Abstract
Chemoresistance is the inevitable outcome of chemotherapy for epithelial ovarian carcinoma (EOC), and its mechanism is still not fully understood. This study explored the role of ribosomal protein L23 (RPL23) in cisplatin resistance of EOC. WGCNA based on TCGA and GEO was used to screen and analyze target genes related to EOC chemotherapy sensitivity. Clinical samples of cisplatin resistance were collected to detect the expression of target genes. Cisplatin resistance was induced in EOC cell lines A2780 and SKOV3. The cell abilities of invasion, migration and adhesion were observed. Western blotting was used to detect protein expressions. Bioinformatics analysis showed that RPL23 may be related to EOC chemotherapy sensitivity, and was highly expressed in clinical samples and cell lines of cisplatin-resistant. After A2780 and SKOV3 were resistant to cisplatin, the inhibitory abilities of therapeutic dose of cisplatin on their invasion, migration and adhesion were significantly attenuated, and N-cadherin and vimentin were significantly up-regulated while E-cadherin was significantly down-regulated. However, above phenomena were significantly reversed after RPL23 knockdown. Taken together, the overexpressed RPL23 may lead to platinum resistance by inducing epithelial-mesenchymal transition (EMT) in EOC. Targeting knockdown RPL23 would restore the sensitivity of EOC cells to cisplatin by inhibiting EMT, suggesting that RPL23 is a potential therapeutic target for EOC after platinum resistance.
Keywords: Epithelial ovarian carcinoma, Platinum resistance, RPL23, Epithelial-mesenchymal transition
Introduction
Epithelial ovarian carcinoma (EOC) accounts for 85–90% of ovarian malignancies and is the seventh most common female cancer in the world, with a 5-year survival rate of 46% (Lheureux et al. 2019). Due to its strong concealment, most of patients are in advanced stage at the time of diagnosis (Lheureux et al. 2019). The conventional treatment is surgery, followed by platinum or taxol chemotherapy, but it will eventually develop into chemoresistance (Freimund et al. 2018). The mechanism of EOC chemoresistance is complex, and optimized treatment after resistance is the focus of research to improve the survival of EOC patients. In the past 40 years, platinum has been the cornerstone of the treatment of EOC. The research on the mechanism of platinum resistance has made some progress, but it is still not completely clear (Bv et al. 2018).
Ribosome biogenesis is necessary for the process of cell growth and proliferation. Therefore, the accelerated biogenesis of ribosomes can be seen in cancer cells to meet the needs of their expansion. The ribosomal protein is the key to ribosomal biogenesis and may be related to chemoresistance (Hassouni et al. 2018). Ribosomal protein L23 (RPL23), a member of ribosomal protein family, is a negative regulator of apoptosis. It is thought to be involved in a variety of tumor chemoresistance processes (Shi et al. 2004; Wu et al. 2012). A recent study based on clinical samples and artificial intelligence prediction model confirmed RPL23 as a prognostic biomarker for advanced-stage high-grade serous ovarian carcinoma (HGSC) (Kang et al. 2021a). In the preliminary work, we predicted that RPL23 might be related to EOC chemoresistance with the help of weighted correlation network analysis (WGCNA) based on The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO). Clinical samples were collected and found that EOC patients with cisplatin resistant had a higher expression level of RPL23 compared with cisplatin sensitive patients. This study demonstrated the possibility of RPL23 as a target for EOC treatment after platinum resistance.
Materials and methods
Data acquisition
The gene expression and clinical data of ovarian carcinoma patients were downloaded from the TCGA database (https://cancergenome.nih.gov). The expression data of 419 patients were obtained, of which 400 patients had overall survival data. The GEO database GSE26712 data set consisted of 185 ovarian carcinoma patients and 10 normal tissue samples (https://ftp.ncbi.nlm.nih.gov/geo/series/GSE26nnn/GSE26712/matrix/).
Identification of differentially expressed genes and survival analysis
The differentially expressed genes were identified by GEO2R. Survival analysis of differentially expressed genes was carried out using R package “survival”, “stats”, “rms”.
Construction of weighted gene co-expression network analysis (WGCNA)
In our research, the TCGA dataset, consisting of 400 ovarian carcinoma patients, has detailed overall survival information and is taken for constructing WGCNA. We can correlate module signature genes with drug sensitivity data of ovarian carcinoma patients (1 = sensitivity, 0 = resistance). The data matrix of gene expression of TCGA ovarian carcinoma patients was constructed, and the first 50% of the coding protein genes with the largest variance in tumor samples were taken as the input data set of subsequent WGCNA. The WGCNA software package in R/Bioconductor software was used. The method of sample hierarchy clustering is used to detect and exclude outliers, and the soft threshold power is selected to realize the standard scale-free network. Further, the gene tree diagram and module identification are achieved through dynamic tree cutting (minimum module size = 100) by constructing adjacency and topological overlap matrix (TOM) and calculating its corresponding difference (1-Tom). Finally, the correlation between module eigengene and clinical phenotype of ovarian carcinoma were calculated and highly similar modules with the dissimilarity of < 0.25 were merged by clustering module eigengenes. Selection included further analysis of the modules most relevant to drug sensitivity traits to obtain gene significance (GS) and module membership (MM) to identify module genes associated with OV clinical characteristics.
Estimation of stromal and immune cells in malignant tumor tissues
Previous research had indicated that the infiltrating stromal and immune cells played a significant role in tumor development. Using the expression data TCGA samples, ESTIMATE achieves score estimation of malignant tissue matrix and immune cells. Our research calculated the ESTIMATE, stromal, and immune scores in OV by adopting the ESTIMATE algorithm from (https://bioinformatics.mdanderson.org/estimate/) (Yoshihara et al. 2013).
Sample acquisition and immunohistochemical staining
This study has been registered on the Chinese Clinical Trial Registry (Registration No.: ChiCTR2100043896). EOC tissues paraffin blocks of cisplatin sensitive [progression-free interval > 6 months (Bv et al. 2018)] and cisplatin resistant (progression-free interval < 6 months) were borrowed from hospital biobank according to an agreement approved by Human Investigation Ethic Committee of the Second Affiliated Hospital of Nanchang University (2021011). The donation procedures were in compliance with the laws of the People's Republic of China, and the specimens obtained have been registered with the relevant government departments of Jiangxi Province. Paraffin sections were prepared and immunohistochemical staining was performed with primary antibody RPL23 (1:500, Proteintech, Rosemont, IL, USA). The paracancerous tissues were stained as control. The staining results were analyzed blindly by two pathologists.
Cell culture
Human ovarian endometrioid adenocarcinoma cell line A2780, human ovarian serous cystadenocarcinoma cell line SKOV3 and human ovarian surface epithelium cell line IOSE80 were all purchased from Shanghai EK-Bioscience Biotechnology Co., Ltd (Shanghai, CHN). The A2780 cells were cultured in DMEM containing 10% fetal bovine serum (FBS). The SKOV3 cells were cultured in McCoy's 5A containing 10% FBS. The IOSE80 cells were cultured in RPMI1640 containing 10% FBS. All cells were cultured at 37 °C in a humidified atmosphere of 5% CO2, fed every 3 days, and sub-cultured at 70–80% confluency. Starting from 0.1 μg/mL, the intervention dose of cisplatin (DDP, Hansoh Pharma, Lianyungang, JS, CHN) was gradually increased to induce the establishment of cisplatin-resistant cells A2780/DDP and SKOV3/DDP, and a cisplatin-containing (0.2 μg/mL) culture system was used to maintain their drug resistance.
siRNA interference
The siRNA oligonucleotides including si-314 (5′-UGCAGGAGUCAUAGUGAACAATT-3′), si-119 (5′-CCUGUAUAUCAUCUCCGUGAATT-3′), and si-198 (5′-GCCACAGUCAAGAAAGGCAAATT-3′), which targeting RPL23 gene, were designed and synthesised by Sangon Biotech (Shanghai, CHN). Lipofectamine™ 3000 Transfection Reagent (Thermo Fisher Scientific, MA, USA) was used to transfect siRNAs into cancer cells as previous (Liu et al. 2021) and the effect of gene knockdown was measured by western blotting and quantitative polymerase chain reaction (qPCR).
Cell counting kit-8 (CCK8) assay
Cell viability was detected using CCK8 (Glpbio, Montclair, CA, USA). Briefly, the cells were inoculated into 96 well plates with 1 × 104 cells per well. After 24 h of culture, different doses of DDP were added. After 24 h, 10 μL of CCK8 reaction solution was add to each well. 4 h later, the OD value of each well was detected at the wavelength of 450 nm and the inhibition rate and half-inhibitory concentration (IC50) value were calculated accordingly.
Cell invasion assay
Briefly, 1 × 105 cells were plated in the upper chamber of transwell chamber (Corning, Corning, NY, USA) which was coated with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) cultured with medium without FBS, while 0.5 mL medium containing 10% FBS was added to the lower chamber. After culturing at 37 °C for 48 h, the cells in lower chamber were washed with phosphate buffered saline (PBS) and fixed with 4% paraformaldehyde for 20 min. Then cells were stained with 0.25% crystal violet (Solarbio, Beijing, CHN), which was dissolved in 20% methanol, for 20–45 min, and washed again with PBS for twice. Light microscope was used to observe and counted for 3 random fields per well. Cell counts are expressed as the mean number of cells per field of view.
Wound healing assay
Briefly, the cells were inoculated into 6-well plates. When they grew until 80–90% confluence, sterilized 200 μL pipette tip was used to scratch the cells vertically to generate wounding across the cell monolayer. Add the therapeutic dose of cisplatin, and observe the migration of cells to the wound 24 h later. Three areas were randomly selected for each well to take images, and Image J v2.1.4.7 software (National Institutes of Health, Bethesda, MD, USA) was used for measurement and analysis.
Cell adhesion assay
As previous report (Liu et al. 2016), the cells transfected siRNA or intervened with cisplatin were inoculated into a 24-well plate at a density of 5 × 104 cells/well. After 1 h, the unattached cells were removed, and the attached cells were counted after trypsinization. The data were presented as a percentage of the attached cells compared to total cells.
Western blotting
Proteins were extracted from cells by using radioimmunoprecipitation assay (RIPA) lysis buffer and size fractionated by SDS polyacrylamide gel electrophoresis. Membranes were incubated with target antibodies of RPL23 (1:1000, Proteintech), E-cadherin (1:1000, Proteintech), N-cadherin (1:1000, Proteintech), Vimentin (1:1000, Proteintech) and β-actin (1:1000, Proteintech) at 4 °C overnight. Then horseradish peroxidase-conjugated secondary antibodies were used to react with membranes for 2 h at room temperature. Finally, the immune complexes were visualized by enhanced chemiluminescence and the band intensity was measured quantitatively by the Image J software (USA).
QPCR assay
Total RNA was extracted from cells using TRIzol reagent (ThermoFisher, Waltham, MA, USA). Premier Primer 5.0 software was used to design the oligonucleotide primer sequences of RPL23 (F: TCCTCTGGTGCGAAATTCCG, R: CGTCCCTTGATCCCCTTCAC). β-Actin was used as an internal control. The synthesized first‑strand cDNA samples were subjected to qPCR using a SYBR Green PCR Master Mix (Toyobo Bio‑Technology, Shanghai, CHN) and the qPCR reaction was performed on an ABI Prism 7700 Sequence Detector (ThermoFisher). Threshold cycle (Ct) values were determined and relative fold changes in mRNA expression were calculated using the formula 2−ΔΔCt.
Statistical analysis
In the section of bioinformatics analysis, the statistical analyses were performed by using R software (version 3.6.1). The “limma” R package was used to perform the differentially expressed analysis. In addition, the "survival" and "survminer" packages were used to perform Cox proportional hazard regression analysis, Kaplan–Meier survival analysis, and C index calculation. The "rms" package was used to build a nomogram that combines the risk scoring model with clinical factors. In the section of in vitro experiments, data was presented as means ± S.E.M and analysed by SPSS version 20.0 (IBM Corp., Armonk, NY, USA) for variance homogeneity test and one-way analysis of variance. P < 0.05 was considered to indicate a statistically significant difference.
Results
Identification of key genes for EOC drug sensitivity diagnosis
WGCNA is a biological analysis method for analyzing the expression patterns of multiple genes in different samples, which can present genes with the same expression pattern in clusters or modules (Beckerman et al. 2017). Genes with the same biological function are clustered in the same module, based on which the relationship between the module and sample characteristics can be analyzed (Tang et al. 2018; Rosen et al. 2011). In the analysis of TCGA ovarian carcinoma data, TCGA.25.1870.01 was excluded as an outlier sample, thus a total of 399 samples with survival data were included in WGCNA (Fig. 1A). The strong gray value (scale-free R2 = 0.950) was chosen as the soft threshold to realize the scale-free network (Fig. 1B, C). After removing the gray module by merging dynamic tree cutting, 6 gene expression modules were identified by consensus (Fig. 1D). The heat map mapped the TOM of the 400 genes selected in the analysis, and the results showed that each module was independently validated (Fig. 1E). Subsequently, it was found that the brown module was the key module with the highest correlation with drug sensitive information of OV treatment (R2 = 0.18, P = 3 × 10–4, Fig. 1F), and the scatter plots of GS and MMIN brown modules for drug sensitive information of OV patients were plotted (correlation = 0.38, P = 3.5 × 10–26).
Fig. 1.
Weighted co-expression network construction and identification of the key module related to drug susceptibility. A Hierarchical dendogram of the top 50% samples of standard deviations in TCGA database. The bottom shows the clinical characteristics of drug sensitivity traits. B-C Analysis of scale-free fit index and the mean connectivity for various soft-thresholding powers. Testing the scale free topology when β = 3. D Hierarchical clustering dendogram of genes with dissimilarity based on topological overlap. Modules are the branches of the clustering tree. E The heatmap describes the TOM among selected 400 genes in WGCNA, and darker colour represents higher overlap and lighter colour corresponds to lower overlap. The gene dendrogram and module assignment are shown along the left side and the top. F Correlation between module characteristic genes and drug sensitivity traits. Each row corresponds to a module characteristic gene and each cell contains correlation and P- values. The brown module with the highest correlation with drug sensitivity traits is selected. G Volcano plots of gene expression profiles in GSE26712. Orange/blue symbols classify the upregulated/downregulated genes according to the criteria: log2FC > 1 and P-value < 0.05. H Volcano map of gene survival analysis in TCGA dataset. Orange/blue symbols classify the risk/protective factors genes according to the criteria: HR > 1 and P-value < 0.05. I Venn diagram shows the intersection of DEGs (log2FC > 1 and P-value < 0.05), survivor-related risk factor genes (HR > 1 and P-value < 0.05), and genes with brown module membership < 0. J Scatter plot of genes in brown module. The vertical line represents cutoff of module membership = 0.5, and the horizontal line represents cutoff of gene significances for drug sensitivity = 0.2. Genes on upper right contains RPL23
Up-regulated differential expression analysis (log2 fold change > 1 and P < 0.05) was performed using GEO data set GSE26712 (Fig. 1G), and univariate Cox proportional hazard regression analysis (P < 0.05 and HR > 1) was performed using TCGA data set to identify risk genes associated with survival (Fig. 1H). There was a total of 16 overlapping genes between the two, and then the overlapping genes were taken with the brown genes, with the cut-off value GS. drug sensitivity < − 0.1, RPL23 was screened out (Fig. 1I, J).
Clinical data and immune infiltration analysis of EOC
We combined clinical factors with RPL23 expression to construct a Nomogram prognostic model. Nome diagram model and calibration curve are shown in Fig. 2A–C. We calculated the C index to evaluate the effect of the Nom graph model. The C-index predicting 5-year OS for EOC patients is 0.405, indicating that this model is a valuable prognostic indicator. Kaplan–Meier analysis of high expression and low expression subgroups of RPL23 (cut off by median) predicting OS in were significantly down-regulated EOC patients based on the TCGA database, the results showed that higher expression predicted poor prognosis (P = 0.03) (Fig. 2D).
Fig. 2.
Relationship among key gene, clinical practice and immune infiltration. A A nomogram model combining RPL23 signature with the clinical factors for predicting the 3,5-year OS of OV patients. B, C The nomogram calibration curve to evaluate the prediction of 3,5-year OS of OV patients. The C index of this model was also calculated. D Kaplan–Meier analysis of high expression and low expression subgroups of RPL23 (cut off by median) predicting OS in OV patients based on the TCGA database. E, F The relationship between key gene and immune infiltration in TCGA database. PRL23 mRNA expression level and Immune Score. Tumor Purity distribution is different. P-values were obtained by Student's t-test
We used TCGA sample expression data to obtain estimates of stromal cells and immune cells in malignant tumor tissues. Using median RPL23 expression as a cutoff value divided the patients into two groups and t test results show that the patients with high RPL23 expression had lower immune scores (P = 2 × 10–5) and higher tumor purity (P = 3.06 × 10–5) (Fig. 2E, F).
RPL23 is highly expressed in cisplatin-resistant EOC
A total of 90 samples of EOC patients were collected. The correlation analysis between RPL23 expression and clinicopathological parameters of the patients is shown in Table 1, including 37 cases of cisplatin resistance and 51 cases of cisplatin sensitivity (2 patients did not receive chemotherapy). Our data showed that high RPL23 expression was closely related to higher pathological grade, positive lymphatic metastasis and cisplatin resistance, whereas no relation was detected with age, tumor diameter or CA125 level. Immunohistochemistry was used to detect the expression of RPL23 in EOC tissues. As shown in Fig. 3A and B, in 37 cisplatin resistant patients, RPL23 was highly expressed in 30 cases and low expressed in 7 cases, and in 51 cisplatin sensitive patients, RPL23 was highly expressed in 13 cases and low expressed in 38 cases, showed a significant difference between the two groups (P < 0.05). Western blotting was used to detect the expression of RPL23 protein in cells of IOSE80, A2780 and SKOV3. As shown in Fig. 3C, significantly up-regulated RPL23 was detected in both A2780 and SKOV3 compared with IOSE80 (P < 0.05). The expression of RPL23 was further up-regulated after cisplatin resistance in A2780 and SKOV3 (P < 0.05 vs. A2780 or SKOV3). The transcriptional level of RPL23 mRNA was detected by qPCR and consistent results were obtained (Fig. 3D, E).
Table 1.
Correlation analysis between RPL23 protein level and the clinicopathological parameters in EOC
| Characteristic | Patients (n) | RPL23 expression | P-value | |
|---|---|---|---|---|
| High | Low | |||
| Age (years) | 0.1527 | |||
| < 50 | 24 | 9 | 15 | |
| > 50 | 66 | 36 | 30 | |
| Tumour diameter | 0.7253 | |||
| < 5 cm | 9 | 4 | 5 | |
| > 5 cm | 81 | 41 | 40 | |
| Pathological stage | 0.4384 | |||
| I-II | 19 | 8 | 11 | |
| III-IV | 71 | 37 | 34 | |
| Grade | < 0.0001 | |||
| G1 | 16 | 2 | 19 | |
| G2-G3 | 29 | 43 | 26 | |
| Lymphatic metastasis | < 0.0001 | |||
| Positive | 45 | 37 | 8 | |
| Negative | 45 | 8 | 37 | |
| CA123 (U/ml) | 0.8315 | |||
| < 500 | 51 | 25 | 26 | |
| 500 | 39 | 20 | 19 | |
| Resistance to cisplatin* | < 0.0001 | |||
| Resistant | 37 | 30 | 7 | |
| Sensitive | 51 | 13 | 38 | |
Fig. 3.
The expression level of RPL23 in cisplatin-resistant EOC. A The representative images of RPL23 immunohistochemistry in clinical samples. B Analysis of the proportion of low/high RPL23 in resistant/sensitive EOC of patients C The representative western blotting bands and its corresponding quantitative analysis histogram of RPL23 in cells of IOSE80, A2780, A2780/DDP, SKOV3/DDP and SKOV3. D, E The transcriptional level of RPL23 mRNA in cells of IOSE80, A2780, A2780/DDP, SKOV3/DDP and SKOV3. Values were expressed as the means ± standard error of the mean (n = 3 for each group). **P < 0.01; *P < 0.05
Effects of cisplatin resistance on cell behavior of EOC
After cisplatin resistance, the morphology of A2780 and SKOV3 cells became irregular significantly (Fig. 4A). The values of IC50 for cisplatin were also significantly increased compared with normal A2780 and SKOV3 cells (Fig. 4B). IC50 values of cisplatin on A2780 and A2780/DDP were 1.097 ± 0.013 and 7.024 ± 0.332 μg/mL, respectively (P < 0.05), while IC50 of cisplatin on SKOV3 and SKOV3/DDP were 1.396 ± 0.030 vs 6.256 ± 0.061 μg/mL, respectively (P < 0.05). Under the therapeutic dose of cisplatin, the abilities levels of attachment, migration and invasion of A2780/DDP or SKOV3/DDP were all significantly increased than those of normal A2780 and SKOV3 cells (Fig. 4C–E, P < 0.05). Western blotting was used to detect the expression levels of epithelial-mesenchymal transition (EMT)—related proteins in the cells. As shown in Fig. 4F, compared with the normal EOC cells, E-cadherin was significantly down-regulated in A2780/DDP and SKOV3/DDP, while N-cadherin and Vimentin were significantly up-regulated (P < 0.05).
Fig. 4.
Effects of cisplatin resistance on cell behavior of EOC. A Representative images of morphological changes of tumor cells after cisplatin induced drug resistance. B Changes of IC50 values in resistant and sensitive cells. C The representative images of cells of A2780, A2780/DDP, SKOV3/DDP and SKOV3 stained by crystal violet in the lower chamber indicating invasive changes. The bar chart at the right represents the count of cells passing through the matrix gel in each group. D The representative images of cells wound healing in sensitive and resistant groups indicating cell migration level and the comparation histogram of percent wound closure. E The comparation histogram of attachment assay. F The representative western blotting bands of E-cadherin, N-cadherin, Vimentin and β-actin in cells of A2780, A2780/DDP, SKOV3 and SKOV3/DDP and the corresponding quantitative analysis histograms, indicating the relationship between drug-resistance and EMT. Values were expressed as the means ± standard error of the mean (n = 3 for each group). **P < 0.01; *P < 0.05
Effects of RPL23 knockdown on cell behavior of cisplatin-resistant EOC
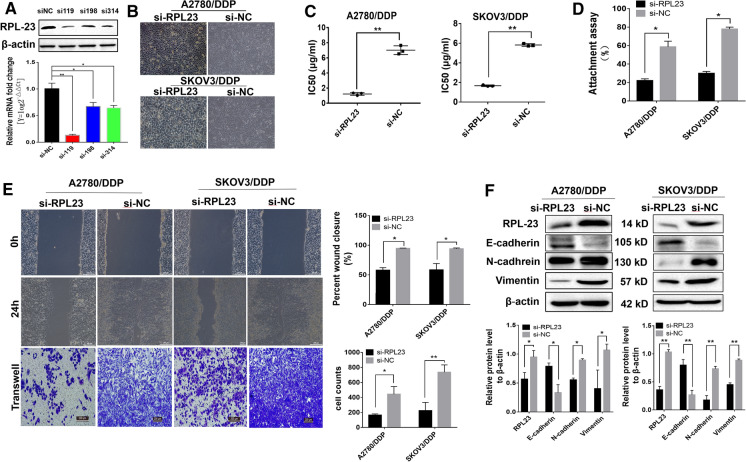
A total of three RPL23 siRNA sequences were designed and synthesized. After transfecting into A2780/DDP for 48 h, the expression of RPL23 in cells was detected by western blotting. The results showed that the interference effect of si119 was the most significant (Fig. 5A, P < 0.05 vs. si198 or si314), and was used in subsequent experiments. After RPL23 was knocked down, the morphology of most A2780/DDP and SKOV3/DDP cells were returned to normal (Fig. 5B). The sensitivity of A2780/DDP and SKOV3/DDP cells to the therapeutic dose of cisplatin increased with the IC50 values decreased obviously (Fig. 5C, P < 0.05 vs. siNC). The abilities of migration, invasion and adhesion of A2780/DDP and SKOV3/DDP cells decreased significantly (Fig. 5D and E, P < 0.05 vs. siNC). In additon, western blotting showed that E-cadherin was significantly up-regulated in RPL23 knockdown cells, while RPL23, N-cadherin and vimentin were significantly down-regulated (Fig. 5F, P < 0.05 vs. siNC).
Fig. 5.
Effects of RPL23 knockdown on cell behavior of cisplatin-resistant EOC. Short interfering RNAs (siRNAs) were used to deplete RPL23 gene expression in vitro, and the silencing effects of siRPL23 were evaluated by western blotting and qPCR. The representative western blotting bands of RPL23 and its mRNA transcription level after A2780/DDP and SKOV3/DDP transfected with siRNAs were showed in (A). B Cell morphological changes after RPL23 gene knockdown in cisplatin-resistant cells. C The comparation of IC50 values of cisplatin on cells of A2780/DDP and SKOV3/DDP after knocking down RPL23. D The comparation histogram of attachment assay. E The representative images of wound healing and crystal violet staining in each group after RPL23 knockdown in cisplatin-resistant cells and the corresponding comparation histograms. F The representative western blotting bands of RPL23, E-cadherin, N-cadherin, Vimentin and and β-actin in each group after RPL23 knockdown in cisplatin-resistant cells and the corresponding quantitative analysis histograms, indicating the relationship between siRNA and EMT. Values were expressed as the means ± standard error of the mean (n = 3 for each group). **P < 0.01; *P < 0.05
Discussion
Previous studies suggest that platinum resistance in ovarian carcinoma is related to p53 and genome-wide mutations, epigenetic changes, and dysfunctional DNA repair (Bv et al. 2018). Various factors work together to cause genome instability, allowing cancer cells to tolerate DNA damage caused by platinum and survive. However, the exact mechanism is still unclear. Ribosome biogenesis plays an important role in regulating cell growth and proliferation by controlling the translation of all proteins in the cells. Ribosomal protein is a ubiquitous RNA tuberculosis protein. It is a component of ribosomal subunits and has a variety of auxiliary extra-ribosomal functions (Kang et al. 2021b). RPL23 is a protein component of the 60S large ribosomal subunit, which can negatively regulate apoptosis by inhibiting the transcriptional activation of the cell cycle inhibitory proteins P15Ink4b and P21Cip1 induced by Myc-associated zinc-finger protein1 (Qi et al. 2017). Some studies believe that the binding of RPL23 and MDM2 inhibits MDM2 from stabilizing the tumor suppressor gene p53, and exerts a tumor suppressor effect (Meng et al. 2016). However, overexpression of RPL23 has been found in many tumors. Meeren et al. observed that RPL23 was generally positive in lymphoma tissues (Meeren et al. 2019). Kang et al. found that the expression of RPL23 in recurrent HGSC patients was 1.6 times higher than that in non-recurrent patients (Kang et al. 2021a). The authors believe that the high expression of RPL23 may lead to the recurrence of HGSC, resulting in a worse prognosis in these patients. Newton et al. analyzed patients with advanced HGSC and showed that RPL23 is one of the genes that are significantly changed in non-responders after first-line chemotherapy, which suggests that RPL23 may be related to chemotherapy sensitivity (Newton et al. 2010).
In this study, with the help of bioinformatics analysis, we identified the close relationship between RPL23 and EOC chemotherapy sensitivity. Based on clinical samples, we have observed that RPL23 was significantly highly expressed in platinum-resistant EOC samples. The abnormally high expression of RPL23 was also observed in platinum resistance cell lines A2780 and SKOV3. These results suggest that RPL23 as a potential biomarker is related to platinum resistance in EOC. By knocking down the expression of RPL23 in cells of A2780/DDP and SKOV3/DDP, we found that the platinum sensitivity of A2780 or SKOV3 was significantly restored, which is reflected in effective inhibition of invasion, migration and adhesion of cells. These results show that RPL23 is a therapeutic target for platinum resistant EOC. Targeting inhibition the expression of RPL23 can improve platinum resistance, which provides an idea for the treatment strategy of EOC after platinum resistance.
EMT is the basic process of embryonic development and tissue repair, which can transform polarized epithelial cells into a mesenchymal phenotype to enhance cell viability (Loh et al. 2019). EMT plays an important regulatory role in the process of cancer, participates in the invasion and metastasis of cancer cells, enhances the cancer stemness of tumor cells, and improves the immune clearance resistance of cancer cells to the host (Lu and Kang 2019). Studies have shown that EMT also contributes to the chemoresistance of a variety of cancers. Early studies have determined the relationship between EMT and drug resistance by evaluating the drug sensitivity of cancer cell lines with altered expression of EMT and transcription factors (Lu and Kang 2019). Fischer and Zheng et al. used genetically engineered mouse models to clarify that the chemoresistance of primary and metastatic tumor cells of breast and prostate were enhanced in an EMT-dependent manner (Fischer et al. 2015; Zheng et al. 2015). Cisplatin can induce EMT by inducing autophagy, nuclear factor kappa B, cell-adhesion molecules and ataxia telangiectasia mutation to reduce the sensitivity of tumor cells, enhance their migration and metastasis ability, and promote the generation of drug resistance (Ashrafizadeh et al. 2020). The positive status of EMT is considered to be a characteristic of malignant ovarian carcinoma and may be the cause of platinum resistance in ovarian carcinoma (Bozhkova and Poryazova-Markova 2019; Zhang et al. 2020). Deng et al. found abundant cells with EMT characteristics in cisplatin-resistant EOC cells. By reversing EMT, drug-resistant cells can be made sensitive to cisplatin (Deng et al. 2019). In this study, we observed that E-cadherin was significantly down-regulated, while N-cadherin and Vimentin were significantly up-regulated in EOC cells after cisplatin resistance, suggesting that EMT was over-activated. Interestingly, the expression of E-cadherin increased while the expression of N-cadherin and Vimentin decreased in cells of A2780/DDP or SKOV3/DDP after knocking down the expression of RPL23, suggesting that the EMT process was effectively inhibited. Above results suggest that overexpression of RPL23 leads to platinum resistance in EOC cells may be achieved by inducing EMT.
In conclusion, in this study, we found that RPL23 may be a biomarker of platinum resistance in EOC. Overexpressed RPL23 could induce EMT, which may be the direct cause of platinum resistance in EOC. Knocking down the expression of RPL23 could restore the sensitivity of EOC cells to cisplatin by inhibiting EMT, suggesting that RPL23 is a potential therapeutic target for EOC after platinum resistance. Further in vivo experiments should be carried out to provide more sufficient evidences.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability
All data generated or analyzed during this study are included in this published article, or available upon reasonable request from the corresponding author.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yujuan Liu, Email: liuyujuan123@163.com.
Yan Jinlong, Email: yjl19880608@126.com.
References
- Ashrafizadeh M, Zarrabi A, Hushmandi K, Kalantari M, Mohammadinejad R, Javaheri T, et al. Association of the epithelial-mesenchymal transition (EMT) with cisplatin resistance. Int J Mol Sci. 2020;21:4002. doi: 10.3390/ijms21114002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckerman P, Qiu C, Park J, Ledo N, Ko Y-A, Park A-SD, et al. Human kidney tubule-specific gene expression based dissection of chronic kidney disease traits. EBioMedicine. 2017;24:267–276. doi: 10.1016/j.ebiom.2017.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozhkova DM, Poryazova-Markova EG. The epithelial-mesenchymal transition, E-cadherin and tumor progression in ovarian serous tumors. Folia Med (plovdiv) 2019;61:296–302. doi: 10.2478/folmed-2018-0082. [DOI] [PubMed] [Google Scholar]
- Bv Z, Tang D, Bowden NA. Biomarkers of platinum resistance in ovarian cancer: what can we use to improve treatment. Endocr Relat Cancer. 2018;25:R303–R318. doi: 10.1530/ERC-17-0336. [DOI] [PubMed] [Google Scholar]
- Deng J, Bai X, Feng X, Ni J, Beretov J, Graham P, et al. Inhibition of PI3K/Akt/mTOR signaling pathway alleviates ovarian cancer chemoresistance through reversing epithelial-mesenchymal transition and decreasing cancer stem cell marker expression. BMC Cancer. 2019;19:618. doi: 10.1186/s12885-019-5824-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong STC, et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527:472–476. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freimund AE, Beach JA, Christie EL, Bowtell DDL. Mechanisms of drug resistance in high-grade serous ovarian cancer. Hematol Oncol Clin N Am. 2018;32:983–996. doi: 10.1016/j.hoc.2018.07.007. [DOI] [PubMed] [Google Scholar]
- Hassouni BE, Sarkisjan D, Vos JC, Giovannetti E, Peters GJ. Targeting the ribosome biogenesis key molecule fibrillarin to avoid chemoresistance. Curr Med Chem. 2018;26:6020–6032. doi: 10.2174/0929867326666181203133332. [DOI] [PubMed] [Google Scholar]
- Kang H, Choi MC, Kim S, Jeong J-Y, Kwon A-Y, Kim T-H, et al. USP19 and RPL23 as candidate prognostic markers for advanced-stage high-grade serous ovarian carcinoma. Cancers (basel) 2021;13:3976. doi: 10.3390/cancers13163976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Brajanovski N, Chan KT, Xuan J, Pearson RB, Sanij E. Ribosomal proteins and human diseases: molecular mechanisms and targeted therapy. Signal Transduct Target Ther. 2021;6:323. doi: 10.1038/s41392-021-00728-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lheureux S, Gourley C, Vergote I, Oza AM. Epithelial ovarian cancer. Lancet. 2019;393:1240–1253. doi: 10.1016/S0140-6736(18)32552-2. [DOI] [PubMed] [Google Scholar]
- Liu Y, Li Y, Wang R, Qin S, Liu J, Su F, et al. MiR-130a-3p regulates cell migration and invasion via inhibition of Smad4 in gemcitabine resistant hepatoma cells. J Exp Clin Cancer Res. 2016;35:19. doi: 10.1186/s13046-016-0296-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Li W, Luo J, Wu Y, Xu Y, Chen T, et al. Cysteine-rich intestinal protein 1 served as an epithelial ovarian cancer marker via promoting Wnt/ β-catenin-mediated EMT and tumour metastasis. Dis Markers. 2021;2021:3566749. doi: 10.1155/2021/3566749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh C-Y, Chai JY, Tang TF, Wong WF, Sethi G, Shanmugam MK, et al. The E-cadherin and N-cadherin switch in epithelial-to-mesenchymal transition: signaling, therapeutic implications, and challenges. Cells. 2019;8:1118. doi: 10.3390/cells8101118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Kang Y. Epithelial-mesenchymal plasticity in cancer progression and metastasis. Dev Cell. 2019;49:361–374. doi: 10.1016/j.devcel.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeren LE, Kluiver J, Rutgers B, Alsagoor Y, Kluin PM, Avd B, et al. A super-SILAC based proteomics analysis of diffuse large B-cell lymphoma-NOS patient samples to identify new proteins that discriminate GCB and non-GCB lymphomas. PLoS ONE. 2019;14:e0223260. doi: 10.1371/journal.pone.0223260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Tackmann NR, Liu S, Yang J, Dong J, Wu C, et al. RPL23 links oncogenic RAS signaling to p53-mediated tumor suppression. Cancer Res. 2016;76:5030–5039. doi: 10.1158/0008-5472.CAN-15-3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton TR, Parsons PG, Lincoln DJ, Cummings MC, Wyld DK, Webb PM, et al. Expression profiling correlates with treatment response in women with advanced serous epithelial ovarian cancer. Int J Cancer. 2010;119:875–883. doi: 10.1002/ijc.21823. [DOI] [PubMed] [Google Scholar]
- Qi Y, Li X, Chang C, Xu F, He Q, Zhao Y, et al. Ribosomal protein L23 negatively regulates cellular apoptosis via the RPL23/Miz-1/c-Myc circuit in higher-risk myelodysplastic syndrome. Rep. 2017;7:2323. doi: 10.1038/s41598-017-02403-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen EY, Wexler EM, Versano R, Coppola G, Gao F, Winden KD, et al. Functional genomic analyses identify pathways dysregulated by progranulin deficiency, implicating Wnt signaling. Neuron. 2011;71:1030–1042. doi: 10.1016/j.neuron.2011.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi O, Zhai H, Wang X, Han Z, Liu C, Lan M, et al. Ribosomal proteins S13 and L23 promote multidrug resistance in gastric cancer cells by suppressing drug-induced apoptosis. Exp Cell Res. 2004;296:337–346. doi: 10.1016/j.yexcr.2004.02.009. [DOI] [PubMed] [Google Scholar]
- Tang J, Kong D, Cui Q, Wang K, Zhang D, Gong Y, et al. Prognostic genes of breast cancer identified by gene co-expression network analysis. Front Oncol. 2018;8:374. doi: 10.3389/fonc.2018.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Li X, Xu F, Chang C, He Q, Zhang Z, et al. Over-expression of RPL23 in myelodysplastic syndromes is associated with apoptosis resistance of CD34+ cells and predicts poor prognosis and distinct response to CHG chemotherapy or decitabine. Ann Hematol. 2012;91:1547–1554. doi: 10.1007/s00277-012-1486-2. [DOI] [PubMed] [Google Scholar]
- Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Cui J-Y, Gao H-F, Yu H, Gao F-F, Chen J-L, et al. Cancer-associated fibroblasts induce epithelial-mesenchymal transition and cisplatin resistance in ovarian cancer via CXCL12/CXCR4 axis. Future Oncol. 2020;16:2619–2633. doi: 10.2217/fon-2020-0095. [DOI] [PubMed] [Google Scholar]
- Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article, or available upon reasonable request from the corresponding author.