Summary
Skeletal muscle stem cells, or satellite cells (SCs), are essential to regenerate and maintain muscle. Quiescent SCs reside in an asymmetric niche between the basal lamina and myofiber membrane. To repair muscle, SCs activate, proliferate, and differentiate, fusing to repair myofibers or reacquiring quiescence to replenish the SC niche. Little is known about when SCs reacquire quiescence during regeneration or the cellular processes that direct SC fate decisions. We find that most SCs reacquire quiescence 5–10 days after muscle injury, following differentiation and fusion of most cells to regenerate myofibers. Single-cell sequencing of myogenic cells in regenerating muscle identifies SCs reacquiring quiescence and reveals that noncell autonomous signaling networks influence SC fate decisions during regeneration. SC transplantation experiments confirm that the regenerating environment influences SC fate. We define a window for SC repopulation of the niche, emphasizing the temporal contribution of the regenerative muscle environment on SC fate.
Subject areas: Cell biology, Stem cells research, Omics, Transcriptomics
Graphical abstract
Highlights
-
•
Satellite cells primarily produce myonuclei in the first 4 days following injury
-
•
Most satellite cells reacquire quiescence 5–7 days following injury
-
•
Asymmetric division pathways are elevated in satellite cells reacquiring quiescence
-
•
The regenerating muscle environment directs satellite cell fate decisions
Cell biology; Stem cells research; Omics; Transcriptomics.
Introduction
Skeletal muscle, essential for locomotion, respiration, and longevity, possesses a remarkable regenerative capacity dependent on resident muscle stem cells, satellite cells (SC) (Lepper et al., 2011; Murphy et al., 2011; Sambasivan et al., 2011). SCs are quiescent, residing in an asymmetric niche bounded by the basal lamina on one side and the myofiber on the other (Mauro, 1961). In response to muscle injury, SCs activate, exit quiescence, and begin proliferating between 24 and 48 h postinjury (Cornelison and Wold, 1997; Siegel et al., 2011). The resulting myoblasts may differentiate and fuse to regenerate the damaged, multinucleated myofibers, continue to proliferate, or reacquire quiescence, replenishing the SC pool.
Replenishing and maintaining a sufficient SC pool is critical to sustaining muscle health and regenerative capacity. Muscle regeneration is abolished when SCs are lost (Lepper et al., 2011; Murphy et al., 2011; Sambasivan et al., 2011), and decreased SC function is implicated in aging (Collins et al., 2007; Sacco et al., 2010; Bernet et al., 2014; Sousa-Victor et al., 2014) and myopathies (Heslop et al., 2000). By returning to the niche and reacquiring quiescence, myoblasts replenish the SC pool, which can occur via symmetric division, giving rise to two quiescent daughter SCs, or asymmetric division where one MyoD positive myoblast daughter cell either proliferates or differentiates and the other MyoD negative daughter reacquires quiescence (Conboy and Rando, 2002; Kuang et al., 2007; Troy et al., 2012). Asymmetric division is regulated by Notch (Conboy and Rando, 2002; Kuang et al., 2007), Wnt7a (Le Grand et al., 2009), EGFR (Wang et al., 2019), and downstream effectors Vangl2 (Le Grand et al., 2009), p38α/β MAPK (Troy et al., 2012; Bernet et al., 2014), and dystrophin (Dumont et al., 2015), which recruit the planar cell polarity pathway to polarize Myf5 and MyoD to establish distinct daughter cell fates. Despite this knowledge, the timing and molecular mechanisms regulating SC replenishment in vivo remain unexplored.
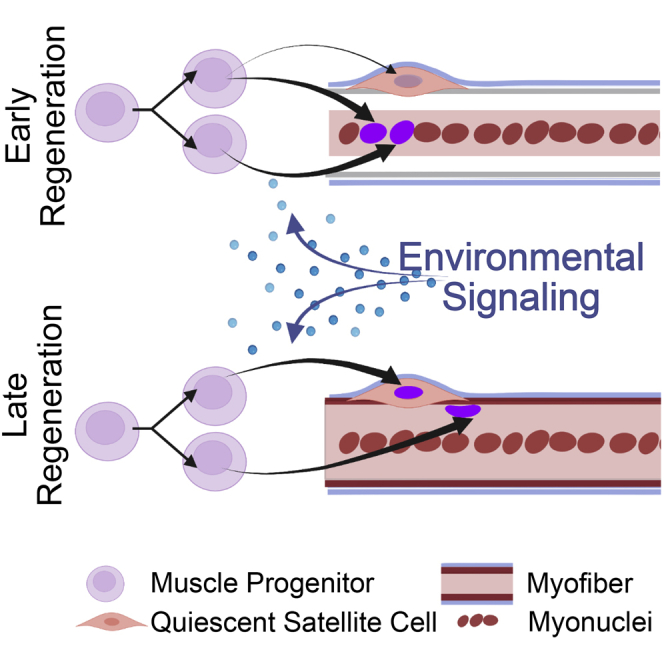
Employing in vivo DNA base-labeling lineage tracing experiments, we found the majority of SCs re-establish quiescence between 5 dpi (days postinjury) and 10 dpi, following the formation of new myofibers, much later than expected from previous work examining asymmetric division in myofiber cultures where asymmetric divisions occur during the initial cell cycle following SC activation (Kuang et al., 2007; Troy et al., 2012; Dumont et al., 2015; Wang et al., 2019). We performed single-cell RNA sequencing (scRNA-Seq) at 4 dpi, when most myonuclear formation is completed, and at 7 dpi, when SCs re-occupy their niche, to identify gene expression changes in myogenic cells and the signaling pathways driving repopulation of the niche by quiescent SCs. We identified signals in nonmyogenic cell populations that may direct SC cell fate via noncell autonomous signaling. Engraftment of donor SCs into the stem cell niche as quiescent SCs is significantly enhanced if transplanted during the time when SCs re-occupy their niche, confirming the regenerative environment as a key driver of SC fate decisions. Thus, the coordinated signaling of the regenerating muscle environment appears to prioritize myofiber reconstruction and then promote SC pool replenishment.
Results
The majority of SCs reacquire quiescence during days 5–10 postinjury
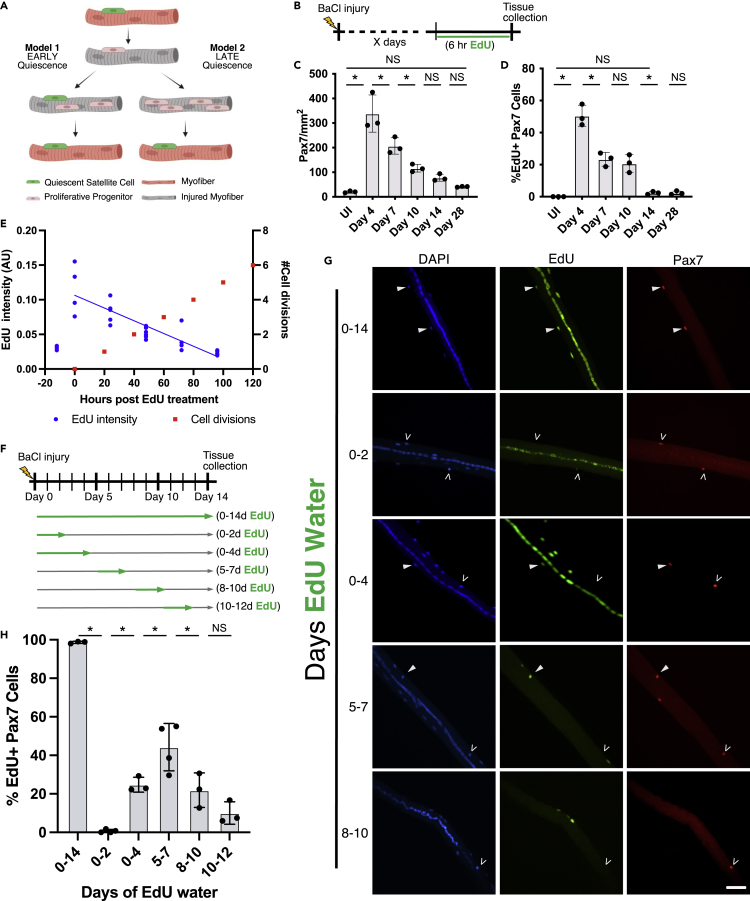
The timing and kinetics of SC expansion, myonuclear generation, and replenishment of the SC niche following a muscle injury are largely unexplored. Based on examination of asymmetric division on cultured myofibers, SC generation is generally accepted to occur via asymmetric division during the initial cell cycle following SC activation (Kuang et al., 2007; Troy et al., 2012; Bernet et al., 2014). If SCs reacquire quiescence during this initial cell division, then the replenished SC pool would undergo fewer cell divisions, suggesting prioritization of replenishing the SC pool early after muscle injury. By contrast, if the majority of SCs reacquire quiescence later in regeneration, then SCs would have undergone more cell divisions, suggesting an initial prioritization for myofiber reconstruction (Figure 1A). To determine when SCs reacquire quiescence in vivo, we lineage-traced SCs with timed pulses of 5-ethynyl-2′-deoxyuridine (EdU) following a muscle injury.
Figure 1.
SCs primarily reacquire quiescence between 5 dpi and 7 dpi
(A) Schematic depicting two simplified models for SC replenishment during regeneration.
(B) Schematic showing 6 h of EdU administration, in three injections 2 h apart, immediately prior to collection on the specified day following injury.
(C) Pax7+ cell numbers quantified per mm2 in tissue sections from uninjured (UI) or injured TA muscle, indicated with the day postinjury.
(D) Percent Pax7+/EdU + cells in tissue sections from injured TA muscle.
(E) Quantification of EdU persistence in proliferating C2C12 cells. The left y axis indicates EdU fluorescence intensity in arbitrary units (AU), and the right y axis indicates number of cell divisions. The x axis indicates hours since EdU withdrawal. Fluorescence intensity is plotted in blue and cell divisions in red. n = technical replicates of coverslips. Analysis by linear regression R2 = 0.7471.
(F) Experimental design schematic for EdU incorporation in DNA-lineage tracing experiments. Green arrows demarcate EdU application timing.
(G) Representative images of extensor digitorum longus (EDL) myofibers collected at 14 dpi after the indicated application of EdU. EdU+/Pax7+ double-positive cells (white arrows) and EdU-/Pax7+ SCs (white carets) are identified. Scale bar: 40 μm.
(H) EdU + SCs quantified on myofibers containing central nuclei exposed to EdU 0–14 days, 0–4 days, 0–2 days, 5–7 days, 8–10 days, or 10–12 days postinjury. All experiments, except EdU persistence in C2C12 cells, were performed as biological triplicates where each n represents an individual mouse and >100 Pax7+ cells were scored per mouse. For all panels, statistical significance tested by ANOVA. ∗p value < 0.05. For all data are represented as mean ± SD. See also Figure S1.
To assess SC and myoblast division kinetics following a muscle injury, we collected tibialis anterior (TA) muscles at various days after BaCl2 injury (days 4, 7, 10, 14, and 28). Because DNA base analogs are rapidly diluted (Matiašová et al., 2014), three injections of EdU 2 h apart were given immediately before tissue collection (Figure 1B). SCs and myoblasts were identified by Pax7 immunoreactivity and tissue sections processed for EdU incorporation (Figure S1). Once activated following injury, myoblasts divide rapidly, increasing over 20-fold by 4 dpi, then gradually decline to within 2-fold of the initial SC number by 28 dpi (Figure 1C). Thus, most myogenic progenitors are produced early in regeneration and then decline, most likely by terminal differentiation and fusion to form myofibers. Without injury, we detected no Pax7+/EdU + cells following the three EdU injections before muscle harvest (Figure 1D). The highest percentage of Pax7+/EdU + cells was detected at 4 dpi (50%), which declined to 23% at 7 dpi, and remained at 21% at 10 dpi (Figure 1D). The lowest percentage of Pax7+/EdU + cells was detected at 14 dpi (2%) with no further decrease by day 28 dpi (Figure 1D). The dramatic decrease in percentage of dividing myogenic cells between 4 dpi and 7 dpi indicates that myogenic cells are leaving the cell cycle either by terminal differentiation or by acquisition of quiescence in the niche. The minimal percentage of myogenic cells dividing at day 14 indicates that the majority of myonuclei and replenished SCs were produced earlier during regeneration.
Because the majority of SCs and myonuclei are produced by 14dpi, we asked precisely when during regeneration SCs reacquire quiescence. Because we rely on EdU incorporation for lineage tracing to estimate when cells exit the cell cycle and EdU signal is diluted each cell cycle, we first performed EdU pulse-chase experiments to determine how long the EdU signal remained detectable in dividing cells. C2C12 cells were grown in the presence of EdU for 12 h, then EdU was removed, and cells were evaluated for EdU fluorescence intensity every 24 h (Figure 1E). EdU intensity decreases linearly and was dimly detectable at 72 h after EdU removal and no longer detectable by 96 h post EdU removal (Figure 1E). Thus, the EdU signal persists for three cell divisions and is not detectable by the fourth cell division. Estimates of muscle precursor division rates, following the first division after activation, range from 8 to 17 h (Zammit et al., 2002; Siegel et al., 2011; Troy et al., 2012), so EdU + nuclei will have exited the cell cycle within 24–51 h following cessation of EdU administration in vivo.
To determine when SCs reacquire quiescence following an injury, mice were given EdU in drinking water in timed pulses during regeneration. Extensor digitorum longus (EDL) muscles were collected at 14 dpi and assessed for EdU incorporation in myofibers with centrally located nuclei (Schematic in Figure 1F). EdU + SCs and myonuclei were quantified from individual regenerated myofibers. SCs were identified by Pax7 immunoreactivity and myonuclei were identified as Pax7 negative nuclei located inside the myofiber. As EdU is detectable for three cell divisions (Figure 1E) before being diluted below detectable levels, EdU + SCs and EdU + myonuclei exited the cell cycle within three cell divisions of EdU removal. All myonuclei in and all SCs on myofibers with central nuclei were EdU + when EdU is provided for 0–14 dpi (Figures 1G and 1H). In contrast, on myofibers from mice provided EdU water for 0–2 dpi, less than 1% of SCs were EdU+ and centrally located myonuclei were labeled with variable intensity (Figures 1G and 1H). Because the initial SC cell division following injury occurs between 24 and 48 h (Jones et al., 2005; Troy et al., 2012; Webster et al., 2016), few, if any, SCs reacquire quiescence during the first cell division following injury in vivo, countering the proposed early SC replenishment model (Kuang et al., 2007; Troy et al., 2012; Dumont et al., 2015; Wang et al., 2019), instead favoring later SC replenishment (Figure 1H).
The variable EdU signal in central myonuclei is consistent with rapidly proliferating myogenic progenitors, diluting EdU following EdU removal at 2 dpi, before terminally differentiating (Figure 1H). When EdU is administered longer, from 0–4 dpi, a minority of SCs (mean 24%) were EdU+, whereas all centrally located myonuclei and some peripheral myonuclei were EdU+ (Figures 1G and 1H). Approximately 75% of SCs reacquire quiescence between 5 dpi and 12 dpi, with 44% acquiring quiescence from 5 dpi to 7 dpi, 22% acquiring quiescence between 8 dpi and 10 dpi, and 6% acquiring quiescence from 10 dpi to 12 dpi, respectively (Figures 1G and 1H). The EdU lineage tracing results define the temporal kinetics of SC fate decisions, with a normal distribution of quiescent SCs centered around 5–7 dpi.
A malleable myogenic transcriptome directs myogenic fate decisions
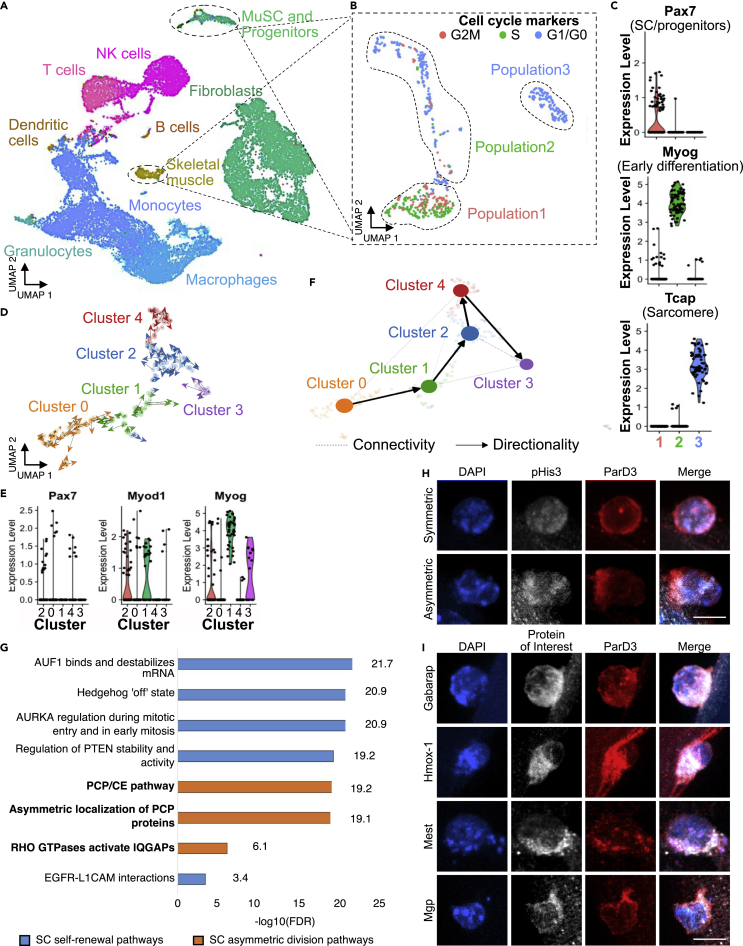
The observation that SCs in vivo do not reacquire quiescence during the initial cell division led us to explore the molecular profiles contributing to SC replenishment. We performed scRNA-Seq at 4 dpi and 7 dpi time points when myonuclear production is at its maximum and when the maximum numbers of SCs reacquire quiescence, respectively. Although a diverse cellular population comprises regenerating muscle, including assorted immune cells, fibroadipose progenitors (FAP), and myogenic cells (Figure 2A), we focused on defining the molecular features of the mononuclear myogenic population within the regenerative environment. Sub-clustering the SCs and myogenic progenitors revealed three dominant populations, which coincide with different stages of myogenesis. In the first population, most cells are Pax7+ and dividing, consistent with a proliferative, activated progenitor population (Figure 2B). In contrast, populations two and three are cells predominately in G1 or G0 expressing transcripts encoding the terminal differentiation commitment protein Myog (population 2) or Tcap, a protein in maturing sarcomeres (population 3) (Figure 2C). Because we were interested in myogenic cells that exit the cell cycle by reacquiring quiescence, we focused subsequent analyses on populations 1 and 2 representing the proliferating and early differentiating myogenic progenitors.
Figure 2.
Identifying and characterizing a myogenic cluster acquiring quiescence
(A) UMAP embedding for single-cell RNA sequencing of regenerating skeletal muscle at 4 dpi and 7 dpi where clusters are colored by cell type (total cells = 25,863).
(B) Subclustering of cells identified as SCs, myogenic progenitors, and muscle with cell-cycle stage indicated in color (total cells = 717).
(C) Gene expression levels of Pax7, Myog, and Tcap are indicated for the clusters presented in B.
(D) RNA velocity analysis of myogenic cells at 4 dpi and 7 dpi combined where distinct clusters are indicated by color and individual cell trajectories are indicated by arrows (total cells = 213).
(E) Gene expression levels of Pax7, Myod1, and Myog are indicated for the clusters presented in D.
(F) Velocity-inferred partition-based graph abstraction of composite myogenic cell fate trajectories between myogenic clusters.
(G) Pathways associated with a return to quiescence identified by Reactome pathway analysis performed on differentially expressed genes identified for cluster 4. Pathways organized by false discovery rate (FDR). Self-renewal pathways are colored blue, and asymmetric division pathways are colored orange. For full list of identified pathways refer to Table S7.
(H) Representative maximum intensity projection images of protein immunoreactivity for phosphorylated histone 3 (white) as a marker of dividing cells, ParD3 (red) as a marker of asymmetric division, as well as DAPI dye (blue) in SCs on isolated myofibers.
(I) Representative maximum intensity projection images of protein immunoreactivity for the protein of interest indicated in each row (white), ParD3 (red) as a marker of asymmetric division, as well as DAPI dye (blue) in SCs on isolated myofibers. Scale bar: 10 μm. See also Figures S2–S4; Tables S1, S2, S3, S4, S5, and S6; and Videos S1, S2, S3, S4, and S5.
We identified five cellular clusters (Figure 2D and S3A), each defined by a unique gene signature (Figure S2A). The expression level of transcripts associated with SC activation (Pax7 and Myod1), SC quiescence (Pax7 and Myf5), and SC differentiation (MyoG and Cdkn1c) were compared for each cluster as a general indication of progression in myogenesis (Figures 2E, S2C, and S2D). Cluster 3 is Pax7-low, Myod1-low, and Myog-high, identifying it as terminally differentiating cells, whereas cluster 0 and cluster 4 are Pax7-high and Myod1-low (Figures 2D, 2E, and S2C), enriched for G1/G0 cells (Figures S2B and S2C), suggesting that clusters 0 and 4 include quiescent SCs or SCs returning to quiescence. Our datasets contain myogenic cells at two ends of a biologic spectrum in injured muscle, cells that have remained quiescent following injury, cells undergoing terminal differentiation, and cells reacquiring quiescence. Satellite cells that did not activate will likely possess transcriptional profiles distinct from those reacquiring quiescence; the time required for transcriptional profiles of satellite cells that reacquired quiescence to become indistinguishable from those that never activated and remained quiescent is not known. Given that cells from injured muscle were sequenced at the height of SC replenishment, we hypothesized that a subset of our captured cells are actively acquiring quiescence.
To infer trajectories between myogenic clusters, we used partition-based graph abstraction (PAGA) to determine collective cellular directionality and assign cell fate trajectories (Bergen et al., 2020), with cells beginning at cluster 0 and progressing through clusters 1, 2, and 4 or exiting through differentiation (cluster 3) (Figures 2F and S3B). Connections between clusters indicate that cells may also transition through paths other than the identified trajectory. An intermediate population (cluster 2) that expresses Myod1 (Figure 2E) may represent cycling myogenic cells poised to commit to terminal differentiation (cluster 3) or return to quiescence (cluster 4) by regulating Myod1 mRNA translation (Hausburg et al., 2015; Morrée et al., 2017). Cluster 4, the cluster putatively reacquiring quiescence, is downstream of the Myod1 high population (cluster 2), supporting that cluster 4 represents SCs returning to quiescence rather than satellite cells that have not activated (Figure 2D).
To interrogate whether cluster 4 contains SCs returning to quiescence and to identify transcripts associated with a return to quiescence, we performed differential gene expression analysis on each myogenic cluster (Figure S4A, Table S1. Differential gene expression analysis of myogenic cluster0, related to Figure 2, Table S2. Differential gene expression analysis of myogenic cluster1, related to Figure 2, Table S3. Differential gene expression analysis of myogenic cluster2, related to Figure 2, Table S4. Differential gene expression analysis of myogenic cluster3, related to Figure 2, Table S5. Differential gene expression analysis of myogenic cluster4, related to Figure 2). We focused our attention on genes differentially expressed in cluster 4, the cluster putatively returning to quiescence (Figure S4B, Table S5). We then performed differential gene expression analysis comparing cluster 4 with cluster 2, the neighbor temporally upstream of cluster 4 (Figure S4C, Table S6). We performed pathway analysis of the differentially expressed genes and found upregulation of genes and pathways involved in myogenic self-renewal. Differential pathway analysis identified EGFR, AURORA A kinase, and AUF1 as significantly upregulated, all components of signaling pathways that regulate SC self-renewal in addition to pathways involved in asymmetric cell division (Table S6, Figure 2G) (Le Grand et al., 2009; Ramilowski et al., 2015; Chenette et al., 2016; Wang et al., 2019).
If the transcripts identified by scRNA-Seq encode proteins upregulated during a return to quiescence, then we expect a subset of proteins identified by scRNA-Seq to be polarized in asymmetrically dividing SCs. A subset of myoblast divisions on isolated myofibers are asymmetric (Kuang et al., 2007; Troy et al., 2012; Bernet et al., 2014), so we selected several of the most overrepresented transcripts from the differential expression analyses and validated expression and subcellular localization of the encoded proteins in myoblasts on isolated myofibers. In asymmetrically dividing cells, identified by phospho-Histone3 (pH3) and asymmetric ParD3 (Troy et al., 2012) (Figure 2H Video S1), we found Gabarap, Hmox-1, Mest, and Mgp asymmetrically partitioned (Figure 2I and Video S1. Hmox-1 symmetric division, related to Figure 2, Video S2. Hmox-1 asymmetric division, related to Figure 2, Video S3. Gabarap asymmetric division, Related to Figure 2, Video S5. Mest asymmetric division, related to Figure 2).
Representative 3D rendering of protein immunoreactivity for DAPI (blue), ParD3 (green) as a marker of asymmetry, and Hmox-1 (red) in SCs on an isolated myofiber
Representative 3D rendering of protein immunoreactivity for DAPI (blue), ParD3 (green) as a marker of asymmetry, and Hmox-1 (red) in SCs on an isolated myofiber
Representative 3D rendering of protein immunoreactivity for DAPI (blue), ParD3 (green) as a marker of asymmetry, and Gabarap (red) in SCs on an isolated myofiber
Representative 3D rendering of protein immunoreactivity for DAPI (blue), ParD3 (green) as a marker of asymmetry, and Mgp (red) in SCs on an isolated myofiber
Representative 3D rendering of protein immunoreactivity for DAPI (blue), ParD3 (green) as a marker of asymmetry, and Mest (red) in SCs on an isolated myofiber
The influence of regenerating muscle environment on SC fate decisions
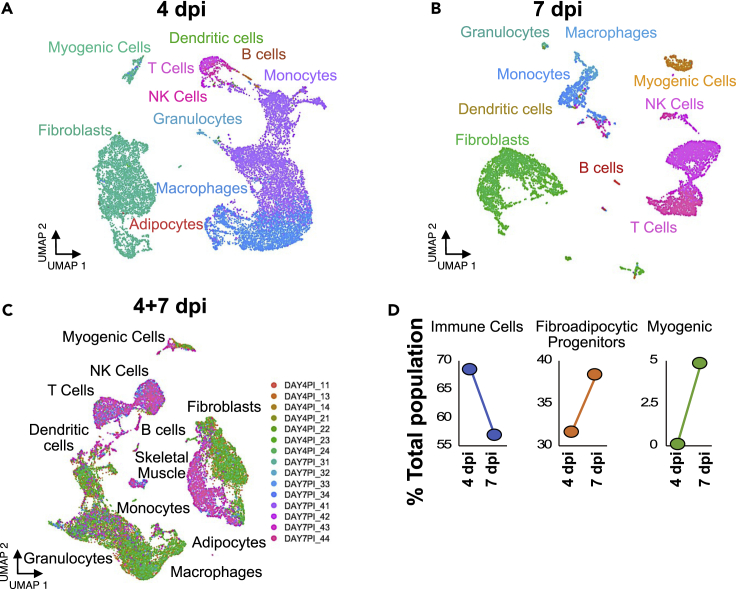
Nonmyogenic cells and myogenic cells undergo dramatic transcriptomic changes between 4 dpi and 7 dpi (when most SCs reacquire quiescence) (Figures 3A and 3B), and thus, it seems likely that the changing regenerative environment may regulate myogenic cell fate decisions. The composition of regenerating muscle is dynamic and changes in entire cellular populations occur over three days (Figure 3C). For example, immune cells globally decrease by 15%, whereas FAPs increase by 10% between day 4 and day 7 postinjury (Figure 3D). Consistent with the DNA lineage tracing experiments (Figure 1B), myogenic cells increase between 4 dpi and 7 dpi, indicative of the terminally differentiated progenitors that underwent DNA replication, exited the cell cycle, and are preparing for fusion (Figure 3D). Dynamic fluctuations in the cellular environment may expose muscle progenitors to a pro-differentiation environment or pro-quiescence environment depending on the signaling milieu, available cellular interactions, and local ligand production.
Figure 3.
Temporal dynamics of myogenic cells in regenerating muscle
(A and B) UMAP embeddings of mononuclear cells in regenerating skeletal muscle at 4 dpi and (B) 7 dpi with clusters colored corresponding to cell type.
(C) Combined UMAP embedding of mononuclear cells in regenerating skeletal muscle at 4 dpi and 7 dpi with clusters colored by sequencing library.
(D) Changes in percentage of the total population of mononuclear cells in regenerating skeletal muscle from 4 dpi to 7 dpi for immune cells, FAPs, and SCs.
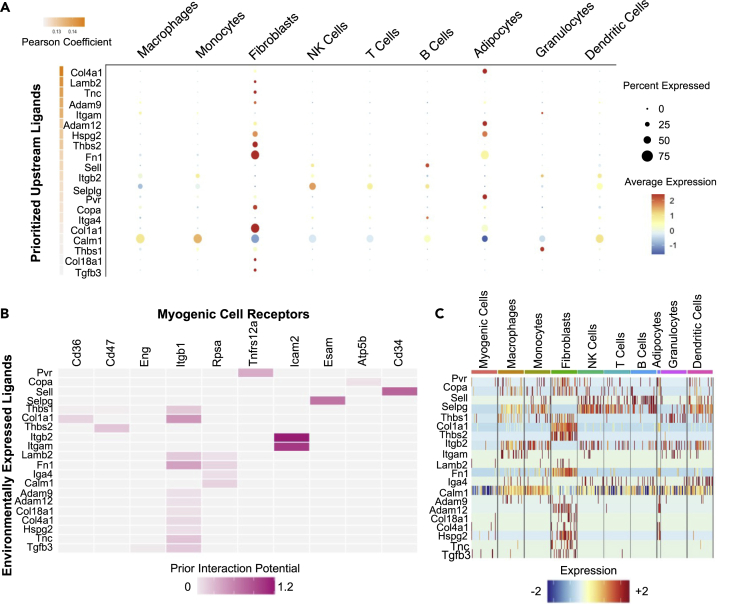
We mapped the differences in ligands during regeneration and examined the impact of ligand-receptor interactions on muscle progenitors at 4 dpi or 7 dpi with NicheNet (Browaeys et al., 2020). NicheNet draws on a priori knowledge of the gene regulatory effects of ligand-receptor interactions on intracellular signaling and gene expression to infer bioactive ligands, identify the cellular source of these ligands, and impute the downstream consequences of ligand-receptor binding on target cell gene expression in an unbiased manner. With stringent cutoffs, we identified differentially expressed ligands predicted to interact with extracellular receptors on muscle progenitors between 4 dpi and 7 dpi (top 20 by differential expression; Figure 4A, Tables S7, and S8). To determine whether NicheNet accurately mapped cell interactions, we queried the NicheNet dataset for described ligand-receptor pairs regulating myogenic cells. Fibronectin 1 (Fn1) is a key adhesion substrate for SCs and an essential extracellular matrix adhesion molecule for muscle regeneration (Lukjanenko et al., 2016). NicheNet accurately maps Fibronectin 1 and Fibronectin 1 binding to the Fn1-receptor, B1-integrin (Itgb1) on myogenic cells as well as perlecan (Hspg2), meltrin alpha (Adam12) and their respective ligands (Figure 4B and Table S7), all of which are involved in muscle regeneration (Galliano et al., 2000, p. 12; Lafuste et al., 2004; Xu et al., 2010; Yamashita et al., 2018). Thus, NicheNet identifies ligand-receptor interactions and provides predictive power for the temporal impact of the niche on muscle regeneration.
Figure 4.
Identifying signaling pathways in a myogenic cluster acquiring quiescence
(A) A Dotplot representing NicheNet-imputed ligands produced by mononuclear cells in regenerating skeletal muscle predicted to influence myogenic cells. The dot size reflects the precent gene expression, and the hue intensity depicts the relative gene expression.
(B) The most significantly expressed genes for cell ligands (by Pearson coefficient) and their potential interactions with genes encoding receptors on SCs determined by NicheNet. Hue indicates interaction potential, a representation of the number and quality of data sources supporting the ligand-receptor interaction.
(C) Expression levels for the most significant genes encoding ligands in mononuclear cells from regenerating skeletal muscle; the heatmap indicates the relative gene expression level. See also Figure S5, Table S7, and S8.
Fine mapping of differential ligand-receptor expression during SC return to quiescence identifies a subset of ligand-receptor interactions between SCs and niche cells. Specific subpopulations of cells show differential ligand-receptor interactions with SCs. For example, Thbs1 and Thbs2 (thrombospondin) ligands produced by niche cells signal to myogenic cells via Cd36 and Cd47, respectively (Figures 4B and S5A). Thbs1 is expressed in granulocytes, whereas Thbs2 is predominately expressed in fibroblasts (Figures 4C and S5B). Granulocytes are most numerous at 4 dpi and fibroblasts at 7 dpi (Figure 3D). Therefore, the availability of ligands or cellular interactions changes as the cellular populations present fluctuate over the course of muscle regeneration. Similarly, several ligands produced by different cell types are each capable of interacting with the SC β1-integrin receptor (Figures 4B, 4C, S5A, and S5B), suggesting that some cellular signaling may remain constant despite shifting cellular composition. Therefore, the changing cellular milieu could provide a complex combinatorial ligand array regulating myogenic cell fate decisions during muscle regeneration.
Quiescence acquisition is determined by cell extrinsic factors
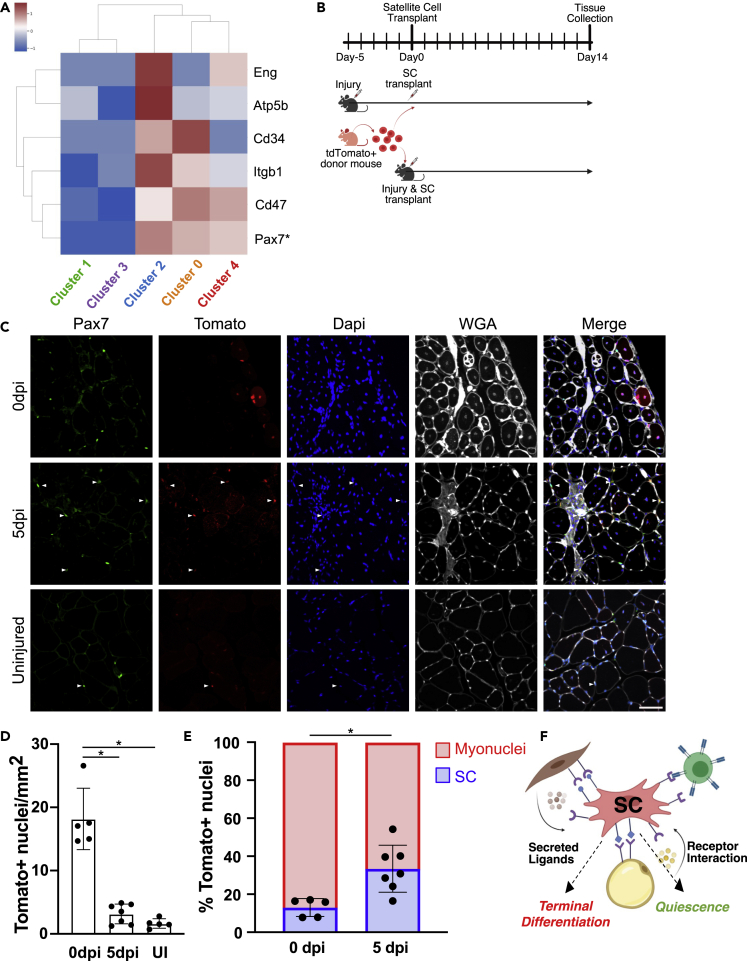
To identify additional signaling pathways potentially involved in quiescence acquisition, we examined the expression of receptors identified by NicheNet in myogenic clusters containing SCs acquiring quiescence. Hierarchical clustering of molecular signatures for myogenic clusters identifies unique gene expression programs contributing to SCs acquiring quiescence (Figure S6), indicating that changes in the signaling environment SCs experience affect the SC transcriptome. Each myogenic cluster expresses distinct cell surface receptor transcripts (Figures 5A and S7A); for example, cluster 4 expresses higher levels of transcripts encoding Cd47 and Eng receptors and much lower levels of Itgb1 and Atp5b receptor transcripts. The selectivity in receptor expression may promote a response in specific SC populations that are poised to acquire quiescence, whereas SC populations undergoing differentiation would be less responsive. Thus, if the environment drives myogenic cell fates, we predict that freshly isolated SCs transplanted into different signaling environments will adopt distinct fates.
Figure 5.
The regenerating muscle environment controls SC cell fate
For a Figure360 author presentation of this figure, see https://doi.org/10.1016/j.isci.2022.104444.
(A) Hierarchical clustering of NicheNet-identified genes encoding receptors on myogenic cell clusters. The heatmap indicates the Z score.
(B) Schematic of experimental design for SC transplantation concomitant with injury, or at 5 dpi (during the peak of SC quiescence acquisition).
(C) Representative images of engrafted tdTomato+/Pax7+ SCs (filled arrowheads) at 14 days posttransplant. Scale bar: 100 μm.
(D) Quantification of tomato+, donor-derived, nuclei identified per mm2 from transplants at 0 dpi, 5 dpi, and into uninjured TA muscle. Statistical significance tested by ANOVA. ∗p value < 0.05.
(E) The percentage of donor cells engrafting either as myonuclei or as SCs. Statistical significance tested by t test with p value = 0.006, For all mean ± SD, n = individual mice, where all tomato + cells in each of three TA muscle sections were quantified.
(F) A model depicting the influence of the regenerating muscle microenvironment on SC cell fate. See also Figures S6 and S7.
We transplanted freshly isolated SCs concomitant with injury, or at 5 dpi, the peak of SC niche repopulation (schematic in Figures 5B ). SCs were isolated from mice harboring a nuclear targeted tdTomato transgene preceded by a lox-stop-lox sequence (Daigle et al., 2018) and a tamoxifen-inducible Cre-recombinase in the Pax7 locus (Murphy et al., 2011), enabling linage tracing of the donor cells. Donor cells were isolated, and the population of cells was divided and transplanted simultaneously into the TA muscles of recipient mice either concomitant with injury or into muscles that had been injured 5 days previously. Muscle tissue was collected at 14 days following transplant and tdTomato + nuclei quantified as Pax7 positive or negative (Figure 5C). In agreement with previous findings (Morton et al., 2019), resuspension in BaCl2 did not negatively affect SCs, as they formed 91% ± 10 of the colonies compared with SCs suspended in normal saline (Figure S7B). Although more tdTomato + nuclei were detected when transplanted concomitant with injury (Figure 5D), a greater proportion of donor-derived cells engrafted as SCs when transplanted into 5 dpi muscle (Figure 5E). When donor SCs were transplanted at 5 dpi, the propotion of engrafted Pax7+ SCs was 3-fold greater than donor SCs transplanted concomitant with injury (Figure 5E), which predominately differentiated and fused, becoming myonuclei. Thus, a single population of donor SCs adopt different fates when introduced to different environments. We conclude that the regenerating muscle environment directs SC fate during skeletal muscle regeneration (Figure 5F).
Discussion
Replenishing the SC pool is essential to maintain long-term muscle health, yet the timing and mechanisms regulating SC replenishment in vivo (generation of quiescent SCs in the niche) remain largely unknown. Because the SC niche is difficult to observe and time-consuming to manipulate in vivo, the majority of information on SC self-renewal is derived from observing asymmetric division of SCs cultured on myofibers (Kuang et al., 2007; Troy et al., 2012; Dumont et al., 2015; Wang et al., 2019). We took advantage of DNA-based lineage tracing and scRNA-Seq to investigate the timing of SC replenishment and identify the signaling pathways likely influencing SC quiescence acquisition during muscle regeneration.
Lineage tracing during muscle regeneration reveals that most SCs reacquire quiescence from 5 dpi to 7 dpi, after the majority of myonuclei have been produced. In contrast, when EdU is provided from 0 dpi to 2 dpi, myonuclei are labeled nearly exclusively, and thus, we identify a critical window when the fate of myogenic cells exiting the cell cycle shifts from terminal differentiation to reacquiring quiescence. Our in vivo findings contrast data from in vitro myofiber cultures showing SCs reacquire quiescence via asymmetric division during the first cell division following activation (Kuang et al., 2007; Troy et al., 2012; Dumont et al., 2015; Wang et al., 2019). In a prior scRNA-Seq dataset of skeletal muscle regeneration, no quiescent SCs were detected until 5 dpi (Oprescu et al., 2020), supporting our finding that the SC pool is replenished relatively late in muscle regeneration. We do, however, detect heterogeneity in the timing of SCs acquiring quiescence. Although more than 75% of quiescent SCs are generated after 5 dpi, a minority of SCs (24%) reacquire quiescence between 2 dpi and 4 dpi. What directs the minority of SCs to reacquire quiescence in an environment promoting proliferation and differentiation is unclear. Because the same population of cells transplanted into two different environments adopted different cell fates, we favor the idea that SC heterogeneity arises from the cellular context an activated SC encounters rather than hierarchical limitation of cell fates. But, as cell intrinsic differences affect SC activation from quiescence following injury (Kuang et al., 2007; Chakkalakal et al., 2012, 2014; Scaramozza et al., 2019), similar differences may also affect the return to quiescence.
We identified when SCs reacquire quiescence in vivo independently from lineage tracing and from scRNA-Seq, where we identified a cluster (cluster 4) of myogenic cells that appear to be reacquiring quiescence. This cluster contains nondividing Pax7-high, Myod1-low cells, and elevated levels of transcripts encoding proteins involved in asymmetric division including Aurora Kinase A, EGFR, GSK, JAK signaling, Auf1, and Pcp-wnt (Le Grand et al., 2009; Chenette et al., 2016; Wang et al., 2019; De Micheli et al., 2020a). Differentially increased transcripts encoding proteins involved in asymmetric division and self-renewal within cluster 4 suggests that SCs may enter quiescence through asymmetric division.
By comparing scRNA-Seq during myonuclear production from 0–4 dpi to 5–7 dpi during SC replenishment, we identified signaling pathways that may regulate the decision to terminally differentiate or to self-renew and reacquire quiescence. We determined significant agreement (p value = 2.5 × 10−18) between our list of ligand-receptor pairs and signaling pairs identified in myogenic progenitors from both mice and human muscle using alternative computational methods (De Micheli et al., 2020a, 2020b). The agreement between these independent datasets emphasizes the robustness of all the experiments and the likely involvement of the signaling pathways we identified in regulating SC return to quiescence. Further studies will be needed to determine the relative contributions and interactions of these signaling pathways in SCs.
In addition to the signaling pathways, we identified differentially expressed transcripts that could be involved in establishing or maintaining quiescence. For example, the myogenic cluster returning to quiescence had significantly increased transcript levels encoding proteins that modulate autophagy and are involved in DNA damage repair, both processes that contribute to SC quiescence (Umansky et al., 2015; Yue et al., 2017; Liu et al., 2018; Gassen et al., 2019). Moreover, we identified several upregulated transcripts encoding proteins that are polarized in asymmetrically dividing SCs, identifying potential new markers of SC asymmetric division and providing additional tools to examine SC behavior in vivo.
The majority of SCs appear to reacquire quiescence and repopulate the SC niche from 5–7 dpi, as demonstrated by lineage tracing and scRNA-Seq. We observed a 3-fold increase in the percentage of engrafted SCs occupying the SC niche when engrafted into skeletal muscle at 5 dpi as opposed to transplantation with injury, demonstrating noncell autonomous signaling plays a role in determining SC fate. These results may provide an explanation for potentially disparate reports for the percentage of SC niche engraftment following transplantation (Sacco et al., 2008; Hall et al., 2010; Lepper et al., 2011; Chan et al., 2018; Incitti et al., 2019). A major hurdle for cell-based therapies is encouraging transplanted cells to engraft both as myonuclei, contributing to healthier, repaired myofibers, and as SCs prepared to maintain the muscle for years to come. By manipulating the signals encountered by SCs upon transplant, a greater percentage of donor cells could be encouraged to adopt an SC fate, ensuring longevity of treatment by manipulating either the response of engrafted SCs to the donor environment or by manipulating the signals in host skeletal muscle tissue.
Limitations of the study
To adequately assess the data, we present the following limitations of our study. We identified a group of genes expressed in a cell population of myogenic cells acquiring quiescence by trajectory analysis. These genes are implicated in stem cell maintenance or stem cell self-renewal in tissues other than skeletal muscle. We validated that a subset of the proteins encoded by these genes are present in SCs and are polarized in asymmetrically dividing SCs associated with explanted myofibers in culture. Because, as we describe in this manuscript, in vivo quiescence acquisition is not fully recapitulated in culture, assessing the functional role of the identified proteins will require either a knockdown or knockout in vivo. Future experiments will require significant experimental manipulation of mice to address whether polarization of the identified proteins regulates SC acquiring quiescence or is a consequence of quiescence.
Trajectory analyses derived from single-cell sequencing allowed us to identify several potential signaling pathways between 4 and 7 days postinjury that may regulate cell fates of transplanted satellite cells. Given the challenging nature of manipulating signaling at specific time points during muscle regeneration, we did not attempt to resolve whether these pathways regulate SC cell fate. Future studies will address the roles of the identified signaling pathways and how they may interact to regulate SC fate, SC asymmetric division, and SC quiescence acquisition.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-Pax7 | Developmental Studies Hybridoma Bank | Cat# PAX7, RRID:AB_2299243 |
| anti-parD3 | Santa Cruz Biotechnology | Cat# Sc-79577; RRID:AB_2159660 |
| anti-phospho histone 3 (ser10) | Sigma-Aldrich | Cat# H9908, RRID:AB_260096 |
| anti-itm2a | Proteintech | Cat# 18306-1-AP, RRID:AB_2249412 |
| anti-HO-1 | Proteintech | Cat# 66743-1-Ig, RRID:AB_2882091 |
| anti-gabarap1 | Abcam | Cat# ab109364, RRID:AB_10861928 |
| anti-mgp | Proteintech | Cat# 10734-1-AP, RRID:AB_2297660 |
| anti-ncoa7 | Novus Biologicals | Cat# NBP1-85200, RRID:AB_11019710 |
| anti-mest | Invitrogen | Cat# PA5-77067, RRID:AB_2720794 |
| anti-mouse IgG1 | Invitrogen | Cat# A-21240, RRID:AB_2535809 |
| anti-rat | Biotium | Cat# 20,027; RRID:AB_2920542 |
| anti-goat | Invitrogen | Cat# A-21432, RRID:AB_2535853 |
| anti-mouse IgG2a | Invitrogen | Cat# A-21241, RRID:AB_2535810 |
| Anti-rabbit | Invitrogen | Cat# A-31573, RRID:AB_2536183 |
| Chemicals, peptides, and recombinant proteins | ||
| 5-Ethynyl-2′-deoxyuridine | Carbosynth | NE08701 |
| Barium chloride | Sigma | B-6394 |
| Collagenase Type 1 | Worthington Biochemical Corporation | CLS-1 Lot: 40C20080 |
| Ham’s F-12C | VWR | 10-080-CV |
| Click-iT EdU cell Proliferation Kit for Imaging, Alexa Fluor 488 dye | Invitrogen | C10337 |
| TritonX-100 | Sigma | T9284 |
| Bovine serum albumin | Sigma | TA9647 |
| Dead Cell Removal kit | Miltenyi Biotec | 130-090-101 |
| LD Columns | Miltenyi Biotec | 130-042-901 |
| Satellite Cell Isolation Kit Mouse | Miltenyi Biotec | 130-104-268 |
| Chromium Next GEM Single Cell 3′ Reagent kits v2 | 10x Genomics | PN-120237 |
| Tamoxifen | Sigma | T5648 |
| Dulbecco’s Modification of Eagle’s medium | VWR | 10-013-CV |
| Deposited data | ||
| Raw 10x scRNA sequencing data | This paper | GEO: GSE63473 |
| Experimental models: Cell lines | ||
| Mouse: C2C12 cells | ATCC | CRL-1772 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | JAX: 000664 |
| Mouse: Pax7-CreERT2 NLS-tdTomato | The Jackson Laboratory | JAX: 025106, JAX: 017763 |
| Software and algorithms | ||
| CellRanger version 3.0.1 | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/release-notes/3-0 |
| R version 3.6.2 | The R Foundation | https://cran.r-project.org/bin/windows/base/old/3.6.2/ |
| R version 4.0.0 | The R Foundation | https://cran.r-project.org/bin/windows/base/old/4.0.0/ |
| Seurat version 3.1.5 | Stuart et al., 2019 | https://github.com/satijalab/seurat/releases?page=2 |
| Doublet Finder | McGinnis et al. 2019 | https://github.com/chris-mcginnis-ucsf/DoubletFinder |
| SingleR version 1.2.3 | Aran et al. 2019 | https://bioconductor.org/packages/release/bioc/html/SingleR.html |
| Seaborn version 0.11.0 (Python version 3.6.3) | N/A | |
| scVelo (Python version 3.6.3) | Bergen et al., 2020 | https://scvelo.readthedocs.io/ |
| Samtools version 1.8 (Python version 3.6.3) | https://github.com/samtools/samtools/releases/ | |
| LoomR version 0.2.0 | https://github.com/mojaveazure/loomR/releases | |
| Nichenetr | Browaeys et al. 2020 | https://github.com/saeyslab/nichenetr |
| Prism version 9.3.1 | GraphPad | https://www.graphpad.com/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Brad Olwin (olwin@colorado.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
Data: scRNA-Seq data have been deposited at GEO: GSE205690 and are publicly available as of the date of publication. Accession numbers are listed in the Key resources table. Microscopy data reported in this paper will be shared by the Lead contact upon request.
-
•
Code: This paper does not report original code
-
•
Any additional information required to reanalyze the data reported in this paper is available from the Lead contact upon request.
Experimental model details
Mice were bred and housed according to National Institutes of Health (NIH) guidelines for the ethical treatment of animals in a pathogen-free facility at the University of Colorado at Boulder. University of Colorado Institutional Animal Care and Use Committee (IACUC) approved animal protocols and procedures. The mice included in experiments were either C57BL6 (Jackson Labs Stock No. 000664) or Pax7-creERT2 NLS-tdTomato (Jackson Labs Stock No. 025106 crossed to No. 017763) between 4 and 8 months old and were a mix of male and female.
Method details
Mouse injuries and EdU delivery
Mice were anesthetized with isoflurane followed by injection with 50μL of 1.2% BaCl2 in normal saline into the TA and EDL muscles. To deliver EdU, mice were either given water containing 0.5 mg/mL EdU (Carbosynth), with 1% glucose or given IP injections of 10 mM EdU (Carbosynth), re-suspended in water, a volume of 100 μL per 25g mouse weight.
Myofiber isolation, immunostaining, and culture
The EDL muscles were dissected and placed into 400 U/mL collagenase at 37°C for 1.5h with rotation and then transferred into Ham’s F-12C and 15% horse serum to inactivate the collagenase. Individual EDL myofibers were separated using a glass pipet and fixed using 4% paraformaldehyde for 10 min at room temperature either immediately, for EdU lineage tracing, or at 36 hours post isolation, for polariztion imaging, and stored in PBS at 4°C. For visualization of EdU incorporation and immunocytochemistry, myofibers were permeabilized with 0.5% Triton-X100 in PBS (phosphate buffered saline without calcium or magnesium pH 7.4), containing 3% bovine serum albumin (Sigma) for 30 min at room temperature. EdU incorporation was visualized using the Click-iT EdU Alexa Fluor 488 imaging kit (ThermoFisher) following the manufacturer’s protocol. Primary antibodies [anti-Pax7, 2 μg/mL (Developmental Studies Hybridoma Bank (DSHB) at the University of Iowa), anti-mgp 1:50 (Proteintech), anti-ncoa7 1:50 (Novus Biologicals), anti-mest 1:100 (Invitrogen), anti-itm2a 1:50 (Proteintech), anti-HO-1 1:100 (Proteintech), anti-gabarap1 1:50 (abcam), anti-parD3 1:200 (Santa Cruz Biotechnology), anti-phospho histone 3 1:500 (Millipore)] were incubated with intact myofibers at room temperature for 1hour followed by three washes in PBS. Myofibers were then incubated with appropriate fluorescently conjugated secondary antibodies [Donkey anti-mouse IgG1 (ThermoFisher), anti-rat (ThermoFisher), anti-goat (ThermoFisher), anti-mouse IgG2a (ThermoFisher)] diluted 1:1000 for 1hour at room temperature. Myofibers were washed 3 times in PBS, incubated with 1 μg/mL DAPI for 10 min at room temperature to label nuclei and then mounted in Mowiol supplemented with DABCO (Sigma-Aldrich) as an anti-fade agent.
TA muscle collections and cell isolations
TA muscles were dissected and placed into 400U/mL collagenase at 37°C for 1h with shaking and then placed into Ham’s F-12C supplemented with 15% horse serum to inactivate the collagenase. Cells were passed through three strainers of 100, 70, and 40μm (BD Falcon) and flow through was centrifuged at 1500×g for 5 min and the cell pellets were re-suspended in Ham’s F-12C. To remove dead cells and debris, cells were passed over the Miltenyi, dead cell removal kit columns (Cat# 130-090-101). To remove red blood cells (RBC), cells were incubated with antiTer119 micro magnetic beads and passed over a Miltenyi column (Cat#130-049-901). For the adult and aged uninjured TAs, 6 TA muscles (from 3 mice) were pooled together. For the injured TA muscles 2 TA muscles from 2 different mice were pooled together. Cells were then counted using a BioRad TC20 automated cell counter and processed with the 10X genomics single cell sequencing kit.
SC transplant
For SC transplants, Pax7CreERT2; R26RNLStdTom mice were injected with 100 μL 20 mg/mL tamoxifen once daily for five days to induce recombination and expression of tdTomato in Pax7+ cells. These tomato positive donor mice were then sacrificed and SCs collected from hindlimb muscles as described above. To remove dead cells and debris, cells were passed over the Miltenyi, dead cell removal kit columns (Cat# 130-090-101). To remove RBCs, cells were incubated with antiTer119 micro magnetic beads and passed over a Miltenyi column (Cat#130-049-901). To deplete non-SCs, cells were incubated with satellite cell isolation kit micro magnetic beads and passed over a Miltenyi column (Cat# 130-104-268). The resulting purified SCs were separated into 100,000 cell aliquots and re-suspended in either normal saline (for 5dpi transplants) or 1.2% BaCl2 in normal saline (for 0dpi transplants) and injected into the TA of an anesthetized mouse. For transplants concomitant with injury or into uninjured muscle, mice were anesthetized with isoflurane and 50μL of SCs suspended in BaCl2 or normal saline, respectively, was injected into the TA muscle. For transplants at 5dpi, 5 days prior to donor cell collection, recipient mice were injected with 50μL of BaCl2 in the TA muscle and the injection site marked. Then, on the day of transplantation, 50μL of SC suspended in normal saline was injected into the same injection site as the initial injury.
C2C12 cell culture and EdU persistence
Muscle cell line C2C12 cells were maintained in 20% FBS DMEM supplemented with 1% penicillin and streptomycin. Cells were plated at clonal density on gelatin coated coverslips the day before beginning the EdU pulse chase. EdU was supplemented to the growth media at a final concentration of 100nM. Cells were maintained in EdU containing media for 12 hours and then the media was replaced with standard growth media. A coverslip was fixed in 4% paraformaldehyde every 24 hours. All coverslips were processed together to detect EdU signal.
Single cell sequencing
To capture, label, and generate transcriptome libraries of individual cells we used the 10X genomics Chromium Single Cell 3′ Library and Gel Bead Kit v2 (Cat #PN-120237) following the manufactures protocols. Barcoded cDNA is used to make sequencing libraries for analysis with Illumina sequencing. We captured 16,868 cells from the 4-dpi and 8,995 cells from the 7-dpi. Sequencing was completed on an Illumina NovaSeq 6000, using paired end 150 cycle 2 × 150 reads by the genomics and microarray core at the University of Colorado Anschutz Medical Campus.
Single cell informatics
CellRanger version 3.0.1 (10X Genomics, Pleasanton, CA) was used for initial processing of sequencing reads in keeping with standardized workflows with reads mapped to the mouse reference transcriptome mm10. For each library, gene expression matrixes generated, and subsequent downstream analysis was conducted using either R version 3.6.2 (2019-12-15) or R version 4.0.0 (2020-04-24). For each library, quality control, filtering, data clustering and visualization was carried out using Seurat version 3.1.5 R package with some custom modifications to the standard pipeline (Stuart et al., 2019). Each dataset was independently analyzed and datasets from the same time points were combined for integrated analysis. Briefly, for each individual dataset, pre-processing was conducted according to standard workflows wherein genes that were expressed in less that 3 cells as well as single cells harboring less than 2500 UMIs, greater than 5% UMIs mapping to mitochondrial genes, or less than 200 genes were removed from the gene expression matrix. Doublets were identified and removed using DoubletFinder (McGinnis et al., 2019). After log-normalizing the data, we performed PCA on the gene expression matrix and used the first 12 principal components for clustering and visualization. Unsupervised shared nearest neighbor (SNN) clustering was performed with a resolution of 0.5 and visualization was done using uniform manifold approximation and projection (UMAP). Cell identities were annotated using SingleR version 1.2.3 (Aran et al., 2019). Differential gene expression analysis was performed using either Seurat version 3.1.5 “FindMarker” or the DESeq2 package. Data was plotted using Seurat enabled ggplot2, custom R ggplot2 scripts, or exported for graphical analysis using Seaborn version 0.11.0 in Python or Tableau visualization software.
RNA velocity and trajectory mapping
RNA velocity was performed using the scVelo package (Bergen et al., 2020). In brief, using Python version 3.6.3, velocyto.py alongside Samtools version 1.8 was used to generate loom files pertaining to spliced and unspliced information from CellRanger version 3.0.1 generated output files. Loom files for each Seurat object from each processed library were created using LoomR version 0.2.0 and combined with velocyto output loom files into an adata object using custom Python scripts. Subsequent RNA velocity analysis along with velocity-inferred trajectory and connectivity mapping were performed using standardized scripts with the outputs superimposed onto the UMAP embeddings.
Ligand-receptor interaction mapping
Ligand-Receptor interaction analysis was conducted using NicheNet using the R-enabled nichenetr plugin (Browaeys et al., 2020) and the Seurat version 3.1.5 R package in R version 4.0.0 (2020-04-24). In brief, a combined processed Seurat object was created for 4 and 7-dpi with embedded cell annotations having been generated using SingleR as above. Metadata was amended to include either a 4-dpi or 7-dpi descriptor for each cell. Previously generated weighted network files were read in and genes were converted to mouse orthologs based on one-to-one orthology. NicheNet analysis and differential expression was performed using standard scripts with “receiver” cells defined as myogenic clusters and “sender” cells defined as all remaining SingleR-annotated cell clusters. Data was plotted using Seurat enabled ggplot2 or exported for graphical analysis using Seaborn version 0.11.0 in Python.
Microscopy and image analysis
Images were captured either on a Nikon inverted spinning disk confocal microscope or on an Olympus IX83 widefield microscope. Objectives used on the Nikon were: 10x/0.45NA Plan Apo, 20x/0.75NA Plan Apo, and 60x/1.4NA Plan Apo. The objective used on the Olympus microscope was 10x/0.040NA PlanSApo. Images were processed using Fiji ImageJ. Confocal stacks were projected as maximum intensity images for each channel and merged into a single image. Brightness and contrast were adjusted for the entire image as necessary.
Quantification and statistical analysis
Statistical analysis was performed in Prism (GraphPad), where statistical significance was assessed using, two-tailed, unpaired Student’s t test or one-way ANOVA with p < 0.05 considered significant.
Acknowledgments
We acknowledge the Biofrontiers Sequencing Core, the Light Microscopy Core Facility, and the Stem Cell Research and Technology Resource Center at the University of Colorado Boulder for use of the facilities and expertise in collecting imaging and sequencing data. We also acknowledge BioRender.com for graphics used in figures. Funding: this work was funded by grants from the ALSAM Foundation (BBO), NIH AR049446 (BBO), and NIH AR070630 (BBO).
Author contributions
BP, AAC, and BBO conceived the experiments. BP, AAC, ND, and TA performed the experiments, analyzed data, and made figures. JW, KJ, and RO performed bioinformatic analysis of single-cell sequencing. AAC, JW, BP, and BBO wrote the manuscript. BBO supervised the research. All authors read and approved the manuscript.
Declaration of interests
BBO discloses a potential conflict of interest as a Scientific Advisory Board Member for Satellos Biosciences.
Published: June 17, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104444.
Supplemental information
Differential gene expression using non-parameteric Wilcoxon rank-sum test performed on myogenic cluster and are ranked by significance according to adjusted p-valve. The gene with significance, average log fold change, the percentage of cells where the gene is detected, the percent of cells where the gene is detected in the comparison group, and adjusted p value are provided
Differential gene expression using non-parameteric Wilcoxon rank-sum test performed on myogenic cluster and are ranked by significance according to adjusted p-valve. The gene with significance, average log fold change, the percentage of cells where the gene is detected, the percent of cells where the gene is detected in the comparison group, and adjusted p-value are provided
Differential gene expression using non-parameteric Wilcoxon rank-sum test performed on myogenic cluster and are ranked by significance according to adjusted p-valve. The gene with significance, average log fold change, the percentage of cells where the gene is detected, the percent of cells where the gene is detected in the comparison group, and adjusted p-value are provided
Differential gene expression using non-parameteric Wilcoxon rank-sum test performed on myogenic cluster and are ranked by significance according to adjusted p-valve. The gene with significance, average log fold change, the percentage of cells where the gene is detected, the percent of cells where the gene is detected in the comparison group, and adjusted p-value are provided
Differential gene expression using non-parameteric Wilcoxon rank-sum test performed on myogenic cluster and are ranked by significance according to adjusted p-valve. The gene with significance, average log fold change, the percentage of cells where the gene is detected, the percent of cells where the gene is detected in the comparison group, and adjusted p-value are provided
Differential gene expression using non-parameteric Wilcoxon rank-sum test performed on myogenic cluster and are ranked by significance according to adjusted p-valve. The gene with significance, average log fold change, the percentage of cells where the gene is detected, the percent of cells where the gene is detected in the comparison group, and adjusted p-value are provided
The pathways, their fold enrichment, the raw p-value, and the false discovery rate are provided in the table. Highlighted pathways indicate pathways that include candidate genes that may regulate SC self-renewal
All NicheNet imputed differentially expressed ligands in the regenerating muscle NicheNet identified ligand with cognate receptor pair inferred from Ramilowski et al. (2015)
References
- Aran D., Looney A.P., Liu L., Wu E., Fong V., Hsu A., Chak S., Naikawadi R.P., Wolters P.J., Abate A.R., et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019;20:163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergen V., Lange M., Peidli S., Wolf F.A., Theis F.J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 2020;38:1408–1414. doi: 10.1038/s41587-020-0591-3. [DOI] [PubMed] [Google Scholar]
- Bernet J.D., Doles J.D., Hall J.K., Kelly Tanaka K., Carter T.A., Olwin B.B. p38 MAPK signaling underlies a cell-autonomous loss of stem cell self-renewal in skeletal muscle of aged mice. Nat. Med. 2014;20:265–271. doi: 10.1038/nm.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browaeys R., Saelens W., Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods. 2020;17:159–162. doi: 10.1038/s41592-019-0667-5. [DOI] [PubMed] [Google Scholar]
- Chakkalakal J.V., Jones K.M., Basson M.A., Brack A.S. The aged niche disrupts muscle stem cell quiescence. Nature. 2012;490:355–360. doi: 10.1038/nature11438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakkalakal J.V., Christensen J., Xiang W., Tierney M.T., Boscolo F.S., Sacco A., Brack A.S. Early forming label-retaining muscle stem cells require p27kip1 for maintenance of the primitive state. Development. 2014;141:1649–1659. doi: 10.1242/dev.100842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S.S.-K., Arpke R.W., Filareto A., Xie N., Pappas M.P., Penaloza J.S., Perlingeiro R.C., Kyba M. Skeletal muscle stem cells from PSC-derived teratomas have functional regenerative capacity. Cell Stem Cell. 2018;23:74–85.e6. doi: 10.1016/j.stem.2018.06.010. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenette D.M., Cadwallader A., Antwine T., Larkin L., Wang J., Olwin B., Schneider R. Targeted mRNA decay by RNA binding protein AUF1 regulates adult muscle stem cell fate, promoting skeletal muscle integrity. Cell Rep. 2016;16:1379–1390. doi: 10.1016/j.celrep.2016.06.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C.A., Zammit P.S., Ruiz A.P., Morgan J.E., Partridge T.A. A population of myogenic stem cells that survives skeletal muscle aging. Stem Cell. 2007;25:885–894. doi: 10.1634/stemcells.2006-0372. [DOI] [PubMed] [Google Scholar]
- Conboy I.M., Rando T.A. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev. Cell. 2002;3:397–409. doi: 10.1016/s1534-5807(02)00254-x. [DOI] [PubMed] [Google Scholar]
- Cornelison D.D., Wold B.J. Single-cell analysis of regulatory gene expression in quiescent and activated mouse skeletal muscle satellite cells. Dev. Biol. 1997;191:270–283. doi: 10.1006/dbio.1997.8721. [DOI] [PubMed] [Google Scholar]
- Daigle T.L., Madisen L., Hage T.A., Valley M.T., Knoblich U., Larsen R.S., Takeno M.M., Huang L., Gu H., Larsen R., et al. A suite of transgenic driver and reporter mouse lines with enhanced brain-cell-type targeting and functionality. Cell. 2018;174:465–480.e22. doi: 10.1016/j.cell.2018.06.035. e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Micheli A.J., Spector J.A., Elemento O., Cosgrove B.D. A reference single-cell transcriptomic atlas of human skeletal muscle tissue reveals bifurcated muscle stem cell populations. Skeletal Muscle. 2020;10:19. doi: 10.1186/s13395-020-00236-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Micheli A.J., Laurilliard E.J., Heinke C.L., Ravichandran H., Fraczek P., Soueid-Baumgarten S., De Vlaminck I., Elemento O., Cosgrove B.D. Single-cell analysis of the muscle stem cell hierarchy identifies heterotypic communication signals involved in skeletal muscle regeneration. Cell Rep. 2020;30:3583–3595.e5. doi: 10.1016/j.celrep.2020.02.067. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont N.A., Wang Y.X., von Maltzahn J., Pasut A., Bentzinger C.F., Brun C.E., Rudnicki M.A. Dystrophin expression in muscle stem cells regulates their polarity and asymmetric division. Nat. Med. 2015;21:1455–1463. doi: 10.1038/nm.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galliano M.-F., Huet C., Frygelius J., Polgren A., Wewer U.M., Engvall E. Binding of ADAM12, a marker of skeletal muscle regeneration, to the muscle-specific actin-binding protein, α-actinin-2, is required for myoblast fusion. J. Biol. Chem. 2000;275:13933–13939. doi: 10.1074/jbc.275.18.13933. [DOI] [PubMed] [Google Scholar]
- Gassen N.C., Niemeyer D., Muth D., Corman V.M., Martinelli S., Gassen A., Hafner K., Papies J., Mosbauer K., Zellner A., et al. SKP2 attenuates autophagy through Beclin1-ubiquitination and its inhibition reduces MERS-Coronavirus infection. Nat. Commun. 2019;10:5770. doi: 10.1038/s41467-019-13659-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J.K., Banks G.B., Chamberlain J.S., Olwin B.B. Prevention of muscle aging by myofiber-associated satellite cell transplantation. Sci. Transl. Med. 2010;2:57ra83. doi: 10.1126/scitranslmed.3001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausburg M.A., Doles J.D., Clement S.L., Cadwallader A.B., Hall M.N., Blackshear P.J., Lykke-Andersen J., Olwin B.B. Post-transcriptional regulation of satellite cell quiescence by TTP-mediated mRNA decay. Elife. 2015;4:e03390. doi: 10.7554/eLife.03390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslop L., Morgan J.E., Partridge T.A. Evidence for a myogenic stem cell that is exhausted in dystrophic muscle. J. Cell Sci. 2000;113:2299–2308. doi: 10.1242/jcs.113.12.2299. [DOI] [PubMed] [Google Scholar]
- Incitti T., Magli A., Darabi R., Yuan C., Lin K., Arpke R.W., Azzag K., Yamamoto A., Stewart R., Thomson J.A., et al. Pluripotent stem cell-derived myogenic progenitors remodel their molecular signature upon in vivo engraftment. Proc. Natl. Acad. Sci. U S A. 2019;116:4346–4351. doi: 10.1073/pnas.1808303116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones N.C., Tyner K.J., Nibarger L., Stanley H.M., Cornelison D.D., Fedorov Y.V., Olwin B.B. The p38α/β MAPK functions as a molecular switch to activate the quiescent satellite cell. J. Cell Biol. 2005;169:105–116. doi: 10.1083/jcb.200408066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S., Kuroda K., Le Grand F., Rudnicki M.A. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafuste P., Sonnet C., Chazaud B., Dreyfus P.A., Gherardi R.K., Wewer U.M., Authier F.J. ADAM12 and α9β1 integrin are instrumental in human myogenic cell differentiation. Mol. Biol. Cell. 2004;16:861–870. doi: 10.1091/mbc.e04-03-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Grand F., Jones A.E., Seale V., Scime A., Rudnicki M.A. Wnt7a activates the planar cell polarity pathway to drive the symmetric expansion of satellite stem cells. Cell Stem Cell. 2009;4:535–547. doi: 10.1016/j.stem.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C., Partridge T.A., Fan C.-M. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development. 2011;138:3639–3646. doi: 10.1242/dev.067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Charville G.W., Cheung T.H., Yoo B., Santos P.J., Schroeder M., Rando T.A. Impaired Notch signaling leads to a decrease in p53 activity and mitotic catastrophe in aged muscle stem cells. Cell Stem Cell. 2018;23:544–556.e4. doi: 10.1016/j.stem.2018.08.019. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukjanenko L., Jung M.J., Hegde N., Perruisseau-Carrier C., Migliavacca E., Rozo M., Karaz S., Jacot G., Schmidt M., Li L., et al. Loss of fibronectin from the aged stem cell niche affects the regenerative capacity of skeletal muscle in mice. Nat. Med. 2016;22:897–905. doi: 10.1038/nm.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matiašová A., Sevc J., Mikes J., Jendzelovsky R., Daxnerova Z., Fedorocko P. Flow cytometric determination of 5-bromo-2ʹ-deoxyuridine pharmacokinetics in blood serum after intraperitoneal administration to rats and mice. Histochem. Cell Biol. 2014;142:703–712. doi: 10.1007/s00418-014-1253-7. [DOI] [PubMed] [Google Scholar]
- Mauro A. Satellite cell of skeletal muscle fibers. J. Biophys. Biochem. Cytol. 1961;9:493–495. doi: 10.1083/jcb.9.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis C.S., Murrow L.M., Gartner Z.J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019;8:329–337.e4. doi: 10.1016/j.cels.2019.03.003. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Morrée A., van Velthoven C.T.J., Gan Q., Salvi J.S., Klein J.D.D., Akimenko I., Quarta M., Biressi S., Rando T.A. Staufen1 inhibits MyoD translation to actively maintain muscle stem cell quiescence. Proc. Natl. Acad. Sci. U S A. 2017;114:E8996–E9005. doi: 10.1073/pnas.1708725114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton A.B., Norton C.E., Jacobsen N.L., Fernando C.A., Cornelison D.D.W., Segal S.S. Barium chloride injures myofibers through calcium-induced proteolysis with fragmentation of motor nerves and microvessels. Skeletal Muscle. 2019;9:27. doi: 10.1186/s13395-019-0213-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M.M., Lawson J.A., Mathew S.J., Hutcheson D.A., Kardon G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development. 2011;138:3625–3637. doi: 10.1242/dev.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oprescu S.N., Yue F., Qiu J., Brito L.F., Kuang S. Temporal dynamics and heterogeneity of cell populations during skeletal muscle regeneration. iScience. 2020;23:100993. doi: 10.1016/j.isci.2020.100993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramilowski J.A., Goldberg T., Harshbarger J., Kloppmann E., Lizio M., Satagopam V.P., Itoh M., Kawaji H., Carninci P., Rost B., Forrest A.R.R. A draft network of ligand–receptor-mediated multicellular signalling in human. Nat. Commun. 2015;6:7866. doi: 10.1038/ncomms8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco A., Doyonnas R., Kraft P., Vitorovic S., Blau H.M. Self-renewal and expansion of single transplanted muscle stem cells. Nature. 2008;456:502–506. doi: 10.1038/nature07384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco A., Mourkioti F., Tran R., Choi J., Llewellyn M., Kraft P., Shkreli M., Delp S., Pomerantz J.H., Artandi S.E., Blau H.M. Short telomeres and stem cell exhaustion model duchenne muscular dystrophy in mdx/mTR mice. Cell. 2010;143:1059–1071. doi: 10.1016/j.cell.2010.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambasivan R., Yao R., Kissenpfennig A., Van Wittenberghe L., Paldi A., Gayraud-Morel B., Guenou H., Malissen B., Tajbakhsh S., Galy A. Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development. 2011;138:3647–3656. doi: 10.1242/dev.067587. [DOI] [PubMed] [Google Scholar]
- Scaramozza A., Park D., Kollu S., Beerman I., Sun X., Rossi D.J., Lin C.P., Scadden D.T., Crist C., Brack A.S. Lineage tracing reveals a subset of reserve muscle stem cells capable of clonal expansion under stress. Cell Stem Cell. 2019;24:944–957.e5. doi: 10.1016/j.stem.2019.03.020. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel A.L., Kuhlmann P.K., Cornelison D. Muscle satellite cell proliferation and association: new insights from myofiber time-lapse imaging. Skeletal Muscle. 2011;1:7. doi: 10.1186/2044-5040-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa-Victor P., Gutarra S., Garcia-Prat L., Rodriguez-Ubreva J., Ortet L., Ruiz-Bonilla V., Jardi M., Ballestar E., Gonzalez S., Serrano A.L., et al. Geriatric muscle stem cells switch reversible quiescence into senescence. Nature. 2014;506:316–321. doi: 10.1038/nature13013. [DOI] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troy A., Cadwallader A., Fedorov Y., Tyner K., Tanaka K., Olwin B. Coordination of satellite cell activation and self-renewal by par-complex-dependent asymmetric activation of p38α/β MAPK. Cell Stem Cell. 2012;11:541–553. doi: 10.1016/j.stem.2012.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umansky K.B., Gruenbaum-Cohen Y., Tsoory M., Feldmesser E., Goldenberg D., Brenner O., Groner Y. Runx1 transcription factor is required for myoblasts proliferation during muscle regeneration. PLoS Genet. 2015;11:e1005457. doi: 10.1371/journal.pgen.1005457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.X., Feige P., Brun C.E., Hekmatnejad B., Dumont N.A., Renaud J.M., Faulkes S., Guindon D.E., Rudnicki M.A. EGFR-aurka signaling rescues polarity and regeneration defects in dystrophin-deficient muscle stem cells by increasing asymmetric divisions. Cell Stem Cell. 2019;24:419–432.e6. doi: 10.1016/j.stem.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster M.T., Manor U., Lippincott-Schwartz J., Fan C.M. Intravital imaging reveals ghost fibers as architectural units guiding myogenic progenitors during regeneration. Cell Stem Cell. 2016;18:243–252. doi: 10.1016/j.stem.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Ichikawa N., Kosaki K., Yamada Y., Sasaki T., Sakai L.Y., Kurosawa H., Hattori N., Arikawa-Hirasawa E. Perlecan deficiency causes muscle hypertrophy, a decrease in myostatin expression, and changes in muscle fiber composition. Matrix Biol. 2010;29:461–470. doi: 10.1016/j.matbio.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y., Nakada S., Yoshihara T., Nara T., Furuya N., Miida T., Hattori N., Arikawa-Hirasawa E. Perlecan, a heparan sulfate proteoglycan, regulates systemic metabolism with dynamic changes in adipose tissue and skeletal muscle. Sci. Rep. 2018;8:7766. doi: 10.1038/s41598-018-25635-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F., Bi P., Wang C., Shan T., Nie Y., Ratliff T.L., Gavin T.P., Kuang S. Pten is necessary for the quiescence and maintenance of adult muscle stem cells. Nat. Commun. 2017;8:14328. doi: 10.1038/ncomms14328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammit P.S., Heslop L., Hudon V., Rosenblatt J.D., Tajbakhsh S., Buckingham M.E., Beauchamp J.R., Partridge T.A. Kinetics of myoblast proliferation show that resident satellite cells are competent to fully regenerate skeletal muscle fibers. Exp. Cell Res. 2002;281:39–49. doi: 10.1006/excr.2002.5653. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative 3D rendering of protein immunoreactivity for DAPI (blue), ParD3 (green) as a marker of asymmetry, and Hmox-1 (red) in SCs on an isolated myofiber
Representative 3D rendering of protein immunoreactivity for DAPI (blue), ParD3 (green) as a marker of asymmetry, and Hmox-1 (red) in SCs on an isolated myofiber
Representative 3D rendering of protein immunoreactivity for DAPI (blue), ParD3 (green) as a marker of asymmetry, and Gabarap (red) in SCs on an isolated myofiber
Representative 3D rendering of protein immunoreactivity for DAPI (blue), ParD3 (green) as a marker of asymmetry, and Mgp (red) in SCs on an isolated myofiber
Representative 3D rendering of protein immunoreactivity for DAPI (blue), ParD3 (green) as a marker of asymmetry, and Mest (red) in SCs on an isolated myofiber
Differential gene expression using non-parameteric Wilcoxon rank-sum test performed on myogenic cluster and are ranked by significance according to adjusted p-valve. The gene with significance, average log fold change, the percentage of cells where the gene is detected, the percent of cells where the gene is detected in the comparison group, and adjusted p value are provided
Differential gene expression using non-parameteric Wilcoxon rank-sum test performed on myogenic cluster and are ranked by significance according to adjusted p-valve. The gene with significance, average log fold change, the percentage of cells where the gene is detected, the percent of cells where the gene is detected in the comparison group, and adjusted p-value are provided
Differential gene expression using non-parameteric Wilcoxon rank-sum test performed on myogenic cluster and are ranked by significance according to adjusted p-valve. The gene with significance, average log fold change, the percentage of cells where the gene is detected, the percent of cells where the gene is detected in the comparison group, and adjusted p-value are provided
Differential gene expression using non-parameteric Wilcoxon rank-sum test performed on myogenic cluster and are ranked by significance according to adjusted p-valve. The gene with significance, average log fold change, the percentage of cells where the gene is detected, the percent of cells where the gene is detected in the comparison group, and adjusted p-value are provided
Differential gene expression using non-parameteric Wilcoxon rank-sum test performed on myogenic cluster and are ranked by significance according to adjusted p-valve. The gene with significance, average log fold change, the percentage of cells where the gene is detected, the percent of cells where the gene is detected in the comparison group, and adjusted p-value are provided
Differential gene expression using non-parameteric Wilcoxon rank-sum test performed on myogenic cluster and are ranked by significance according to adjusted p-valve. The gene with significance, average log fold change, the percentage of cells where the gene is detected, the percent of cells where the gene is detected in the comparison group, and adjusted p-value are provided
The pathways, their fold enrichment, the raw p-value, and the false discovery rate are provided in the table. Highlighted pathways indicate pathways that include candidate genes that may regulate SC self-renewal
All NicheNet imputed differentially expressed ligands in the regenerating muscle NicheNet identified ligand with cognate receptor pair inferred from Ramilowski et al. (2015)
Data Availability Statement
-
•
Data: scRNA-Seq data have been deposited at GEO: GSE205690 and are publicly available as of the date of publication. Accession numbers are listed in the Key resources table. Microscopy data reported in this paper will be shared by the Lead contact upon request.
-
•
Code: This paper does not report original code
-
•
Any additional information required to reanalyze the data reported in this paper is available from the Lead contact upon request.