Abstract
In vivo expression technology (IVET) was employed to study colonization of Phytophthora parasitica by a biological control bacterium, Pseudomonas putida 06909, based on a new selection marker. The pyrB gene, which encodes aspartate transcarbamoylase, an enzyme used for pyrimidine biosynthesis, was cloned from P. putida 06909. A pyrB-disrupted mutant did not grow in pyrimidine-deficient media unless it was complemented with pyrBC′ behind an active promoter. Thirty clones obtained from P. putida 06909 that were expressed on fungal hyphae but not on culture media were isolated by IVET based on the promoterless transcriptional fusion between pyrBC′ and lacZ. Nineteen of these clones were induced during late-stage bacterial growth in vitro, while 11 of the clones were expressed only when they were inoculated onto fungal hyphae. Restriction analysis of these 11 clones revealed that there were five unique clones. Sequence analyses of three of the five unique clones showed that the 3′ ends of the clones fused to pyrB were similar to genes encoding diacylglycerol kinase (DAGK), bacterial ABC transporters, and outer membrane porins. The sequences of the two other clones were not similar to the sequences of any of the genes in the database used. A LuxR family response regulator was found upstream of DAGK, and a LysR family response regulator was found upstream of the ABC transporter. The location of the inducible promoter of two clones suggested that DAGK and the ABC transporter are induced and may play a role in colonization of the fungus P. parasitica by P. putida 06909.
Pseudomonas putida 06909 suppresses populations of the root-rotting fungus Phytophthora parasitica in the citrus rhizosphere (50). This strain does not produce antibiotics in culture media. Mutagenesis has shown that both adherence to fungal hyphae and siderophore production by P. putida 06909 are important in the inhibition of P. parasitica (54). However, little else is known about the interaction between P. putida 06909 and P. parasitica during hyphal colonization. Continued interest in the use of this strain for biological control of citrus root rot (48) has prompted a more thorough analysis of this bacterium-fungus interaction.
We used in vivo expression technology (IVET) (25) to study the colonization of P. parasitica by P. putida 06909. IVET is a strategy for selecting cloned bacterial genes that are specifically induced in vivo. The in vivo-induced genes can be identified by their ability to express a promoterless selection marker gene that is essential for survival in vivo. Expression in vitro can then be monitored by studying the expression of another promoterless reporter gene cloned downstream from the selection marker as a transcriptional fusion. The IVET strategy has advantages over traditional mutagenesis techniques since there is positive selection for genes that are specifically induced by environmental cues in vivo and the procedure does not disrupt genes that may be essential in vivo.
In the original IVET system used for Salmonella typhimurium, a purine requirement for bacterial survival or antibiotic resistance was used as the selection criterion during bacterial infection of the host cells (25, 26). Other genes essential for bacterial survival were also used as selection markers to use the IVET system during various bacterial infections of animal cells (6, 13, 14, 24). In our study, since the target fungus, P. parasitica, is very sensitive to several antibiotics, antibiotic resistance genes were not useful as selection markers. In addition, we wanted to use the IVET strategy in rhizosphere and soil studies, in which antibiotic selection would not be feasible. A growth factor requirement, the pyrimidine requirement for bacterial growth, was used as a selection marker in our Phytophthora-Pseudomonas system.
The pyrB gene encodes aspartate transcarbamoylase (ATCase), an enzyme that is essential for biosynthesis of pyrimidines, and pyrB mutants cannot grow in pyrimidine-deficient culture media (42). Because the supply of pyrimidines is limited in many natural environments, the pyrB gene could provide selection in those environments. lacZ was used as a reporter gene for in vitro expression studies (9). To our knowledge, this is the first study in which the nondisruptive IVET strategy was used to identify bacterial genes induced during interaction with a plant-pathogenic fungus.
Our hypothesis is that bacterial genes that are specifically induced during fungal colonization are directly involved in the colonization of fungal hyphae and that some of these genes are important in the biocontrol activity of P. putida. These genes may ultimately be useful in novel biocontrol approaches. In this paper we describe application of the IVET strategy based on the pyrimidine requirement for bacterial growth of a mutant strain and identification of several genes specifically induced during colonization of P. parasitica by P. putida 06909. Five chromosomal loci of P. putida 06909 that are specifically expressed during colonization of P. parasitica were isolated. The sequences of two of the chromosomal loci of P. putida 06909 and potential promoter regions in these two loci induced during colonization of fungal hyphae were determined in this study.
MATERIALS AND METHODS
Microorganisms, plasmids, and culture conditions.
All of the organisms and plasmids used in this study are listed in Table 1. Escherichia coli cultures were grown at 37°C on Luria-Bertani agar or in Luria-Bertani broth supplemented with the appropriate antibiotics (27). The following antibiotic concentrations were used for E. coli strains: tetracycline, 15 μg/ml; kanamycin, 50 μg/ml; and ampicillin, 100 μg/ml. P. putida strains were grown at 28°C on mannitol-glutamate medium (MG medium) (19) supplemented with yeast extract (0.25 g/liter) (MGY medium) or in MGY broth. The following antibiotic concentrations were used in MGY medium: tetracycline, 20 μg/ml; kanamycin, 30 μg/ml; and ampicillin, 200 μg/ml. MG medium or MGY medium was supplemented with uracil (50 μg/ml) for growth of pyrimidine auxotrophs of P. putida 06909. When it was necessary for bacterial growth in rich medium, P. putida strains were cultured on Pseudomonas agar F under the same conditions. P. parasitica M191 was grown at 28°C on V8C agar or in V8C broth (30). Plasmids pHRP309 and pHSK828 were kindly provided by Caroline Harwood, University of Iowa, Iowa City, and by Noel Keen, University of California, Riverside, respectively.
TABLE 1.
Microorganisms and plasmids used
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F−recA1 Δ(lacZYA-argF)U169 hsdR17 thi-1 gyrA66 supE44 endA1 relA1 φ80dlac Δ(lacZ)M15 | 40 |
| HB101 | F−hsdA20 (r− m−) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 supE44 λ− | 2 |
| P. putida strains | ||
| 06909 | Wild type; Ampr | 54 |
| 06909u1 | pyrB; uracil auxotroph; Ampr Kanr | This study |
| 06909u2 | pyrB; uracil auxotroph; Ampr Kanr | This study |
| P. parasitica M191 | 54 | |
| Plasmids | ||
| pUC119 | Apr; cloning vector | 55 |
| pRK415 | Tcr; RK2-derived broad-host-range cloning vector | 20 |
| pHRP309 | Gmr; RSF1010-based broad-host-range transcriptional fusion vector | 34 |
| pHSK728 | Apr Smr; Tn7 delivery plasmid | 44 |
| pHSK828 | Apr Smr; pHSK728 derivative; a lux operon in pHSK728 was replaced with an ice nucleation gene | Unpublished data |
| pDSK509 | Kmr; RSF1010-derived broad-host-range cloning vector | 20 |
| pRK2013 | Kmr; mobilization helper | 10 |
| pDAC41 | Apr; pyrB cloned in pUC119 at HincII site | This study |
| pDAC42 | Apr Kmr; 1.2-kb kanamycin cassette inserted in pDAC41 | This study |
| pDAC43 | Apr; pyrBC′ cloned in pUC119 at HincII site | This study |
| pDAC54 | Apr; Φ(pyrBC′-lacZ) cloned in pHSK828 | This study |
| pRIV11 | Tcr; pRK415 carrying a 1.8-kb SphI-EcoRI fragment (blunt ended) from pDAC42 containing the pyrB::km fragment | This study |
| pRIV13 | Tcr; pRK415 carrying a 2.4-kb HindIII-BamHI fragment from pDAC43 containing pyrBC′ | This study |
| pRIV16 | Tcr; pRK415 carrying a 6.1-kb HindIII-BamHI fragment from pDAC54 containing a promoterless fusion; Φ(pyrBC′-lacZ) in opposite orientation compared to lac promoter | This study |
| pRIV16C | Tcr; pRK415 carrying a promoterless fusion; Φ(pyrBC′-lacZ) in same orientation as lac promoter | This study |
| pRIVA1 | Tcr; pRIV16 carrying a constitutive promoter | This study |
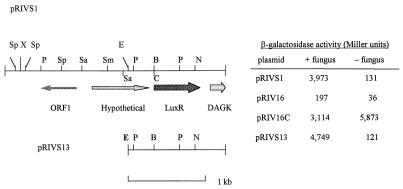
| pRIVS1 | Tcr; pRIV16 carrying a clone expressed on fungal plates | This study |
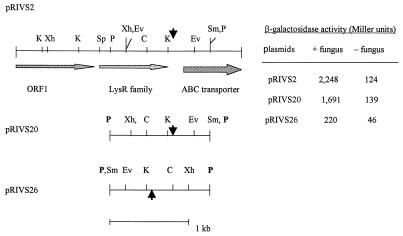
| pRIVS2 | Tcr; pRIV16 carrying a clone expressed on fungal plates | This study |
| pRIVS3 | Tcr; pRIV16 carrying a clone expressed on fungal plates | This study |
| pRIVS4 | Tcr; pRIV16 carrying a clone expressed on fungal plates | This study |
| pRIVS5 | Tcr; pRIV16 carrying a clone expressed on fungal plates | This study |
| pRIVS13 | Tcr; 3.9-kb EcoRI fragment from pRIVS1 transferred into pRIV16 | This study |
| pRIVS20 | Tcr; pRIV16 carrying a 1.4-kb PstI fragment from pRIVS2; the fragment has the same orientation as Φ(pyrBC′-lacZ) of pRIV16 | This study |
| pRIVS26 | Tcr; pRIV16 carrying a 1.4-kb PstI fragment from pRIVS2; the fragment has the opposite orientation compared to Φ(pyrBC′-lacZ) of pRIV16 | This study |
| pUIVS1 | Apr; pUC119 carrying a 5.4-kb KpnI fragment from pRIVS1 | This study |
| pUIVS11 | Apr; pUC119 carrying a 1.4-kb EcoRI-HindIII fragment from pUIVS1 | This study |
| pUIVS2 | Apr; pUC119 carrying a 5.5-kb EcoRI fragment from pRIVS2 | This study |
| pUIVS21 | Apr; pUC119 carrying a 2-kb SphI fragment from pUIVS2 | This study |
| pUIVS3 | Apr; pUC119 carrying a 4.2-kb EcoRI fragment from pRIVS5 | This study |
| pUIVS4 | Apr; pUC119 carrying a 2-kb EcoRI-SphI fragment from pRIVS4 | This study |
| pUIVS5 | Apr; pUC119 carrying a 4.5-kb EcoRI fragment from pRIVS5 | This study |
Ampr, chromosomal ampicillin resistance; Kanr, chromosomal kanamycin resistance; Apr, ampicillin resistance; Tcr, tetracycline resistance; Smr, streptomycin resistance; Gmr, gentamicin resistance; Kmr, kanamycin resistance.
Recombinant DNA techniques and DNA sequencing.
Plasmid preparation, restriction endonuclease cleavage, ligation, agarose gel electrophoresis, and other standard recombinant DNA techniques were carried out as described previously (40). Total DNA of P. putida wild-type strain 06909 and its mutants was prepared by the standard cetyltrimethylammonium bromide method described elsewhere (1). Southern blot analysis of digested genomic DNA from P. putida strains was performed with nylon membranes (MSI, Westboro, Mass.). A 1.2-kb AvaII fragment carrying a kanamycin resistance cassette and a 700-bp PstI fragment of pyrB were used as probes to confirm the correct replacement of the pyrB locus of P. putida 06909 with pyrB::km. Probes were labeled with a digoxigenin-dUTP DNA labeling kit (Boehringer GmbH, Mannheim, Germany) and were detected with a chemiluminescent substrate, disodium 3-(4-methoxyspiro-{1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.13,7]decan}-4-yl)phenyl phosphate (Boehringer), as described in the manufacturer's instructions.
DNA sequencing was carried out at the DNA Sequencing Facility of the University of California, Berkeley. When automated sequencing was not successful for a segment containing a palindromic sequence, DNA sequencing was carried out manually by using the chain termination method (41) and a Sequenase version 2.0 DNA sequencing kit (U.S. Biochemicals, Braunschweig, Germany). The DNA sequences were analyzed by using a software package obtained from the Genetics Computer Group of the University of Wisconsin and the Blast programs provided by the National Center for Biotechnology Information. The primers used in this study were synthesized commercially (Genosys Biotechnologies, Inc., The Woodlands, Tex.).
Cloning of pyrB and marker exchange mutagenesis.
Degenerate primers D1 (5′-dATGACGCCNATHGAYGCNAAR-3′) and D2 (5′-TCAYTGNGCRTTYTCYTGRTC-3′), based on the PyrB sequence from P. putida PPN and Pseudomonas aeruginosa PAO (42), were designed to amplify pyrB from P. putida 06909 (Y = T or C; N = A, T, G, or C; R = A or G; H = A, T, or C). DNA was amplified in a 100-μl (total volume) reaction mixture which contained 10 μl of Vent DNA polymerase 10× buffer (New England BioLabs, Inc., Beverly, Mass.), each deoxynucleoside triphosphate (New England BioLabs) at a concentration of 200 μM, 5 mM MgSO4, 5% dimethyl sulfoxide, each primer at a concentration of 1 μM, and 0.5 U of Vent DNA polymerase. The template DNA added was 200 ng of total P. putida 06909 DNA. PCR amplification was performed with an automated thermocycler (EasyCycler Series; Ericomp, Inc., San Diego, Calif.) by using the following program: an initial DNA template denaturation step consisting of 95°C for 5 min; 30 cycles consisting of denaturation at 95°C for 1 min, annealing at 52°C for 30 s, and extension at 72°C for 1 min; and a final extension step consisting of 72°C for 5 min. The 1-kb PCR product was cloned at the HincII site of pUC119 to generate pDAC41. The identity of the PCR product was confirmed by DNA sequencing. The PCR product was used for marker exchange mutagenesis. To disrupt the wild-type pyrB gene, a marker exchange plasmid was constructed as follows. The 400-bp internal HincII fragment of pDAC41 was deleted, and a 1.2-kb AvaII fragment carrying a kanamycin resistance cassette from pDSK509 (20) was filled with the Klenow DNA polymerase and ligated at the HincII site of pDAC41. The resulting plasmid was pDAC42. A 1.8-kb SphI-EcoRI fragment carrying pyrB in pDAC42 with the kanamycin resistance cassette was filled with the Klenow DNA polymerase and ligated into a blunt-end EcoRI site of a broad-host-range plasmid, pRK415 (20), to generate plasmid pRIV11. The plasmid was introduced into P. putida 06909 by triparental mating in which pRK2013 was used as a helper plasmid. Marker exchange mutagenesis was carried out as described previously (23). Plasmid pRK415 is unstable in the absence of selection, and a cycloserine enrichment culture was used to enhance recovery of homologous recombinants. The growth rates of the mutants and the wild-type strain were compared by monitoring A600 in MG broth supplemented with 50 μg of uracil per ml.
Complementation of the pyrB mutant.
To complement the pyrB mutation in P. putida, pyrBC′ was amplified from wild-type P. putida 06909 with one specific primer, primer P3, and one degenerate primer, primer D3. Primer P3 (5′-dTAGGAGAACCCCGCGATGAGCCGATCGACGCCAAG-3′) was designed based on the PCR product of pyrB, and primer D3 (5′-dTTACTCAGGCCCRTGRCAGACRTGNCCRTCNAC-3′) was designed based on the 3′ ends of pyrC′ of P. putida PPN and P. aeruginosa PAO (in the primer P3 sequence the boldface type indicates a conserved Shine-Dalgarno sequence, and the underlined portion is the translation start codon of the pyrB gene). The pyrC′ gene is a pyrC homolog downstream of pyrB that is required to form a functional ATCase in P. putida PPN, as previously described (42). The reaction mixture was the same as the reaction mixture used for pyrB amplification except that the concentration of each deoxynucleoside triphosphate was 400 μM and the concentration of MgSO4 was 6 mM. DNA amplification was performed by using the following program: an initial DNA template denaturation step consisting of 95°C for 5 min; 24 cycles consisting of denaturation at 95°C for 1 min, annealing at 70°C for 30 s, and extension at 72°C for 2 min 20 s; and a final extension step consisting of 72°C for 5 min. The 2.4-kb amplified fragment was cloned into pUC119 at the HincII site as a blunt-end fragment to obtain pDAC43. The 2.4-kb HindIII-EcoRI fragment in pDAC43, carrying promoterless pyrBC′, was subcloned behind the lac promoter of pRK415 to generate pRIV13.
Bacterial growth on fungal cultures.
To study the potential use of the pyrB gene as a selection marker for IVET, we compared growth of P. putida 06909u2 (pyrB) and growth of wild-type strain P. putida 06909 on V8C agar covered by fully grown P. parasitica. P. parasitica M191 was grown at 28°C on V8C agar plates until the plates were completely covered by the fungus. Wild-type P. putida and the pyrB mutant were grown in MG medium and MG medium supplemented with uracil and kanamycin, respectively, and then they were resuspended in sterile water. Each bacterial suspension was diluted with sterile water to obtain a concentration of 4 × 103 CFU/ml. A 1.5-ml portion of each dilution was applied to a fungal plate covered by P. parasitica. The plates were air dried briefly in a laminar flow hood and incubated at 28°C for 20 to 29 h. The bacteria were harvested from the fungal plates by adding 5 ml of sterile water to each plate, loosening the bacteria with a small glass rod, and removing the suspension with a pipette. The number of CFU per milliliter in each final suspension was determined by plating serial dilutions onto MG medium containing ampicillin for the wild-type strain, MG medium containing kanamycin and uracil for mutant strain 06909u2, and MG medium containing kanamycin and tetracycline for mutant strain 06909u2 carrying pRIV13.
Bacterial growth in the presence of P. parasitica was also studied by using small agar discs covered by the fungus. V8C agar discs containing actively growing P. parasitica were transferred to fresh V8C agar plates, and a bacterial inoculum was applied to the surface of each disc. Bacterial cultures were grown on MG medium and transferred with toothpicks. The plates were incubated overnight, and the fungal agar discs were dropped into 1-ml portions of sterile water to release the bacteria. The bacterial titer was determined by measuring the A600 of each final suspension. A fungal column assay was also used to determine whether the pyrB mutation affected the initial adhesion of P. putida 06909 to P. parasitica hyphae. Preparation of the fungal column and quantification of bacterial adhesion to fungal hyphae were performed as previously described (54).
Plasmid construction.
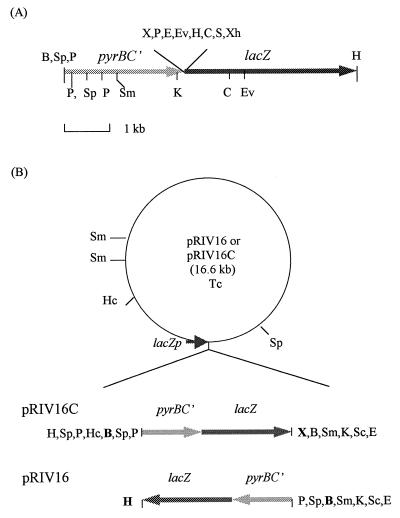
Broad-host-range plasmid pRIV16, a derivative of pRK415, was constructed and used as a basic plasmid to deliver the IVET construct to P. putida 06909u2 (Fig. 1). A 3.6-kb BamHI-HindIII fragment containing the promoterless lacZ gene was cut from pHRP309 and filled with the Klenow enzyme to generate blunt ends. This fragment was cloned into a blunt-end XhoI site of pHSK828, a derivative of pHSK728 (44). The lux operon of pHSK728 was replaced with an ice nucleation gene to create pHSK828 (unpublished data). A 2.4-kb HindIII-BamHI fragment carrying the promoterless pyrBC′ genes in pDAC43 was blunt ended with Klenow DNA polymerase and cloned into the SmaI site in front of the promoterless lacZ gene to generate pDAC54. Plasmid pDAC54 was first digested with BamHI and then partially digested with HindIII to obtain a 6.1-kb fragment containing a promoterless transcriptional fusion between pyrBC′ and lacZ. This fragment was cloned into the BamHI-HindIII site of pRK415 to generate pRIV16. In addition, this fragment was filled with the Klenow enzyme and cloned at the blunt-end XbaI site of pRK415 to generate pRIV16C. Plasmid pRIV16C was used as a positive control to check the expression of the transcriptional fusion in pyrB mutant strain 06909u2, since the promoterless transcriptional fusion was located behind the lac promoter, which is constitutive in pseudomonads. The nucleotide sequences at the 5′ end of pyrBC′, including polylinker sites in pRIV16, were confirmed by DNA sequencing.
FIG. 1.
Construction of IVET plasmids pRIV16 and pRIV16C. (A) Restriction map of a promoterless transcriptional fusion between pyrBC′ and lacZ. (B) pRIV16 and pRIV16C carry pyrBC′-lacZ in the reverse orientation. Promoterless pyrBC′-lacZ digested with BamHI and partially digested with HindIII was cloned into pRK415 to generate pRIV16 and was cloned into a XbaI site as a blunt-end ligation to generate pRIV16C. The restriction enzyme sites in the pyrBC′-lacZ genes are not shown for pRIV16 and pRIV16C. The SphI (Sp) and PstI (P) sites in the upstream region of pyrBC′ were derived from pUC119 in the subcloning process. The polylinker sites between pyrBC′ and lacZ were originally from pHSK728. The boldface BamHI (B), XbaI (X), and HindIII (H) sites are cloning sites in pRK415. The boldface BamHI (B) and XbaI (X) sites in pRIV16C were retained after blunt-end ligation. The enzyme sites located outside the boldface letters were derived from the polylinker of pRK415. Abbreviations: B, BamHI; C, ClaI; E, EcoRI; Ev, EcoRV; H, HindIII; Hc, HincII; K, KpnI; P, PstI; S, SalI; Sc, SacI; Sm, SmaI; Sp, SphI; X, XbaI; Xh, XhoI.
Isolation of clones expressed on P. parasitica.
A genomic library of P. putida 06909 was constructed in pRIV16 by using the standard recombinant DNA protocol (40). Briefly, total genomic DNA of P. putida 06909 was partially digested with Sau3AI and dephosphorylated with calf intestinal phosphatase to prevent self-ligation between small insert fragments. DNA fragments were fractionated in a sucrose density gradient (10 to 40% sucrose) for 24 h at 22,000 rpm. Fractions containing DNA fragments between 2 and 7 kb long were collected. The fragments were ligated into the unique BamHI site of pRIV16 to generate the library. The recombinant clones were introduced into pyrB mutant strain P. putida 06909u2 by triparental mating on yeast extract-dextrose-calcium carbonate agar (53) supplemented with 50 μg of uracil per ml. The transconjugants carrying recombinant clones were applied to V8C agar plates covered by P. parasitica at levels of 50,000 to 70,000 CFU per plate. The fungal plates were air dried briefly in a laminar flow hood and incubated for 24 h at 28°C. The cells were harvested with 2 ml of sterile water and plated onto MG agar supplemented with uracil, tetracycline, and kanamycin. The cells that grew on MG agar were resuspended, diluted with sterile water, and reapplied to V8C agar fungal plates at levels of 50,000 to 70,000 CFU per plate to enrich the growing cells. This process was repeated at least three times to enrich for positive clones. Finally, cells were spread onto MG agar supplemented with uracil, tetracycline, kanamycin, and 80 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) per ml. Expression of the promoterless transcriptional fusion, pyrBC′-lacZ in pRIV16, was expected to be dependent on expression of the upstream library insert DNA. A P. putida 06909u2 transconjugant which carried putative clones expressed specifically during colonization of P. parasitica should have grown in the presence of P. parasitica or in media containing uracil. When the isolates were grown on V8C agar supplemented with uracil in the absence of P. parasitica, the colonies should have been white on X-Gal-containing medium due to a lack of expression of the upstream gene. A total of 30 white colonies were selected, and 11 plasmids were isolated and used for further characterization. The remaining 19 colonies turned blue at a late stage of bacterial growth and therefore were not considered as specific in their expression patterns. The plasmids isolated were analyzed by determining the restriction enzyme digestion patterns obtained with EcoRI, BamHI, HindIII, SphI, KpnI, and XbaI. The plasmids were reintroduced into pyrB mutant strain 06909u2 by triparental mating. The transconjugants carrying selected plasmids were applied to fungal agar discs, as described above, to confirm that bacterial growth occurred in the presence of P. parasitica. Growth of the transconjugants in the absence of P. parasitica was also examined. P. putida 06909 genomic DNA fragments in induced clones pRIVS1, pRIVS2, pRIVS3, pRIVS4, and pRIVS5 were subcloned into pUC119 for sequencing. A 5.4-kb KpnI fragment from pRIVS1, a 5.5-kb EcoRI fragment from pRIVS2, a 4.2-kb EcoRI fragment from pRIVS3, a 4.4-kb EcoRI fragment from pRIVS5, and a 2-kb SphI-EcoRI fragment from pRIVS4 were subcloned into pUC119 that was digested with the same restriction enzymes. The subcloned fragments from pRIVS1, pRIVS2, pRIVS3, and pRIVS5 also contained the pyrBC′ genes, because convenient restriction enzymes to subclone the inserts without pyrBC′ were not found except for the pRIVS4 clone. The final constructs were designated pUIVS1, pUIVS2, pUIVS3, pUIVS4, and pUIVS5.
β-Galactosidase assays.
P. putida 06909u2 transconjugants carrying selected plasmids were grown overnight in V8C broth supplemented with uracil and were subcultured in 5 ml of fresh V8C broth supplemented with uracil. The resulting subcultures were then grown for 6 h. The cells were harvested by centrifugation. Finally, the cells were resuspended in sterile water and assayed for β-galactosidase activity, as described by Miller (27), by using o-nitrophenyl-β-d-galactopyranoside as the substrate. The same transconjugants were also grown on V8C agar supplemented with 50 μg of uracil per ml or on V8C agar discs covered by P. parasitica for 20 to 24 h. The cells grown on a V8C agar plate containing uracil and appropriate antibiotics were resuspended in water and assayed for β-galactosidase activity by using the same procedure. The V8C agar fungal discs colonized by P. putida strains were dropped into 1 ml of sterile water and resuspended to release the bacterial cells grown on the fungal discs. The β-galactosidase activities of the cells obtained from fungal discs were determined by the same procedure. The controls included a fungal agar disc that was not inoculated with bacteria and a fungal agar disc that was inoculated with P. putida 06909u2(pRIV13). Plasmid pRIV13 complemented the uracil auxotrophy of P. putida 06909u2, but this plasmid does not contain a lacZ gene.
Defining potential inducible promoter areas.
Potential fungus-induced clones were subcloned into pRIV16 to determine the location of promoters induced during colonization of P. parasitica M191 by P. putida 06909. Figures 2 and 3 show restriction maps of two clones that were completely sequenced and subclones with restriction enzyme sites. Sometimes fragments were first subcloned into pUC119 to have enough restriction sites to transfer into pRIV16, which had only a few unique restriction sites. The negative control plasmid used was pRIV16.
FIG. 2.
Organization of the potential ORFs in pRIVS1 and various restriction enzyme sites. The promoterless pyrBC′ and lacZ genes are located downstream of the DAGK gene as a transcriptional fusion in pRIVS1. ORF1 is a potential secreted protein. pRIVS13 restores induction of the clone during colonization of fungal hyphae at the level of pRIVS1. The levels of β-galactosidase activity in P. putida 06909u2 containing different plasmids were determined in the presence and in the absence of P. parasitica. Plasmids pRIV16 and pRIV16C were the negative and positive controls, respectively. The boldface EcoRI (E) site was used to subclone the 1.35-kb fragment to generate pRIVS13. Abbreviations: B, BamHI; C, ClaI; E, EcoRI; N, NcoI; P, PstI; Sa, SalI; Sm, SmaI; Sp, SphI; X, XbaI.
FIG. 3.
Organization of the potential ORFs in pRIVS2 and various restriction enzyme sites. The promoterless pyrBC′ and lacZ genes are located downstream of the ABC transporter as a transcriptional fusion. ORF1 is similar to heavy-metal-transporting ATPases. pRIVS20 restores induction of pRIVS2 during colonization of fungal hyphae at the level of pRIVS2. The levels of β-galactosidase activity in P. putida 06909u2 containing different plasmids were determined in the presence and in the absence of P. parasitica. pRIVS26 carries the reverse-oriented PstI fragment. The arrows indicate the positions of the transcriptional terminator-bearing palindromic sequences. Abbreviations: C, ClaI; Ev, EcoRV; K, KpnI; P, PstI; Sm, SmaI; Sp, SphI; Xh, XhoI.
Nucleotide sequence accession numbers.
The DNA sequences determined in this study have been deposited in the GenBank database under accession no. AF249735 (clone in pRIVS1), AF249736 (clone in pRIVS2), and AF249737 (clone in pRIVS3)].
RESULTS
Cloning of pyrB from P. putida 06909.
ATCase is an enzyme that is essential for pyrimidine biosynthesis. We cloned a gene encoding the P. putida 06909 ATCase to determine whether it could be used as a selectable marker in the IVET strategy. An ATCase gene, pyrB, was amplified from P. putida 06909 by PCR by using a pair of degenerate primers designed on the basis of the amino acid sequence of PyrB from P. putida PPN and P. aeruginosa PAO. The PCR product, which was approximately 1 kb long, was cloned in pUC119 to generate pDAC41. The partial 5′ and 3′ nucleotide sequences of the cloned PCR product were very similar to the pyrB nucleotide sequences of P. putida PPN and P. aeruginosa PAO (data not shown). A restriction map of the PCR product of pyrB was determined by using restriction endonuclease cleavage patterns (data not shown).
Generation of pyrB mutant.
To determine whether the pyrB gene would be a good selectable marker, it was necessary to create a mutation in the pyrB gene of P. putida 06909. pyrB mutants should be auxotrophic for pyrimidine. Therefore, a pyrimidine requirement for growth or survival of an auxotroph should be a good selection criterion in pyrimidine-deficient environments. To create a mutation in the pyrB gene of P. putida 06909, marker exchange mutagenesis was used. Plasmid pRIV11 carrying the marker exchange construct was introduced into wild-type strain P. putida 06909, and the wild-type pyrB gene was replaced with the marker exchange construct by homologous recombination. Two kanamycin-resistant but tetracycline-sensitive mutants were obtained, and they did not grow on minimal medium (MG medium) unless uracil was supplied. Growth of the two mutants in minimal medium (MG medium) supplemented with uracil was comparable to growth of the wild-type strain in the same medium (data not shown). This indicated that the mutational process did not impair bacterial growth except for the uracil requirement. The mutation was further confirmed by Southern blot hybridization with two different probes (data not shown). When a pyrB probe was used, the expected size difference (800 bp) between the wild-type and mutant pyrB loci was observed. This difference was due to removal of the 400-bp internal HincII fragment and insertion of the 1.2-kb kanamycin resistance gene. In addition, the kanamycin resistance gene hybridized only with DNA isolated from the mutant strains. No differences between the two pyrB mutants were found. Therefore, only one of the pyrB mutant strains (P. putida 06909u2) was used in the experiments described below.
Complementation of pyrB.
The pyrB gene with a complete Shine-Dalgarno sequence, driven by the lac promoter, did not complement mutants P. putida 06909u1 and P. putida 06909u2 in trans. A similar observation was made by Schurr et al. (42), who proposed that another open reading frame (ORF) (pyrC′) downstream of pyrB was required for complementation of the pyrB mutation in trans. Even though PyrC′ is similar to PyrC at the amino acid level, the PyrC′ polypeptide does not exhibit the dihydroorotase activity that is associated with PyrC. The PyrC′ polypeptide appears to be required for ATCase folding. Since six polypeptides of PyrB and six polypeptides of PyrC′ form a dodecameric functional ATCase in P. putida PPN, these polypeptides may have to be cotranslated to fold correctly (42). Therefore, we amplified the 2.4-kb pyrBC′ fragment with specific primer P3, which was based on the 5′ nucleotide sequence of the pyrB PCR product, and degenerate primer D3, which was based on the 3′ end of pyrC′ from P. putida PPN. Primer P3 has a conserved Shine-Dalgarno sequence, GGAGAA. The 2.4-kb pyrBC′ PCR product was cloned into pRK415 to generate pRIV13. pyrB mutant strain P. putida 06909u2 carrying pRIV13 grew in minimal medium. This result confirmed that the pyrBC′ gene was required for complementation of pyrimidine auxotrophy in P. putida 06909u2. The promoterless pyrBC′ gene was therefore used as a selection marker in our IVET construction.
Bacterial growth on fungal plates.
A fungal plate assay was developed to determine whether the pyrimidine requirement provided a good selection criterion for clones expressed during colonization of P. parasitica by P. putida 06909. P. parasitica was grown so that it completely covered a V8C agar plate. Since V8C agar did not contain sufficient pyrimidine to support the growth of P. putida 06909u2 (a pyrB mutant strain), the only way for the mutants to grow on this fungal plate was to acquire pyrimidine from P. parasitica. Therefore, if pyrimidine was not available from P. parasitica, the pyrimidine requirement would provide a good criterion to select between wild-type strain P. putida 06909 and pyrB mutant strain P. putida 06909u2.
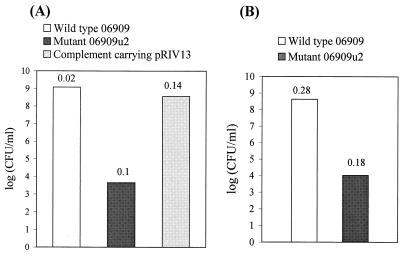
Bacterial growth on the fungal plate revealed that there was significant selection between the wild type and the pyrB mutant (Fig. 4). P. putida 06909u2 did not grow on the fungal plate after 24 h, while the wild-type strain grew rapidly. There was a more-than-4-log difference between the sizes of the wild-type and pyrB mutant bacterial populations (Fig. 4A). The size of the pyrB mutant population did not increase above the size of the initial population applied (approximately 3 × 103 to 6 × 103 CFU per plate). In addition, the pyrB mutant carrying pyrBC′ behind the lac promoter (pRIV13) could grow and complement pyrimidine auxotrophy on the fungal plate (Fig. 4A). The mixed-application experiment in which both the wild type and the mutant (50:50) were used resulted in a similar selection (Fig. 4B). The results obtained after mixed application of the two strains indicated that there was no cross-feeding on fungal hyphae by the strains during the fungal colonization process. Additional experiments in which we used different ratios of the wild-type strain and the pyrB mutant strain on fungal plates revealed that there was a significant selection for the wild-type bacteria (data not shown). We concluded that the pyrB gene was a good selection marker with which to study colonization of P. parasitica by P. putida 06909.
FIG. 4.
Bacterial growth on P. parasitica-covered V8C agar plates. (A) Individual application of the wild type, the pyrB mutant, and the pyrB mutant carrying pRIV13. (B) Mixed application of both the wild type and the pyrB mutant. The P. putida 06909 inoculum contained 4 × 103 CFU/ml. The numbers above the bars are the standard errors based on three replications.
Adhesion of the wild-type strain and the pyrB mutant strain to fungal hyphae was determined by using a fungal column assay (54). The fungal column assay showed that the initial adhesion to the fungal hyphae was not affected by the pyrB mutation (data not shown).
Selection of clones expressed on P. parasitica.
Putative clones that were specifically expressed during colonization of fungal hyphae were selected as described in Materials and Methods. A total of 30 clones were selected. Only 11 of these clones were used for further characterization because the other 19 clones exhibited increases in gene expression in culture media when they were cultured for a prolonged period (data not shown). It was hard to determine whether these 19 clones were expressed during colonization of P. parasitica or induced at the late bacterial growth stage. However, we could not rule out the possible involvement of these genes in colonization of fungal hyphae. The 11 clones which we studied were analyzed by comparing restriction enzyme digestion patterns obtained with different restriction enzymes. Some of the clones were identical, and five unique clones were characterized further.
In addition to the clones that were induced during colonization of P. parasitica, we identified one plasmid (pRIVA1) that was constitutively expressed. pRIVA1 was isolated from a blue colony of P. putida on V8C agar supplemented with X-Gal. This plasmid carried an unknown insert in front of the promoterless pyrBC′-lacZ genes. Plasmid pRIVA1 was used as a positive control.
To confirm that the clones selected were specifically expressed during colonization of P. parasitica, we measured the bacterial growth and β-galactosidase activity of the P. putida 06909u2 pyrB mutant carrying each of the selected clones (Table 2). The pyrB mutant carrying pRIV16C or pRIVA1 grew in the absence of the fungus due to constitutive expression of downstream pyrBC′. In contrast, the pyrB mutant carrying one of five selected clones (pRIVS1, pRIVS2, pRIVS3, pRIVS4, or pRIVS5) did not grow in the absence of the fungus but grew quickly in the presence of P. parasitica. The pyrB mutant carrying pRIV16, a negative control, did not grow in the presence of the fungus or in the absence of the fungus unless uracil was present in the medium.
TABLE 2.
Bacterial growth and β-galactosidase activity of P. putida 06909u2 containing different plasmids
| Introduced plasmid or prepn | Bacterial growth on V8C agar
|
β-Galactosidase activity (Miller units)
|
|||
|---|---|---|---|---|---|
| Without fungusa | With fungusb | V8CU brothc | V8CU agard | V8C agar fungal disc | |
| pRIV13 | + | + | NDe | ND | ND |
| pRIV16 | − | − | 49 | 26 | ND |
| pRIV16C | + | + | 1,570 | 4,375 | 2,166 |
| pRIVA1 | + | + | 3,009 | 933 | 3,005 |
| pRIVS1 | − | + | 237 | 189 | 3,297 |
| pRIVS2 | − | + | 201 | 142 | 2,597 |
| pRIVS3 | − | + | 371 | 3,957 | |
| pRIVS4 | − | + | 166 | 144 | 731 |
| pRIVS5 | − | + | 306 | 141 | 1,834 |
| Fungal disc | ND | ND | ND | ||
Bacterial growth on a V8C agar plate. −, no visible bacterial colonies after 2 days; +, visible bacterial growth after 1 day.
Bacterial growth on a fungal disc as determined by turbidity. −, A600 less than 0.075; +, A600 more than 0.15.
V8CU broth, V8C broth supplemented with uracil (50 μg/ml).
V8CU agar, V8C agar supplemented with uracil.
ND, not detected.
Gene expression was quantified by measuring β-galactosidase activity in the bacterial suspensions. Expression of pRIVS1, pRIVS2, pRIVS3, pRIVS4, and pRIVS5 increased at least 5- to 20-fold during colonization of fungal hyphae (Table 2). The enhanced level of lacZ expression was equivalent to the level of expression by the lac promoter (pRIV16C) or the other constitutive promoter (pRIVA1). In contrast, expression of pRIV16 was not detected in the presence of the fungus. A control V8C agar disc covered by P. parasitica was also dropped into 1 ml of sterile water to monitor the β-galactosidase activity in the fungal mycelium alone. The suspension did not become turbid even when it was vortexed vigorously, and there was no β-galactosidase activity in the suspension, indicating that the fungal mycelium itself did not exhibit detectable β-galactosidase-like enzyme activity.
Another control experiment was conducted by applying the pyrB mutant strain carrying pRIV13, which expressed pyrBC′ from the lac promoter and lacked the lacZ reporter gene, to a fungal disc. The bacteria grew actively on the fungal disc, but the bacterial suspension released from the fungal hyphae did not exhibit β-galactosidase activity. Therefore, we concluded that the clones selected, clones pRIVS1, pRIVS2, pRIVS3, pRIVS4, and pRIVS5, were specifically induced in the presence of the fungus.
DNA sequence analysis.
Two of the clones selected were completely sequenced, and three of them were partially sequenced at the 3′ ends of the inserts. Two of the clones were not significantly similar to any genes in the GenBank database, and three of them exhibited high levels of similarity to previously described bacterial genes.
We determined putative ORFs and the organization of these putative ORFs in a 2.9-kb DNA fragment spanning most of the insert in front of pyrBC′ in pUIVS1 (Fig. 2). The 3′ end of the genomic DNA insert of pUIVS1 fused to promoterless pyrBC′ was a truncated ORF encoding only 63 amino acids. This truncated ORF was similar to the corresponding 5′ regions of genes encoding the diacylglycerol kinases (DAGK) of Escherichia coli (52% amino acid sequence identity; GenBank accession no. K00127) (22), Haemophilus influenzae (43% identity; GenBank accession no. U32718) (49), and Pseudomonas denitrificans (44% identity; GenBank accession no. M62868) (5). Intact E. coli DAGK is 122 amino acids long, and Pseudomonas denitrificans DAGK has 137 residues. Thus, the amino acid sequence of the truncated DAGK of P. putida 06909 corresponds to one-half of the DAGK coding region. The membrane-spanning domains of E. coli DAGK (47) were conserved in P. putida 06909 DAGK (data not shown).
Upstream of the DAGK homolog, a LuxR family response regulator homolog was found. The deduced 216-amino-acid sequence exhibited high levels of similarity to the sequences of a large number of response regulators from various bacteria. High levels of similarity were found with AgmR of P. aeruginosa (42% amino acid sequence identity; GenBank accession no. M60805) (43), which is involved in glycerol metabolism; FlcA of Azospirillum brasilense (38% identity; GenBank accession no. Y12363) (35), which is involved in colonization of the wheat rhizosphere and flocculation of bacteria; FlhR of Paracoccus denitrificans (GenBank accession no. AJ223460) (unpublished data), which is responsible for methanol, methyamine, and formaldehyde oxidation; and NarL of E. coli (34% identity; GenBank accession no. X14884) (38). An alignment of the sequence of the response regulator with the sequences of AgmR of P. aeruginosa and FlcA of A. brasilense showed that these proteins exhibit high levels of identity in a conserved area corresponding to residues 168 to 191 for a DNA-binding, helix-turn-helix motif (3, 17).
The 3-kb insert of pUIVS2 was subcloned into pUC119 as two SphI fragments. The 2-kb SphI fragment was completely sequenced, and it consisted of one complete ORF and one truncated ORF (Fig. 3). The truncated ORF was located at the fusion with the promoterless pyrBC′. The deduced amino acid sequence encoded by the truncated ORF was very similar to the sequences of many ABC transporters from various bacteria. The 282-amino-acid sequence encoded by this truncated ORF was very similar to the sequences of sulfate transporters from Synechococcus sp. (53% amino acid sequence identity; GenBank accession no. J04512) (12), Synechocystis sp. (52% identity; GenBank accession no. D90916) (18), and E. coli (48% identity; GenBank accession no. M32101) (46). It was also very similar to the sequences of the ferric transporter of E. coli (44% identity; GenBank accession no. P37009) (51), the putrescine transporter of E. coli (49% identity; GenBank accession no. M93239) (36), and the maltose transporter of Pseudomonas fluorescens (42% identity; GenBank accession no. U39468) (4). Other transporters, such as the transporters for polyamines, sugars, peptides, etc., from various bacteria were also very similar to the putative ABC transporter from P. putida 06909 (data not shown).
A second complete ORF was located upstream of the ABC transporter. Since there is 229 bp between the stop codon of this ORF and the start codon of the ABC transporter, it is likely that a potential promoter is located in this space. The deduced 289-amino-acid sequence encoded by the ORF was similar to the sequences of the ribulose bisphosphate carboxylase operon transcriptional regulators of various photosynthetic bacteria, such as Synechocystis sp. (31% amino acid sequence identity; GenBank accession no. D90910) (18; see reference 45 for a review). The ORF-encoded protein was also similar to the LysR family response regulators, such as OxyR from E. coli, which regulates the oxidative stress response, a specific colony morphology switch, bacterial aggregation, and piliation (8, 52). The highly conserved DNA-binding motif, a helix-turn-helix, is located in the N-terminal region of this response regulator and aligns with the DNA-binding motifs of other LysR family response regulators (3) (data not shown). Our DNA sequence comparison of the clone in pRIVS2 and the incomplete P. aeruginosa genomic DNA sequence showed that the similar sequences were scattered through different contigs. This indicated that the gene organization in this locus in P. putida 06909 is different from the gene organization in P. aeruginosa (data not shown).
One of the clones (pRIVS3) was partially sequenced, and it exhibited high levels of similarity to a few outer membrane porin proteins. The deduced amino acid sequence encoded by the truncated ORF was similar to the sequence of outer membrane porin OprD2 of P. aeruginosa, which forms the imipenem-permeable porin (56), and was also similar to the sequence of PhaK of P. putida, which transports phenylacetic acid (33).
Potential promoter areas in pUIVS1 and pUIVS2.
Potential promoters induced during colonization of P. parasitica were subcloned into pRIV16. The 1.35-kb portion of the pUIVS1 EcoRI fragment spanning the truncated dgk homolog and the luxR family response regulator (nucleotides 1547 to 2915) retained inducible promoter activity (Fig. 2). In the presence of P. parasitica, the level of β-galactosidase activity of this subclone was the same as the level of activity observed with the original pRIVS1 clone. Thus, the inducible promoter could be located in front of the LuxR family response regulator gene or between the response regulator gene and dgk.
The 1.4-kb PstI fragment of pUIVS2 exhibited inducible promoter activity when the fragment was oriented correctly with respect to the promoterless pyrBC′-lacZ genes (pRIVS20). No inducible promoter activity was observed when the fragment was placed in the opposite orientation (pRIVS26) (Fig. 3). DNA sequence analysis of the genomic DNA fragment in pUIVS2 revealed a strong palindromic sequence with a 33-bp stem and a 24-bp loop between the stop codon for the LysR family response regulator and the start codon for the ABC transporter (Fig. 3). It is likely that the palindromic sequence is a transcriptional terminator of the LysR family response regulator gene. Therefore, we concluded that the inducible promoter is probably located in front of the ABC transporter gene and that the ABC transporter may play a role in colonization of P. parasitica by P. putida 06909.
DISCUSSION
P. parasitica hyphal colonization and subsequent inhibition of fungal growth by P. putida 06909 are important for the biocontrol activity of P. putida (54). The colonization of hyphae by P. putida is a biologically interesting phenomenon. However, little is known about bacterial colonization of fungal hyphae. In this study, we developed an IVET system based on a pyrimidine requirement for growth of a mutant bacterial strain in vivo to genetically investigate bacterial colonization of P. parasitica. It has been shown previously that the thymidylate synthase gene is a good alternative selection marker that can replace antibiotic resistance genes in Rhizobium meliloti and Lactococcus lactis (32, 39). Our data suggested that the pyrimidine requirement for bacterial growth was a good selection marker during colonization of fungal hyphae by P. putida. The fungal hyphae did not produce sufficient pyrimidine to support growth of the pyrB mutant strain. The pyrimidine requirement also provided a good selection criterion for studying bacterial gene expression in the citrus rhizosphere, especially on actively growing citrus root tips (21).
We selected five clones that were specifically induced in the presence of the fungus P. parasitica. Truncated ORFs were found to be fused to the promoterless pyrBC′ genes at the 3′ end of each these clones. These truncated ORFs, or upstream genes, should be induced during colonization of P. parasitica by P. putida 06909. The gene fused to the promoterless pyrBC′ genes in pRIVS1 was a dgk homolog encoding DAGK. The function of DAGK in E. coli is to recycle diacylglycerol generated during membrane-derived oligosaccharide biosynthesis (16). The same enzyme is involved in recycling of bacterial diacylglycerol generated during cyclic β-1,2-glucan biosynthesis in Rhizobium meliloti (29). It has been suggested that the glucans of members of the Rhizobiaceae may function during plant infection and adaptation to osmotic stress (7, 28, 37). We do not know if the DAGK of P. putida 06909 is linked to specific cell surface carbohydrate biosynthesis to recycle diacylglycerol. Since the DNA sequence of the dgk gene that we obtained was only 190 bp long, the complete ORF of the dgk should be cloned to conduct mutational studies to test the direct involvement of the gene in colonization of P. parasitica.
A LuxR family response regulator was found upstream of dgk. Since subclone pRIVS13, which carried luxR-dgk, restored the inducible promoter activity of pRIVS1, the inducible promoter expressed during colonization of fungal hyphae should be either in front of this LuxR family response regulator or in front of dgk. Since 1% glycerol in the culture medium slightly induced expression of pRIVS1, the chemical nature of the inducer of pRIVS1 may be similar to glycerol. It will be interesting to investigate whether the LuxR family response regulator regulates expression of the DAGK gene. No sensor kinase-like DNA sequence was found in the vicinity of the LuxR family response regulator. However, this finding does not rule out the possibility that the LuxR family response regulator is a response regulator of a two-component regulatory system. Further definition of the inducible promoter area is required to determine if the response regulator-like gene is induced or if only dgk is induced during colonization of the fungus by P. putida 06909.
Another clone in pRIVS2 had an ABC transporter gene at the fusion with pyrBC′, and its promoter was induced during colonization of P. parasitica. The truncated ABC transporter gene encodes a 282-amino-acid protein that includes most of the conserved domains in different ABC transporters. The sizes of ABC transporters are variable but usually are more than 300 amino acids. The C-terminal portions of the ABC transporters are not highly conserved compared with the N-terminal conserved regions (15). The ABC transporters import or export many different molecules by using ATP, and each transporter is highly specific for its substrate (31). Therefore, the ABC transporter induced during colonization of P. parasitica may be involved in uptake of a specific compound from P. parasitica or in secretion of a compound onto P. parasitica to inhibit its growth. Since the ABC transporter was similar to transporters for sulfate, ferric ions, polyamine, maltose, etc., we added several compounds to V8C medium individually to examine ABC transporter induction specificity. None of the substrates induced ABC transporter gene expression. High levels of similarity to the ABC transporter were observed for sulfate transporters from various bacteria that are induced during sulfate depletion (11, 12). However, the ABC transporter was not induced under sulfate-deficient conditions. We did not observe any induction of this ABC transporter on various culture media, including minimal medium and rich medium, except in the presence of P. parasitica. Each locus induced during colonization of P. parasitica may cooperate to inhibit fungal growth in natural environments. Further work is necessary to define the role of the ABC transporter either in specific chemical uptake from P. parasitica or in specific chemical export to P. parasitica.
The inducible promoter area of pRIVS2 has a potential transcriptional terminator and three direct repeats (7 to 9 bp) (data not shown) between the LysR family response regulator and ABC transporter genes. It is likely that the transcriptional terminator blocks transcription of the LysR type of response regulator found in front of the ABC transporter. It is not clear if the LysR family response regulator is induced during colonization of P. parasitica by P. putida 06909. Since the promoter area between the LysR family response regulator and the ABC transporter alone restores original pRIVS2 induction activity, the LysR family response regulator may be the last ORF of a separately transcribed operon.
One of the partially sequenced clones encodes a putative outer membrane porin protein at its 3′ end, and it is likely that the porin gene is induced during colonization of P. parasitica. It may be involved in uptake of specific chemicals from the fungus; together with the ABC transporter, this suggests that P. putida 06909 may acquire multiple compounds from P. parasitica.
Adding various chemicals as possible inducers did not induce pRIVS1 or pRIVS2. Only 1% glycerol in V8C agar resulted in a slight induction of pRIVS1 carrying dgk. Therefore, the nature of the inducing substances during colonization of P. parasitica by P. putida 06909 is not known. Further work to define the roles of other genes selected by using the IVET strategy should help us understand the nature of colonization of P. parasitica by P. putida 06909. It would be interesting to determine whether the genes are induced by the chemicals produced by P. parasitica or by the presence of P. parasitica hyphae. In the future we will confirm the potential involvement of selected clones during fungal colonization by P. putida 06909 and biological control on citrus by analyzing mutations at specific loci of selected clones in the wild-type bacterial chromosome.
ACKNOWLEDGMENTS
We thank Linda L. Walling for critically reading the manuscript, Hamid R. Azad for technical assistance, and Noel T. Keen and Caroline S. Harwood (University of Iowa) for providing plasmids and helpful information.
S.-W. Lee was supported by a Korean Government Overseas Scholarship. This work was supported by U.S. Department of Agriculture/National Research Initiative competitive grant 93-37303-9227 to D. A. Cooksey.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley Interscience; 1987. [Google Scholar]
- 2.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 3.Brennan R G, Matthews B W. The helix-turn-helix DNA binding motif. J Biol Chem. 1989;264:1903–1906. [PubMed] [Google Scholar]
- 4.Brunker P, Altenbuchner J, Mattes R. Structure and function of the genes involved in mannitol, arabitol and glucitol utilization from Pseudomonas fluorescens DSM50106. Gene. 1998;206:117–126. doi: 10.1016/s0378-1119(97)00574-x. [DOI] [PubMed] [Google Scholar]
- 5.Cameron B, Blanche F, Rouyez M-C, Bisch D, Famechon A, Couder M, Cauchois L, Thibaut D, Debussche L, Crouzet J. Genetic analysis, nucleotide sequence, and products of two Pseudomonas denitrificans cob genes encoding nicotinate-nucleotide:dimethylbenzimidazole phosphoribosyltransferase and cobalamin (5′-phosphate) synthase. J Bacteriol. 1991;173:6066–6073. doi: 10.1128/jb.173.19.6066-6073.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camilli A, Mekalanos J J. Use of recombinase gene fusion to identify Vibrio cholerae genes induced during infection. Mol Microbiol. 1995;18:671–683. doi: 10.1111/j.1365-2958.1995.mmi_18040671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cangelosi G A, Martinetti G, Nester E W. Osmosensitivity phenotypes of Agrobacterium tumefaciens mutants that lack periplasmic β-1,2-glucan. J Bacteriol. 1990;172:2172–2174. doi: 10.1128/jb.172.4.2172-2174.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christman M F, Storz G, Ames B N. OxyR, a positive regulator of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella typhimurium, is homologous to a family of bacterial regulatory proteins. Proc Natl Acad Sci USA. 1989;86:3484–3488. doi: 10.1073/pnas.86.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drahos D J, Hemming B C, McPherson S. Tracking recombinant organisms in the environment: β-galactosidase as a selectable non-antibiotic marker from fluorescent pseudomonads. Bio/Technology. 1986;4:439–444. [Google Scholar]
- 10.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green L S, Grossman A R. Changes in sulfate transport characteristics and protein composition of Anacystis nidulans R2 during sulfur deprivation. J Bacteriol. 1988;170:583–587. doi: 10.1128/jb.170.2.583-587.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green L S, Laudenbach D E, Grossman A R. A region of a cyanobacterial genome required for sulfate transport. Proc Natl Acad Sci USA. 1989;86:1949–1953. doi: 10.1073/pnas.86.6.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Handfield M, Schweizer H P, Mahan M J, Sanschagrin F, Hoang T, Levesque R C. ASD-GFP vectors for in vivo expression technology in Pseudomonas aeruginosa and other Gram-negative bacteria. BioTechniques. 1998;24:261–264. doi: 10.2144/98242st02. [DOI] [PubMed] [Google Scholar]
- 14.Heithoff D M, Conner C P, Hanna P C, Julio S M, Hentschel T, Mahan M J. Bacterial infection as assessed by in vivo gene expression. Proc Natl Acad Sci USA. 1997;94:934–939. doi: 10.1073/pnas.94.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higgins C F. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 16.Jackson B J, Kennedy E P. The biosynthesis of membrane-derived oligosaccharides: a membrane-bound phosphoglycerol transferase. J Biol Chem. 1983;258:2394–2398. [PubMed] [Google Scholar]
- 17.Kahn D, Ditta G. Modular structure of FixJ: homology of the transcriptional activator domain with the −35 binding domain of sigma factors. Mol Microbiol. 1991;5:987–997. doi: 10.1111/j.1365-2958.1991.tb00774.x. [DOI] [PubMed] [Google Scholar]
- 18.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 19.Keane P J, Kerr A, New P B. Crown gall of stone fruit. II. Identification and nomenclature of Agrobacterium isolates. Aust J Biol Sci. 1970;23:585–595. [Google Scholar]
- 20.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 21.Lee S-W, Menge J A, Cooksey D A. Cloning genes expressed during colonization of fungal hyphae or citrus root tips by Pseudomonas putida. Phytopathology. 1998;88:S52. [Google Scholar]
- 22.Lightner V A, Bell R M, Modrich P. The DNA sequences encoding plsB and dfkA loci of Escherichia coli. J Biol Chem. 1983;258:10856–10861. [PubMed] [Google Scholar]
- 23.Lorang J M, Shen H, Kobayashi D, Cooksey D A, Keen N T. avrA and avrB in Pseudomonas syringae pv. tomato PT23 play a role in virulence on tomato plants. Mol Plant-Microbe Interact. 1994;7:508–515. [Google Scholar]
- 24.Lowe A M, Beattie D T, Deresiewicz R L. Identification of novel staphylococcal virulence genes by in vivo expression technology. Mol Microbiol. 1998;27:967–976. doi: 10.1046/j.1365-2958.1998.00741.x. [DOI] [PubMed] [Google Scholar]
- 25.Mahan M J, Slauch J M, Mekalanos J J. Selection of bacterial virulence genes that are specifically induced in host tissues. Science. 1993;259:686–688. doi: 10.1126/science.8430319. [DOI] [PubMed] [Google Scholar]
- 26.Mahan M J, Tobias J W, Slauch J M, Hanna P C, Collier R J, Mekalanos J J. Antibiotic-based selection for bacterial genes that are specifically induced during infection of a host. Proc Natl Acad Sci USA. 1995;92:669–673. doi: 10.1073/pnas.92.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N. Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 28.Miller K J, Kennedy E P, Reinhold V N. Osmotic adaptation by gram-negative bacteria: possible role for periplasmic oligosaccharides. Science. 1986;231:48–51. doi: 10.1126/science.3941890. [DOI] [PubMed] [Google Scholar]
- 29.Miller K J, McKinstry M W, Hunt W P, Nixon B T. Identification of the diacylglycerol kinase structural gene of Rhizobium meliloti 1021. Mol Plant-Microbe Interact. 1992;5:363–371. doi: 10.1094/mpmi-5-363. [DOI] [PubMed] [Google Scholar]
- 30.Miller P M. V-8 juice agar as a general purpose medium for fungi and bacteria. Phytopathology. 1955;45:461–462. [Google Scholar]
- 31.Nikaido H, Hall J A. Overview of bacterial ABC transporters. Methods Enzymol. 1998;292:3–21. doi: 10.1016/s0076-6879(98)92003-1. [DOI] [PubMed] [Google Scholar]
- 32.O'Flaherty S, Moenne-Loccoz Y, Boesten B, Higgins P, Dowling D N, Condon S, O'Gara F. Greenhouse and field evaluations of an autoselective system based on an essential thymidylate synthase gene for improved maintenance of plasmid vectors in modified Rhizobium meliloti. Appl Environ Microbiol. 1995;61:4051–4056. doi: 10.1128/aem.61.11.4051-4056.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olivera E R, Minambres B, Garcia B, Muniz C, Moreno M A, Ferrandez A, Diaz E, Garcia J L, Luengo J M. Molecular characterization of the phenylacetic acid catabolic pathway in Pseudomonas putida U: the phenylacetyl-CoA catabolon. Proc Natl Acad Sci USA. 1998;95:6419–6424. doi: 10.1073/pnas.95.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parales R E, Harwood C S. Construction and use of a new broad-host-range lacZ transcriptional fusion vector, pHRP309, for gram negative bacteria. Gene. 1993;133:23–30. doi: 10.1016/0378-1119(93)90220-w. [DOI] [PubMed] [Google Scholar]
- 35.Pereg-Gerk L, Paquelin A, Gounon P, Kennedy I R, Elmerich C. A transcriptional regulator of the LuxR-UhpA family, FlcA, controls flocculation and wheat root surface colonization by Azospirillum brasilense Sp7. Mol Plant-Microbe Interact. 1998;11:177–187. doi: 10.1094/MPMI.1998.11.3.177. [DOI] [PubMed] [Google Scholar]
- 36.Pistocchi R, Kashiwagi K, Miyamoto S, Nukui E, Sadakata Y, Kobayashi H, Igarashi K. Characteristics of the operon for a putrescine transport system that maps at 19 minutes on the Escherichia coli chromosome. J Biol Chem. 1993;268:146–152. [PubMed] [Google Scholar]
- 37.Puvanesarajah V, Schell F M, Stacey G, Douglas C J, Nester E W. Role for 2-linked-β-d-glucan in the virulence of Agrobacterium tumefaciens. J Bacteriol. 1985;164:102–106. doi: 10.1128/jb.164.1.102-106.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rabin R S, Stewart V. Dual response regulators (NarL and NarP) interact with dual sensors (NarX and NarQ) to control nitrate- and nitrite-regulated gene expression in Escherichia coli K-12. J Bacteriol. 1993;175:3259–3268. doi: 10.1128/jb.175.11.3259-3268.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ross P, O'Gara F, Condon S. Thymidylate synthase gene from Lactococcus lactis as a genetic marker: an alternative to antibiotic resistance genes. Appl Environ Microbiol. 1990;56:2164–2169. doi: 10.1128/aem.56.7.2164-2169.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 41.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schurr M J, Vickrey J F, Kumar A P, Campbell A L, Cunin R, Benjamin R C, Shanley M S, O'Donovan G A. Aspartate transcarbamoylase genes of Pseudomonas putida: requirement for an inactive dihydroorotase for assembly into the dodecameric holoenzyme. J Bacteriol. 1995;177:1751–1759. doi: 10.1128/jb.177.7.1751-1759.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schweizer H P. The agmR gene, an environmentally responsive gene, complements defective glpR, which encodes the putative activator for glycerol metabolism in Pseudomonas aeruginosa. J Bacteriol. 1991;173:6798–6806. doi: 10.1128/jb.173.21.6798-6806.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen H, Gold S E, Tamaki S J, Keen N T. Construction of a Tn7-lux system for gene expression studies in Gram-negative bacteria. Gene. 1992;122:27–34. doi: 10.1016/0378-1119(92)90028-n. [DOI] [PubMed] [Google Scholar]
- 45.Shively J M, van Keulen G, Meijer W G. Something from almost nothing: carbon dioxide fixation in chemoautotrophs. Annu Rev Microbiol. 1998;52:191–230. doi: 10.1146/annurev.micro.52.1.191. [DOI] [PubMed] [Google Scholar]
- 46.Sirko A, Hryniewicz M M, Hulanicka D, Bock A. Sulfate and thiosulfate transport in Escherichia coli K-12: nucleotide sequence and expression of the cysTWAM gene cluster. J Bacteriol. 1990;172:3351–3357. doi: 10.1128/jb.172.6.3351-3357.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith R L, O'Toole J F, Maguire M E, Sanders C R., II Membrane topology of Escherichia coli diacylglycerol kinase. J Bacteriol. 1994;176:5459–5465. doi: 10.1128/jb.176.17.5459-5465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steddom K C, Menge J A. Continuous application of the biocontrol bacterium, Pseudomonas putida 06909, improves biocontrol of Phytophthora parasitica on citrus. Phytopathology. 1999;89:S75. doi: 10.1094/PHYTO.2002.92.8.850. [DOI] [PubMed] [Google Scholar]
- 49.Tatusov R L, Mushegian A R, Bork P, Brown N P, Hayes W S, Borodovsky M, Rudd K E, Koonin E V. Metabolism and evolution of Haemophilus influenzae deduced from a whole-genome comparison with Escherichia coli. Curr Biol. 1996;6:279–291. doi: 10.1016/s0960-9822(02)00478-5. [DOI] [PubMed] [Google Scholar]
- 50.Turney J K. The biological control of Phytophthora root rot of citrus using rhizobacteria. Ph.D. thesis. Riverside: University of California; 1995. [Google Scholar]
- 51.Volkert M R, Loewen P C, Switala J, Crowley D, Conley M. The Δ(argF-lacZ)205(U169) deletion greatly enhances resistance to hydrogen peroxide in stationary-phase Escherichia coli. J Bacteriol. 1994;176:1297–1302. doi: 10.1128/jb.176.5.1297-1302.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Warne S R, Varley J M, Boulnois G J, Norton M G. Identification and characterization of a gene that controls colony morphology and auto-aggregation in Escherichia coli K12. J Gen Microbiol. 1990;136:455–462. doi: 10.1099/00221287-136-3-455. [DOI] [PubMed] [Google Scholar]
- 53.Wilson E E, Zeitoun F M, Fredrickson D L. Bacterial phloem canker, a new disease of Persian walnut trees. Phytopathology. 1967;57:618–621. [Google Scholar]
- 54.Yang C-H, Menge J A, Cooksey D A. Mutations affecting hyphal colonization and pyoverdine production in pseudomonads antagonistic toward Phytophthora parasitica. Appl Environ Microbiol. 1994;60:473–481. doi: 10.1128/aem.60.2.473-481.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 56.Yoneyama H, Yoshihara E, Nakae T. Nucleotide sequence of the protein D2 gene of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1992;36:1791–1793. doi: 10.1128/aac.36.8.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]