin>GCCATCAGGCAGTTTC-3′) and DIG-ApisP (5′-<digoxigenin>TCTTTTGGGTAGATCC-3′) were designed to specifically detect the S- and P-symbionts, respectively. BIO-EUB338 and DIG-EUB338, which generally recognize eubacterial 16S rRNA (2, 26), were used to visualize both the S- and P-symbionts. About 200 μl of hybridization buffer (20 mM Tris-HCl [pH 8.0], 0.9 M NaCl, 0.01% sodium dodecyl sulfate, 30% formamide) containing 50 pmol of probe per ml was applied to a tissue section, and the preparation was covered with a coverslip and incubated in a humidified chamber at room temperature overnight. To eliminate nonspecific binding of the probe, the tissue section was rinsed in washing buffer (20 mM Tris-HCl [pH 8.0], 0.9 M NaCl, 0.01% sodium dodecyl sulfate, 30% formamide) for 10 min at 37°C. After the tissue section was washed with 1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), bound probe was detected as previously described (26). Biotin-labeled probes were visualized by using a Vectastain Elite ABC kit (Vector). Digoxigenin-labeled probes were detected by using a DIG nucleic acid detection kit (Boehringer Mannheim). To confirm the specificity of the hybridization procedure, the following control experiments were conducted: no-probe control experiment, RNase digestion control experiment, and competitive suppression control experiment performed with excess unlabeled probe (26).
Examination of hemolymph.
The abdominal dorsa of adult aphids, the surfaces of which had been sterilized and washed with 70% ethanol and sterile water, were fixed onto glass slides with Scotch tape carefully so that the insects were not damaged. The legs were removed with forceps, and the hemolymph coming from the injury was collected with a glass capillary. About 1 μl of hemolymph was subjected to either DNA extraction with a QIAamp tissue kit or direct microscopic observation. For the latter procedure, the hemolymph was applied to microscopic immersion oil on a glass slide, covered with a coverslip, and observed with a light microscope.
Electron microscopy.
The embryos of unwinged adult aphids were removed under a dissecting microscope in the presence of 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4), prefixed in the fixative at 4°C overnight, postfixed in 2% osmium tetroxide in 0.1 M phosphate buffer (pH 7.4) at 4°C for 60 min, and subjected to block staining with 0.5% uranyl acetate for 60 min. The embryos were dehydrated with an ethanol series and embedded in Epon 812. Ultrathin sections were cut with an ultramicrotome (Ultracut-N; Leichert-Nissei), mounted on collodion-coated copper mesh, stained with uranyl acetate and lead citrate, and observed with a transmission electron microscope (model H-7000; Hitachi) at 75 kV.
Nucleotide sequence accession numbers.
The 16S rDNA sequences of the P- and S-symbionts of the A. pisum strains described in this paper have been deposited in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession numbers AB033772 through AB033779 (Table 1).
RESULTS
Analysis of the 16S rDNA of the P- and S-symbionts of strain IS.
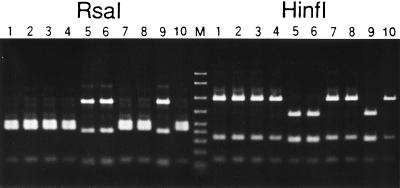
Almost the entire length of eubacterial 16S rDNA in a strain IS adult was amplified by PCR, and the products were subjected to cloning. When the clones obtained were examined to determine their RFLP patterns, two major types of clones were identified (Fig. 1). Three clones of the first type and five clones of the second type were sequenced. The three sequences of the first type were identical and exhibited a very high level of similarity to the sequence of the P-symbiont of A. pisum in the database. Only one substitution and two indels were found among 1,473 aligned nucleotide sites. The five sequences of the second type were identical except for two nucleotide sites. At sites 137 and 232 the sequences of three clones contained G, while the sequences of the other two clones contained A. These sequences exhibited very high levels of similarity to the sequence of the S-symbiont in the database. Out of 1,462 aligned nucleotide sites not including the two polymorphic sites, only two substitutions were found.
FIG. 1.
RFLP analysis of bacterial 16S rDNA amplified and cloned from the total DNA of A. pisum IS. Lanes 1 through 10 contained cloned 16S rDNA fragments digested by RsaI (left) or HinfI (right) and resolved in a 2% agarose gel. Lanes 1 through 4, 7, 8, and 10, clones containing the P-symbiont sequence; lanes 5, 6, and 9, clones containing the S-symbiont sequence. Lane M contained DNA size markers (2,000, 1,500, 1,000, 700, 500, 400, 300, 200, and 100 bp, from top to bottom).
Diagnostic PCR detection of the S-symbiont in various strains of A. pisum.
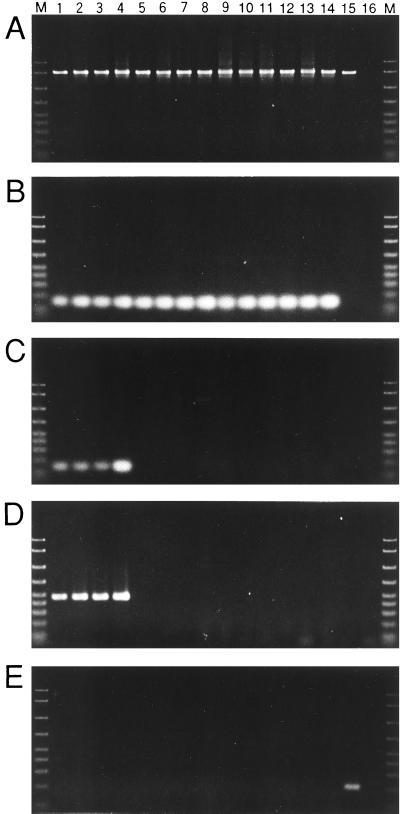
To examine the presence of the S-symbiont in various strains of A. pisum, we performed diagnostic PCR experiments with specific reverse primers in combination with universal forward primer 16SA1 (Fig. 2). When specific primer ApisP was used, the P-symbiont was detected in all 14 strains examined (Fig. 2B). When specific primer ApisS was used, on the other hand, the S-symbiont was detected only in four strains, strains IS, MR88, TKC93, and HG99 (Fig. 2C); these findings were supported by the results of PCR performed with specific primer PASScmp (Fig. 2D). Although Chen et al. (9) identified a Rickettsia species in many strains of A. pisum, PCR performed with specific primer Rick16SR demonstrated that all 14 strains were Rickettsia negative (Fig. 2E).
FIG. 2.
Diagnostic PCR detection of endosymbionts in A. pisum strains. (A) Universal detection of eubacterial endosymbionts with primers 16SA1 and 16SB1. (B) Specific detection of the P-symbiont with primers 16SA1 and ApisP. (C) Specific detection of the S-symbiont with primers 16SA1 and ApisS. (D) Specific detection of the S-symbiont with primers 16SA1 and PASScmp. (E) Specific detection of Rickettsia spp. with primers 16SA1 and Rick16SR. Lane 1, strain IS; lane 2, TKC93; lane 3, MR88; lane 4, HG99; lane 5, EF99; lane 6, SM; lane 7, AIST99; lane 8, LL01; lane 9, LL02; lane 10, LL04; lane 11, LL05; lane 12, LC06; lane 13, LF08; lane 14, LC09; lane 15, bruchid beetle Kytorhinus sharpianus containing Rickettsia sp. (24); lane 16, no-template control; lane M, DNA size markers (2,000, 1,500, 1,000, 700, 500, 400, 300, 200, and 100 bp, from top to bottom). Although the results for only one individual of each strain are shown, the reproducibility of the results was confirmed by examining more than 12 individuals of each strain.
16S rDNA sequences of the P- and S-symbionts of various strains.
In addition to the 16S rDNA sequences of strain IS symbionts, we determined the 16S rDNA sequences of the P- and S-symbionts of four strains. Both the P- and S-symbiont sequences were found in disymbiotic strains MR88 and TKC93, whereas only the P-symbiont sequence was found in monosymbiotic strains AIST99 and LL01. The sequences of the P-symbionts of different strains were almost identical, as were the sequences of the S-symbionts. Molecular phylogenetic analysis showed that, as previously reported, the P-symbionts clustered with Buchnera strains obtained from other species belonging to the Aphidinae, whereas the S-symbionts were related to enteric bacteria, such as Serratia, Enterobacter, Erwinia, and Escherichia strains (data not shown).
In situ hybridization of the S-symbiont.
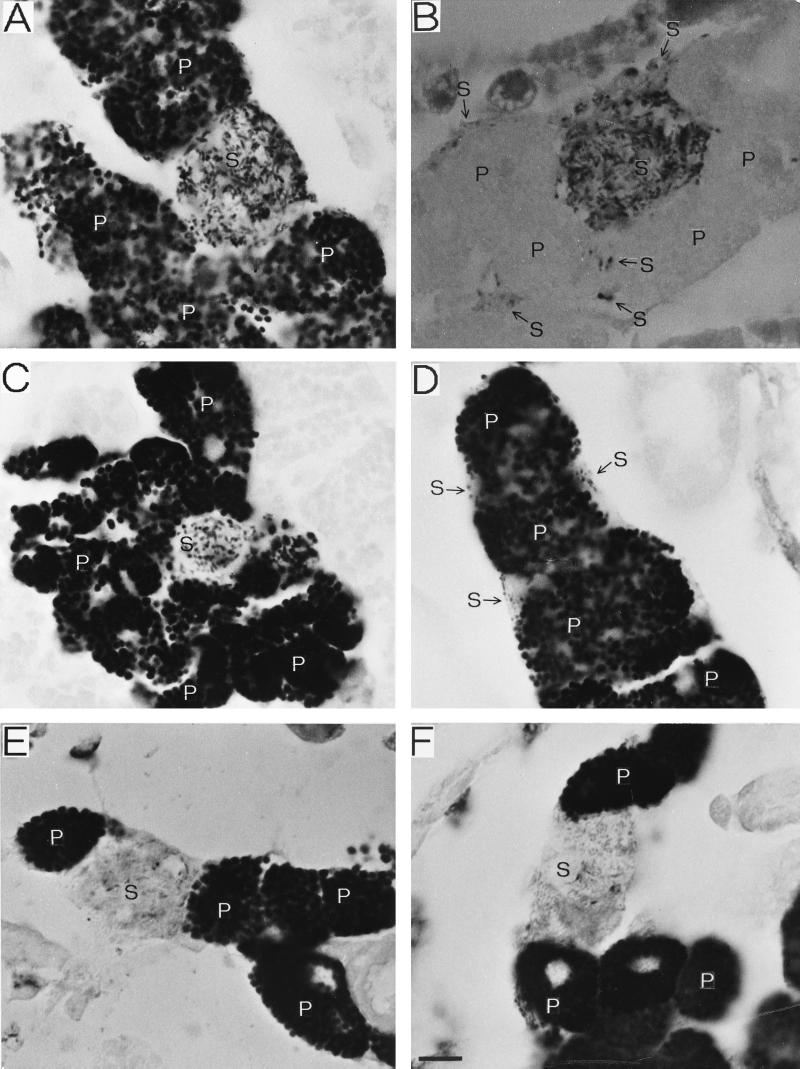
In order to determine the morphology and localization of the S-symbiont in vivo, tissue sections of the insects were subjected to in situ hybridization with oligonucleotide probes that target bacterial 16S rRNA (Fig. 3). In disymbiotic strain IS, two types of intracellular bacteria were detected with probe BIO-EUB338, which recognizes eubacteria universally (Fig. 3A, C, and D). One type of bacteria was globular and predominant and was harbored in round uninucleate mycetocytes that constituted a huge mycetome in the abdomen of each embryo. On the basis of these histological and morphological features this bacterium was identified as the P-symbiont, Buchnera sp.; this identification was confirmed by in situ hybridization with probe DIG-ApisP, which recognizes the P-symbiont specifically (data not shown). In addition to the P-symbionts, intracellular bacteria that had a different shape and localization were also found. When preparations were probed with BIO-EUB338, small portions of cytoplasm containing tiny short rods were observed (Fig. 3D). The rod-shaped cells were closely associated with the P-mycetocytes on the periphery of each embryonic mycetome. These histological features corresponded to the features of sheath cells previously reported to harbor S-symbionts. In addition, the S-symbionts were found in special cells that have not been found previously in A. pisum (Fig. 3A through C). These cells, designated secondary mycetocytes (S-mycetocytes), were similar in size to the P-mycetocytes. In embryos, the mycetomes were usually composed of tens of P-mycetocytes and one S-mycetocyte or a few S-mycetocytes. The S-mycetocytes were normally full of tubes or long rods (Fig. 3A), although the bacteria appeared to be shorter rods in some cases (Fig. 3C). The bacteria in the S-mycetocytes were larger and longer than the bacteria in the sheath cells. However, when tissue sections were probed with BIO-PASScmp, which specifically recognizes the S-symbiont, the bacteria were visualized in both the S-mycetocytes and the sheath cells (Fig. 3B). Hybridization with probe BIO-ApisS gave the same results (data not shown). Therefore, we concluded that in A. pisum the S-symbionts are harbored in two types of cells, the S-mycetocytes and the sheath cells. In other disymbiotic strains, including strains MR88 and TKC93, S-mycetocytes containing tubular S-symbionts were also present (Fig. 3E and F). The S-symbiont-specific probes certainly detected the bacteria in the S-mycetocytes and sheath cells, as shown in Fig. 3B (data not shown). In the monosymbiotic strains, in contrast, the S-symbionts were not detected by in situ hybridization (data not shown).
FIG. 3.
In situ hybridization of the P- and S-symbionts of A. pisum. (A) Mycetome of a strain IS embryo probed with BIO-EUB338. Tubular S-symbionts in a S-mycetocyte and globular P-symbionts in many P-mycetocytes are present. (B) Mycetome of a strain IS embryo probed with BIO-PASScmp. Tubular S-symbionts in a S-mycetocyte and small S-symbionts in sheath cells are specifically visualized in the mycetome. (C) Mycetome of a strain IS embryo probed with BIO-EUB338. In this S-mycetocyte, the S-symbionts are short rods. (D) Mycetome of a strain IS embryo probed with BIO-EUB338. Sheath cells containing small S-symbionts are associated with the P-mycetocytes. (E) Mycetome of a strain MR88 embryo probed with BIO-EUB338. A S-mycetocyte harboring tubular S-symbionts is located between P-mycetocytes. (F) Mycetome of a strain TKC93 embryo probed with BIO-EUB338. The same disymbiotic organization is observed. Bar = 10 μm. The arrows indicate the locations of sheath cells. Abbreviations: P, P-symbiont: S, S-symbiont.
Disordered localization of the S-symbiont.
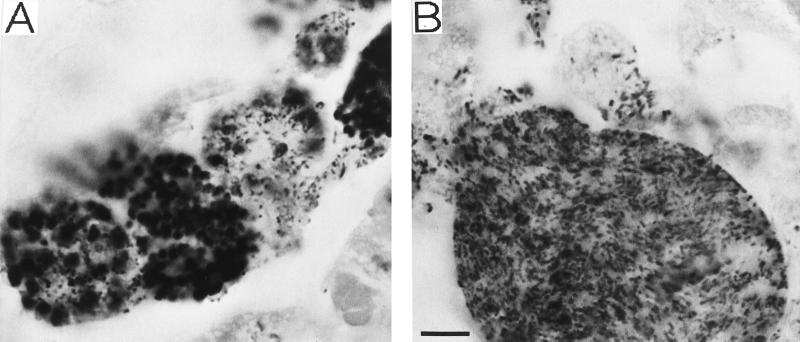
In adults of strain IS, we occasionally observed abnormal localization of the S-symbionts (Fig. 4). In these insects, S-symbionts were detected not only in the S-mycetocytes but also in other cells and tissues. In embryos, normally the P- and S-symbionts are specifically harbored in different types of cells. However, Fig. 4A shows an embryonic mycetome in which some of the P-mycetocytes were also infected by S-symbionts. Although some mycetocytes looked normal, some were infected with a mixture of cells, and others were predominantly occupied by S-symbionts. In addition, other embryonic tissues around the mycetome were also sporadically infected. In maternal tissues, normally huge P-mycetocytes are the only cells that contain P-symbionts, and no special cells harboring S-symbionts are found. However, Fig. 4B shows a maternal mycetocyte filled with S-symbionts in place of P-symbionts; around this mycetocyte fat body cells and hemocoel also contained S-symbionts. We have infrequently encountered insects exhibiting such symptoms; none or a few of the tens of individuals sampled and processed at the same time for histological studies have shown them. So far, however, such insects have been found in at least three samples that were independently collected.
FIG. 4.
Disordered localization of the S-symbiont found in several unwinged adults of A. pisum IS. (A) Embryonic P-mycetocytes infected by S-symbionts. Abnormally, the P- and S-symbionts coexist in several cells. (B) Maternal P-mycetocyte filled with S-symbionts. The P-symbionts are almost completely replaced by S-symbionts. The symbionts were visualized by in situ hybridization with probe BIO-EUB338. Bar = 10 μm.
Examination of hemolymph.
We examined the hemolymph of Japanese strains of A. pisum to determine whether symbionts were present by using diagnostic PCR and light microscopy. As shown in Fig. 2, in the disymbiotic strains both P- and S-symbionts were detected by PCR while in the monosymbiotic strains only P-symbionts were detected (data not shown); these findings indicated that at least a small number of symbionts were present in the hemolymph of A. pisum. When the hemolymph was directly observed by light microscopy, we found many tubular or rod-shaped particles which were morphologically reminiscent of the S-symbionts observed by in situ hybridization only in disymbiotic strains (data not shown).
Electron microscopy of the S-symbiont.
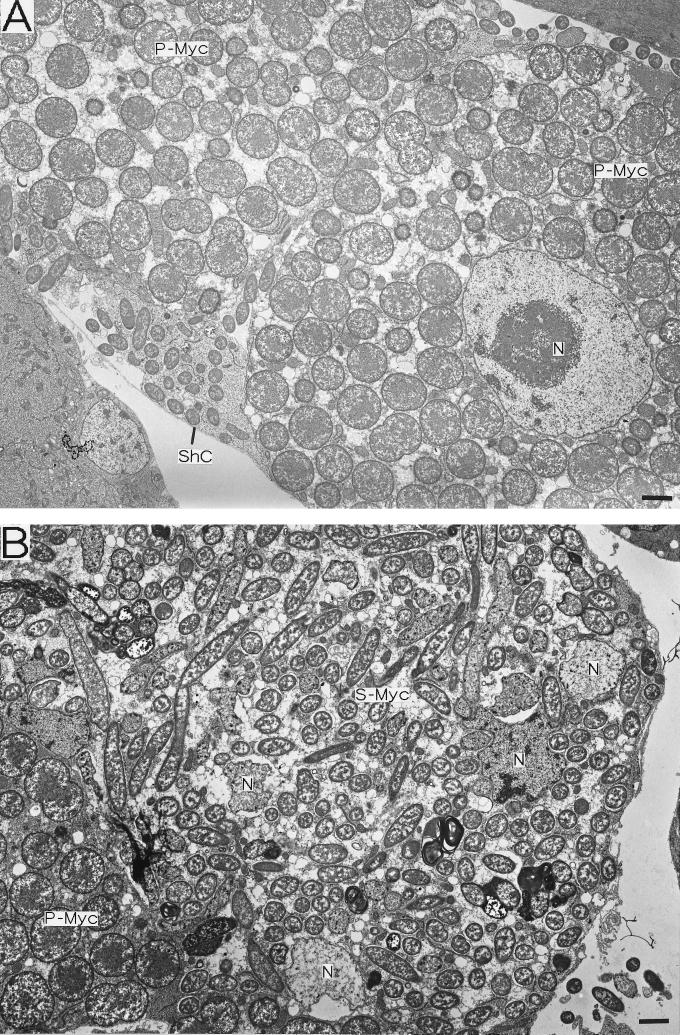
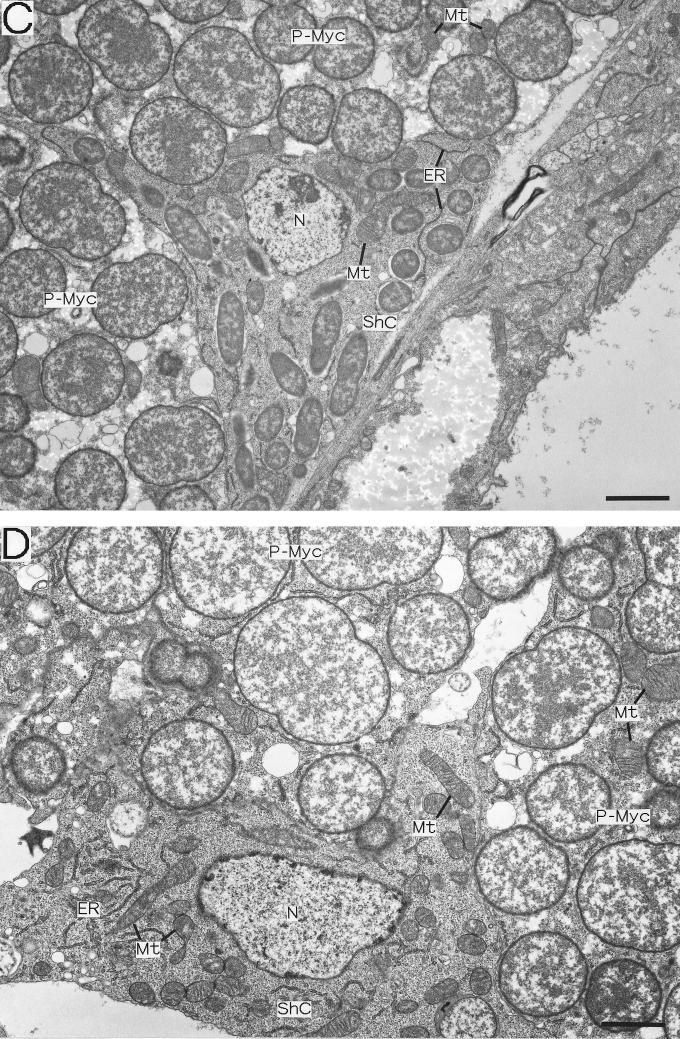
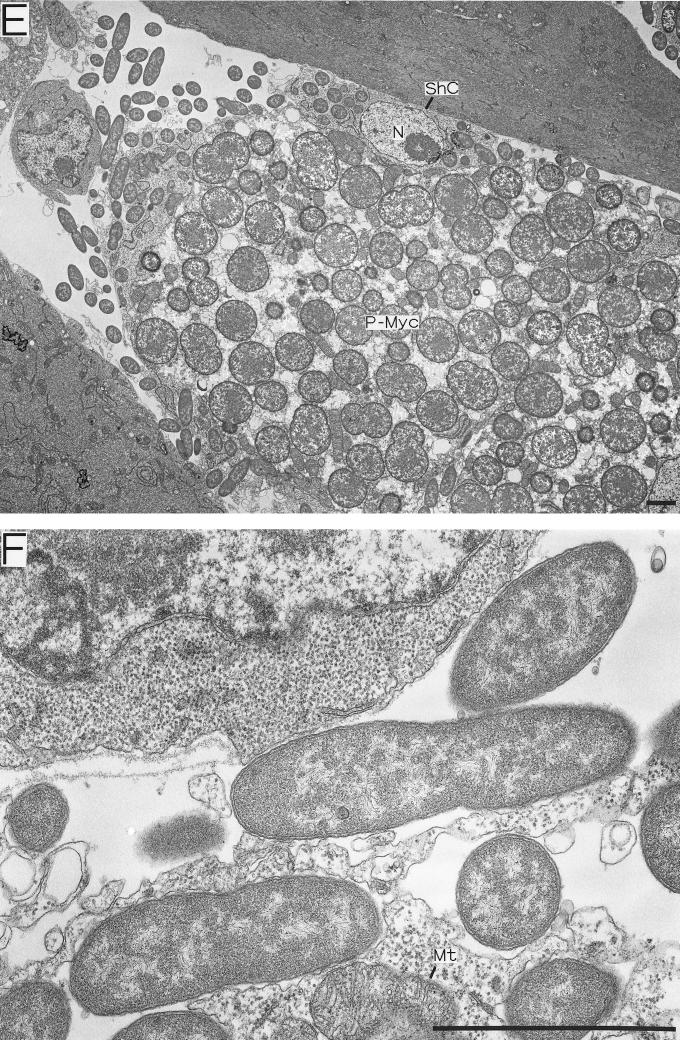
The fine structure and localization of the S-symbiont in strain IS embryos were investigated by transmission electron microscopy (Fig. 5). Sheath cells harboring many rod-shaped S-symbionts were found throughout the mycetome and were closely associated with the P-mycetocytes (Fig. 5A). In addition to the S-symbionts, the cytoplasm of the sheath cells contained mitochondria, endoplasmic reticulum, and ribosomes (Fig. 5C), as described previously. Notably, in the monosymbiotic strains the sheath cells had similar ultrastructural features, although they did not contain S-symbionts (Fig. 5D). The large S-mycetocytes harbored a number of S-symbionts, which were generally larger than the S-symbionts in the sheath cells, although the overall shape of the bacteria was difficult to determine with ultrathin sections. The cytoplasm of the S-mycetocytes was highly vacuolated and in this respect differed from that of the sheath cells (Fig. 5B). We found that even in embryos a number of S-symbionts were present in extracellular locations, although they were still closely associated with the sheath cells and S-mycetocytes (Fig. 5B, E, and F).
FIG. 5.
Electron microscopy of the endosymbiosis in A. pisum embryos. (A through C, E, and F) Strain IS containing both P- and S-symbionts. (D) Strain SM lacking the S-symbiont. (A) Strain IS sheath cell located between large P-mycetocytes. Small rod-shaped S-symbionts are located intracellularly. (B) Strain IS S-mycetocyte harboring many tubular S-symbionts. The S-symbionts are apparently larger than the S-symbionts in sheath cells. The cytoplasm of the S-mycetocyte is highly vacuolated. (C) Magnified image of a strain IS sheath cell containing S-symbionts, mitochondria, and endoplasmic reticulum. (D) Magnified image of a sheath cell of a strain SM aphid without the S-symbiont. The sheath cell contains developed mitochondria and endoplasmic reticulum but no S-symbionts. (E) Extracellular S-symbionts associated with strain IS sheath cells on the periphery of a mycetome. (F) Magnified image of S-symbionts on the periphery of a strain IS sheath cell. Bars = 2 μm. Abbreviations: ER, endoplasmic reticulum; Mt, mitochondrion; N, nucleus; P-Myc, primary mycetocyte; ShC, sheath cell; S-Myc, secondary mycetocyte.
DISCUSSION
Since the first electron microscopic observations of endosymbiosis in pea aphids (30, 40), it has been repeatedly claimed and widely accepted that the S-symbionts in A. pisum are harbored by the sheath cells (4, 10, 11, 34, 42). In the present study, however, we demonstrated that in Japanese strains of A. pisum, the S-symbionts are also found in large special cells, the S-mycetocytes in the mycetomes (Fig. 3 and 5). Although it has been found for many aphids that secondary intracellular bacteria are harbored in the cytoplasm of large mycetocytes that are cytologically distinct from the P-mycetocytes (7, 18, 20, 22, 26), this is the first description of this type of mycetocyte in A. pisum. The morphology and ultrastructure of the S-mycetocytes were clearly distinguishable from the morphology and ultrastructure of the sheath cells. Thus, the S-symbionts are harbored in two different types of host cells specialized for endosymbiosis.
Our results indicated that in A. pisum there are three types of cells involved in endosymbiosis: the P-mycetocytes, S-mycetocytes, and sheath cells. The developmental and evolutionary origins of these cells are very interesting but poorly understood. The size, shape, and location of the S-mycetocytes were reminiscent of the size, shape, and location of the P-mycetocytes (Fig. 3), and we observed that the S-symbionts occasionally invaded the P-mycetocytes (Fig. 4). These facts suggest that the S-mycetocytes are developmentally homologous to the P-mycetocytes, although the ultrastructure of the S-mycetocytes was not similar to the ultrastructure of the P-mycetocytes (Fig. 5). Cytologically, it appeared that the sheath cells were quite different from the P- and S-mycetocytes. The mechanisms which enable S-symbionts to target different types of cells are also intriguing.
Are S-mycetocytes found in disymbiotic strains in general, or are they restricted to Japanese strains? To answer this question, a more extensive histological study to determine the presence of S-symbionts in A. pisum strains is needed. However, it is quite likely that S-mycetocytes are present at least in an American disymbiotic strain. When McLean and Houk (40) observed a smear preparation of mycetocytes of a California A. pisum strain by light microscopy, they found a large cell containing aggregations of bacilliform bacteria (40). Judging from its size and cytological traits, the cell was probably a S-mycetocyte rather than a sheath cell, although McClean and Houk did not examine the cell by electron microscopy. The mycetome of an A. pisum embryo is normally composed of tens of P-mycetocytes and sheath cells and only one S-mycetocyte or a few S-mycetocytes. It is conceivable that in previous electron microscopic studies the researchers failed to obtain images of S-mycetocytes because of the scarcity of these cells in the material. In fact, we had to examine a large number of ultrathin sections to observe an S-mycetocyte, while sheath cells were found in most ultrathin sections containing mycetomes.
Intracellular bacteria in the sheath cells and intracellular bacteria in the S-mycetocytes differed in size and morphology (Fig. 3 and 5). In the sheath cells the bacteria were small rods, whereas in the S-mycetocytes the bacteria were larger and longer. However, in situ hybridization experiments in which specific oligonucleotide probes were used unequivocally showed that the bacteria in the two types of cells are genetically identical (Fig. 3B). The differences in morphology were attributed to the pleomorphism of S-symbiont cells induced under different environmental conditions, a trait commonly found in microorganisms (48).
In addition to the sheath cells and the S-mycetocytes, the hemocoel was identified as a third location of S-symbiont cells. A considerable population of S-symbionts was present in the hemocoel. Electron microscopic examination clearly showed that even in embryos extracellular S-symbionts were closely associated with the sheath cells and the S-mycetocytes (Fig. 5). These results suggest that the hemocoel is a normal location of S-symbionts. Although the presence of S-symbionts in the hemolymph has been suggested previously by the results of PCR and injection experiments (8), this is the first study in which the morphology and localization of extracellular S-symbionts have been determined microscopically. Insects have humoral and cellular immune systems that effectively attack and kill bacterial and fungal intruders in the body fluid (27, 39). Thus, the extracellular location of S-symbionts suggests that these cells are able to avoid the host immune system in some way. In addition, S-symbionts are able to target the sheath cells and the S-mycetocytes specifically. To investigate endosymbiotic mechanisms at a molecular level, the S-symbiont of A. pisum might provide a good experimental system because manipulation and artificial transmission of the S-symbiont are possible (8).
16S rDNA sequence analyses strongly suggested that the S-symbionts of Japanese A. pisum strains identified in this study and the S-symbionts of American strains described in previous works (8, 51) are the same bacterium. The S-symbiont of A. pisum was closely related to enteric bacteria, such as Serratia, Enterobacter, Erwinia, and Escherichia strains, which may suggest that its evolutionary origin was a gut bacterium. In fact, various gut bacteria have been isolated from A. pisum; one such predominant bacterium was identified as an Erwinia species (31–33).
In strain IS adults examined histologically, we occasionally observed surprising localization of the S-symbiont (Fig. 4). In these adults, the S-symbionts, which infected various tissues and cells, appeared to be out of control. Unfortunately, we did not establish whether these insects exhibited pathological symptoms when they were alive. Since we examined only a small number of cases, it is not clear whether such disordered behavior is exceptional and observed only sometimes or is induced by particular physiological and environmental conditions. Our observations may reflect imperfect coadaptation in the endosymbiosis involving the S-symbiont and A. pisum, presumably due to the short history of coevolution of these organisms.
With the disymbiotic strains, PCR experiments showed that all of the individuals contained the S-symbiont after several years of maintenance in the laboratory (Fig. 2). As previously reported (8), the offspring of disymbiotic mothers all inherited the S-symbiont through parthenogenesis. Therefore, the S-symbiont is passed to the next generation by vertical transmission with high fidelity, at least under laboratory conditions. Although the transmission process has not been studied in detail, it must occur at an early embryonic stage in the body of the mother, because S-symbionts were found in the S-mycetocytes and sheath cells of young embryos (Fig. 3 and 5). Considering that injection of hemolymph can establish a heritable infection of the S-symbiont (8; T. Fukatsu, unpublished data), it is conceivable that the S-symbiont is transmitted to embryos via the hemolymph.
On the other hand, both Chen and Purcell (8) and we have demonstrated that the S-symbiont does not infect all individuals in populations of A. pisum, suggesting that vertical transmission of the S-symbiont may not be perfect under natural conditions. If the transmission rate is not 100%, the rate of S-symbiont infection must decline, and eventually the S-symbiont must disappear from populations as host generations proceed. However, S-symbiont infection is certainly maintained in natural populations, which suggests that some processes may counter imperfect vertical transmission. One possibility is that the S-symbiont makes the host slightly more fit, while another possibility is horizontal transmission of the S-symbiont.
To date, there has been no evidence of horizontal transmission of the S-symbiont from disymbiotic A. pisum to monosymbiotic A. pisum under laboratory mixed-rearing conditions (8; Fukatsu, unpublished data). Interestingly, however, it has been reported that a 16S rDNA sequence identical to that of the S-symbiont was found in an aphid belonging to different genus, Macrosiphum rosae (8), suggesting that interspecific horizontal transmission of the S-symbiont may have occurred under natural conditions. The S-symbiont of A. pisum was stably maintained and vertically transmitted when it was injected into a different aphid species, Acyrthosiphon kondoi (8), suggesting that the S-symbiont is not strictly host specific and thus may be transmitted horizontally. Although speculative, several horizontal transmission routes are conceivable. Parasitoid wasps might be vectors for the S-symbiont and perform microinjection in the wild. Considering its phylogenetic affinity to gut bacteria, the S-symbiont might sometimes be excreted with honeydew. If oral ingestion can occasionally establish an infection, honeydew, squashed aphids, and phloem sap of plants heavily populated by aphids could be sources of infection.
At this stage, the biological effects of the S-symbiont on the host aphid are not known; however, it is known that the S-symbiont is not essential for the host because there are monosymbiotic strains and populations. Nutritional, physiological, and population dynamics studies on A. pisum strains with and without the S-symbiont should be performed in order to determine whether the S-symbiont is almost neutral, parasitic, or slightly advantageous for the host under various environmental conditions. Considering the spatial proximity and integrity of the P- and S-symbionts in the endosymbiotic system, it is conceivable that the S-symbiont may interact with and modify the established mutualism between the aphid and Buchnera sp. in some way.
The S-symbiont of A. pisum is very interesting because it can be considered an intermediate between facultative endosymbiotic bacteria, such as Wolbachia spp., and highly specialized mutualistic intracellular symbionts, such as Buchnera spp. (41). Like Wolbachia spp., the S-symbiont is not essential for the host and sometimes is horizontally transmitted across lineages and species. Like Buchnera spp., the S-symbiont is harbored by specialized host cells for endosymbiosis. In future studies on the S-symbiont, we may be able to gain insight into the evolutionary transition from facultative guest microbe to obligately mutualistic symbiont.
We identified two intracellular symbiotic bacteria, the P- and S-symbionts, in the A. pisum strains examined in this study. Frequent occurrence of Rickettsia sp. has been reported for American strains (9), although this organism was not detected in this study. As far as we know, these three microbes are the major endosymbiotic bacteria of the pea aphid that have been described so far. However, it is not certain that this is the complete picture of the endosymbiotic microbiota. Further investigations may reveal other types of interesting endosymbiotic associates in A. pisum.
ACKNOWLEDGMENTS
We thank K. Honda, H. Ishikawa, Y. Narai, and Y. Rahbe for aphid samples, A. Sugimura, S. Kumagai, and K. Sato for technical and secretarial assistance, and T. Wilkinson and Y. Rahbe for reading the manuscript.
This research was supported by the Industrial Science and Technology Frontier Program “Technological Development of Biological Resources in Bioconsortia” of the Ministry of International Trade and Industry of Japan and by the Program for Promotion of Basic Research Activities for Innovation Biosciences (ProBRAIN) of the Bio-Oriented Technology Research Advancement Institution.
REFERENCES
- 1.Adachi J, Hasegawa M. MOLPHY version 2.3: programs for molecular phylogenetics based on maximum likelihood. Comput Sci Monogr Inst Stat Math Tokyo. 1996;28:1–150. [Google Scholar]
- 2.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumann P, Moran N A. Non-cultivable microorganisms from symbiotic associations of insects and other hosts. Antonie Leeuwenhoek. 1997;72:38–48. doi: 10.1023/a:1000239108771. [DOI] [PubMed] [Google Scholar]
- 4.Baumann P, Baumann L, Lai C-Y, Rouhbakhsh D, Moran N A, Clark M A. Genetics, physiology and evolutionary relationships of the genus Buchnera: intracellular symbionts of aphids. Annu Rev Microbiol. 1995;49:55–94. doi: 10.1146/annurev.mi.49.100195.000415. [DOI] [PubMed] [Google Scholar]
- 5.Blackman R L, Eastop V F. Aphids on the world's trees. Wallingford, United Kingdom: CAB International; 1994. [Google Scholar]
- 6.Buchner P. Eine neue Form der Endosymbiose bei Aphiden. Zool Anz. 1958;160:222–230. [Google Scholar]
- 7.Buchner P. Endosymbiosis of animals with plant microorganisms. New York, N.Y: Interscience; 1965. [Google Scholar]
- 8.Chen D Q, Purcell A H. Occurrence and transmission of facultative endosymbionts in aphids. Curr Microbiol. 1997;34:220–225. doi: 10.1007/s002849900172. [DOI] [PubMed] [Google Scholar]
- 9.Chen D Q, Campbell B C, Purcell A H. A new Rickettsia from a herbivorous insect, the pea aphid Acyrthosiphon pisum (Harris) Curr Microbiol. 1996;33:123–128. doi: 10.1007/s002849900086. [DOI] [PubMed] [Google Scholar]
- 10.Dixon A F G. Aphid ecology. London, United Kingdom: Chapman & Hall; 1998. [Google Scholar]
- 11.Douglas A E. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu Rev Entomol. 1998;43:17–37. doi: 10.1146/annurev.ento.43.1.17. [DOI] [PubMed] [Google Scholar]
- 12.Douglas A E, Dixon A F G. The mycetocyte symbiosis of aphids: variation with age and morph in virginoparae of Megoura viciae and Acyrthosiphon pisum. J Insect Physiol. 1987;33:109–113. [Google Scholar]
- 13.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 14.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 15.Feng D F, Doolittle R F. Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J Mol Evol. 1987;25:351–360. doi: 10.1007/BF02603120. [DOI] [PubMed] [Google Scholar]
- 16.Fukatsu T. Acetone preservation: a practical technique for molecular analysis. Mol Ecol. 1999;8:1935–1945. doi: 10.1046/j.1365-294x.1999.00795.x. [DOI] [PubMed] [Google Scholar]
- 17.Fukatsu T, Ishikawa H. A novel eukarytoic extracellular symbiont in an aphid, Astegopteryx styraci (Homoptera, Aphididae, Hormaphidinae) J Insect Physiol. 1992;38:765–773. [Google Scholar]
- 18.Fukatsu T, Ishikawa H. Soldier and male of an eusocial aphid Colophina arma lack endosymbiont: implications for physiological and evolutionary interaction between host and symbiont. J Insect Physiol. 1992;38:1033–1042. [Google Scholar]
- 19.Fukatsu T, Ishikawa H. Synthesis and localization of symbionin, an aphid endosymbiont protein. Insect Biochem Mol Biol. 1992;22:167–174. [Google Scholar]
- 20.Fukatsu T, Ishikawa H. Occurrence of chaperonin 60 and chaperonin 10 in primary and secondary bacterial symbionts of aphids: implications for the evolution of an endosymbiotic system in aphids. J Mol Evol. 1993;36:568–577. doi: 10.1007/BF00556361. [DOI] [PubMed] [Google Scholar]
- 21.Fukatsu T, Ishikawa H. Phylogenetic position of yeast-like symbiont of Hamiltonaphis styraci (Homoptera, Aphididae) based on 18S rDNA sequence. Insect Biochem Mol Biol. 1996;26:383–388. doi: 10.1016/0965-1748(95)00105-0. [DOI] [PubMed] [Google Scholar]
- 22.Fukatsu T, Ishikawa H. Differential immunohistochemical visualization of the primary and secondary intracellular symbiotic bacteria of aphids. Appl Entomol Zool. 1998;33:321–326. [Google Scholar]
- 23.Fukatsu T, Nikoh N. Two intracellular symbiotic bacteria from the mulberry psyllid Anomoneura mori (Insecta, Homoptera) Appl Environ Microbiol. 1998;64:3599–3606. doi: 10.1128/aem.64.10.3599-3606.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fukatsu T, Shimada M. Molecular characterization of Rickettsia sp. in a bruchid beetle, Kytorhinus sharpianus (Cleoptera: Bruchidae) Appl Entomol Zool. 1999;34:391–397. [Google Scholar]
- 25.Fukatsu T, Aoki S, Kurosu U, Ishikawa H. Phylogeny of Cerataphidini aphids revealed by their symbiotic microorganisms and basic structure of their galls: implications for host-symbiont coevolution and evolution of sterile soldier castes. Zool Sci. 1994;11:613–623. [Google Scholar]
- 26.Fukatsu T, Watanabe K, Sekiguchi Y. Specific detection of intracellular symbiotic bacteria of aphids by oligonucleotide-probed in situ hybridization. Appl Entomol Zool. 1998;33:461–472. [Google Scholar]
- 27.Gillespie J P, Kanost M R. Biological mediators of insect immunity. Annu Rev Entomol. 1997;42:611–643. doi: 10.1146/annurev.ento.42.1.611. [DOI] [PubMed] [Google Scholar]
- 28.Gotoh O. Optimal alignment between groups of sequences and its application to multiple sequence alignment. Comput Applic Biosci. 1993;9:361–370. doi: 10.1093/bioinformatics/9.3.361. [DOI] [PubMed] [Google Scholar]
- 29.Grenier A M, Nardon C, Rahbe Y. Observations on the microorganisms occurring in the gut of the pea aphid Acyrthosiphon pisum. Entomol Exp Appl. 1994;70:91–96. [Google Scholar]
- 30.Griffiths G W, Beck S D. Intracellular symbiotes in the pea aphids, Acyrthosiphon pisum. J Insect Physiol. 1973;19:75–84. [Google Scholar]
- 31.Harada H, Ishikawa H. Gut microbe of aphid closely related to its intracellular symbiont. BioSystems. 1993;31:185–191. doi: 10.1016/0303-2647(93)90048-h. [DOI] [PubMed] [Google Scholar]
- 32.Harada H, Oyaizu H, Ishikawa H. A consideration about the origin of aphid intracellular symbiont in connection with gut bacterial flora. J Gen Appl Microbiol. 1996;42:17–26. [Google Scholar]
- 33.Harada H, Oyaizu H, Kosako Y, Ishikawa H. Erwinia aphidicola, a new species isolated from pea aphid, Acyrthosiphon pisum. J Gen Appl Microbiol. 1997;43:349–354. doi: 10.2323/jgam.43.349. [DOI] [PubMed] [Google Scholar]
- 34.Houk E J. Symbionts. In: Minks A K, Harrewijn P, editors. Aphids: their biology, natural enemies and control. 2A. Amsterdam, The Netherlands: Elsevier; 1987. pp. 123–129. [Google Scholar]
- 35.Houk E J, Griffiths G W. Intracellular symbiotes of Homoptera. Annu Rev Entomol. 1980;25:161–187. [Google Scholar]
- 36.Ishikawa H, Yamaji M. Symbionin, an aphid endosymbiont-specific protein. I. Production of insects deficient in symbiont. Insect Biochem. 1985;15:155–163. [Google Scholar]
- 37.Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 38.Kolb G. Die Endosymbiose der Thelaxiden unter besonderer Berücksichtigung der Hormaphidinen und ihrer Embryonalentwicklung. Z Morphol Oekol Tiere. 1963;53:185–241. [Google Scholar]
- 39.Marmaras V J, Charalambidis N D, Zervas C G. Immune response in insects: the role of phenoloxidase in defense reactions in relation to melanization and sclerotization. Arch Insect Biochem Physiol. 1996;31:119–133. doi: 10.1002/(SICI)1520-6327(1996)31:2<119::AID-ARCH1>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 40.McLean D L, Houk E J. Phase contrast and electron microscopy of the mycetocytes and symbiotes of the pea aphid, Acyrthosiphon pisum. J Insect Physiol. 1973;19:625–633. [Google Scholar]
- 41.Moran N, Baumann P. Phylogenetics of cytoplasmically inherited microorganisms of arthropods. Trends Ecol Evol. 1994;9:15–20. doi: 10.1016/0169-5347(94)90226-7. [DOI] [PubMed] [Google Scholar]
- 42.Moran N A, Telang A. Bacteriocyte-associated symbionts of insects. BioScience. 1998;48:295–304. [Google Scholar]
- 43.Moran N A, Munson M A, Baumann P, Ishikawa H. A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. Proc R Soc London B. 1993;253:167–171. [Google Scholar]
- 44.Munson M A, Baumann P, Kinsey M G. Buchnera gen. nov. and Buchnera aphidicola sp. nov., a taxon consisting of the mycetocyte-associated, primary endosymbionts of aphids. Int J Syst Bacteriol. 1991;41:566–568. [Google Scholar]
- 45.Munson M A, Baumann P, Clark M A, Baumann L, Moran N A. Evidence for the establishment of aphid-eubacterium endosymbiosis in an ancestor of four aphid families. J Bacteriol. 1991;173:6321–6324. doi: 10.1128/jb.173.20.6321-6324.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohtaka C, Ishikawa H. Effects of heat treatment on the symbiotic system of an aphid mycetocyte. Symbiosis. 1991;11:19–30. [Google Scholar]
- 47.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 48.Stanier R Y, Ingraham J L, Wheelis M L, Painter P R. The microbial world. Englewood Cliffs, N.J: Prentice-Hall; 1986. [Google Scholar]
- 49.Swofford D L. PAUP*: phylogenetic analysis using parsomony version 4.0b2. Sunderland, Mass: Sinauer Associates; 1999. [Google Scholar]
- 50.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Unterman B M, Baumann P, McLean D L. Pea aphid symbiont relationships established by analysis of 16S rRNAs. J Bacteriol. 1989;171:2970–2974. doi: 10.1128/jb.171.6.2970-2974.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]