Graphical abstract

Keywords: Hepatocellular Carcinoma, mRNAsi, Cancer stem cell, Molecular subtype, Prognosis
Abstract
Background
Recent studies have shown that the mRNA expression-based stemness index (mRNAsi) can accurately quantify the similarity of cancer cells to stem cells, and mRNAsi-related genes are used as biomarkers for cancer. However, mRNAsi-driven tumor heterogeneity is rarely investigated, especially whether mRNAsi can distinguish hepatocellular carcinoma (HCC) into different molecular subtypes is still largely unknown.
Methods
Using OCLR machine learning algorithm, weighted gene co-expression network analysis, consistent unsupervised clustering, survival analysis and multivariate cox regression etc. to identify biomarkers and molecular subtypes related to tumor stemness in HCC.
Results
We firstly demonstrate that the high mRNAsi is significantly associated with the poor survival and high disease grades in HCC. Secondly, we identify 212 mRNAsi-related genes that can divide HCC into three molecular subtypes: low cancer stemness cell phenotype (CSCP-L), moderate cancer stemness cell phenotype (CSCP-M) and high cancer stemness cell phenotype (CSCP-H), especially over-activated ribosomes, spliceosomes and nucleotide metabolism lead to the worst prognosis for the CSCP-H subtype patients, while activated amino acids, fatty acids and complement systems result in the best prognosis for the CSCP-L subtype. Thirdly, we find that three CSCP subtypes have different mutation characteristics, immune microenvironment and immune checkpoint expression, which may cause the differential prognosis for three subtypes. Finally, we identify 10 robust mRNAsi-related biomarkers that can effectively predict the survival of HCC patients.
Conclusions
These novel cancer stemness-related CSCP subtypes and biomarkers in this study will be of great clinical significance for the diagnosis, prognosis and targeted therapy of HCC patients.
1. Introduction
Hepatocellular Carcinoma (HCC) is one of most common and aggressive human malignancies with the 5-years survival rate less than 5% [1], [2]. Especially, the lack of reliable early diagnostic markers and effective methods to distinguish molecular subtypes results in the poor prognosis for HCC patients [3], [4], [5]. Of note, many recent studies demonstrate that HCC can be divided into different subtypes by using distinct molecular characteristics [3], [4], [5], [6]. HCC can be subdivided into seven groups by 591 variable CpG sites [7]. HCC can be distinguished into three molecular subtypes:cell proliferation, metabolic disorder and immune disorder based on the whole protein expression profile [8]. HCC can be also divided into three molecular subtypes:iCluster1, iCluster2, iCluster3 by integrating multi-omics data [9], [10]. Even so, the high heterogeneity of HCC makes it difficult to accurately classify HCC into molecular subtypes up to now. Obviously, similar to the classic molecular subtypes of breast cancer [11], establishing a reliable model for distinguishing molecular subtypes is very necessary for effective diagnosis and treatment of HCC patients.
Cancer progression involves the loss of a differentiated phenotype and acquisition of progenitor and stem-cell-like features, particularly cancer stemness is often used to assess how similar cancer cells (especially cancer stem cells, CSC) in tumor tissue are to stem cells [12], [13]. CSCs is an important component in the complex tumor microenvironment and has the ability to self-renew and differentiate from cell origin, which can produce a variety of tumor cells through its own stem cell characteristics [14], [15]. Remarkably, these undifferentiated cell populations with stem cell-like properties have been identified as the main factors affecting recurrence and progression in HCC [16], [17]. However, due to the complexity of the tumor microenvironment, CSCs cannot be quantified well. Fortunately, a recent study indicated that CSCs can be well quantified by the mRNA expression-based stemness index (mRNAsi) and the mRNAsi can effectively quantify the degree of oncogenic differentiation of tissues [18]. This mRNAsi is an cancer stemness score to measure the similar degree between tumor cells and stem cells, and can quantify the CSC in tumor tissue. The value of mRNAsi is between 0 and 1. The closer to 1, the lower the degree of differentiation of tumor cells and the stronger the characteristics of CSCs [18]. The mRNAsi has been confirmed to be significantly related to the level of tumor dedifferentiation and the biological process of cancer stem cells [18]. Interestingly, multiple mRNAsi-related genes have been proved to widely participate in the occurrence of tumors and act as prognosis markers of patients [19], [20], [21]. However, most studies are mainly focused on the identification of mRNAsi-related prognostic genes, but not tumor heterogeneity. More importantly, the relationship between tumor heterogeneity and mRNAsi in HCC patients is still unknown to date. Therefore, further revealing the cancer stemness-driven heterogeneity is of great significance for the accurate classification and targeted treatment of HCC patients.
In this study, we used the weighted gene co-expression network analysis (WGCNA) to screen 212 mRNAsi-related genes that can subdivide HCC patients into three subtypes: CSCP-L, CSCP-M, and CSCP-H, especially patients with the CSCP-L subtype have the best prognosis, but the worst prognosis for patients with the CSCP-H subtype. Interestingly, our study has demonstrated that three CSCP subtypes have different mutation characteristics, immune infiltration microenvironment and immune checkpoint expression. Overall, we first reveal three molecular subtypes associated with cancer stemness in HCC, which will be very helpful for further promoting the diagnosis and treatment of HCC patients.
2. Materials and methods
2.1. Data collection and preprocessing
Gene expression profiles (FPKM) and corresponding clinical information of 341 HCC tumor tissues and 50 para-cancerous samples of TCGA were originated from UCSC Xena database (https://xena.ucsc.edu/). The mRNAsi of TCGA samples was obtained from the study of Tathiane et al [18], and the mRNAsi of verification dataset was obtained by running the source code of the one class linear regression (OCLR) algorithm [18]. The somatic mutation profiles of HCC patients were downloaded from the GDC (https://portal.gdc.cancer.gov/). The verification data numbered GSE14520 came from Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/) [22], [23]. The Japan HCC samples came from the International Cancer Genome Consortium (ICGC) database (https://icgc.org/).
2.2. Calculation of the mRNAsi of tumor samples
The OCLR machine learning algorithm was used to calculate the mRNAsi of tumor tissue [18]. The OCLR can use the expression data of various stem cells generated by the Progenitor Cell Biology Consortium (PCBC) as a training set to build a predictive model to predict the mRNAsi of new samples [18]. These main code steps are as follows (https://github.com/dxsbiocc/learn/tree/main/R/CSCs). First, register and download these expression data of the stem cell training set of PCBC. Second, mean and normalize these data. Third, construct a prediction model by the gelnet function of the gelnet package and use one-class logistic regression to obtain the weights for each gene. Fourth, map these gene names of the new tissue and PCBC data, and extract the expression matrix and weights of these shared genes. Fifth, use spearman to calculate the correlation between weights and expression values to measure the mRNAsi of new samples and normalize it to fall between 0 and 1.
2.3. Differential expression analysis and WGCNA analysis
Differentially expressed genes (DEGs) were screened using the limma R package [24] with filtering criteria |log2FC| > 1 and FDR < 0.05. These DEGs were used for the WGCNA algorithm to identify mRNAsi-related gene modules by using the WGCNA R package [25]. The WGCNA algorithm is a systems biology tool to describe the correlation pattern of gene expression in samples, particularly it can use the expression correlation coefficient between genes to measure their co-expression relationships [25]. Genes with similar expression patterns may be involved in the same biological process or pathway, thereby simplifying complex omics data into several functional modules. These biologically meaningful modules can be discovered by correlating these modules with phenotypic information. These phenotype-related module genes identified by the WGCNA are closely related and may jointly affect the phenotype, which coheres with the biological significance of functional modules. In addition, this WGCNA adopts a soft threshold to construct a co-expression network, which enables the network model to be more in line with the scale-free network distribution and be closer to the biological network.
2.4. Identification of CSCP molecular subtypes in HCC
The ConsensusClusterPlus R package [26] was used to perform the consistent unsupervised clustering of HCC samples to identify different molecular subtypes. According to the Consensus Cumulative Distribution Function (CDF) and Delta Area Plot, the optimal cluster number K value was determined to be 3. Principal component analysis was used to verify whether mRNAsi-related genes can effectively distinguish HCC patients into different subtypes. The Pheatmap R package (https://cran.r-project.org/web/packages/pheatmap/index.html) was used to analyze these expression patterns among different molecular subtypes. According to the default distance algorithm built in Pheatmap, these 212 mRNAsi-related genes are clustered into 2 clusters in CSCP subtypes.
2.5. Functional enrichment and mutation data analyses
Use Pheatmap to extract two clusters of mRNAsi-related genes for subsequent functional analysis. Both Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were conducted by the clusterProfiler R package [27]. The R package maftools was used to analyze these HCC mutation data [28]. The built-in somaticInteractions and mafCompare function was used to investigate co-mutations and mutually exclusive mutations as well as differential mutations, respectively.
2.6. Tumor immune infiltration cell (TIICs) and tumor purity analysis
The single sample gene set enrichment analysis (ssGSEA) in this GSVA package [29] was used to evaluate relative abundance of 28 kinds of TIICs based on their 782 marker genes [30]. The estimate R package was used to calculate the tumor purity of HCC samples [31].
2.7. Survival analysis and prognostic model construction
The Kaplan–Meier survival analysis was used to compare the survival rate between different groups. These independent prognostic marker genes were identified through the following steps. First, the univariate cox hazard analysis was applied to 212 mRNAsi-related genes to identify potential markers (p-value < 0.05). Next, the batch survival analysis was performed to further filter these prognostic genes (p-value < 0.05). Finally, the multivariate stepwise regression analysis was used to identify robust independent markers (p-value < 0.05). These robust markers were further used to establish a prognostic model and predict the patient's risk score. Of note, the model was constructed by executing the coxph function, and the patient's risk score was calculated by the predict function of the survival R package. The mathematical formula is: Riskscore = h0(t) * exp (β1X1 + β2X2 + … + βnXn). Herein, Xn represents 10 prognostic genes, βn represents the regression coefficient of the gene, exp represents the expression level of the gene, and h0(t) is a constant. The receiver operating characteristic curve (ROC) was used to verify the model reliability. The rms R package (https://cran.r-project.org/web/packages/rms/index.html) was used to construct nomograms and transform complex multivariate cox regression equations into clinically available visualization model.
2.8. Data statistics and visualization
Statistical analysis of all data was performed using R (version 4). The Mann-Whitney-Wilcoxon test was used to calculate the statistical significance of mRNAsi scores and immune cell infiltration. The fisher's exact test or chi-square test was used to calculate the statistical significance of tumor grade and mutation significance of different subtypes. The log-rank test was used to calculate the statistical significance of survival curves.
3. Result
3.1. High mRNAsi is associated with the poor prognosis of HCC patients
Here, we systematically examined whether mRNAsi is related to the survival and the disease progression of HCC patients from TCGA dataset (Table S1). Our results demonstrated that mRNAsi in tumor tissues is significantly higher than that in normal tissues (p < 0.001) (Fig. 1A), and patients of the high mRNAsi group have lower survival rate than ones of the low mRNAsi group (p = 0.003) (Fig. 1B), as well as the mRNAsi in these dead patients is higher that one in the alive population (p = 0.043) (Fig. 1C), implying that the increase of tumor stem characteristics is not conducive to the survival of patients. Similarly, HCC patients with higher T stage, G grade, American Joint Committe on cancer (AJCC) stage and tumor burden have higher mRNAsi (Fig. 1D ∼ 1G), but no significant difference between patients of different ages and genders (Fig. S1). Notably, although mRNAsi is generally higher in higher T and AJCC stages, it is decreased in T4 and AJCC stage IV (Fig. 1D and Fig. 1F). This cause may be due to the small number of patient samples for T4 and AJCC stage IV. Furthermore, we found that the tumor purity of tumor tissues is significantly positively correlated with the value of mRNAsi (R = 0.47, p < 0.001) (Fig. 1H), particularly a significant positive correlation exists between mRNAsi and AFP (the most commonly used clinical detection marker for HCC) (R = 0.23, p < 0.001) (Fig. 1I). Taken together, our results indicated that the increase of tumor stem characteristics is closely associated with the poor prognosis and the disease progression of HCC patients.
Fig. 1.
mRNAsi is associated with the prognosis and disease progression of HCC patients. A: Differences in mRNAsi between HCC adjacent tissues and tumor tissues of TCGA cohort. B: The overall survival rate of HCC patients in the high and low mRNAsi groups. According to the median mRNAsi value of HCC patients, patients were divided into high and low mRNAsi groups. C: Differences in mRNAsi between living and dead HCC patients. D: Differences in mRNAsi of HCC patients with different T stages. E: Differences in mRNAsi of HCC patients with different G grades. F: Differences in mRNAsi of HCC patients with different AJCC stages. G: Differences in mRNAsi of HCC patients with different tumor burdens. H: Correlation between mRNAsi and tumor purity. I: Correlation between mRNAsi and HCC clinical detection marker AFP (alpha-fetoprotein). The Mann-Whitney-Wilcoxon test was used to calculate the significance of mRNAsi difference between the two groups. Analysis of variance (ANOVA) was used to calculate the significance of mRNAsi differences between multiple groups.
3.2. Identifing HCC molecular subtypes
Herein, we further used this WGCNA to analyze the relationship between mRNAsi and 1,527 differentially expressed genes in tumor and para-cancerous tissues of TCGA cohort. Notably, we used the soft threshold (β = 10) to realize the scale-free topology criterion of the network (Fig. S2A∼S2B). Our findings showed that under this threshold, the number of non-scale topological structure connected genes changes exponentially, and the linear fitting result further proves that this data network conforms to the non-scale network distribution with R2 > 0.9 (Fig. 2A). We then constructed a cluster dendrogram and used the hybrid dynamic cutting tree algorithm to divide these co-expressed genes into multiple gene modules (GM) in different colors (Fig. 2B), finding that 7 modules are significantly related to mRNAsi (Fig. 2C). Especially, the blue module has the strongest positive correlation with mRNAsi (r = 0.41, p < 0.001), whereas the yellow module has the strongest negative correlation with mRNAsi (r = -0.7, p < 0.001) (Fig. 2C). Herein, therefore, we further chose the blue gene module (containing 212 DEGs) for subsequent analysis (Table S2).
Fig. 2.
WGCNA analysis identifies mRNAsi-related gene modules in HCC of TCGA. A: The linear fitting curve when the soft threshold β = 10. This is used to determine whether the gene network identified by WGCNA conforms to the scale-free network distribution. The closer the fitted value R2 is to 1, the more consistent it is. B: Clustering dendrogram of mRNAsi-related genes, based on the difference in topological overlap, and the assigned merged module color and original module color. C: The correlation between gene modules and mRNAsi calculated based on WGCNA. D: Consensus clustering of HCC patients based on 212 mRNAsi-related genes. When the clustering matrix parameter is 3, the patients are effectively clustered into three subtypes. E: The heat map shows the differences in the expression of 212 mRNAsi-related genes in the three subtypes. F: Principal component analysis verifies and visualizes that HCC patients are divided into three groups.
Interestingly, we found that 212 mRNAsi-related genes from the blue gene module can precisely divide HCC patients into three different subtypes, and HCC patients of group1, group2 and group3 subtype account for 32.8%, 21.1% and 46.0%, respectively (Fig. 2D and Fig. S2C∼S2D). Surprisingly, 212 mRNAsi-related genes can be further subdivided into two large clusters (Fig. 2E and Fig. S3). The first cluster of 45 genes only highly expressed in the group1 subtype (Table S2), and the second cluster of 167 genes just highly expressed in the group2 subtype (Table S2), whilst certain genes of the two clusters are moderately expressed simultaneously in the group3 subtype (Fig. 2E and Fig. S3). Correspondingly, these results from the principal component analysis further revealed that 212 mRNAsi-related genes can effectively distinguish HCC patients into three subtypes (Fig. 2F).
3.3. Prognostic value of three CSCP subtypes
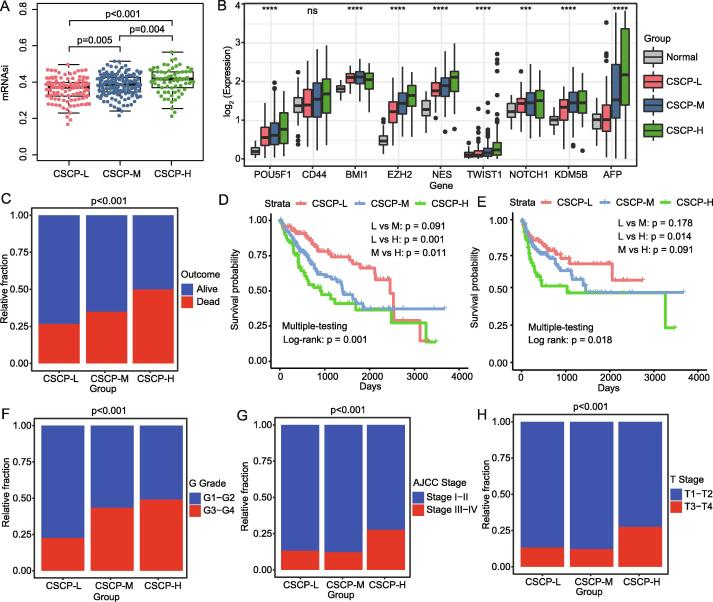
We here found that significant mRNAsi difference exists among three HCC subtypes, and the order of the mRNAsi value is group2 > group3 > group1 (p < 0.001) (Fig. 3A). Therefore, we further named group1, group2 and group3 subtype as low cancer stemness cell phenotype (CSCP-L), high cancer stemness cell phenotype (CSCP-H) and moderate cancer stemness cell phenotype (CSCP-M) respectively (Fig. 3A and Table S1). Interesting, we found that some classical CSC markers, such as POU5F1, CD44, BMI1, EZH2, NES, TWIST1, NOTCH1, KDM5B, are more higher expressed in patients with CSCP-H and CSCP-M subtype than those in patients with CSCP-L subtype, in particular the tumor detection marker AFP is also highest expressed in CSCP-H subtype patients (Fig. 3B). As expected, our results demonstrated that patients with CSCP-H subtype have a higher proportion of deaths (p < 0.001) (Fig. 3C) and a worse overall survival (p = 0.001) and a disease-free survival rate (p = 0.018) (Fig. 3D∼3E). In contrast, patients with CSCP-L subtype have a better survival outcome (Fig. 3D ∼ 3E). Similarly, patients with CSCP-H subtype have a more severe disease progression accompanied by a higher proportion of G grade, AJCC stage and T stage (Fig. 3F∼3H). Remarkably, patients with CSCP-M subtype have a moderate overall survival and tumor progression, which intermediates between patients with CSCP-L and CSCP-H subtypes (Fig. 3C∼Fig. 3H). As a whole, our findings implied that three CSCP subtypes may have an important clinical significance for the diagnosis and prognosis of HCC patients.
Fig. 3.
Differences in clinical characteristics of the three subtypes. A: Differences in mRNAsi among three subtypes. B: Differences in the expression levels of CSC markers and AFP in the three subtypes. C: Stacked histogram showing the proportion of survival outcomes in different subtypes. D: The overall survival rate curve of different subtypes. E: The disease-free survival rate curve of different subtypes. F: Stacked histogram showing the proportion of G grade in different subtypes. G: Stacked histogram showing the proportion of AJCC stage in different subtypes. H: Stacked histogram showing the proportion of T stage in different subtypes. Analysis of variance (ANOVA) was used to calculate the significance of mRNAsi differences between multiple groups. The tumor grade significance of different subtypes were statistically calculated by chi-square test. The log-rank test was used to calculate the significance of survival curves.
3.4. Functional roles of mRNAsi-related genes
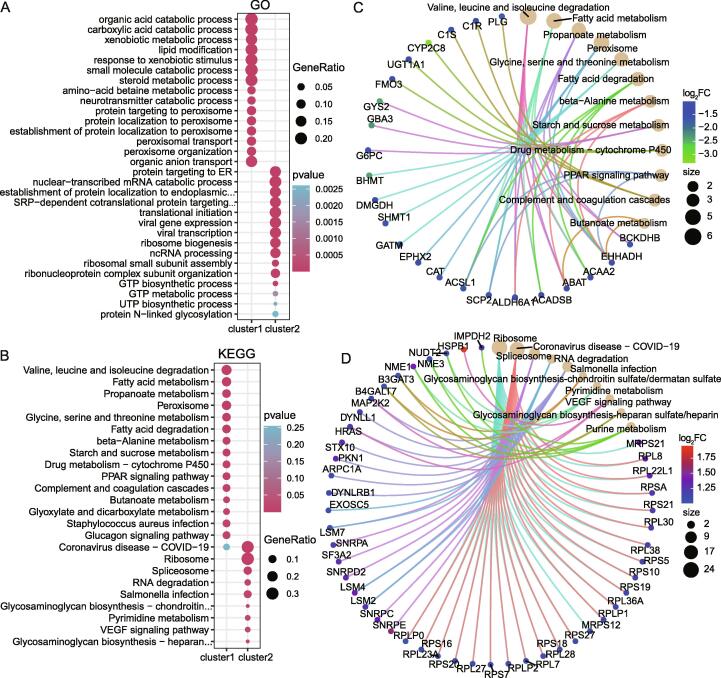
We further carried out both GO annotation and KEGG pathway enrichment analysis on these 212 mRNAsi-related genes, and found that 45 mRNAsi-related genes of the CSCP-L subtype not only can be functionally annotated as organic acid metabolism, carboxylic acid metabolism, heterologous metabolism, organic acid transport (Fig. 4A), but also can be enriched in these signaling pathways such as amino acids, fatty acids, propanoate and P450 drug metabolism and coagulation complement system (Fig. 4B). Interestingly, based on the network analysis, we found that ABAT, ACAA2, CAT, G6PC, CYP2C8, C1S and C1R are involved in the metabolic and complement system pathway (Fig. 4C and Fig. S5A). Especially, highly expressed CAT, G6PC and ABAT and so on can significantly promote the survival of HCC patients, respectively (Fig. S6A∼S6C). These results implied that 45 highly expressed mRNAsi-related genes can enhance the survival of HCC patients with CSCP-L subtype.
Fig. 4.
Differences in molecular characteristics of three subtypes. A: Functional enrichment analysis shows the biological processes (BP) involved in two clusters of genes. B: Functional enrichment analysis shows the KEGG signaling pathway involved in two clusters of genes. C: KEGG network analysis of the first cluster of genes enriched by CSCP-L subtype. The cluster genes are mainly enriched in pathways such as metabolism and complement system. Most of them are down-regulated in tumors and may act as potential tumor suppressors such as CAT, G6PC and ABAT (refer to Fig. S6). D: KEGG network analysis of the second cluster of genes enriched by CSCP-H subtype. The cluster genes are mainly enriched in the ribosome, spliceosome and pyrimidine metabolism pathways. Most of them are up-regulated in tumors and may act as potential oncogenic factors such as RPL8, RPS21, RPL223A, RPL27, RPL38, NME1, SNRPA, SNRPC and SNRPE, etc. (refer to Fig. S6). The gene names included in Cluster 1 and Cluster 2 refer to table S2.The size of the circle in the KEGG network represents the number of genes enriched in the pathway, and the color of the circle represents the difference fold of the gene. For space constraints in the figure, GO or KEGG descriptions that are too long in the figure are replaced by “…”, see supplementary table S3 and table S4 for full names.
In contrast, 167 mRNAsi-related genes of the CSCP-H subtype not only can participate in nuclear transcription, endoplasmic reticulum protein localization, ribosomal subunit assembly, and pyrimidine (Fig. 4A and Fig. S4A and Fig. S4C), but also can involve in ribosomes, spliceosomes, RNA degradation, pyrimidine metabolism and VEGF pathway (Fig. 4B and Fig. S4B and Fig. S4D). These ribosomal genes include RPL8, RPL38, RPS7 and RPS27 and so on, and the spliceosome genes consist of SNRPA, SNRPC, SNRPE, as well as the pyrimidine metabolism genes are NME1, NME2 and NME3 (Fig. 4D and Fig. S5B). In particular, these high expressions of RPL8, RPS21, RPL223A, RPL27, RPL38, NME1, SNRPA, SNRPC and SNRPE significantly reduce the survival rate of HCC patients, respectively (Fig. S6D∼S6L). These above results revealed that these highly expressed genes may result in the worst prognosis of patients with CSCP-H subtype.
3.5. Three CSCP subtypes are verified by using other HCC data sets
Here, we further used three other independent data sets to verify whether mRNAsi-related genes can also divide HCC into three CSCP subtypes. Interestingly, HCC patients from ICGC dataset can be clearly clustered into three different subtypes by 212 mRNAsi-related genes (Fig. 5A). The heat map clustering showed that three subtypes have same molecular characteristics as three CSCP subtypes of TCGA, respectively (Fig. 5B, Fig. S3 and Fig. S7). For example, some metabolism and complement system-related genes, such as ACAA2, CAT, G6PC, CYP2C8, C1R, C1S, are highly expressed in the group3 patients, which is similar to the CSCP-L subtype of TCGA. Many ribosome-related genes (e.g. RPL8, RPSA, RPS7, RPL27) and spliceosome genes (e.g. SNRPA, SNRPC, SNRPE) as well as pyrimidine metabolism related genes (e.g. NME1, NME2, NUDT2) are significantly up-regulated in the group2 patients, which is agreement with the CSCP-H subtype of TCGA (Fig. 5B, Fig. S3 and Fig. S7). Interestingly, patients with the group1 subtype moderately expressed all subtype genes, suggesting a similar transition state to the CSCP-M subtype in the TCGA cohort (Fig. 5B, Fig. S3 and Fig. S7). Similar to the TCGA dateset, patients with group3 subtype have a higher survival rate and a fewer proportion of patients with severe tumors progression (Stage 3 ∼ 4) (Fig. 5C∼5D), but patients with group2 subtype have the worse survival rate (p = 0.038) and a higher proportion of patients with severe stage (p < 0.001) (Stage 3 ∼ 4) (Fig. 5C∼5D). Remarkably, patients with group2 subtype have the highest tumor stemness mRNAsi (p < 0.001) (Fig. 5E) and the highest expression level of CSC marker such as POU5F1, KLF4, CD44, EZH2, NES, HIF1A, NOTCH1 and KDM5 (p < 0.001) (Fig. 5F), which is consistent with the result from the TCGA dateset. Of note, other two datasets (GPL571 and GPL3921) of GSE14520 also confirmed the result from the TCGA dateset (Fig. S8). Collectively, three CSCP subtypes are robust for the identification of HCC patients.
Fig. 5.
Validation of CSCPs subtypes in other HCC datasets. A: Consensus clustering of HCC patients in the ICGC dataset based on 212 mRNAsi-related genes. When the clustering matrix parameter is 3, patients are effectively clustered into three subtypes. B: The heat map shows the expression differences of 212 mRNAsi-related genes in the 3 subtypes of the ICGC data set. C: The overall survival rate curve of different subtypes in ICGC data set. D:Stacked histogram showing the proportion of Liver Cancer Study Group of Japan (LCSGJ) stage in different subtypes in ICGC data set. E: Differences in mRNAsi among three subtypes in ICGC data set. F: Differences in the expression levels of CSC markers in the three subtypes ICGC data set. Analysis of variance (ANOVA) was used to calculate the significance of mRNAsi differences between multiple groups. The tumor grade significance of different subtypes were statistically calculated by chi-square test. The log-rank test was used to calculate the significance of survival curves.
3.6. Gene mutation characteristics of three CSCP subtypes
To reveal the underlying mechanism of leading to the different prognosis among three CSCP subtype patients, we here further dectected these gene mutation features of these three subtypes. Our results showed that the CSCP-H subtype has a higher gene mutation rate of 37.1% (Fig. 6A), in particular these higher gene mutation types are nonsense mutation, in frame del, frame shift del and missense mutation, while the lower mutation type is in frame ins (Fig. 6A). Especially, we identified some high frequency mutation genes such as TP53, CTNNB1, TTN, MUC16 (Fig. 6B), which have been proved to be dysregulated in HCC patients [10]. Interestingly, compared with CSCP-L, both CSCP-H and CSCP-M subtypes have a higher proportion of TP53 mutations, respectively (p < 0.01 and p < 0.05) (Fig. 6B and Fig. S9A ∼ S9C), revealing that the mutation of the classic tumor suppressor TP53 may promote their poorer prognosis than patients with CSCP-L subtype. On the contrary, the MUC4 mutation in the CSCP-H subtype is significantly lower than that of CSCP-L and CSCP-M subtypes (Fig. 6B and Fig. S9A ∼ S9C), suggesting that MUC4 may be a novel marker for HCC patients. Remarkably, these co-mutated gene pairs in the CSCP-L subtype include ABCA12_TTN, ABCA12_CTNNB1, ABCA12_MUC16, and KMT2D_APOB, but the mutually exclusive mutation gene pair only includs TP53_CTNNB1 (Fig. 6C). Whereas these co-mutated gene pairs in the CSCP-H subtype include CUBN_USH2A, FLG_OBSCN, etc., and the mutually exclusive mutation pair is TP53_BAP1 (Fig. 6D). In contrast to CSCP-L and CSCP-M subtypes, the CSCP-M subtype has fewer co-mutations and mutually exclusive mutations (Fig. S9D). Generally, genes with cooperative mutations will jointly drive the development of tumors, while genes with mutually exclusive mutations may be potential synthetic lethality [32], [33]. Therefore, our results seemed to suggest that the different prognosis between CSCP-L and CSCP-H subtypes may be caused by these different combinations of mutations. In particular, CTNNB1 and BAP1 genes, which are mutually exclusive with TP53, may be potential synthetic lethal genes and they are expected to become potential therapeutic targets for HCC patients with TP53 mutations.
Fig. 6.
Differences in genome mutations in different CSCP subtypes. A: The histogram shows the proportion of different types of mutations in different CSCP subtypes. B: The waterfall chart shows high-frequency mutations of different CSCP subtypes. C: Analysis of cooperative mutations and mutually exclusive mutations in CSCP-L subtypes. D: Analysis of cooperative mutations and mutually exclusive mutations in CSCP-H subtypes. Co-mutated genes mean that these genes are often mutated simultaneously in tumor tissues or cells, and they tend to synergistically promote tumor initiation and progression such as ABCA12_TTN and ABCA12_CTNNB1 of CSCP-L subtype as well as FLG_OBSCN and CUBN_USH2A of CSCP-H subtype. In contrast, mutually exclusive genes mean that these genes do not co-mutate in tumor tissues or cells, and they may play antagonistic functions in promoting tumor progression such as TP53_CTNNB1 of CSCP-L subtype and BAP1_TP53 of CSCP-H subtype. Different combinations of mutations may have affected tumor progression. Use fisher's exact test to analyze co-occurring or exclusiveness between mutant genes.
3.7. The comparison of tumor immune microenvironment of three CSCP subtype patients
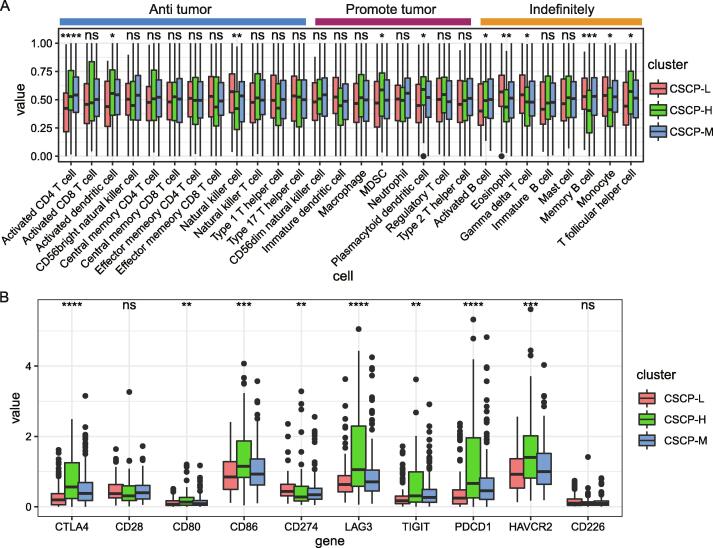
Herein, we further explored immune cell differences within three CSCP subtypes. Our results indicated that the ratio of MDSC (p < 0.05), plasmacytoid dendritic cell (p < 0.05) and T follicular helper cell (p < 0.05) in the CSCP-H subtype is respectively significant higher than other two CSCP subtypes (Fig. 7A). Of note, MDSC and plasmacytoid dendritic cells usually play a role in promoting cancer [30], [34], thereby their increase may lead to the worst prognosis of CSCP-H subtype patients. Additionally, we found that eosinophil (p < 0.01), gamma delta T cell (p < 0.05), memory B cell (p < 0.001) and monocyte (p < 0.05) in the CSCP-H subtype are respectively significantly decreasing (Fig. 7A). Interestingly, we found that the CSCP-H subtype has a high proportion of activated CD4 T cells (p < 0.001) and activated dendritic cells (p < 0.05) (Fig. 7A), which are inconsistent with their roles of anti-tumor by presenting tumor antigens. Moreover, CD8 T cells, as tumor-killing effector cells, have similar proportions among CSCP-L, CSCP-M and CSCP-H subtype, indicating that CD8 T cells are not the main cause of promoting the prognosis difference among three CSCP subtypes. These findings implied that the poorer prognosis of CSCP-H patients may be related to immune escape. We thus further detected these expression levels of multiple immune checkpoint molecules in three CSCP subtype patients. Surprisingly, we found that CTLA4, CD274 (PDL1), TIGIT, LAG3 and PDCD1 (PD-1) involved in inhibiting the immune activity of T cells are significantly highly expressed in the CSCP-H subtype (p < 0.01) (Fig. 7B). Especially, CD80 and CD86 are also significantly highly expressed in the CSCP-H subtype (p < 0.01) (Fig. 7B). Previous studies revealed that the B7 molecular ligand (CD80/CD86) is usually expressed in antigen presenting cells and can simultaneously bind to CD28 (p = ns) to activate T cell immunity or CTLA4 to inhibit T cell immunity [35], [36]. Taken together, our results suggested that CTLA4-mediated immune escape may exist in patients with CSCP-H subtype accompanied by a significant upregulation of CTLA4 instead of CD28, in particular this result can explain the incompetence of the increase of activated CD4 T cells and activated dendritic cells.
Fig. 7.
Differences of immune microenvironment in different CSCP subtypes. A:Differences of tumor immune infiltrating cells in different CSCP subtypes. B: Differences of immune checkpoint molecules in different CSCP subtypes. The abundance of immune infiltrating cells in tumor samples was calculated by single-sample gene set enrichment analysis (ssGSEA). The Mann-Whitney-Wilcoxon test is used to calculate the significance of immune cell abundance and immune checkpoints.
3.8. Construction of the prognostic model based on mRNAsi-related genes
Herein, we identified 10 robust prognostic markers (EIF3B, G6PC, SAC3D1, DYNLL1, PSMG3, TMEM147, SNRPA, SNRPD2, CYTOR, CPEB3) from 212 mRNAsi-related genes through univariate cox regression and multivariate cox regression analysis (Fig. S10). Among them, highly expressed G6PC and CPEB3 can act as protective factors to promote the survival of HCC patients, while these high expressions of remaining risk factors significantly reduce the survival rate of HCC patients (Fig. S10). Similarly, these 10 markers are closely associated to HCC patient's disease progression and tumor burden (Fig. 8A). We further used the prognostic model consisting of 10 markers to score the risk of patients and found that the survival rate of the high-risk group is significantly lower than that of the low-risk group with about 3 times cumulative deaths within 5 years (p < 0.0001) (Fig. 8B). These receiver operating characteristic curves (ROC) also proved that the prognostic model has a good accuracy and sensitivity with 1-year, 3-year and 5-year AUC value for 0.794, 0.733 and 0.753, respectively (Fig. 8C). Besides, the model risk score can still act as an independent prognostic factor by including clinical indicators G grade, T stage, AJCC stage and tumor burden as a covariate correction (p < 0.001) (Table 1). Of note, similar results were also verified in the ICGC cohort (Fig. 8D∼8F). Interestingly, we found strong associations between high and low risk groups and CSCP subtypes (Fig. S11). The high-risk group of the TCGA cohort had a higher proportion of patients with CSCP-H and CSCP-M subtypes and a lower proportion of patients with CSCP-L subtype, while the low-risk group had a higher proportion of patients with CSCP-L subtype and a lower proportion of patients with CSCP-H subtype (p < 0.001) (Fig. S11A)., Interestingly, this result was validated again in the ICGC cohort (p < 0.001) (Fig. S11B). Finally, to provide a clinically usable practical model, we constructed a nomogram model containing 10 markers (Fig. 8G). Clinicians can obtain the individual score of each marker and the total score according to their expression levels and the nomogram, which can be directly used to predict the survival rate of this patient in different years (Fig. 8G).
Fig. 8.
Identifing robust prognostic markers in mRNAsi-related genes. A: The relationship between robust markers in 10 mRNAsi-related genes and clinical facor of HCC in TCGA. B: Survival curve of HCC patients in high and low risk groups in TCGA. The risk score of patients is predicted by the model composed of 10 markers. C: The ROC curve is used to evaluate model reliability in TCGA. D: Survival curve of HCC patients in high and low risk groups in ICGC cohort. E: Use the ROC curve to evaluate the reliability of the model in the ICGC. F: Differences in risk scores of different AJCC stages in the ICGC cohort. G: A nomogram constructed based on 10 markers predicts the survival rate of HCC patients. Draw a vertical line between the expression value of each gene and points to get the corresponding score. The total risk score of HCC patients is obtained by adding the scores of all genes. Draw the vertical line between the patient's total risk score and the risk probability to obtain the 1-year, 3-year, and 5-year survival probabilities of HCC patients.
Table 1.
Univariate and multivariate analysis of model risk value and other clinical indicators.
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Hazard ratio (95%CI) | pvalue | Hazard ratio (95%CI) | pvalue | |
| Grade | 1.115(0.858–1.454) | 0.422 | 1.107(0.830–1.479) | 0.489 |
| T stage | 1.76(1.430–2.165) | <0.001 | 1.456(0.680–3.119) | 0.334 |
| AJCC stage | 1.789(1.436–2.229) | <0.001 | 1.010(0.447–2.287) | 0.98 |
| Status | 2.717(1.786–4.133) | <0.001 | 2.246(1.451–3.477) | <0.001 |
| Riskscore | 3.293(2.138–5.074) | <0.001 | 3.635(1.714–7.710) | <0.001 |
Note: The abbreviations in the table are as follows, which are derived from the guidelines of the American Joint Committee on Cancer (AJCC). Grade: A numerical value expressing the degree of abnormality of cancer cells. It is an indicator of differentiation and invasiveness. T stage: Extent of the primary cancer when the patient was first diagnosed. AJCC Stage: The extent of a cancer, that whether the disease has spread from the original site to other parts of the body. Status: The neoplasm cancer status when the patient was first diagnosed. Risk scores were predicted by a multivariate cox regression model constructed from 10 prognostic genes. The model was constructed by executing the coxph function and the patient's risk score was calculated by the predict function of the survival R package. The mathematical formula is: Riskscore = h0(t) * exp (β1X1 + β2X2 + … + βnXn). Xn represents 10 prognostic genes, βn represents the regression coefficient of the gene, exp represents the expression level of the gene, and h0(t) is a constant.
4. Disscusion
The mRNAsi has been used as a digital phenotype for the identification of CSC-related genes and diagnostic and prognostic markers for different cancer patients [19], [20], [21], but studies on mRNAsi-driven tumor heterogeneity are still sorely lacking. A previous report has demonstrated that the CSC-driven tumor heterogeneity can cause the differences of prognosis and treatment for HCC patients [37]. Obviously, systematically revealing the mechanism of CSC-driven tumor heterogeneity is helpful for further accurately distinguishing HCC patients into different subtypes and providing effectively targeted treatment. Remarkably, the tumor stemness related methylated locus has been applied to distinguish these molecular subtypes of prostate cancer [38]. Interestingly, our present works have identified 2 gene modules significantly associated with mRNAsi through the WGCNA (Fig. 2C). Among them, the positively correlated blue modules include 212 mRNAsi-related genes that can divide HCC patients into three molecular subtypes: CSCP-L, CSCP-M and CSCP-H (Fig. 2). Especially, HCC patients with CSCP-H subtype have the worst survival rate and tumor status, while patients with CSCP-L subtype have the best prognosis (Fig. 3). Importantly, three CSCP subtypes can be well validated in three other cohorts (Fig. 5 and Fig. S8), indicating that these three CSCP subtypes may have an important clinical application value for effectively monitoring and treating HCC patients. In contrast, this negatively correlated yellow module containes 52 mRNAsi-related genes. Although these 52 mRNAsi-related genes can separate patients into two subtypes with different expression patterns in the TCGA cohort, there is no significant difference in the survival rate between two subtypes (Fig. S12A∼S12C), particularly these results from the ICGC cohort are also inconsistent with those of the TCGA cohort, either in terms of survival curves or molecular signature expression patterns (Fig. S12D∼Fig. S12F), suggestting that 52 mRNAsi-related genes fo this yellow module cannot be used as a stable and effective typing tool for HCC patients.
The liver is an important metabolic organ, and its normal metabolism is necessary to ensure the good prognosis of patients [39]. The complement system has also been proved to be essential for maintaining human normal immunity [40]. Of note, our findings demonstrate that 45 mRNAsi-related genes in the CSCP-L subtype are mainly involved in amino acids and fatty acids metabolism as well as coagulation complement system (Fig. 4 and Fig. S3), and highly expressed CAT, G6PC and ABAT significantly promote the survival of HCC patients (Fig. S6), implying that 45 highly expressed mRNAsi-related genes are responsible for the good prognosis of HCC patients with CSCP-L subtype. In contrast, 167 mRNAsi-related genes in the CSCP-H subtype are significantly enriched in ribosomes, spliceosomes and pyrimidine metabolism-related pathways. Previous studies revealed that ribosomes are important protein synthesis organelles, in partular ribosomes can synthesize many certain proteins to induce the metastasis of cancer cells [41], [42], [43]. For example, ribosomes can synthesize some certain proteins to promote the migratation of epithelial-mesenchymal transition (EMT) during the tumor metastasis [42]. Remarkably, the EMT transition state can promote the production of circulating tumor cells and tumor stem cells, which can promote tumor cells to invade and infect surrounding cells, thereby helping them to acquire drug resistance [44], [45]. In our work, we find that these highly expressed ribosome-related genes RPL8, RPS21, RPL23A, RPL27, RPL38 do significantly reduce the survival rate of HCC patients (Fig. S6). Similarly, many spliceosome- and pyrimidine metabolism-related genes, such SNRPA, SNRPE, NME1, and NME2, have also been reported to be involved in the tumorigenesisprocess of various cancers [46], [47], [48], [49]. Interestingly, our study has shown that highly expressed NME1, SNRPA, SNRPC and SNRPE singnificatly reduce the survival rate of HCC patients (Fig. S6), indicating that 167 highly expressed mRNAsi-related genes may result in the poor prognosis for patients with CSCP-H subtype. Remarkably, these mRNAsi-related genes do not appear to be clearly stratified in the CSCP-M subtype, but they do present a transitional state of moderate expression (Fig. 2E), which may explain that the survival rate of patients with CSCP-M subtype is between CSCP-L and CSCP-H subtypes. Additionally, we attempted to identify specific DEGs of patients with CSCP-M subtype. Unfortunately, we did not find these feature genes that are significantly enriched in the CSCP-M subtype at the transcriptome level (Fig. S13A). This reason may be that CSCP-M is the transition state of CSCP-L and CSCP-H at the transcriptome level (Fig. S13B), thereby these specific characteristics of CSCP-M need to be explored from the proteome, DNA methylation or copy number variation. Especially, our works have verified the accuracy of three CSCP subtypes in three other independent data sets (Fig. 5 and Fig. S8), which means that three CSCP subtypes are widely presented in HCC patients and have real clinical significance for diagnosis and prognosis of HCC patients.
Remarkably, our works reveal that the CSCP-H subtype has a higher overall mutation rate than two other subtypes, in particular TP53 (Fig. 6). Studies indicated that the disorder of TP53 can lead to the detunings of ribosomal biosynthesis and protein synthesis. For example, TP53 can inhibit RNA Pol I-mediated transcriptions of ribosomal genes by preventing the interaction between SL1 and UBF [50]. TP53 can bind to the core initiation factor TF-IIIB of RNA Pol III to interfere the combination of other components (such as TF-IIIC2), thereby significantly inhibiting RNA Pol III-mediated ribosomal biosynthesis and translation [51], [52]. These results further support that the high-frequency mutation of TP53 in the CSCP-H subtype causes the abnormal activation of ribosome genes and accelerates tumor progression. Additionally, the proportion of cancer-promoting MDSC and plasmacytoid dendritic cell (pDC) in the CSCP-H subtype is significant higher than two other subtypes (Fig. 7A). Previous studies reported that the accumulation of MDSCs is significantly related to the decrease of tumor infiltrating lymphocytes and the increase of tumorigenicity in HCC [53], while myeloid LAMP3 + DC cells are related to tumor migration to lymph nodes [54]. Interestingly, our findings indicate that some neutral cell types, such as eosinophils, memory B cells and monocytes, are also significantly reduced in the CSCP-H subtype (Fig. 7A). Many works have revealed that eosinophil-mediated anti-tumor response is necessary for DPP4 inhibitor to treat HCC and breast cancer [55], and eosinophils activated by IL5 and eotaxin have anti-tumor activity in HCC [56], as well as B cells in the tertiary lymphatic structure are found to be closely related to the patient's response to immune checkpoint inhibitor therapy [57]. Herein, we do find that activated CD4 T cells and activated dendritic cells with anti-tumor activity are enriched in the CSCP-H subtype. Activated CD4 T cell and activated dendritic cell are the main cell types for antigen presentation [58], so their increases should not promote the prognosis of the CSCP-H subtype patients. Of note, these high expressions of multiple immune checkpoint genes have been suggested to inhibit the immune activity of T cells and further result in the immune escape [59], [60], [61], [62], [63]. We thus suggest that highly expressed CTLA4, CD274 (PDL1), TIGIT, HAVCR2 (TIM3), LAG3 and PDCD1(PD-1) may lead to the immune escape and be responsible for the worst prognosis of the CSCP-H subtype patients (Fig. 7B). Especially, these highly expressed immune checkpoints and the higher mutation load of the CSCP-H subtype mean that they are likely to benefit for immunotherapy [64].
In this work, we establish the prognostic model, which consists of 10 mRNAsi-related genes including down-regulated G6PC and CPEB3 and up-regulated EIF3B, SAC3D1, SNRPA, DYNLL1, CYTOR, PSMG3, TMEM147 and SNRPD2 (Fig. 8 and Fig. S10). Previous studies have shown that the inactivating mutation or down-regulation of G6PC gene can cause glycogen accumulation to induce liver cancer occurrence [65], [66]. The CPEB3-mediated translational inhibition of MTDH can inhibit the progression of HCC and be used as a prognostic marker of liver cancer [67]. EIF3B can induce C-MET protein synthesis to promote cell proliferation and invasion of HCC [68]. SAC3D1 can act as a new prognostic marker for HCC [69]. SNRPA has been found to be differentially expressed in other cohorts [70], [71], but its relationship with HCC has to be further studied. The up-regulation of methylation-driven DYNLL1 is associated with HCC mortality and higher tumor stages [72]. LncRNA CYTOR can affect the proliferation, cell cycle and apoptosis of liver cancer cells [73], and it can also promote colon cancer EMT and metastasis [74]. However, the relationship between PSMG3, TMEM147, SNRPD2 and HCC is still rarely reported, so we suggest that they may serve as new markers and therapeutic targets for HCC patients in future studies.
In this study, we have constructed three molecular subtypes CSCP-L, CSCP-M and CSCP-H associated with mRNAsi in HCC. Our results demonstrated that patients with the CSCP-H subtype have the worst prognosis, while patients with the CSCP-L subtype have the best prognosis, and patients with the CSCP-M subtype have a moderate prognosis, implying that 212 mRNAsi-related genes might act as a potential molecular typing for clinical application of HCC. Whilst our findings suggested that developing diagnostic kits targeting these 212 genes should be also a good option for HCC patient diagnosis. Additionally, HCC patients are classified into three CSCP subtypes, which will be helpful for judging and predicting the prognosis and treatment plan of HCC patients. For example, clinicians may employ more conservative treatment based on the favorable molecular profile of patients with the CSCP-L subtype. Conversely, clinicians may need to give patients with the CSCP-H subtype more monitoring and more aggressive treatment regimens due to their own malignant molecular profile. In particular, HCC patients with CSCP-H subtype may be considered as a candidate for immunotherapy (e.g. anti-CTLA4) due to their high expressions of multiple immune checkpoints and a higher mutational load. However, our present work belongs to the category of basic research, and the clinical significance and practicability of CSCPs still need to be tried and tested clinically.
In summary, our study provides important enlightenments for molecular typing and prognostic prediction of HCC patients.
Statement of ethics
This article titled “mRNAsi-related genes can effectively distinguish hepatocellular carcinoma into new molecular subtypes” was written by Canbiao Wang, Shijie Qin, Wanwan Pan, Xuejia Shi, Hanyu Gao, Ping Jin, Xinyi Xia and Fei Ma.
The main data of this study were obtained from public databases, and no ethical permission was involved or required.
This research was funded by grants from the National Natural Science Foundation of China (No. 31970477) and the Natural Science Foundation of Jiangsu Province (No. BK20191368).
We promise that there will be no plagiarism and has not been published in any journal or platform. All authors have read and approved the manuscript and we declare that there is no conflict of interest.
Acknowledgments
Acknowledgments
We thank our colleagues for their suggestions and criticisms on the manuscript.
Funding
This research was funded by grants from the National Natural Science Foundation of China (No. 31970477) and the Natural Science Foundation of Jiangsu Province (No. BK20191368).
Author contributions
Shijie Qin and Fei Ma conceived the study. Canbiao Wang, Shijie Qin and Xuejia Shi collected omics data and conducted analysis. Xuejia Shi and Wanwan Pan visualized diagrams. Shijie Qin and Canbiao Wang wrote the draft. Fei Ma, Xinyi Xia and Ping Jin revised the draft. Ping Jin and Wanwan Pan supervised the project progress. Hanyu Gao, Wanwan Pan and Xinyi Xia participated in the commentary of the manuscript.
Conflicts of interest
We declare that we have no conflict of interest.
Availability of data and materials
All the data supporting the findings of this study are available within the article and its supplementary information files.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.06.011.
Contributor Information
Ping Jin, Email: jinping8312@163.com.
Xinyi Xia, Email: xinyixia@nju.edu.cn.
Fei Ma, Email: mafei01@tsinghua.org.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Huang X., Gan G., Wang X., Xu T., Xie W. The HGF-MET axis coordinates liver cancer metabolism and autophagy for chemotherapeutic resistance. Autophagy. 2019;15(7):1258–1279. doi: 10.1080/15548627.2019.1580105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Y., Chen L., Gu J., Zhang H., Yuan J., Lian Q., et al. Recurrently deregulated lncRNAs in hepatocellular carcinoma. Nat Commun. 2017;8:14421. doi: 10.1038/ncomms14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bidkhori G, Benfeitas R, Klevstig M, Zhang C, Nielsen J, Uhlen M, Boren J, Mardinoglu A: Metabolic network-based stratification of hepatocellular carcinoma reveals three distinct tumor subtypes. Proc Natl Acad Sci U S A 2018, 115(50):E11874-E11883. [DOI] [PMC free article] [PubMed]
- 4.Calderaro J., Couchy G., Imbeaud S., Amaddeo G., Letouze E., Blanc J.F., et al. Histological subtypes of hepatocellular carcinoma are related to gene mutations and molecular tumour classification. J Hepatol. 2017;67(4):727–738. doi: 10.1016/j.jhep.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 5.Chaisaingmongkol J., Budhu A., Dang H., Rabibhadana S., Pupacdi B., Kwon S.M., et al. Common molecular subtypes among Asian hepatocellular carcinoma and cholangiocarcinoma. Cancer Cell. 2017;32(1):57–70 e53. doi: 10.1016/j.ccell.2017.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li W., Wang H., Ma Z., Zhang J., Ou-Yang W., Qi Y., et al. Multi-omics analysis of microenvironment characteristics and immune escape mechanisms of hepatocellular carcinoma. Front Oncol. 2019;9:1019. doi: 10.3389/fonc.2019.01019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng J., Wei D., Ji Y., Chen L., Yang L., Li G., et al. Integrative analysis of DNA methylation and gene expression reveals hepatocellular carcinoma-specific diagnostic biomarkers. Genome Med. 2018;10(1):42. doi: 10.1186/s13073-018-0548-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao Q., Zhu H., Dong L., Shi W., Chen R., Song Z., et al. Integrated proteogenomic characterization of HBV-related hepatocellular carcinoma. Cell. 2019;179(5):1240. doi: 10.1016/j.cell.2019.10.038. [DOI] [PubMed] [Google Scholar]
- 9.Zhao Y, Zhang L, Zhang Y, Meng B, Ying W, Qian X: Identification of hedgehog signaling as a potential oncogenic driver in an aggressive subclass of human hepatocellular carcinoma: A reanalysis of the TCGA cohort. (1869-1889 (Electronic)). [DOI] [PubMed]
- 10.Cancer Genome Atlas Research Network. Electronic address wbe, Cancer Genome Atlas Research N: Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017, 169(7):1327-1341 e1323. [DOI] [PMC free article] [PubMed]
- 11.Roulot A., Hequet D., Guinebretiere J.M., Vincent-Salomon A., Lerebours F., Dubot C., et al. Tumoral heterogeneity of breast cancer. Ann Biol Clin (Paris) 2016;74(6):653–660. doi: 10.1684/abc.2016.1192. [DOI] [PubMed] [Google Scholar]
- 12.Lian H., Han Y.P., Zhang Y.C., Zhao Y., Yan S., Li Q.F., et al. Integrative analysis of gene expression and DNA methylation through one-class logistic regression machine learning identifies stemness features in medulloblastoma. Mol Oncol. 2019;13(10):2227–2245. doi: 10.1002/1878-0261.12557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang K., Che S., Pan C., Su Z., Zheng S., Yang S., et al. The SHH/Gli axis regulates CD90-mediated liver cancer stem cell function by activating the IL6/JAK2 pathway. J Cell Mol Med. 2018;22(7):3679–3690. doi: 10.1111/jcmm.13651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyssiotis C.A., Kimmelman A.C. Metabolic Interactions in the Tumor Microenvironment. Trends Cell Biol. 2017;27(11):863–875. doi: 10.1016/j.tcb.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin S., Long X., Zhao Q., Zhao W. Co-Expression network analysis identified genes associated with cancer stem cell characteristics in lung squamous cell carcinoma. Cancer Invest. 2020;38(1):13–22. doi: 10.1080/07357907.2019.1697281. [DOI] [PubMed] [Google Scholar]
- 16.Nio K., Yamashita T., Kaneko S. The evolving concept of liver cancer stem cells. Mol Cancer. 2017;16 doi: 10.1186/s12943-016-0572-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamashita T., Wang X.W. Cancer stem cells in the development of liver cancer. J Clin Invest. 2013;123(5):1911–1918. doi: 10.1172/JCI66024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malta T.M., Sokolov A., Gentles A.J., Burzykowski T., Poisson L., Weinstein J.N., et al. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell. 2018;173(2):338–354 e315. doi: 10.1016/j.cell.2018.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang YA-O, Tseng JT, Lien IC, Li F, Wu W, Li H: mRNAsi index: machine learning in mining lung adenocarcinoma stem cell biomarkers. LID - 10.3390/genes11030257 [doi] LID - 257. (2073-4425 (Electronic)). [DOI] [PMC free article] [PubMed]
- 20.Lian H, Han YP, Zhang YC, Zhao Y, Yan S, Li QF, Wang BC, Wang JJ, Meng W, Yang J et al: Integrative analysis of gene expression and DNA methylation through one-class logistic regression machine learning identifies stemness features in medulloblastoma. (1878-0261 (Electronic)). [DOI] [PMC free article] [PubMed]
- 21.Zhang M, Wang X, Chen X, Guo F, Hong J: Prognostic value of a stemness index-associated signature in primary lower-grade glioma. (1664-8021 (Print)). [DOI] [PMC free article] [PubMed]
- 22.Roessler S., Jia H.L., Budhu A., Forgues M., Ye Q.H., Lee J.S., et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70(24):10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roessler S., Long E.L., Budhu A., Chen Y., Zhao X., Ji J., et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology. 2012;142(4):957–966 e912. doi: 10.1053/j.gastro.2011.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilkerson M.D., Hayes D.N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayakonda A., Lin D.C., Assenov Y., Plass C., Koeffler H.P. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28(11):1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Charoentong P., Finotello F., Angelova M., Mayer C., Efremova M., Rieder D., et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18(1):248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 31.Yoshihara K., Shahmoradgoli M., Martinez E., Vegesna R., Kim H., Torres-Garcia W., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sinha S., Thomas D., Chan S., Gao Y., Brunen D., Torabi D., et al. Systematic discovery of mutation-specific synthetic lethals by mining pan-cancer human primary tumor data. Nat Commun. 2017;8:15580. doi: 10.1038/ncomms15580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dobzhansky T: Genetics of natural populations; recombination and variability in populations of Drosophila pseudoobscura. (0016-6731 (Print)). [DOI] [PMC free article] [PubMed]
- 34.Jia Q., Wu W., Wang Y., Alexander P.B., Sun C., Gong Z., et al. Local mutational diversity drives intratumoral immune heterogeneity in non-small cell lung cancer. Nat Commun. 2018;9(1):5361. doi: 10.1038/s41467-018-07767-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greene J.L., Leytze G.M., Emswiler J., Peach R., Bajorath J., Cosand W., et al. Covalent dimerization of CD28/CTLA-4 and oligomerization of CD80/CD86 regulate T cell costimulatory interactions. J Biol Chem. 1996;271(43):26762–26771. doi: 10.1074/jbc.271.43.26762. [DOI] [PubMed] [Google Scholar]
- 36.Slavik J.M., Hutchcroft J.E., Bierer B.E. CD28/CTLA-4 and CD80/CD86 families: signaling and function. Immunol Res. 1999;19(1):1–24. doi: 10.1007/BF02786473. [DOI] [PubMed] [Google Scholar]
- 37.Chang JC: Cancer stem cells: Role in tumor growth, recurrence, metastasis, and treatment resistance. Medicine (Baltimore) 2016, 95(1 Suppl 1):S20-S25. [DOI] [PMC free article] [PubMed]
- 38.Li S., Yue D., Chen X., Wang L., Li J., Ping Y., et al. Epigenetic regulation of CD271, a potential cancer stem cell marker associated with chemoresistance and metastatic capacity. Oncol Rep. 2015;33(1):425–432. doi: 10.3892/or.2014.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ding H.R., Wang J.L., Ren H.Z., Shi X.L. Lipometabolism and glycometabolism in liver diseases. Biomed Res Int. 2018;2018:1287127. doi: 10.1155/2018/1287127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Defendi F., Thielens N.M., Clavarino G., Cesbron J.Y., Dumestre-Perard C. The immunopathology of complement proteins and innate immunity in autoimmune disease. Clin Rev Allergy Immunol. 2020;58(2):229–251. doi: 10.1007/s12016-019-08774-5. [DOI] [PubMed] [Google Scholar]
- 41.Madan B., Harmston N., Nallan G., Montoya A., Faull P., Petretto E., et al. Temporal dynamics of Wnt-dependent transcriptome reveal an oncogenic Wnt/MYC/ribosome axis. J Clin Invest. 2018;128(12):5620–5633. doi: 10.1172/JCI122383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prakash V., Carson B.B., Feenstra J.M., Dass R.A., Sekyrova P., Hoshino A., et al. Ribosome biogenesis during cell cycle arrest fuels EMT in development and disease. Nat Commun. 2019;10(1):2110. doi: 10.1038/s41467-019-10100-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou W., Ouyang J., Li J., Liu F., An T., Cheng L., et al. MRPS17 promotes invasion and metastasis through PI3K/AKT signal pathway and could be potential prognostic marker for gastric cancer. J Cancer. 2021;12(16):4849–4861. doi: 10.7150/jca.55719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katsuno Y., Lamouille S., Derynck R. TGF-β signaling and epithelial-mesenchymal transition in cancer progression. Curr Opin Oncol. 2013;25(1):76–84. doi: 10.1097/CCO.0b013e32835b6371. [DOI] [PubMed] [Google Scholar]
- 45.Saitoh M. Involvement of partial EMT in cancer progression. J Biochem. 2018;164(4):257–264. doi: 10.1093/jb/mvy047. [DOI] [PubMed] [Google Scholar]
- 46.Dou N, Yang D, Yu S, Wu B, Gao Y, Li YA-OX: SNRPA enhances tumour cell growth in gastric cancer through modulating NGF expression. (1365-2184 (Electronic)). [DOI] [PMC free article] [PubMed]
- 47.Jia D, Wei L Fau - Guo W, Guo W Fau - Zha R, Zha R Fau - Bao M, Bao M Fau - Chen Z, Chen Z Fau - Zhao Y, Zhao Y Fau - Ge C, Ge C Fau - Zhao F, Zhao F Fau - Chen T, Chen T Fau - Yao M et al: Genome-wide copy number analyses identified novel cancer genes in hepatocellular carcinoma. (1527-3350 (Electronic)). [DOI] [PubMed]
- 48.Hindupur SK, Colombi M, Fuhs SR, Matter MS, Guri Y, Adam K, Cornu M, Piscuoglio S, Ng CKY, Betz C et al: The protein histidine phosphatase LHPP is a tumour suppressor. (1476-4687 (Electronic)). [DOI] [PMC free article] [PubMed]
- 49.Yamaguchi A, Urano T Fau - Fushida S, Fushida S Fau - Furukawa K, Furukawa K Fau - Nishimura G, Nishimura G Fau - Yonemura Y, Yonemura Y Fau - Miyazaki I, Miyazaki I Fau - Nakagawara G, Nakagawara G Fau - Shiku H, Shiku H: Inverse association of nm23-H1 expression by colorectal cancer with liver metastasis. (0007-0920 (Print)). [DOI] [PMC free article] [PubMed]
- 50.Zhai W., Comai L. Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol Cell Biol. 2000;20(16):5930–5938. doi: 10.1128/mcb.20.16.5930-5938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.White R.J., Trouche D., Martin K., Jackson S.P., Kouzarides T. Repression of RNA polymerase III transcription by the retinoblastoma protein. Nature. 1996;382(6586):88–90. doi: 10.1038/382088a0. [DOI] [PubMed] [Google Scholar]
- 52.Cairns CA, White RJ: p53 is a general repressor of RNA polymerase III transcription. (0261-4189 (Print)). [DOI] [PMC free article] [PubMed]
- 53.Liu Y.T., Tseng T.C., Soong R.S., Peng C.Y., Cheng Y.H., Huang S.F., et al. A novel spontaneous hepatocellular carcinoma mouse model for studying T-cell exhaustion in the tumor microenvironment. J Immunother Cancer. 2018;6(1):144. doi: 10.1186/s40425-018-0462-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Q., He Y., Luo N., Patel S.J., Han Y., Gao R., et al. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell. 2019;179(4):829–845 e820. doi: 10.1016/j.cell.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 55.Hollande C., Boussier J., Ziai J., Nozawa T., Bondet V., Phung W., et al. Inhibition of the dipeptidyl peptidase DPP4 (CD26) reveals IL-33-dependent eosinophil-mediated control of tumor growth. Nat Immunol. 2019;20(3):257–264. doi: 10.1038/s41590-019-0321-5. [DOI] [PubMed] [Google Scholar]
- 56.Kataoka S., Konishi Y., Nishio Y., Fujikawa-Adachi K., Tominaga A. Antitumor activity of eosinophils activated by IL-5 and eotaxin against hepatocellular carcinoma. DNA Cell Biol. 2004;23(9):549–560. doi: 10.1089/dna.2004.23.549. [DOI] [PubMed] [Google Scholar]
- 57.Cabrita R., Lauss M., Sanna A., Donia M., Skaarup Larsen M., Mitra S., et al. Author Correction: Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature. 2020;580(7801):E1. doi: 10.1038/s41586-020-2155-6. [DOI] [PubMed] [Google Scholar]
- 58.Gardner A., Ruffell B. Dendritic cells and cancer immunity. Trends Immunol. 2016;37(12):855–865. doi: 10.1016/j.it.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han Y., Liu D., Li L. PD-1/PD-L1 pathway: current researches in cancer. Am J Cancer Res. 2020;10(3):727–742. [PMC free article] [PubMed] [Google Scholar]
- 60.Chen D.S., Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541(7637):321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 61.Meng F., Li L., Lu F., Yue J., Liu Z., Zhang W., et al. Overexpression of TIGIT in NK and T cells contributes to tumor immune escape in myelodysplastic syndromes. Front Oncol. 2020;10:1595. doi: 10.3389/fonc.2020.01595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Das M., Zhu C., Kuchroo V.K. Tim-3 and its role in regulating anti-tumor immunity. Immunol Rev. 2017;276(1):97–111. doi: 10.1111/imr.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Woo S.R., Turnis M.E., Goldberg M.V., Bankoti J., Selby M., Nirschl C.J., et al. Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T-cell function to promote tumoral immune escape. Cancer Res. 2012;72(4):917–927. doi: 10.1158/0008-5472.CAN-11-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Osipov A., Murphy A., Zheng L. From immune checkpoints to vaccines: the past, present and future of cancer immunotherapy. Adv Cancer Res. 2019;143:63–144. doi: 10.1016/bs.acr.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 65.Liu Q., Li J., Zhang W., Xiao C., Zhang S., Nian C., et al. Glycogen accumulation and phase separation drives liver tumor initiation. Cell. 2021 doi: 10.1016/j.cell.2021.10.001. [DOI] [PubMed] [Google Scholar]
- 66.Resaz R, Vanni C, Segalerba D, Sementa AR, Mastracci L, Grillo F, Murgia D, Bosco MC, Chou JY, Barbieri O et al: Development of hepatocellular adenomas and carcinomas in mice with liver-specific G6Pase-α deficiency. (1754-8411 (Electronic)). [DOI] [PMC free article] [PubMed]
- 67.Zhang H., Zou C., Qiu Z., Li Q., Chen M., Wang D., et al. CPEB3-mediated MTDH mRNA translational suppression restrains hepatocellular carcinoma progression. Cell Death Dis. 2020;11(9):792. doi: 10.1038/s41419-020-02984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fang Q.L., Zhou J.Y., Xiong Y., Xie C.R., Wang F.Q., Li Y.T., et al. Long non-coding RNA RP11-284P20.2 promotes cell proliferation and invasion in hepatocellular carcinoma by recruiting EIF3b to induce c-met protein synthesis. Biosci Rep. 2020;40(3) doi: 10.1042/BSR20200297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Han M.E., Kim J.Y., Kim G.H., Park S.Y., Kim Y.H., Oh S.O. SAC3D1: a novel prognostic marker in hepatocellular carcinoma. Sci Rep. 2018;8(1):15608. doi: 10.1038/s41598-018-34129-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dou N., Yang D., Yu S., Wu B., Gao Y., Li Y. SNRPA enhances tumour cell growth in gastric cancer through modulating NGF expression. Cell Prolif. 2018;51(5):e12484. doi: 10.1111/cpr.12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yuan M., Yu C., Chen X., Wu Y. Investigation on potential correlation between small nuclear ribonucleoprotein polypeptide A and lung cancer. Front Genet. 2020;11 doi: 10.3389/fgene.2020.610704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Berkel C., Cacan E. DYNLL1 is hypomethylated and upregulated in a tumor stage- and grade-dependent manner and associated with increased mortality in hepatocellular carcinoma. Exp Mol Pathol. 2020;117 doi: 10.1016/j.yexmp.2020.104567. [DOI] [PubMed] [Google Scholar]
- 73.Hu B., Yang X.B., Yang X., Sang X.T. LncRNA CYTOR affects the proliferation, cell cycle and apoptosis of hepatocellular carcinoma cells by regulating the miR-125b-5p/KIAA1522 axis. Aging (Albany NY) 2020;13(2):2626–2639. doi: 10.18632/aging.202306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yue B., Liu C., Sun H., Liu M., Song C., Cui R., et al. A positive feed-forward loop between LncRNA-CYTOR and Wnt/β-catenin signaling promotes metastasis of colon cancer. Mol Ther. 2018;26(5):1287–1298. doi: 10.1016/j.ymthe.2018.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data supporting the findings of this study are available within the article and its supplementary information files.