Abstract
This study evaluated the antibiotic resistance characteristics and virulence genes of enterococci isolated from raw and processed seafood sold in the Marmara Region, Turkey. In this context, the enterococcal load was determined as between 1.0 and 2.5 log CFU/g in 39 of a total of 397 samples. It was determined that 117 strains isolated from the samples belonged to Enterococcus gallinarum, E. casseliflavus, E. durans, E. faecium, and E. faecalis species. Erythromycin, tetracycline, streptomycin, and gentamicin resistance was observed, whereas the tetM, ermB, aac(6')-aph(2'')-la genes were found in a majority of the isolates. It was also determined that the isolates carried the agg2 and gelE virulence genes. When all these results are evaluated, the presence of these isolates in aquatic products may pose a risk in terms of food safety and public health.
Keywords: Antibiotic resistance, Enterococci, Seafood, Virulence genes
Introduction
Seafood, particularly those having bacterial and/or chemical qualities, may carry high potential health risks for human beings. Pathogenic microorganisms may contaminate seafood during the breeding, handling, processing, or preparation of products (Iwamoto et al. 2010). During the breeding of seafood products, using antibiotics as therapeutic and/or prophylactic agents to prevent bacterial diseases leads to the development of antibiotic resistance in bacterial populations in aquaculture environments (Muñoz-Atienza et al. 2013).
Enterococci are the member of the commensal microbiota of the intestinal tract of animals and humans and are also found in soil and water, domestic sewage and animal waste (Facklam et al. 2002; Hancock and Gilmore 2006). Sewage and wastes can contaminate certain surfaces, which are used by the public, and groundwater (Korajkic et al. 2020). Because of their widespread presence in animal and human faeces, they are indicated as a faecal indicator for water quality (Di Cesare et al. 2014).
Studies have reported enterococcal species can be isolated from particular aquatic ecosystem, including coastal marine environments and aquaculture, because of their tolerance to high salt conditions. Seafood may carry enterococci obtained from several aquatic sources, such as lakes, rivers, and seawater, which contaminate urban and aquaculture wastewater (Ben Said et al. 2017). Enterococci have also been isolated from seafood such as shellfish, the intestine of fish, traditionally processed fish products, shrimp, salmon, turbot, etc. In some studies, it has been reported that the Enterococcus faecium, E. faecalis, E. durans, E. raffinosus, E. avium, E. malladoratus, E. casseliflavus, E. gallinarum, E. phoeniculicola, E. saccharolyticus, and E. gilvus strains were isolated from aquatic habitats and traditional or industrially processed seafood products (Hammad et al. 2014; Klibi et al. 2013; Psoni et al. 2006; Pinto et al. 2009; Valenzuela et al. 2010).
Although E. gallinarum was described originally from chickens, this species was rarely found, suggesting that these species may not belong to the normal intestinal flora of poultry (Lebreton et al. 2014). Badgley et al. (2010) identified enterococcal species isolated from water, sediment, and submerged aquatic vegetation such as E. casseliflavus, E. faecalis, E. faecium, E. hirae, and E. mundtii. It was suggested that the persistence of certain strains was not directly related to pollution events.
Enterococcal species can tolerate improper environmental conditions such as high salt concentrations, high or low pH and temperatures. (Byappanahalli et al. 2012). Enterococci are also isolated from intestinal fish microflora or fish environments (Valenzuela et al. 2010). Certain enterococcal strains are considered as opportunistic pathogens which have transferable antibiotic resistance characteristics and may cause multi-resistant nosocomial infections (Clewell et al. 2014). Some enterococcal strains may also carry virulence determinants that are responsible for infections in humans such as gelatinase, cytolysin, aggregation substances, and extracellular surface proteins (Aslam et al. 2012). Certain virulence characteristics in the genotype and/or phenotype of enterococci isolated from seafood were reported. Igbinosa and Beshiru (2019) detected S-layer, gelatinase production, and β-hemolysis reaction in phenotype and high number of virulence genes in the Enterococcus species from ready-to-eat seafood samples. Hammad et al. (2014) mentioned that the virulence determinants such as gelE (gelatinase), asa1 (aggregation substance), hyl (hyaluronidase), and esp (enterococcal surface protein) were related to pathogenicity proved by animal models. They also reported that gelE and asa1 genes were detected in E. faecalis and E. faecium strains isolated from ready-to-eat raw fish. Chajecka-Wierzchowska et al. (2017) determined that all tested enterococcal strains isolated from shrimp most frequently carried esp, gelE, efaA cpd, cob, ccf virulence genes.
The release of antimicrobial drugs in agricultural, and industrial applications and in the environment used in the medical treatment of humans and animals may cause an increase in the spreading of resistant enterococcal biota in water, soil, food, and wastewater. The virulent strains may be transferred to humans through sediment and seawater (Di Cesare et al. 2012; Pasquaroli et al. 2014; Citterio et al. 2017). It was reported that antibiotic-resistant enterococcal clinic isolates originated from hospital and urban wastewater and marine environmental isolates were found to be closely related. Antibiotic resistant bacterial isolates in the environment may be formed by the genetic transfer of resistance genes from clinical strains (Vignaroli et al. 2018). In humans, the enterococcal species, such as E. gallinarum, E. durans, E. casseliflavus, E. avium, E. hirae, may cause infection occasionally. Enterococcus gallinarum and E. casseliflavus sourced infections are considered because of their intrinsic resistance to vancomycin used to treat aminoglycoside-resistant enterococcal infections which caused a health risk for the public (Lebreton et al. 2014).
Enterococci are widely spread in the marine environment, despite the stress conditions such as salinity and sunlight. This habitat is affected by farming activities, marine sediments, and urban and hospital sewage. Because of this, it becomes a reservoir of the enterococcal strains of humans. The link between the high prevalence of enterococci in seawater and an increased ratio of human illness, the enterococcal strains carrying virulence genes and/or antibiotic resistance characteristic thought to be a threat to human health (Di Cesare et al. 2014).
Citterio et al. (2017) isolated enterococci from fish, shellfish, seawater, marine sediment, and beach sand. It was informed that bivalve shellfish may collect a high number of microorganisms in their body which could cause infection for humans and animals and which may also be a source for horizontal gene transfer, including antibiotic resistance genes. The use of antibiotics in aquaculture production may cause damaging environmental effects such as the occurrence of antibiotic resistance among the bacterial community in cultured aquatic animals and ponds. Concerns regarding the fact that antibiotic resistance characteristics in phenotype and genotype and also antimicrobial drug residues may be transferred to humans via consumption of seafood such as fish and shrimps (Ellis-Iversen et al. 2020).
Clinical and environmental enterococcal epidemics were reported in the literature (Gassiep et al. 2015) with limited information regarding outbreaks originated from food-borne enterococci. However, it was mentioned that the consumption of ready-to-eat seafood carrying virulence genes was an important way of transfer. Chajecka-Wierzchowska et al. (2017) characterized virulent enterococcal isolates from seafood samples.
Although it is widely known that antibiotic-resistant strains are a potential problem for humans and ecosystem health, there are a few studies regarding the investigation of antibiotic-resistant and pathogenic enterococci in the seafood of Turkey. The objective of this study was to do research on the occurrence and distribution of antibiotic-resistant characteristics and virulence genes of enterococci isolated from processed and raw seafood in the Marmara Region, Turkey.
Material and methods
Sample collection and isolation
A total of 397 samples were obtained from supermarkets, convenience stores, delicatessens, and traditional and fish markets in the Marmara Region, Turkey, including raw seafood (n = 290) and processed seafood samples (n = 107) such as frozen (n = 61), salted (n = 50), smoked (n = 1), canned (n = 5), marinated (n = 7) and surimi foods (n = 3) (Table 1).
Table 1.
Analyzed raw and processed seafood samples and the distribution of isolated enterococcal species
| Product type (Raw) | Number of samples (n = 290) | The species isolated from the samples | Product type (processed) | Number of samples (n = 107) | The species isolated from the samples |
|---|---|---|---|---|---|
| Sardinella (Sardina pilchardus) | 51 | E. casseliflavus, E. gallinarum, E. faecalis, E. Faecium | Fish Kroket (Frozen) | 3 | –* |
| Mackerel (Scomber scombru) | 3 | – | Salmon Fillet (Frozen) | 1 | – |
| Blotched picarel (Spicara maena) | 32 | E. gallinarum, E. faecium | Cod Fish Fillet (Frozen) | 1 | – |
| Horse mackerel (Trachurus trachurus) | 28 | E. gallinarum, E. Durans, E. casseliflavus | Anchovy Fillet (Frozen) | 10 | – |
| Sea bream (Diplodus vulgaris) | 11 | E. gallinarum, E. faecium | Boiled Shrimp (Frozen) | 3 | – |
| Red mullet (Mullus barbatus) | 33 | E. durans, E. gallinarum, E. casseliflavus | Calamary (Frozen) | 13 | – |
| Shrimp (Parapenaeus longirostris) | 22 | E. casseliflavus, E. gallinarum | Shrimp (Frozen) | 8 | – |
| Striped red mullet (Mullus surmuletus) | 12 | E. casseliflavus, E. gallinarum | Clam (Frozen) | 20 | E. gallinarum, E. casseliflavus, E. hirae |
| Baracuda (Sphyraena sphyrena) | 6 | – | Mixed Seafood (Frozen) | 2 | E. faecium |
| Bouge (Boops boops) | 10 | – | Sardinella (Salted) | 3 | – |
| Bluefish (Pomatomus saltator) | 11 | E. gallinarum, E. casseliflavus | Dried macherel (Salted) | 2 | – |
| Mediterranean horse mackerel (Trachurus mediterraneus) | 20 | E. durans, E. gallinarum | Salted bonito (Salted) | 22 | E. gallinarum, E. faecium |
| Squid (Loligo vulgaris) | 17 | E. faecium, E. durans, E. Faecalis, E. gallinarum | Anchovy (Salted) | 22 | E. casseliflavus |
| Anchovy (Engraulis encrasicolus) | 25 | E. gallinarum | Pate (Salted) | 1 | – |
| Gilthead seabream (Sparus aurata) | 9 | – | Mackerel Fillet (Smoked) | 1 | E. gallinarum |
| Sardinella (Canned) | 5 | E. casseliflavus, E. gallinarum | |||
| Anchovy (Marinated) | 3 | – | |||
| Seafood salads (Marinated) | 2 | – | |||
| Clam (Marinated) | 2 | E. faecalis | |||
| Crab (Surimi) | 3 | – |
* Not isolated any enterococcal strains
The whole seafood sample was cut into pieces and mixed individually in aseptic conditions. Then the analytical unit of seafood sample was weighed out (10 g) and homogenized with 90 mL of a physiological saline (PS) solution (0.85% NaCl) using a laboratory stomacher. The decimal dilutions of the samples were conducted in sterile PS and inoculated on Kanamycin Aesculin Azide (KAA, Fluka) agar and then incubated at 37 °C for 24- 48 h. After incubation, the black colonies around the black zone considered to be “suspicious” were purified on Trypticase Soy agar (Merck, Germany). The strain was maintained in stock culture at -80 °C in Brain Heart Infusion broth (Oxoid, Canada) containing 20% (v/v) glycerol.
Control strains
In the study, E. faecalis NCIMB 700584 (The National Collection of Industrial, Marine and Food Bacteria, UK) and Enterococcus hirae FM 2.16 were used as positive control strains for virulence genes and ermB gene, respectively (Eaton and Gasson 2001; Pasquaroli et al. 2014).
Identification and antibiotic susceptibility testing
The isolates were identified using Gram-staining and GP (Gram-positive cocci) cards in the VITEK 2 Compact 30 automated micro identification system (Biomereux, France). (Abele-Horn et al. 2006).
A VITEK 2 compact system (bioMérieux, France) and disc diffusion test method were used to analyze the antibiotic susceptibility of the isolates. Antimicrobial susceptibility testing (AST) was applied by VITEK 2 Compact (Biomérieux, Marcy l'Etoile, France) using AST card P592 (EUCAST 2015) and double-checked by means of the disk diffusion method. The strains were evaluated for antibiotic resistance against streptomycin (10 µg), chloramphenicol (30 µg), erythromycin (15 µg), tetracycline (30 µg), gentamycin (10 µg), and vancomycin (30 µg) using the disc diffusion method on Muller–Hinton agar (MHA, Oxoid, UK), and the results were evaluated through the consideration of the CLSI guidelines (CLSI 2016). The isolates displaying MIC > 1 μg/mL for streptomycin were thought to be resistant as suggested by the European Committee on Antimicrobial Susceptibility Testing (EUCAST 2015).
In this study, the multiple antibiotic resistance (MAR) index was also calculated for enterococcal isolates through the MAR formula (“a/b”). The “a” refers to the number of antimicrobials, to which an isolate was resistant, whereas “b” refers to the total number of antimicrobials to which the isolate was exposed. The MAR index can be calculated when there are more than three antibiotics showing resistance (Krumperman 1983).
Determination of antibiotic resistance and virulence genes
The vancomycin (vanA and vanB), tetracycline (tetM), erythromycin (ermB), and amyloglucosidase (aac(6')-aph(2'')-la) resistance genes and virulence genes (agg2, gelE, cylM, cylB, cylA) of the enterococcal strains were determined by polymerase chain reaction (PCR). The genomic DNAs of the enterococcal strains were extracted by using a commercial DNA isolation kit (Qiagen). PCR primers for the antibiotic resistance and virulence genes (Table 2) were choosen by considering Eaton and Gasson 2001, Reviriego et al. 2005; Pasquaroli et al. 2014).
Table 2.
The primers for virulence and antibiotic resistance genes
| Genes | Primer sequence (5’-3’) | Product size (bp) |
|---|---|---|
| agg2 | F-5’ GTT GTT TTA GCA ATG GGG TAT | 1210 |
| R-5’ TCC TGT CAC TCC TCT TCT CAG | ||
| gelE | F-5’ ACC CCG TAT CAT TGG TTT | 419 |
| R-5’ ACG CAT TGC TTT TCC ATC | ||
| cylM | F-5’ TGC TTC TCC ACT GTG ACC T | 742 |
| R-5’ ATC TAG TAA ATG TTA AGA AAT ACA | ||
| cylB | F-5’ TGG AAG CAT TAC TTC CAG CT | 843 |
| R-5’ AAC TGC AAC CTC AAG ATT GG | ||
| cylA | F-5’ AAT CCT ATC GGT TAC TGC TTA | 517 |
| R-5’ AGC ATC ACA ACC ATC CTA AC | ||
| vanA | F-5’ GTA CAA TGC GGC CGT TA | 732 |
| R-5’ GGG ACA GTT ACA ATT GC | ||
| vanB | F-5’ GTG CTG CGA GAT ACC ACA GA | 1145 |
| R-5’ CGA ACA CCA TGC AAC ATT TC | ||
| tetM | F-5’ GTT AAA TAG TGT TCT TGG AG | 657 |
| R-5’ CTA AGA TAT GGC TCT AAC AA | ||
| aac(6’)-aph(2’’)-la | F-5’ GAG CAA TAA GGG CAT ACC AAA AAT C | 505 |
| R-5’ CCG TGC ATT TGT CTT AAA AAA CTG G | ||
| ermB | F-5’ CAT TTA ACG ACG AAA CTG GC | 425 |
| R-5’ GGA ACA TCT GTG GTA TGG CG |
PCR amplifications were performed in 25 µL reaction mixtures using dNTP mix (1 mM) (Promega, USA), Go Taq Flexi DNA polymerase (1 U) (Promega), DNA (1 µL), and primers (10 pmol) obtained from IDT (Integrated DNA Technologies, USA). The samples were subjected to an initial cycle of denaturation (95 °C for 2 min), which was followed by 35 cycles of denaturation (94 °C for 45 s), annealing (53 °C for 30 s) and elongation (72 °C for 45 s) (Eaton and Gasson 2001; Reviriego et al. 2005).
Results and discussion
Distribution of enterococcal isolates
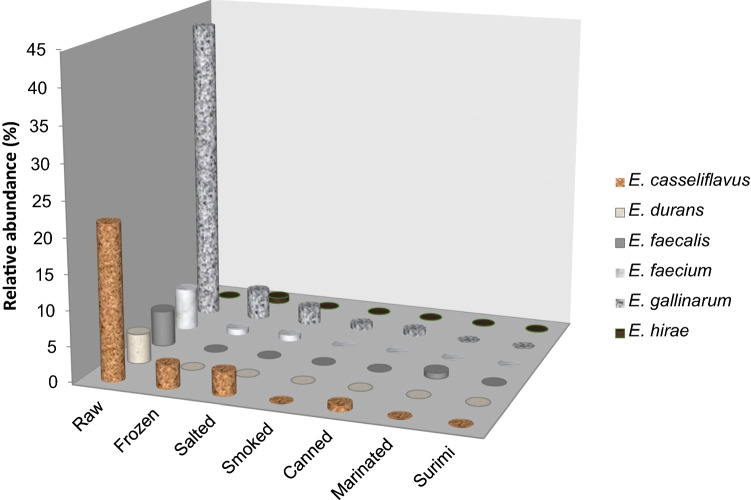
In the study, the enterococcal load was determined as between 1.0—2.5 log CFU/g in 39 of 397 (9.82%) total raw and processed seafood samples, and 117 strains were identified as Enterococcus spp. Among the enterococci, E. gallinarum (50.43%) was the most prevalent species, followed by E. casseliflavus (29.06%), E. faecium (7.70%), E. durans (6.84%), E. faecalis (5.13%), and E. hirae (0.85%) (Table 3). E. gallinarum (43.22%) and E. casseliflavus (22.03%) species were mostly isolated from raw fish samples. E. gallinarum strains were dominantly found in frozen (4.24%), salted (2.54%), smoked (0.85%), and canned (0.85%) seafood samples. E. faecalis strains were frequently isolated from marinated (0.85%) samples (Fig. 1) (Table 1).
Table 3.
Species distribution of antibiotic-resistant enterococci isolated from seafood samples
| Antimicrobial agent | Number and percentages (%) of enterococcal isolates distributed by species | |||||
|---|---|---|---|---|---|---|
| E. gallinarum (n = 59) | E. casseliflavus (n = 34) | E. faecium (n = 9) | E. durans (n = 8) | E. faecalis (n = 6) | E. hirae (n = 1) | |
| Streptomycin | 55 (93.22) | 33 (97.06) | 9 (100) | 7 (87.5) | 6 (100) | 1 (100) |
| Chloramphenicol | 25 (42.37) | 16 (47.06) | 3 (33.33) | 4 (50) | 1 (16.66) | 0 |
| Erythromycin | 44 (74.58) | 33 (97.06) | 7 (77.77) | 4 (50) | 5 (83.33) | 0 |
| Tetracycline | 34 (57.63) | 22 (64.71) | 6 (66.66) | 5 (62.5) | 5 (83.33) | 0 |
| Vancomycin | 4 (6.78) | 2 (5.89) | 0 | 4 (50) | 0 | 0 |
| Gentamycin | 29 (49.15) | 25 (73.53) | 4 (44.44) | 2 (25) | 4 (66.66) | 0 |
Fig. 1.
Distribution of Enterococcus species in raw and prossed seafood samples
There are certain studies that reported different enterococcal species isolated from raw and processed seafood samples in literature. It was thought that seafood contamination has occurred naturally from the environment during fish harvesting or cross-contamination during processing or preparation where bacteria were transferred from raw fish, contaminated surfaces or from equipment (Chajecka-Wierzchowska et al. 2016). Campista-Leon et al. (2021) had isolated E. olivae, E. saigonensis, E. hirae, E. avium, E. pseudoavium, E. termitis, and E. gallinarum species from twenty samples of seafood cocktails containing raw and/or cooked shrimp, scallop, ax callus (Atrina maura), octopus, oyster, fish, and vegetables. They emphasized that enterococci can be found as a result of faecal contamination or cross contamination at the time of preparation in seafood. Chajecka-Wierzchowska et al. (2017) reported that E. faecalis (62.9%), E. faecium (28.6%), E. casseliflavus (5.7%) and E. gallinarum (2.9%) species isolated from retail shrimps. Boss et al. (2016) found the enteroccal distribution as E. faecalis (n = 55), E. faecium (n = 6), Enterococcus casseliflavus (n = 8) Enterococcus gilvus (n = 6), Enterococcus thailandicus (n = 5), Enterococcus hirae (n = 4), Enterococcus mundtii (n = 3), Enterococcus phoenoculicola (n = 2), Enterococcus avium (n = 1), Enterococcus malodoratus (n = 1) isolated from salmon (n = 11), pangasius (shark catfish; n = 12), shrimp (n = 11), and oysters (n = 10). They mentioned that enterococci might be an indicator of faecal pollution of the aquaculture environment or could be acquired during processing. Although dominant species are different from the mentioned research, similarly it was thought that the sources of enterococcal microbiota might be transferred from human or animal pollution of the marine environment where hunting the seafood samples as well as during processing and handling of foods. Besides this, it should be considered that the persistence of certain strains of enterococci in fish intestine, sediment and submerged aquatic vegetation was not directly related to pollution events (Badgley et al. 2010).
Enterococcal load and species diversity were found higher in raw seafood samples than processed types in this study. It was thought that the raw seafood samples might be contaminated during fish evisceration and environmental sources during processing and handling by humans as mentioned by Hammad et al. (2014). However, the processing technologies such as smoking and canning may reduce more the bacterial load of foods than salting and freezing because of applied high process temperature in addition to antimicrobial and water activity reducing characteristics of smoking. So, that’s the reason of low prevalence of enterococci species in smoked and canned seafood in comparison with the raw, frozen and salted seafood samples. It was also thought that the low prevalence of enterococci in marinated seafood samples caused by relatively low resistance of this bacteria to high acidic conditions.
Hammad et al. (2014) reported that 96 enterococcal isolates were obtained from 90 (45%, 90/200) ready-to-eat raw fish samples. E. faecalis species were predominantly identified, followed by E. faecium and E. casseliflavus as well as E. gallinarum, E. raffinosus, E. phoeniculicola, E. gilvus, and E. saccharolyticus. They also mentioned that seafood products could be a reservoir for unusual Enterococcus spp. such as E. phoeniculicola,. E. raffinosus, E. saccharolyticus, and E. gilvus.
Igbinosa and Beshiru found that the prevalence of enterococci was 8.19% (59/720) in ready-to-eat shrimps samples. They identified mostly E. faecalis and E. faecium species from the samples following E. gallinarum, E. casseliflavus, E. durans, and E. hirae. Several studies have reported that seafood cross-contamination may develop as a result of food preparation or processing. Accordingly, Sánchez Valenzuela et al. (2010) isolated 24 enterococcal strains from uncooked fish fillets, fish intestine, clams and molluscs samples, and all strains were identified as E. faecium. They also mentioned that seafoods may be contaminated by enterococci during fish evisceration from fish intestine, and during handling and processing from the environment of the plant.
Antibiotic resistance profiles of enterococcal isolates
The results of the study showed that there was a high percentage (> 50%) of streptomycin, erytromycin, and tetracycline resistance along with gentamycine, chloramphenicol, and vancomycin resistance in the enterococcal strains isolated from the raw and processed seafood samples. In all the isolates, the streptomycin resistance prevalence had the highest ratio. Except streptomycin resistance, the highest antibiotic-resistance distribution was seen in E. casseliflavus for erythromycin, in E. durans, and E. casseliflavus for chloramphenicol, in E. faecalis for tetracycline, in E. durans for vancomycin, and in E. casseliflavus for gentamycin (Table 3).
The results also indicated that enterococcal strains isolated from seafood samples were found as commonly resistant against tested antibiotics. The multidrug resistance characteristics of the isolates demonstrated by Multiple Antibiotic Resistance (MAR) index and it was indicated the strain number that showed antibiotic resistance in the risk zone and showed the isolates obtained from the environment which exposed to overused antibiotics (Paul et al. 1997) (Fig. 2).
Fig. 2.
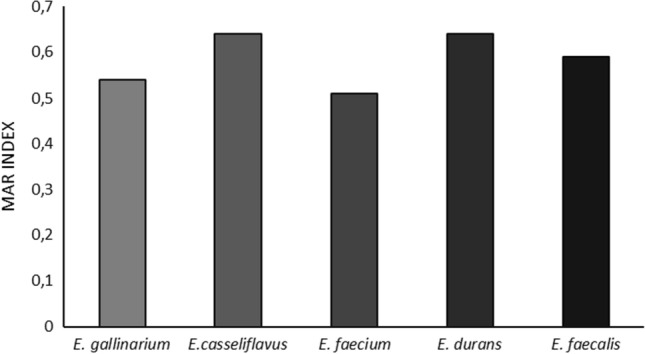
Multiple antibiotic resistance (MAR) indices of enterococcal isolates
All of the enterococcal isolates were determined as intermediate- or high-level resistant against at least one of the tested antibiotics. According to the antibiotic resistance profiles of the isolates; 59% of the isolates were resistant to three and more antibiotics, 19.4% were resistant to five and more antibiotics, and 2.8% were resistant to all of the antibiotics, whereas 55.5% of the MAR isolates were resistant to vancomycin. All of the enterococcal isolates exhibited multiple antibiotic resistance with the average MAR index (Fig. 2).
There were limited reports of antibiotic-resistant enterococcal strains isolated from seafood. Ben Said et al. (2017) found that E. faecalis, E. faecium and E. hirae strains of enterococci isolated from fish and shellfish in the Tunisian region were resistant to streptomycin, tetracycline, and erythromycin, as well as to ciprofloxacin, gentamycin, and chloramphenicol. Valenzuela et al. (2010) reported that most of the enterococci strains isolated from uncooked mollusca, fish, and fish fillets were resistant to nitrofurantoin, erythromycin, and rifampicin. Hammad et al. (2014) reported that clindamycin, erythromycin, kanamycin, gentamycin, streptomycin, and tetracycline resistance were detected in enterococcal isolates from raw fish samples.
In Turkey, Savaşan et al. (2008) reported that streptomycin-, gentamicin-, and ciprofloxacin-resistant E. faecalis strains were found as prevalent in fishes from the Aegean Region. Despite its low proportion, the presence of a vancomycin-resistance in E. faecalis strains, the fish samples must be considered as a source of vancomycin-resistant enterococci.
The rate of the MAR index (0.200) differentiates the low and high risk. If the rate is between 0.200–0.250, it means a very risky phase in which there are equal chances that the MAR may fall in the high-risk and low-risk phases (Krumperman 1983). The MAR is accepted as a good instrument for risk assessment. The MAR of the enterococcal strains isolated from seafood was reported in certain studies. Igbinosa and Beshiru (2019) determined that 73.1%, 100%, and 44.4% of enterococcal strains as MAR were isolated from sauced, boiled, and smoked shrimp samples, respectively. Citterio et al. (2017) also reported that 24.2% of enterococcal strains (E. casseliflavus, E. faecium, and E. hirae) isolated from clam were found as the MAR. These strains were found as resistant to up to 5 antibiotics, and the resistance profile detected more frequently were ampicillin-streptomycin-erythromycin and ampicillin-gentamicin-levofloxacin. They mentioned that coastal marine areas were contaminated with enteric bacteria at various levels, by both humans and animals, generally because of insufficient sewage treatment and wastewater flow. The high filtration rates of clams lead to the accumulation of small environmental pollution particles such as bacteria, including enterococci. Therefore, this environment may be a source for horizontal gene transfer and the emergence of multidrug-resistant strains.
Antibiotic resistance and virulence genes in enterococcal isolates
The phenotypic vancomycin resistance was detected in some E. gallinarum, E. casseliflavus, and E. durans isolates (Table 3). The vanA and/or vanB genes were not found in the vancomycin-resistant isolates of enterococci.
The tetM gene was detected in the fifty-nine tetracycline-resistant isolates of enterococci, whereas the ermB gene was found in the fifty-seven erythromycin-resistant isolates of enterococci. The thirty-eight aminoglycosides-resistant isolates of enterococci harbored the aac(6’)-aph(2’’)-la gene. It was determined that the tetM and ermB genes were mostly found in the E. casseliflavus, E. faecalis, and E. gallinarum strains isolated from the sardinella, squid, and red mullet samples. Besides, the aac(6’)-aph(2’’)-la gene was detected in the E. gallinarum, E. casseliflavus, E. faecalis, and E. durans strains isolated from the Mediterranean horse mackerel, sardinella, shrimp, and squid samples. Erythromycin-, tetracycline-, and aminoglycosides-resistant characteristics were usually found in the E. gallinarum and E. casseliflavus strains isolated from sardinella in phenotype and genotype.
The virulence genes agg2, gelE, cylM, cylB, and cylA were detected in the Enterococcus strains. Some isolates carried the gelE (43.58%) and agg2 (39.31%) genes which are important in pathogenesis (Fig. 3). Two of E. durans (horse mackerel and red mullet), one of E. casseliflavus (red mullet), E. faecalis (sardinella), and E. faecium (sardinella) strains were found to carry all the tested virulence genes (agg2, gelE, cylM, cylB, cylA).
Fig. 3.
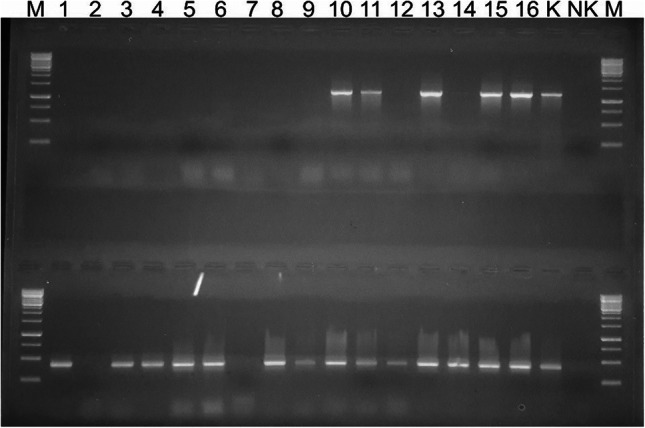
Amplification of virulence genes agg2 (upper) and gelE (below) by PCR. (Product size for agg2; 1210 base pair and for gelE; 419 base pair, M; marker (Thermo Fisher Scientific), the numbers 10, 11, 13, 15 and16 were positive isolates for agg2 and the numbers 1–16 except 2 and 7 were positive isolates for gelE, negative control; NK, positive control; K)
The researchers mentioned that acquired vanA and vanB genes were detected mostly in E. faecalis and E. faecium species. Vancomycin resistance may also be seen intrinsically in certain rare species of enterococci, resulting in low-level vancomycin-resistant strains. The vanC ligase genes, which encode this type of resistance, are not acquired or transferable and are specific for Enterococcus casseliflavus, Enterococcus gallinarum, Enterococcus flavescens, and Enterococcus mundtii (Toye et al. 1997). It is a need to investigate the presence of vanC gene and susceptibility to teicoplanin in the vancomycin-resistant E. casseliflavus and E. gallinarum isolates in the study. Enterococci enhanced and acquired resistance genes on plasmids or transposons from other bacteria that provided an increasing level of resistance to enterococci. It is possible that the antibiotic-resistant fecal bacteria of urban sewage, animal wastewater, or fish farming discharged into the sea might have transferred their antibiotic-resistant genes to fish microflora (Valenzuela et al. 2010).
There is limited information regarding the virulence characteristics of seafood-borne enterococci. Valenzuela et al. (2010) reported that hemolysin/cytolysin virulence genes were not found in any of the enterococci isolated from uncooked aquaculture consisting of mollusk, fish, and fish fillet. Hammad et al. (2014) reported that E. faecium isolates from seafood had gelE virulence gene. Migaw et al. (2014) reported that clyA virulence gene was detected only in one enterococcal isolate from the fish gastrointestinal tract. Muñoz-Atienza et al. (2013) mentioned that enterococcal virulence genes were found more frequently in E. faecalis (95%) isolated from seafood and that the strains harbored at least one of the most frequent virulence genes such as efaAfs, gelE, and agg.
Conclusions
This is the first study in terms of the distribution of enterococcal microbiota at species level isolated from raw or processed seafoods that are hunted and consumed in Turkey and showing that enterococcal isolates from seafoods may carry some virulence genes (agg2 and gelE) as well as high antibiotic resistance profile (tetM, ermB and aac(6')-aph(2'')-la genes). At this point, considering that enterococci can have pathogenicity potential at isolate level and transfer their antibiotic resistance properties to other bacteria, the consumption of these products may pose a serious risk in terms of public health and food safety. The way to produce and consume healthy and safe raw and processed seafood products will be possible by ensuring the hygienic safety of the aquatic environments where these products are hunted, as well as by reducing the unconscious use of antibiotics in the triangle of humans, animals and the environment, and thus limiting the antibiotic resistance rate of the bacteria in the ecosystem microbiota.
Acknowledgements
This research was supported by The Scientific and Technological Research Council of Turkey (Project no. 215O374). The authors are also grateful to Prof. Dr. Gülşen Altuğ from the Department of Marine Biology, Faculty of Aquatic Sciences, İstanbul University, Turkey, for supplying the VITEK 2 Compact System.
Authors' contributions
All authors have participated the experimental and writing parts of the study. Spesifically, Dr. Mine Çardak, Dr. Sine Özmen Toğay, Onur Karaalioğlu and Dr. Ufuk Bağcı were included in the study about sampling, isolation, identification and determination of antibiotic resistance characteristics of enterococcal isolates. Dr. Mustafa Ay and Dr. Özlem Erol Tınaztepe worked out the molecular experiments for screening of antibiotic resistance and virulence genes in enterococcal strains.
Funding
This research was supported by The Scientific and Technological Research Council of Turkey (Project no. 215O374).
Data availability
The data of the study are available and transparent.
Declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Consent for publication
All authors have approved the manuscript and all are aware of the submission to JFST.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mine Çardak, Email: mine_bio98@hotmail.com.
Sine Özmen Toğay, Email: sineozmen@gmail.com, Email: sinetogay@uludag.edu.tr.
Mustafa Ay, Email: may@comu.edu.tr.
Onur Karaalioğlu, Email: karaaliogluonur94@gmail.com.
Özlem Erol, Email: ozlemerolbio@gmail.com.
Ufuk Bağcı, Email: ufukbagci@gmail.com.
References
- Abele-Horn M, Stoy K, Frosch M, Reinert RR. Comparative evaluation of a new Vitek 2 system for identification and antimicrobial susceptibility testing of Streptococcus pneumoniae. Eur J Clin Microbiol. 2006;25:55–57. doi: 10.1007/s10096-005-0071-1. [DOI] [PubMed] [Google Scholar]
- Aslam M, Diarra MS, Checkley S, Bohaychuk V, Masson L. Characterization of antimicrobial resistance and virulence genes in Enterococcus spp. isolated from retail meats in Alberta. Canada Int J Food Microbiol. 2012;156:222–230. doi: 10.1016/j.ijfoodmicro.2012.03.026. [DOI] [PubMed] [Google Scholar]
- Badgley BD, Nayak BS, Harwood VJ. The importance of sediment and submerged aquatic vegetation as potential habitats for persistent strains of enterococci in a subtropical watershed. Water Res. 2010;44(20):5857–5866. doi: 10.1016/j.watres.2010.07.005. [DOI] [PubMed] [Google Scholar]
- Ben Said L, Hamdaoui M, Klibi A, Ben Slama K, Torres C, Klibi N. Diversity of species and antibiotic resistance in enterococci isolated from seafood in Tunisia. Annal Microbiol. 2017;67:135–141. doi: 10.1007/s13213-016-1246-y. [DOI] [Google Scholar]
- Boss R, Overesch G, Baumgartner A. Antimicrobial resistance of Escherichia coli, enterococci, Pseudomonas aeruginosa, and staphylococcus aureus from raw fish and seafood imported into Switzerland. J Food Prot. 2016;79(7):1240–1246. doi: 10.4315/0362-028X.JFP-15-463. [DOI] [PubMed] [Google Scholar]
- Byappanahalli MN, Nevers MB, Korajkic A, Staley ZR, Harwood VJ. Enterococci in the environment. Microbiol Mol Biol R. 2012;76(4):685–706. doi: 10.1128/MMBR.00023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campista-León S, Rivera-Serrano BV, Garcia-Guerrero JT, Peinado-Guevara LI. Phylogenetic characterization and multidrug resistance of bacteria isolated from seafood cocktails. Arch Microbiol. 2021;203:3317–3330. doi: 10.1007/s00203-021-02319-1. [DOI] [PubMed] [Google Scholar]
- Chajecka-Wierzchowska W, Zadernowska A, Łaniewska-Trokenheim Ł. Virulence factors of Enterococcus spp. presented in food. LWT-Food Sci Technol. 2017;75:670–676. doi: 10.1016/j.lwt.2016.10.026. [DOI] [Google Scholar]
- Chajecka-Wierzchowska W, Zadernowska A, Łaniewska-Trokenheim Ł. Virulence factors, antimicrobial resistance and biofilm formation in Enterococcus spp. isolated from retail shrimps. LWT-Food Sci Technol. 2016;69:117–122. doi: 10.1016/j.lwt.2016.01.034. [DOI] [Google Scholar]
- Citterio B, Pasquaroli S, Mangiaterra G, Vignaroli C, Di Sante L, Leoni F, Chierichetti S, Ottaviani D, Rocchi M, Biavasco F. Venus clam (Chamelea gallina): a reservoir of multidrug-resistant enterococci. Food Control. 2017;82:184–189. doi: 10.1016/j.foodcont.2017.06.022. [DOI] [Google Scholar]
- Clewell DB, Weaver KE, Dunny GM, Coque TM, Francia MV, Hayes F (2014) Extrachromosomal and mobile elements in enterococci: transmission, maintenance, and epidemiology. In: Gilmore MS, Clewell DB, Ike Y (ed). Enterococci: from commensals to leading causes of drug resistant ınfection, [Internet]. Boston: Massachusetts Eye and Ear Infirmary; 2014-. Available from: https://www.ncbi.nlm.nih.gov/books/NBK190430/ [PubMed]
- Clinical and Laboratory Standards Institute (CLSI), Performance Standards for Antimicrobial Susceptibility Testing, 2016 M100S, M02-A12, M07-A10, and M11-A8. 26th Edition.
- Di Cesare A, Vignaroli C, Luna GM, Pasquaroli S, Biavasco F. Antibiotic-resistant enterococci in seawater and sediments from a coastal fish farm. Microb Drug Resist. 2012;18(5):502–509. doi: 10.1089/mdr.2011.0204. [DOI] [PubMed] [Google Scholar]
- Di Cesare A, Pasquaroli S, Vignaroli C, Paroncini P, Luna GM, Manso E, Biavasco F. Environmental vs. clinical enterococci. Env Microbiol Rep. 2014;6:184–190. doi: 10.1111/1758-2229.12125. [DOI] [PubMed] [Google Scholar]
- Eaton TJ, Gasson MJ. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl Environ Microb. 2001;67(4):1628–1635. doi: 10.1128/AEM.67.4.1628-1635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis-Iversen J, Seyfarth AM, Korsgaard H, Bortolaia V, Munck N, Dalsgaard A. Antimicrobial resistant E. coli and enterococci in pangasius fillets and prawns in Danish retail imported from Asia. Food Control. 2020;114:106958. doi: 10.1016/j.foodcont.2019.106958. [DOI] [Google Scholar]
- EUCAST, 2015 European Committee on Antimicrobial Susceptibility Testing, Breakpoint tables for interpretation of MICs and zone diameters, Version 7.1, valid from 2017-03-10
- Facklam RR, da Gloria M, Lucia SC, Teixeira M (2002) History, taxonomy, biochemical characteristics, and antibiotic susceptibility testing of enterococci (Pages: 1–54) In: Gilmore MS, Clewell DB, Courvalin P, Dunny GM, Murray BE, Rice LB (ed), The Enterococci: Pathogenesis, Molecular Biology, and Antibiotic Resistance, ISBN:9781119738732, Online ISBN:9781683672302. Copyright © 2002 ASM Press. 10.1128/9781555817923
- Gassiep I, Armstrong M, Van Havre Z, Schlebusch S, McCormack J, Griffin P. Acute vancomycin-resistant enterococcal bacteraemia outbreak analysis in haematology patients: a case-control study. Healthc Infect. 2015;20(3–4):115–123. doi: 10.1071/HI15013. [DOI] [Google Scholar]
- Hammad AH, Shimamoto T. Genetic characterization of antibiotic resistance and virulence factors in Enterococcus spp. from Japanese retail ready-to-eat raw fish. Food Microbiol. 2014;38:62–66. doi: 10.1016/j.fm.2013.08.010. [DOI] [PubMed] [Google Scholar]
- Hancock LE, Gilmore MS (2006) Pathogenicity of enterococci. In: Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Rood JI (ed), Gram-Positive Pathogens, 2nd edition. ASM Press. pp. 299–311.
- Igbinosa EO, Beshiru A. Antimicrobial resistance, virulence determinants, and biofilm formation of Enterococcus species from ready-to-eat seafood. Front Microbiol. 2019;10:728. doi: 10.3389/fmicb.2019.00728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto M, Ayers T, Mahon BE, Swerdlow DL. Epidemiology of seafood-associated infections in the United States. Clin Microbiol Rev. 2010;23(2):399–411. doi: 10.1128/CMR.00059-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klibi N, Said LB, Jouini A, Slama KB, López M, Sallem RB, Boudabous A, Torres C. Species distribution, antibiotic resistance and virulence traits in enterococci from meat in Tunisia. Meat Sci. 2013;93(3):675–680. doi: 10.1016/j.meatsci.2012.11.020. [DOI] [PubMed] [Google Scholar]
- Korajkic A, McMinn BR, Staley ZR, Ahmed W, Harwood VJ. Antibiotic-resistant Enterococcus species in marine habitats: a review. Curr Opin Environ Sci Health. 2020;16:92–100. doi: 10.1016/j.coesh.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumperman PH. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl Environ Microbiol. 1983;46(1):165–170. doi: 10.1128/AEM.46.1.165-170.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebreton F, Willems RJL, Gilmore MS. Enterococcus diversity, origins in nature, and gut colonization. In: Gilmore MS, Clewell DB, Ike Y, Shankar N, editors. Enterococci: from commensals to leading causes of drug resistant infection [Internet] Boston: Massachusetts Eye and Ear Infirmary; 2014. [PubMed] [Google Scholar]
- Migaw S, Ghrairi T, Belguesmia Y, Choiset Y, Berjeaud JM, Chobert JM, Hani K, Haertle T. Diversity of bacteriocinogenic lactic acid bacteria isolated from Mediterranean fish viscera. World J Microbiol Biotechno. 2014;30:1207–1217. doi: 10.1007/s11274-013-1535-6. [DOI] [PubMed] [Google Scholar]
- Muñoz-Atienza E, Gómez-Sala B, Araújo C, Campanero C, del Campo R, Hernández PE, Herranz C, Cintas LM. Antimicrobial activity, antibiotic susceptibility and virulence factors of lactic acid bacteria of aquatic origin intended for use as probiotics in aquaculture. BMC Microbiol. 2013;13:15. doi: 10.1186/1471-2180-13-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquaroli S, Di Cesare A, Vignaroli C, Conti G, Citterio B, Biavasco F. Erythromycin- and copper-resistant Enterococcus hirae from marine sediment and co-transfer of erm(B) and tcrB to human Enterococcus faecalis. Diagn Microbiol Infect Dis. 2014;80:26–28. doi: 10.1016/j.diagmicrobio.2014.06.002. [DOI] [PubMed] [Google Scholar]
- Paul S, Bezbaruah RL, Roy MK, Ghosh AC. Multiple antibiotic resistance (MAR) index and its reversion in Pseudomonas aeruginosa. Letters in Applied Microbiology. 1997;24(3):169–71. doi: 10.1046/j.1472-765X.1997.00364.x. [DOI] [PubMed] [Google Scholar]
- Pinto AL, Fernandes M, Pinto C, Albano H, Castilho F, Teixeira P, Gibbs PA. Characterization of anti-Listeria bacteriocins isolated from shellfish: potential antimicrobials to control non-fermented seafood. Int J Food Microbiol. 2009;129:50–58. doi: 10.1016/j.ijfoodmicro.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Psoni L, Kotzamanides C, Andrighetto C, Lombardi A, Tzanetakis N, Litopoulou-Tzanetaki E. Genotypic and phenotypic heterogeneity in Enterococcus isolates from Batzos, a raw goat milk cheese. Int J Food Microbiol. 2006;109(1–2):109–120. doi: 10.1016/j.ijfoodmicro.2006.01.027. [DOI] [PubMed] [Google Scholar]
- Reviriego C, Eaton T, Martín R, Jiménez E, Fernández L, Gasson MJ, Rodríguez JM. Screening of virulence determinants in Enterococcus faecium strains isolated from breast milk. J Hum Lact. 2005;21(2):131–138. doi: 10.1177/0890334405275394. [DOI] [PubMed] [Google Scholar]
- Savaşan S, Kaya O, Kirkan Ş, Çiftci A. Balık kökenli Enterococcus faecalis suşlarının antibiyotik dirençlilikleri. Ankara Üniv Vet Fak Derg. 2008;55:107–110. [Google Scholar]
- Toye B, Shymanski J, Bobrowska M, Woods W, Ramotar K. Clinical and epidemiological significance of enterococci intrinsically resistant to vancomycin (possessing the vanC genotype) J Clin Microbiol. 1997;35(12):3166–3170. doi: 10.1128/jcm.35.12.3166-3170.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela AS, Benomar N, Abriouel H, Cañamero MM, Gálvez A. Isolation and identification of Enterococcus faecium from seafoods: antimicrobial resistance and production of bacteriocin-like substances. Food Microbiol. 2010;27:955–961. doi: 10.1016/j.fm.2010.05.033. [DOI] [PubMed] [Google Scholar]
- Vignaroli C, Pasquaroli S, Citterio B, Di Cesare A, Mangiaterra G, Fattorini D, Biavasco F. Antibiotic and heavy metal resistance in enterococci from coastal marine sediment. Environ Poll. 2018;237:406–413. doi: 10.1016/j.envpol.2018.02.073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data of the study are available and transparent.