Abstract
Introduction
Chlamydia psittaci pneumonia has been a global public health hotspot in recent years. Although some scattered cases of C. psittaci pneumonia have been reported, there is a lack of large case studies worldwide.
Methods
In this multicenter, observational study, we recruited all consecutive patients with confirmed C. psittaci pneumonia from October 4, 2018, to October 23, 2020, in nine tertiary general hospitals in Central-South China. Epidemiologic and clinical data from patients’ electronic medical records were collected and analyzed.
Results
One hundred and sixteen patients with C. psittaci pneumonia were included in the study. The mean age was 59.7 years. Fever (96.6%) and cough (65.5%) were the most common clinical symptoms. Most patients presented with an increase in the proportion of neutrophils, neutrophil to lymphocyte ratio, LDH, alanine aminotransferase (ALT) and aspartate aminotransferase (AST) and a significant decrease in lymphocytes. The main CT lung findings were consolidation (81%) and pleural effusion (35.3%), and bilateral lung consolidation was mainly found in severe patients. Chlamydia psittaci DNA was detected in BALF (bronchoalveolar lavage fluid) or blood samples by metagenomic next-generation sequencing (mNGS) in all patients. Use of quinolone was associated with shorter length of hospital stay and fever duration after antibiotic use. Multivariate logistic regression analysis indicated that respiratory support was associated with both severe pneumonia and in-hospital death.
Conclusions
The clinical phenotype of C. psittaci pneumonia is complex and variable. mNGS is helpful in the diagnosis and treatment of C. psittaci pneumonia, and early treatment with quinolone may benefit patients.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40121-022-00662-4.
Keywords: Central-South China, Chlamydia psittaci pneumonia, Metagenomic next-generation sequencing, Observational study, Quinolone
Key Summary Points
| Why carry out this study? |
| Chlamydia psittaci (C. psittaci) pneumonia has been a global public health hotspot in recent years. It was estimated that the incidence of C. psittaci accounts for at least 1% of the incidence of CAP. Although some scattered cases of C. psittaci pneumonia have been reported, there is a lack of large case studies worldwide. |
| This study is the largest descriptive study to date describing the clinical and epidemiological characteristics of Chlamydia psittaci pneumonia. We retrospectively collected data from 116 patients with Chlamydia psittaci diagnosed by mNGS (metagenomic next-generation sequencing) in nine tertiary hospitals in Central-south China. |
| What was learned from the study? |
| Contrary to previous studies, our study found that metagenomes are helpful in the diagnosis and treatment of C. psittaci and that quinolone can effectively reduce the hospital stay and fever duration. |
| Since the previous studies on the efficacy of C. psittaci drugs were mainly case reports, clinical studies on the effectiveness of quinolones against C. psittaci are not yet available. |
Introduction
Psittacosis, which is also known as parrot fever or ornithosis, is a zoonotic bacterial infectious disease caused by the obligate intracellular gram negative organism Chlamydia psittaci (C. psittaci). C. psittaci can not only infect humans through the respiratory tract in the form of air or aerosols, leading to the occurrence of atypical community-acquired pneumonia (CAP), but also infect human skin, mucous membranes and digestive tract by excreta [1]. Parrot fever was endemic in 12 countries in the 1930s and led directly to the creation of the National Institutes of Health [2]. The flow of C. psittaci from parrots spread from person to person in a limited way [3–6], and over the past few decades, sporadic outbreaks of C. psittaci have spread from place to place [7–10]. Hogerwerf estimated that the incidence of C. psittaci is at least 1% of the incidence of CAP [11]. In fact, unlike traditional microbiologic diagnosis, C. psittaci pneumonia in humans is often mis- and underdiagnosed.
C. psittaci mainly causes respiratory infections in humans, with variable clinical symptoms. After the initial replication in the epithelial cells and macrophages of the respiratory system, the bacteria may spread throughout the body and affect different organs [1]. The severity of C. psittaci pneumonia ranges from mild flu-like symptoms to severe life-threatening pneumonia [1]. Due to the variable clinical characteristics and limited human-to-human transmission of C. psittaci pneumonia, it is more likely to be misdiagnosed in the context of the global overlap of influenza and COVID-19 [6]. Therefore, there is an urgent need for clinical analysis with large sample size to further understand the epidemiology and clinical characteristics of patients with C. psittaci pneumonia.
As we known, the culture of C. psittaci is too sensitive and complex to be routinely conducted [12]. The performance of other laboratory tests in the diagnosis of parrot fever is poor, for example, serologic analysis has both low specificity and sensitivity, while polymerase chain reaction (PCR) has low sensitivity [12]. Given the limitations of traditional diagnostic methods, non-targeted metagenomic next-generation sequencing (mNGS) has been increasingly applied for the diagnosis of infectious diseases [13]. Here, by collecting data from 116 confirmed mNGS cases, we attempt to provide an up-to-date description of the clinical characteristics of patients with C. psittaci pneumonia in China.
Methods
Study Design and Subjects
In this retrospective, multi-center study, all consecutive patients who were diagnosed with C. psittaci pneumonia and admitted to nine major tertiary hospitals in Central-South China from October 4, 2018, to October 23, 2020, were enrolled. The inclusion criteria were: first, patients met with the diagnostic criteria for community-acquired pneumonia [14]; second, metagenomic next-generation sequencing (mNGS) from an airway sample or blood revealed a specific DNA fragment in C. psittaci. Diagnostic criteria, clinical classification, treatment and discharge criteria for C. psittaci pneumonia cases were based on the Guidelines of the American Thoracic Society on CAP [15]. This study was approved by the institutional review board of the Second Xiangya Hospital (no. luohong201906). Informed consent was obtained from all included patients.
Data Collection
Epidemiologic and clinical data were extracted from electronic medical records. Clinical outcomes were followed up to October 31, 2020. The date of disease onset was defined as the day when the symptoms were noticed by the patient. A high fever was defined as a body temperature > 39.1 °C. Fever duration day after antibiotic use referred to the time from first use of quinolones or tetracyclines to recover from fever for patients who had fever during hospitalization.
Laboratory Confirmation
Bronchoalveolar lavage fluid (BALF) or blood was collected based on the standard clinical procedure [16]; 0.3 ml BALF or blood samples were separated in a new micro-centrifuge tube, and DNA was extracted using the TIANamp Micro DNA Kit (DP316, Tiangen Biotech) according to the manufacturer’s recommendation. The DNA library was constructed according to the protocol of the BGISEQ-50 sequencing platform. Constructed library was qualified by Agilent2100 (Agilent Technologies, Santa Clara, CA) and Qubit 2.0 (Invitrogen, USA), and a qualified double-strand DNA library was transformed into a single-strand circular DNA library. DNA nanoballs (DNBs) were generated from single-strand circular DNA using rolling circle amplification. DNBs were qualified using Qubit 2.0. Qualified DNBs were loaded into the flow cell and sequenced (50 bp, single end) on the BGISEQ-50 platform.
Statistical Analysis
Patients were grouped into severe/non-severe pneumonia groups according to the ATS guideline on CAP [15]. Furthermore, patients in the severe group were grouped into survival/non-survival groups according to their prognosis. Continuous variables were tested for normality using the skewness-kurtosis test and expressed as mean ± standard deviation (SD) or median (interquartile range [IQR]) depending on normal/non-normal distribution; continuous variables were compared using Student’s t-test or Mann-Whitney U test, as appropriate. Categorical variables were expressed as number and percentage and compared using the chi-square test. Univariate and multivariate logistic regression models were performed to explore the risk factors for severe cases and hospital death. To build a multivariate regression model, variables with p < 0.05 in the univariate model and known related factors based on clinical consideration were included in the original model and screened by backward method, which removed the most insignificant variable with a significance level > 0.05 stepwise. All data were analyzed by Stata (16.0) and Python (3.7). A two-tailed p value < 0.05 was considered statistically significant in all tests.
Results
Epidemiologic and Baseline Characteristics of 116 Patients with C. psittaci Pneumonia
One hundred and sixteen patients with C. psittaci pneumonia were included in this study. Their demographics and baseline characteristics are shown in Table 1. For all 116 patients, the mean age was 59.7 (SD 12.5; range 26.0–81.0) years, and 79.3% patients were older than 50 years; 72.4% patients were male. Only 18 patients had a history of exposure to parrots or poultry. C. psittaci pneumonia was diagnosed throughout the whole spectrum of seasons, but was more likely to occur in autumn (38.8%) and winter (33.6%). Forty-seven patients had at least one underlying disease including hypertension (19%), diabetes (18.1%), cardiovascular disease (8.6%), digestive disease (7.8%), cerebrovascular diseases (6.9%), etc. The most common symptoms included fever (96.6%), high fever (78.4%), cough (65.5%), dyspnea (46.6%) and fatigue (44.8%). Time from illness onset to first hospital admission was 7.0 (IQR 6.0–9.2; range 1.0–20.0) days. Among them, four medical staff were associated with clusters.
Table 1.
Demographics and baseline characteristics of 116 patients with Chlamydia psittaci pneumonia
| Variables | All patients (N = 116) | Non-severe group (N = 63) | Severe group (N = 53) | p values (< 0.050) | Survival group (N = 43) | Non-survival group (N = 10) | p values (< 0.050) |
|---|---|---|---|---|---|---|---|
| Age, years | 59.7 (12.5) | 56.7 (13.0) | 63.2 (11.1) | 0.005 | 64.4 (10.2) | 58.0 (13.7) | 0.100 |
| 18–49 | 24 (20.7%) | 20 (31.7%) | 4 (7.5%) | 0.001 | 4 (7.5%) | 2 (20.0%) | 0.098 |
| 50–64 | 45 (38.8%) | 22 (34.9%) | 23 (43.4%) | 0.351 | 18 (41.9%) | 5 (50.0%) | 0.640 |
| Above 65 | 47 (40.5%) | 21 (33.3%) | 26 (49.1%) | 0.086 | 23(53.5%) | 3 (30.0%) | 0.181 |
| Sex | 0.796 | 0.574 | |||||
| Male | 84 (72.4%) | 45 (71.4%) | 39 (73.6%) | 31(72.1%) | 8(80%) | ||
| Female | 32(27.6%) | 18(28.6%) | 14(26.4%) | 12(27.9%) | 2(20%) | ||
| Smoke history | 22 (19.0%) | 12 (19.0%) | 10 (18.9%) | 0.980 | 7 (16.3%) | 1 (10.0%) | 0.449 |
| Expose history | 18 (15.5%) | 9 (14.3%) | 9 (17.0%) | 0.690 | 2 (4.7%) | 2 (20.0%) | 0.682 |
| Onset seasons | 0.222 | 0.827 | |||||
| Spring | 14 (12.1%) | 11 (17.5%) | 3 (5.7%) | 3 (7.0%) | 1 (10.0%) | ||
| Summer | 18 (15.5%) | 10 (15.9%) | 8(15.1%) | 8 (18.6%) | 2 (20.0%) | ||
| Autumn | 45 (38.8%) | 23 (36.5%) | 17 (32.1%) | 17 (39.5%) | 4 (40.0%) | ||
| Winter | 39 (33.6%) | 19 (30.2%) | 25 (47.6%) | 25(58.1%) | 3 (30.0%) | ||
| Coexisting conditions | |||||||
| Any | 47 (40.5%) | 22 (34.9%) | 25 (47.2%) | 0.181 | 18 (41.9%) | 7 (70.0%) | 0.047 |
| Respiratory diseases | 3 (2.6%) | 1 (1.6%) | 2 (3.8%) | 0.460 | 0 (0%) | 2 (20.0%) | < 0.001 |
| Diabetes | 21 (18.1%) | 10 (15.9%) | 11 (20.8%) | 0.496 | 9 (20.9%) | 2 (20.0%) | 0.871 |
| Cardiovascular diseases | 10 (8.6%) | 3 (4.8%) | 7 (13.2%) | 0.106 | 7 (16.3%) | 0 (0%) | 0.310 |
| Cerebrovascular diseases | 8 (6.9%) | 4 (6.3%) | 4 (7.5%) | 0.800 | 3 (7.0%) | 1 (10.0%) | 0.685 |
| Hypertension | 22 (19.0%) | 8 (12.7%) | 14 (26.4%) | 0.060 | 12 (27.9%) | 2 (20.0%) | 0.930 |
| Tumor | 1 (0.9%) | 1 (1.6%) | 0 (0%) | 0.357 | 0 (0%) | 0 (0%) | 0.758 |
| Liver diseases | 5 (4.3%) | 2 (3.2%) | 3 (5.7%) | 0.511 | 2 (4.7%) | 1 (10.0%) | 0.354 |
| Kidney diseases | 3 (2.6%) | 2 (3.2%) | 1 (1.9%) | 0.663 | 1 (2.3%) | 0 (0%) | 0.590 |
| Digestive diseases | 9 (7.8%) | 5 (7.9%) | 4 (7.5%) | 0.938 | 3 (7.0%) | 1 (10.0%) | 0.782 |
| Surgery history | 15 (12.9%) | 10 (15.9%) | 5 (9.4%) | 0.303 | 3 (7.0%) | 2 (20.0%) | 0.486 |
| Time from illness onset to first hospital admission | 7.0 (6.0–9.2) | 7.0 (4.5–9.0) | 7.0 (7.0–9.0) | 0.268 | 7.0 (7.0–9.0) | 6.7 (2.9) | 0.588 |
| Signs and symptoms | |||||||
| High fever | 91 (78.4%) | 51 (81.0%) | 40 (75.5%) | 0.475 | 31 (72.1%) | 9 (90.0%) | 0.236 |
| Fever | 112 (96.6%) | 61 (96.8%) | 51 (96.2%) | 0.860 | 41 (95.3%) | 10 (100.0%) | 0.487 |
| Cough | 76 (65.5%) | 42 (66.7%) | 34 (64.2%) | 0.776 | 26 (60.5%) | 8 (80.0%) | 0.246 |
| Dyspnea | 54 (46.6%) | 26 (41.3%) | 28 (52.8%) | 0.214 | 22 (51.2%) | 6 (60.0%) | 0.614 |
| Muscle ache | 14 (12.1%) | 10 (15.9%) | 4 (7.5%) | 0.170 | 4 (9.3%) | 0 (0%) | 0.316 |
| Headache | 27 (23.3%) | 18 (28.6%) | 9 (17.0%) | 0.141 | 8 (18.6%) | 1 (10.0%) | 0.514 |
| Dizzyness | 27 (23.3%) | 13 (20.6%) | 14 (26.4%) | 0.463 | 10 (23.3%) | 4 (40.0%) | 0.279 |
| Stomach ache | 3 (2.6%) | 1 (1.6%) | 2 (3.8%) | 0.460 | 2 (4.7%) | 0 (0%) | 0.487 |
| Diarrhea | 6 (5.2%) | 4 (6.3%) | 2 (3.8%) | 0.533 | 1 (2.3%) | 1 (10.0%) | 0.251 |
| Fatigue | 52 (44.8%) | 29 (46.0%) | 23 (43.4%) | 0.776 | 16 (37.2%) | 7 (70.0%) | 0.059 |
| Vomit | 14 (12.1%) | 8 (12.7%) | 6 (11.3%) | 0.821 | 4 (9.3%) | 2 (20.0%) | 0.336 |
| Runny nose | 3 (2.6%) | 1 (1.6%) | 2 (3.8%) | 0.460 | 2 (4.7%) | 0 (0%) | 0.487 |
| Severity of illness scores | |||||||
| APACHE II | 11.0 (9.0–13.2) | 9.4 (3.8) | 13.0 (11.0–18.0) | < 0.001 | 11.0 (11.0–16.5) | 19.4 (6.9) | 0.028 |
| SOFA | 2.0 (1.0–3.0) | 2.0 (0.5–2.0) | 3.0 (2.0–6.0) | < 0.001 | 3.0 (2.0–6.0) | 5.4 (2.2) | 0.041 |
| PSI | 2.9 (1.1) | 3.0 (1.5–3.0) | 3.0 (3.0–4.0) | < 0.001 | 3.4 (0.8) | 4.2 (0.9) | 0.013 |
APACHE II Acute Physiology and Chronic Health Evaluation II, SOFA Sequential Organ Failure Assessment, PSI Pneumonia Severity Index
After admission, 63 and 53 patients were categorized into non-severe/severe subgroups as mentioned above. The severe group were of a significantly older age than the non-severe group (63.2 vs. 56.7 years, p < 0.001). There was no difference in gender, exposure history, season or coexisting conditions between the two groups. Subsequently, we divided the 53 patients in the severe subgroup into survival/non-survival groups according to their prognosis. We found that the non-survival group had a higher proportion of having at least one comorbidity (70% vs. 41.9%, p < 0.05), especially for respiratory diseases (20% vs. 0, p < 0.001), compared with the survival group.
Laboratory Examinations
Table 2 shows the radiologic and laboratory findings on admission. Chest computed tomography (CT) on admission was performed in 96 patients. Among these 96 patients with available CT results, 53.1% showed bilateral pulmonary involvement on lung CT, and the most common imaging findings on CT at admission included consolidation (99.0%) and pleural effusion (43.8%), while interstitial changes (5.2%) were rare. Figure 1 demonstrates the representative CT findings of four patients with non-severe or severe C. psittaci pneumonia. Severe cases yielded more prominent bilateral pulmonary involvement on CT than non-severe cases (68.2% vs. 40.4%, p < 0.01).
Table 2.
Radiographic and laboratory findings of 116 patients with Chlamydia psittaci pneumonia
| Variables | All patients (N = 116) | Non-severe group (N = 63) | Severe group (N = 53) | p values (< 0.050) | Survive group (N = 43) | Non-survive group (N = 10) | p values (< 0.050) | Normal range |
|---|---|---|---|---|---|---|---|---|
| Computed tomography images | N = 96 | N = 52 | N = 44 | N = 34 | N = 10 | |||
| Left pulmonary involvement | 20 (20.8%) | 11 (21.2%) | 9 (20.5%) | 0.933 | 9 (26.5%) | 0 (0%) | 0.068 | / |
| Right pulmonary involvement | 27 (28.1%) | 22 (42.3%) | 5 (11.4%) | 0.001 | 2 (5.9%) | 3 (30.0%) | 0.035 | / |
| Bilateral pulmonary involvement | 51 (53.1%) | 21 (40.4%) | 30 (68.2%) | 0.007 | 23 (67.6%) | 7 (70.0%) | 0.888 | / |
| Consolidation | 95 (99.0%) | 51 (98.1%) | 44 (100.0%) | 0.355 | 34 (100.0%) | 10 (100.0%) | 1 | / |
| Interstitial change | 5 (5.2%) | 3 (5.8%) | 2 (4.5%) | 0.788 | 2 (5.9%) | 0 (0%) | 0.432 | / |
| Pleural effusion | 42 (43.8%) | 20 (38.5%) | 22 (50.0%) | 0.256 | 17 (50.0%) | 5 (50.0%) | 1 | / |
| Metagenomic next-generation sequencing | ||||||||
| Airway sample sent for detection | 106 (91.4%) | 58 (92.1%) | 48 (90.6%) | 0.775 | 39 (90.7%) | 9 (90.0%) | 0.946 | / |
| Blood sample sent for detection | 17 (14.7%) | 8 (12.7%) | 9 (17.0%) | 0.516 | 6 (14.0%) | 3 (30.0%) | 0.223 | / |
| Airway and blood samples sent for detection | 7 (6.0%) | 3 (4.8%) | 4 (7.5%) | 0.530 | 2 (4.7%) | 2 (20.0%) | 0.098 | / |
| Airway/blood DNA sequence number of C. psittaci | 100.0 (23.8–372.8) | 56.0 (9.5–202.5) | 113.0 (57.0–531.0) | 0.015 | 100.0 (31.5–433.0) | 593.9 (641.4) | 0.165 | 0.0 |
| Airway DNA sequence number of C. psittaci | 107.0 (30.2–285.2) | 107.0 (23.5–191.5) | 107.0 (76.0–412.0) | 0.066 | 107.0 (34.5–348.0) | 55.0 (107.0–859.2 | 0.303 | 0.0 |
| Combine detection of bacteria | 19.0 (16.4%) | 11.0 (17.5%) | 8.0 (15.1%) | 0.732 | 7 (16.3%) | 1 (10.0%) | 0.617 | 0.0% |
| Combine detection of fungal infection | 21.0 (18.1%) | 12.0 (19.0%) | 9.0 (17.0%) | 0.773 | 7 (16.3%) | 2 (20.0%) | 0.778 | 0.0% |
| Combine detection of virus | 12.0 (10.3%) | 6.0 (9.5%) | 6.0 (11.3%) | 0.752 | 5 (11.6%) | 1 (10.0%) | 0.884 | 0.0% |
| Blood routine | ||||||||
| White blood cell count (× 109/L) | 7.8 (5.6–10.4) | 7.6 (5.8–9.5) | 9.3 (4.8) | 0.228 | 9.1 (4.9) | 10.1 (4.5) | 0.552 | 4.00–10.00 |
| Neutrophil percentage (%) | 87.1 (82.0–90.8) | 86.8 (80.7–89.2) | 89.0 (85.1–93.7) | 0.003 | 88.6 (84.5–92.5) | 90.7 (5.0) | 0.172 | 50.00–70.00 |
| Lymphocyte percentage (%) | 7.3 (5.1–10.0) | 7.4 (7.3–11.0) | 6.5 (3.5–7.4) | < 0.001 | 6.8 (3.9) | 4.3 (2.4) | 0.043 | 20.00–40.00 |
| Neutrophil-to-lymphocyte ratio | 12.1 (8.2–17.1) | 11.9 (7.6–12.1) | 13.7 (12.1–26.4) | < 0.001 | 12.6 (10.8–21.3) | 27.4 (14.8) | 0.046 | 1.25–3.5 |
| Hemoglobin (g/L) | 115.0 (106.8–121.0) | 115.0 (110.5–127.5) | 109.4 (17.3) | 0.041 | 108.7 (18.0) | 112.5 (14.4) | 0.537 | 110–160 |
| Platelet count (× 109/L) | 190.0 (166.0–207.5) | 190.0 (168.0–217.5) | 182.9 (66.6) | 0.257 | 190.4 (66.7) | 150.7 (58.6) | 0.089 | 100–300 |
| Biochemistry tests | ||||||||
| Albumin (g/L) | 28.2 (4.4) | 29.3 (4.5) | 26.9 (3.8) | 0.003 | 26.7 (3.8) | 27.7 (3.8) | 0.483 | 35.0–55.0 |
| Globulin (g/L) | 26.9 (26.4–27.4) | 27.6 (3.8) | 26.0 (3.8) | 0.018 | 26.2 (3.4) | 25.4 (5.5) | 0.676 | 20.2–29.5 |
| Albumin-to-globulin ratio | 1.1 (1.1–1.1) | 1.1 (1.1–1.1) | 1.1 (1.1–1.1) | 0.749 | 1.1 (1.1–1.1) | 1.1 (0.3) | 0.617 | 1.5–2.5 |
| Alanine aminotransferase (U/L) | 63.5 (47.8–99.3) | 63.5 (44.0–97.2) | 63.5 (48.2–98.9) | 0.644 | 63.5 (51.0–99.7) | 68.1 (28.3) | 0.600 | 0.0–42.0 |
| Aspartate aminotransferase (U/L) | 100.1 (57.9–146.1) | 100.1 (50.2–138.9) | 100.1 (77.6–156.0) | 0.109 | 100.1 (63.4–153.6) | 143.0 (78.8) | 0.500 | 0.0–37.0 |
| Total bilirubin (μmol/L) | 12.7 (12.7–12.8) | 12.7 (12.1–12.7) | 12.7 (12.7–14.8) | 0.422 | 12.7 (12.7–15.8) | 12.2 (9.7–12.7) | 0.052 | 3.4–17.1 |
| Direct bilirubin (μmol/L) | 6.3 (6.2–6.8) | 6.3 (6.0–6.3) | 6.3 (6.3–9.5) | 0.228 | 6.3 (6.3–10.8) | 6.2 (5.0–6.3) | 0.077 | 0.0–6.0 |
| Blood urea nitrogen (mmol/L) | 5.8 (5.7–6.3) | 5.3 (1.7) | 5.8 (5.8–9.4) | < 0.001 | 5.8 (5.8–8.4) | 10.5 (7.0) | 0.674 | 2.9–7.14 |
| Creatinine (μmol/L) | 74.8 (69.8–80.0) | 73.4 (17.0) | 74.8 (74.8–95.2) | 0.035 | 74.8 (74.8–88.0) | 74.8 (62.8–122.8) | 0.575 | 44.0–133.0 |
| Creatine kinase (U/L) | 237.0 (227.4–274.0) | 237.0 (160.5–237.0) | 237.0 (237.0–326.0) | 0.296 | 237.0 (237.0–295.5) | 296.7 (122.6) | 0.634 | 10.0–190.0 |
| Lactate dehydrogenase (U/L) | 394.7 (393.4–396.0) | 394.7 (307.1–394.7) | 394.7 (394.7–482.6) | 0.006 | 394.7 (394.7–464.8) | 508.8 (210.8) | 0.487 | 80.0–245.0 |
| Infection biomarkers | ||||||||
| Erythrocyte sedimentation rate (mm/h) | 86.0 (81.8–89.0) | 86.0 (86.0–90.5) | 86.0 (75.0–87.0) | 0.256 | 86.0 (77.0–88.5) | 75.1 (20.0) | 0.277 | 0.0–15.0 |
| High-sensitivity C-reactive protein (mg/L) | 162.0 (107.2–240.1) | 149.0 (79.3–198.2) | 197.6 (95.3) | 0.011 | 192.0 (95.9) | 221.7 (93.9) | 0.380 | 0.0–8.0 |
| Procalcitonin (mg/L) | 1.1 (0.3–4.9) | 0.5 (0.2–1.1) | 3.9 (1.1–9.8) | < 0.001 | 3.4 (1.1–10.0) | 7.2 (5.6) | 0.488 | 0.0–0.05 |
| D-dimer (mg/L) | 1.8 (1.4–3.2) | 1.8 (1.0–1.8) | 1.8 (1.8–4.8) | < 0.001 | 1.8 (1.8–4.7) | 2.5 (1.8–8.2) | 0.773 | 0.0–0.05 |
| Blood gas analysis | ||||||||
| Hydrogen ion concentration | 7.5 (7.5–7.5) | 7.5 (7.5–7.5) | 7.5 (7.5–7.5) | 0.234 | 7.5 (7.5–7.5) | 7.4 (0.1) | 0.049 | 7.35–7.45 |
| Partial pressure of carbon dioxide (mmHg) | 29.1 (29.1–29.1) | 29.1 (29.1–29.1) | 29.1 (29.1–32.0) | 0.062 | 29.1 (29.1–30.6) | 35.0 (9.8) | 0.095 | 35.0–45.0 |
| Oxygenation index (mmHg) | 241.0 (241.0–241.0) | 253.0 (68.5) | 241.0 (166.7–241.0) | 0.009 | 241.0 (198.6–241.0) | 194.8 (129.5) | 0.186 | 400.0–500.0 |
Values are numbers (percentages) or median (IQR) unless stated otherwise. Percentages do not total to 100% owing to missing data
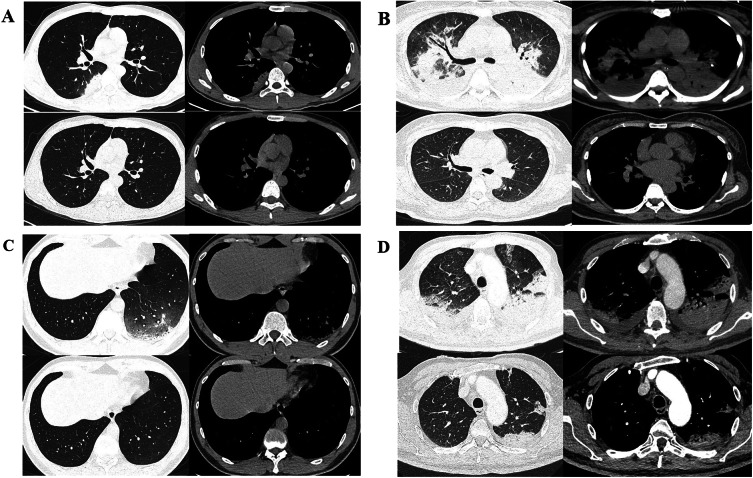
Fig. 1.
Typical CT findings of patients with Chlamydia psittaci pneumonia at admission and discharge. A A 26-year-old male with fever, cough, muscle pain and headache for 2 days, diagnosed with non-severe C. psittaci pneumonia. Axial CT on admission indicated unilateral lung consolidation and pleural effusion, and re-examination suggested complete absorption. B A 48-year-old female with fever, cough, shortness of breath and headache for 15 days was diagnosed with severe C. psittaci pneumonia. Axial CT at admission showed bilateral pulmonary exudation and bilateral pleural effusion, and re-examination showed complete absorption. C A 36-year-old male presented with fever, cough, muscle pain and headache for 4 days and was diagnosed with non-severe Chlamydia psittaci pneumonia. Axial CT scan at admission showed unilateral interstitial lung lesion, and re-examination showed complete absorption. D A 66-year-old male presented with fever, cough and shortness of breath for 13 days and was diagnosed with severe Chlamydia psittaci pneumonia. Axial CT scan at admission showed bilateral interstitial lesions, and re-examination showed most of the lesions had been completely absorbed
Laboratory examination data are also given in Table 2. On admission, all patients underwent metagenomic sequencing with airway sample (91.4%) or/and blood sample (14.7%). The median DNA sequence number of airway/blood samples for C. psittaci was 100.0 (IQR 23.8–372.8). This number was higher in the severe and non-survival groups compared with the non-severe and survival groups (113.0 vs. 56.0, p < 0.05; 593.9 vs. 100.0, p = 0.165). The combined detection rates of bacteria, fungi and virus were 16.4%, 18.1% and 10.3%, respectively, and there was no difference in the detection rates for these pathogens between different groups.
Interestingly, most patients presented with increased neutrophils and significantly decreased lymphocyte percentage. In general, compared with the non-severe and surviving groups, the severe group and non-survive group had a higher neutrophil-to-lymphocyte ratio and fewer lymphocytes (all p < 0.05). Biochemistry tests showed elevated alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels in most patients, with median ALT and AST values of 63.5 and 100.1 U/l (IQR 47.8–99.3; 57.9–146.1). However, no obvious difference in ALT and AST values was found between severe and non-severe subgroup patients. For infection biomarkers, erythrocyte sedimentation rate (ESR) and high-sensitivity C-reactive protein (h-CRP) were increased in > 90% of patients. Moreover, compared with the non-severe group, the severe group had a higher level of h-CRP, procalcitonin, d-dimer and lactate dehydrogenase and lower oxygenation index (all p < 0.05).
Treatment and Clinical Outcomes
Treatment and clinical outcomes in 116 patients with C. psittaci pneumonia are shown in Table 3. During hospitalization, 68 (58.6%) of 116 patients received respiratory support therapy, including oxygen (35.3%), high-flow nasal cannula (7.8%), non-invasive ventilation (6.9%) and invasive ventilation (18.1%). More severe cases received mechanical ventilation (non-invasive: 15.1% vs. 0%; invasive: 39.6% vs. 0%, p < 0.01) compared with non-severe cases. During hospitalization, the percentages of patients being admitted to the ICU, requiring mechanical ventilation and death were 50.0%, 20.7% and 8.6%, respectively. The length of hospital stay ranged from 4 to 57 days in discharged patients. Comparison between survival and non-survival in all 106 included patients are shown in Table S2.
Table 3.
Treatments and clinical outcomes in 116 patients with Chlamydia psittaci pneumonia
| Variables | All patients (N = 116) |
Non severe group (N = 63) |
Severe group (N = 53) |
p values (< 0.050) |
Survive group (N = 43) |
None survive group (N = 10) |
p values (< 0.050) |
|---|---|---|---|---|---|---|---|
| Respiratory support | |||||||
| Oxygen | 41 (35.3%) | 34 (54.0%) | 7 (13.2%) | < 0.001 | 6 (14.0%) | 1 (10.0%) | 0.739 |
| High-flow nasal cannula | 9 (7.8%) | 4 (6.3%) | 5 (9.4%) | 0.536 | 5 (11.6%) | 0 (0%) | 0.257 |
| Non-invasive ventilation | 8 (6.9%) | 0 (0%) | 8 (15.1%) | 0.001 | 5 (11.6%) | 3 (30.0%) | 0.144 |
| Invasive ventilation | 21 (18.1%) | 0 (0%) | 21 (39.6%) | < 0.001 | 14 (32.6%) | 7 (70.0%) | 0.029 |
| Respiratory therapy except oxygen | 31 (26.7%) | 4 (5.6%) | 27 (60.0%) | < 0.001 | 19 (44.2%) | 8 (80.0%) | 0.041 |
| Other supportive treatment | |||||||
| CRRT | 4 (3.4%) | 0 (0%) | 4 (7.5%) | 0.026 | 2 (4.7%) | 2 (20.0%) | 0.098 |
| ECMO | 2 (1.7%) | 0 (0%) | 2 (3.8%) | 0.120 | 1 (2.3%) | 1 (10.0%) | 0.251 |
| Length of stay | |||||||
| Hospital day | 14.0 (10.0–17.0) | 14.0 (8.0–14.0) | 14.0 (14.0–20.0) | < 0.001 | 14.0 (14.0–19.5) | 13.5 (8.4) | 0.112 |
| ICU day | 0.0 (0.0–4.0) | 0.0 (0.0–0.0) | 3.0 (0.0–14.0) | < 0.001 | 0.0 (0.0–12.0) | 10.2 (8.2) | 0.032 |
| Outcomes | |||||||
| Death | 10 (8.6%) | 0 (0%) | 10 (18.9%) | < 0.001 | 0 (0%) | 10 (100.0%) | < 0.001 |
| ICU | 58 (50.0%) | 15 (23.8%) | 43 (81.1%) | < 0.001 | 34 (79.1%) | 9 (90.0%) | 0.426 |
| Survive | 106 (91.4%) | 63(100%) | 43(81%) | < 0.001 | 43 | 0 (0%) | < 0.001 |
Values are numbers (percentages) or median (IQR) unless stated otherwise. Percentages do not total to 100% owing to missing data
As for antibiotics treatment, 98 patients with detailed antibiotic usage data were further analyzed, and the results are shown in Table 4. Ninety-eight patients were sub-grouped according to antibiotic usage. The group with use of tetracycline or quinolones only had no statistically significant association with severe pneumonia, ICU admission or hospital death. However, length of hospital stay (13.8 vs. 18.8 day, p < 0.001) and fever duration days after antibiotic use (3 vs. 4, p < 0.05) were significantly reduced in the quinolone group compared with the non-quinolone group, which was not observed in the tetracycline group.
Table 4.
Efficacy of antibiotics for treatment in 98 patients with Chlamydia psittaci pneumonia
| Variables | All patients (N = 98) |
Quinolones group (N = 52) |
Non-quinolones group(N = 46) |
p values (< 0.050) |
Tetracyclines group(N = 24) |
Non-tetracyclines group(N = 74) |
p values (< 0.050) |
|---|---|---|---|---|---|---|---|
| ICU | 47(48.0%) | 21(40.4%) | 26(56.5%) | 0.111 | 15(62.5%) | 32(43.2%) | 0.101 |
| Severe | 44(44.9%) | 20(38.5%) | 24(52.2%) | 0.173 | 11(45.8%) | 33(44.6%) | 0.916 |
| Death | 7(7.1%) | 3(5.77%) | 4(8.7%) | 0.575 | 2(8.3%) | 5(6.8%) | 0.794 |
| Hospital day | 16.3(0.8) | 13.8(1.0) | 18.8(1.1) | 0.001 | 19.2(1.4) | 15.3(0.9) | 0.014 |
| Fever duration day after antibiotics use | 3(2–5) | 3(1–4) | 4(2–7.5) | 0.019 | 3(1–5) | 3(2–5) | 0.579 |
Risk Factors for Severe Pneumonia, ICU Admission and Hospital Death
Univariate logistic analysis was performed to explore the risk factors for severe pneumonia and hospital death, respectively, and the results are shown in Table S1 in Supplementary Appendix. To adjust the confounding effect, multivariate logistic regression models were built for severe pneumonia and hospital death following the previously mentioned method, and the results are shown in Table 5 and 6. In the logistic model for severe pneumonia, we initially included the following variables: age, APACHE II, PSI, WBC, neutrophil percentage, lymphocyte percentage, neutrophil-to-lymphocyte ratio, hemoglobin, albumin, globulin, total/direct bilirubin, lactate dehydrogenase, ESR, procalcitonin, d-dimer, partial carbon dioxide pressure, cardiovascular diseases, any complications, bilateral pulmonary involvement and respiratory therapy except oxygen. The final model revealed that higher globulin concentration (OR 0.838, 95% CI 0.704–0.998, p = 0.048) was a protective factor for survival, and higher APACHE II score (OR 38.342, 95% CI 1.118–1.59, p = 0.001) and respiratory therapy except oxygen (OR 8.304, 95% CI 1.874–36.783, p = 0.005) were risk predictors of severe pneumonia. Respiratory therapy except oxygen refers to all other respiratory therapy except oxygen therapy, including high-flow nasal cannula, non-invasive ventilation and invasive ventilation. In the regression model for hospital death, the included variables were APACHE II, PSI, neutrophil percentage, lymphocyte percentage, platelet count, d-dimer, partial pressure of carbon dioxide, cardiovascular diseases, any complications, bilateral pulmonary involvement and respiratory therapy except oxygen. The final model showed that higher lymphocyte percentage (OR 0.696, 95% CI 0.514–0.942, p = 0.019) was the only protective factor for survival, and higher PSI (OR 5.984, 95% CI 1.812–19.759, p = 0.003) and respiratory therapy except oxygen (OR 10.077, 95% CI 1.361–74.579, p = 0.002) were risk predictors of hospital death.
Table 5.
Risk factors for severe pneumonia in multivariate logistic regression model
| Risk factor | SE | Z | OR | 95% CI | p value |
|---|---|---|---|---|---|
| APACHE II | 0.119 | 3.2 | 1.333 | 1.118–1.590 | 0.001 |
| Globulin | 0.075 | − 1.98 | 0.838 | 0.704–0.998 | 0.048 |
| Respiratory therapy except oxygen | 10.291 | 2.26 | 8.304 | 1.874–36.783 | 0.005 |
SE standard error, OR odds ratio, CI confidence interval
Table 6.
Risk factors for hospital death in multivariate logistic regression model
| Risk factor | SE | Z | OR | 95% CI | p value |
|---|---|---|---|---|---|
| Lymphocyte percentage | 0.108 | − 2.34 | 0.696 | 0.514–0.942 | 0.019 |
| PSI | 3.647 | 2.94 | 5.984 | 1.812–19.759 | 0.003 |
| Respiratory therapy except oxygen | 10.291 | 2.26 | 10.077 | 1.361–74.579 | 0.024 |
SE standard error, OR odds ratio, CI confidence interval
Discussion
This report, to our knowledge, is the largest case series of hospitalized patients with C. psittaci pneumonia to date. In this nine-center observational study, epidemiology and clinical characteristics of 116 C. psittaci pneumonia patients in Central-South China were described and compared between severe/non-severe and survival/non-survival patients. Effectiveness of quinolone and tetracycline treatment and risk factors for severe pneumonia and hospital death were also analyzed. Our study found that metagenomes are helpful in the diagnosis and treatment of C. psittaci, and quinolone is associated with fewer hospital days and fever duration days after antibiotic use.
The host species of C. psittaci are diverse, including birds with psittacosis, the main host, and poultry such as chickens and ducks. Previous studies have indicated that C. psittaci has limited human-to-human transmission ability [5, 6, 17–19]. Many countries have listed C. psittaci as a notifiable infectious disease for surveillance [20]. In our research, only one cluster of disease was found, which was caused by a patient with C. psittaci who infected four medical staff members, but the exact route of infection was unclear [6]. In the past, the incidence of C. psittaci pneumonia has been underestimated because of limited testing techniques for diagnosis. To date, there are no data on the incidence of C. psittaci pneumonia in China. Therefore, the route and details of C. psittaci infection need more epidemiologic investigation.
The incidence of C. psittaci pneumonia could be related to variations in ecologic and geographic factors such as temperature, rainfall and landscape [21]. Central South China is a place of bird migration [22]. Autumn and winter are the migratory seasons for migratory birds [22, 23]. During the migration season, migratory birds easily get sick, and cross infection occurs when they mix with poultry during migration. Our research found that C. psittaci pneumonia occurs mainly in autumn and winter. Therefore, during the migratory season, the public should pay attention to hygiene when raising live poultry or holding live poultry markets.
Previous studies have shown that C. psittaci pneumonia mainly occurs in the middle-aged and elderly population, with male preponderance, and more than half have a history of bird exposure [8, 10, 24]. Similar to previous studies, our study found that 93.1% of C. psittaci pneumonia patients were > 40 years old, and male patients accounted for the most cases. However, inconsistent with previous studies, only 15.5% of them could be traced back to a clear exposure history in this study. Due to the retrospective nature of our study, and the long time from illness onset to first hospital admission and the diversity of patients, it was difficult to trace back the exposure history of all patients.
Chlamydia psittaci is a zoonotic pathogen parasitizing strictly in eukaryotic cells. After infection, C. psittaci first invades epithelial columnar cells and then multiplies in the inclusion bodies of mononuclear macrophages, avoiding the immune defense response of host cells and phagocytosis by lysosomes [1]. Human infection with C. psittaci can lead to systemic symptoms due to the extensive presence of epithelial and mononuclear macrophage systems [1]. Previous studies have shown that people infected with C. psittaci generally present with influenza-like atypical symptoms such as fever, chills, headache, myalgia and fatigue, with or without respiratory symptoms. Our study seems to be similar to previous studies, with nearly 50% of patients presenting with headache, dizziness, stomach ache, diarrhea, frequent urination or urgent urination as the first symptoms [1, 25, 26]. However, as the disease progresses, almost all patients develop respiratory symptoms, with high fever, cough, dyspnea and fatigue being the most common clinical symptoms. The symptoms of C. psittaci are variable, and the diagnosis is difficult. In the current situation of the COVID-19 epidemic, it is more important to raise people’s attention and vigilance, and medical staff should be carefully trace the source of exposure when recording the medical history.
Chlamydia psittaci infection is easily ignored because its symptoms are not typical and it is difficult to diagnose. Clinicians have limited knowledge of C. psittaci infection. Laboratory testing for C. psittaci includes culture, serologic assay and PCR. Chlamydia psittaci culturing is complex and time-consuming and requires a P3 laboratory. The sensitivity and specificity of serologic detection and PCR diagnosis are limited, so most clinical laboratories cannot carry out routine testing. Because acute and convalescent sera are both required, serologic tests are only useful for retrospective diagnosis. PCR-based nucleic acid detection is the fastest and most specific method, but is sensitive only in the acute phase. mNGS is increasingly recommended for the diagnosis of infectious diseases when the empirical use of conventional anti-infective therapy is not effective. All the patients in our study were confirmed by mNGS with blood and/or airway samples, and consistently with the clinical situation, the number of sequences was significantly higher in severe patients. Our study shows that mNGS in both blood and BALF samples is effective in the detection of C. psittaci, so we may choose to send blood samples for mNGS in primary hospitals without bronchoscopic equipment or when patients cannot tolerate bronchoscopy.
The recommended treatment for C. psittaci pneumonia includes tetracyclines, macrolides and quinolones, and the recommended first-line drugs are tetracyclines [1, 11, 26]. Since the previous studies on the efficacy of C. psittaci drugs were mainly case reports, clinical studies on the effectiveness of quinolones against C. psittaci are not yet available [1]. Contrary to previous studies, our study found that use of quinolones rather than tetracyclines was associated with fewer hospital days and fever duration days after antibiotic use. Currently, the efficacy of moxifloxacin against C. psittaci is controversial, although Turkish researchers conducted drug sensitivity tests on doxycycline and ciprofloxacin in vitro and found that the MIC of doxycycline was lower than that of ciprofloxacin [27]. Due to the widespread use of tetracycline drugs in the poultry and pet bird industries [1], many cases of tetracycline-resistant chlamydia strains have been reported in recent years [28, 29]. Moreover, as a new generation of quinolones, moxifloxacin has a stronger antibacterial effect than ciprofloxacin against atypical pathogens such as Chlamydia [30]. Prospective randomized controlled trial (RCT) studies on the efficacy of these two drugs against C. psittaci are urgently needed.
There are several limitations that should be mentioned in this study. First, due to the retrospective multi-center study design, there is information bias in tracing the exposure history. Second, all patients were diagnosed by mNGS and did not undergo traditional gold standard laboratory tests, including C. psittaci culture and PCR, for diagnosis. Third, multiple factors related to medicine use such as drug combination, different drug types, different manufacturers and inconsistent duration of drug use were missing and might lead to bias in the analysis of drug efficacy. A prospective study is needed to further study the therapeutic effect of the drugs on psittacosis.
Conclusion
In summary, this multi-center case series of 116 hospitalized patients with confirmed C. psittaci pneumonia in Central-South China provides clinicians with not only a greater basis for diagnosis and treatment, but also some useful information for governments and communities to help them carry out effective epidemiologic control measures.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank all the patients for participating in the present study. We thank Prof. Ding-Ding Deng, Xiang-Long Kong and Jian-Bo Chen for collecting clinical data.
Funding
This study was supported by the National Natural Science Foundation of China (82070003 to Hong Luo), the Natural Science Foundation of Hunan Province (2021JJ30943 to Hong Luo), the Science and Technology Program of Changsha, China (kq1901120 to Hong Luo), Xiangya Medical Big Data of Central South University (Pulmonary Inflammatory Disease) and the National Key Clinical Specialty Construction Projects of China ((2012) No.650). The Second Xiangya Hospital of Central South University and Central South University funded the journal’s Rapid Service fees.
Author Contributions
Conception and design of the experiments: Hong Luo and Ping Chen. Collection of clinical data: Min Yang, Huan Yang, Cai-Hong Liu, Hui-Ming Yin, Dan Liu. Analysis of the data: Shui-Zi Ding, Dan-Hui Yang. Writing the text of the main manuscript: Dan-Hui Yang, Min Yang. Preparation of the tables and figures: Dan-Hui Yang. All authors reviewed and revised the manuscript.
Disclosures
Min Yang, Dan-Hui Yang, Shui-Zi Ding, Huan Yang, Cai-Hong Liu, Hui-Ming Yin, Dan Liu, Ping Chen and Hong Luo confirm that they have no competing interests to declare.
Compliance with Ethics Guidelines
This study was approved by the institutional review board of the Second Xiangya Hospital (no. luohong201906). Informed consent was obtained from all included patients.
Data Availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Min Yang, Dan-Hui Yang, Ping Chen and Hong Luo contributed equally to this work.
Contributor Information
Ping Chen, Email: pingchen0731@csu.edu.cn.
Hong Luo, Email: luohonghuxi@csu.edu.cn.
References
- 1.Beeckman DS, Vanrompay DC. Zoonotic Chlamydophila psittaci infections from a clinical perspective. Clin Microbiol Infect. 2009;15(1):11–17. doi: 10.1111/j.1469-0691.2008.02669.x. [DOI] [PubMed] [Google Scholar]
- 2.Lepore J. It’s spreading: outbreaks, media scares, and the parrot panic of 1930. The New Yorker American Chronicles; June 1,2009 Issue:46–50.
- 3.Gelfand MS, Cleveland KO. Family outbreak of psittacosis with an exhumation-based diagnosis: following in the footsteps of Dr. House. Am J Med Sci. 2013;345(3):252–253. doi: 10.1097/MAJ.0b013e31826e366a. [DOI] [PubMed] [Google Scholar]
- 4.Laroucau K, Aaziz R, Meurice L, et al. Outbreak of psittacosis in a group of women exposed to Chlamydia psittaci-infected chickens. Euro Surveill. 2015 doi: 10.2807/1560-7917.es2015.20.24.21155. [DOI] [PubMed] [Google Scholar]
- 5.Wallensten A, Fredlund H, Runehagen A. Multiple human-to-human transmission from a severe case of psittacosis, Sweden, January-February 2013. Euro Surveill. 2014 doi: 10.2807/1560-7917.es2014.19.42.20937. [DOI] [PubMed] [Google Scholar]
- 6.Lei JH, Xu Y, Jiang YF, Shi ZH, Guo T. Clustering cases of Chlamydia psittaci pneumonia in COVID-19 screening ward staff. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arenas-Valls N, Chacón S, Pérez A, Del Pozo R. Atypical Chlamydia psittaci pneumonia. Four related cases. Arch Bronconeumol. 2017;53(5):277–279. doi: 10.1016/j.arbres.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 8.Gu L, Liu W, Ru M, et al. The application of metagenomic next-generation sequencing in diagnosing Chlamydia psittaci pneumonia: a report of five cases. BMC Pulm Med. 2020;20(1):65. doi: 10.1186/s12890-020-1098-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Gier B, Hogerwerf L, Dijkstra F, van der Hoek W. Disease burden of psittacosis in the Netherlands. Epidemiol Infect. 2018;146(3):303–305. doi: 10.1017/s0950268817003065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong CY, Zhu J, Fau-Lu J-J, Lu J, Fau-Xu Z-H, Xu ZH. Clinical characteristics of Chlamydia psittaci pneumonia. Chin Med J Engl. 2021;134(3):353–355. doi: 10.1097/CM9.0000000000001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hogerwerf L, de Gier B, Baan B, Van der Hoek W. Chlamydia psittaci (psittacosis) as a cause of community-acquired pneumonia: a systematic review and meta-analysis. Epidemiol Infect. 2017;145(15):3096–3105. doi: 10.1017/s0950268817002060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nieuwenhuizen AA, Dijkstra F, Notermans DW, van der Hoek W. Laboratory methods for case finding in human psittacosis outbreaks: a systematic review. BMC Infect Dis. 2018;18(1):442. doi: 10.1186/s12879-018-3317-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao L, Rong X, He M, et al. Metagenomic analysis of potential pathogens from blood donors in Guangzhou, China. Transfus Med. 2020;30(1):61–69. doi: 10.1111/tme.12657. [DOI] [PubMed] [Google Scholar]
- 14.Mandell LA, Wunderink RG, Anzueto A, et al. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis. 2007;44(Suppl 2):S27–S72. doi: 10.1086/511159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Metlay JP, Waterer GW, Long AC, et al. Diagnosis and treatment of adults with community-acquired pneumonia. An official clinical practice guideline of the American Thoracic Society and Infectious Diseases Society of America. Am J Respir Crit Care Med. 2019;200(7):e45–e67. doi: 10.1164/rccm.201908-1581ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X, Ding S, Lei C, et al. Blood and bronchoalveolar lavage fluid metagenomic next-generation sequencing in pneumonia. Can J Infect Dis Med Microbiol. 2020;2020:6839103. doi: 10.1155/2020/6839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li N, Li S, Tan WA-O, Wang H, Xu H, Wang D. Metagenomic next-generation sequencing in the family outbreak of psittacosis: the first reported family outbreak of psittacosis in China under COVID-19. Emerg Microbes Infect. 2021;10(1):1418–1428. doi: 10.1080/22221751.2021.1948358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Boeck C, Dehollogne C, Dumont A, et al. Managing a cluster outbreak of psittacosis in Belgium linked to a pet shop visit in The Netherlands. Epidemiol Infect. 2016;144(8):1710–1716. doi: 10.1017/s0950268815003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ciftçi B, Güler ZM, Aydoğdu M, Konur O, Erdoğan Y. Familial outbreak of psittacosis as the first Chlamydia psittaci infection reported from Turkey. Tuberk Toraks. 2008;56(2):215–220. [PubMed] [Google Scholar]
- 20.McGovern OI, Kobayashi M, Shaw KA, Szablewski C, et al. Use of real-time PCR for Chlamydia psittaci detection in human specimens during an outbreak of psittacosis—Georgia and Virginia, 2018. MMWR Morb Mortal Wkly Rep. 2021;70(14):505–509. doi: 10.15585/mmwr.mm7014a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cong W, Huang SY, Zhang XX, et al. Chlamydia psittaci exposure in pet birds. J Med Microbiol. 2014;63(Pt 4):578–581. doi: 10.1099/jmm.0.070003-0. [DOI] [PubMed] [Google Scholar]
- 22.Zou YA, Zhang PY, Zhang SQ, et al. Crucial sites and environmental variables for wintering migratory waterbird population distributions in the natural wetlands in East Dongting Lake, China. Sci Total Environ. 2019;655:147–157. doi: 10.1016/j.scitotenv.2018.11.185. [DOI] [PubMed] [Google Scholar]
- 23.Lei J, Jia Y, Zuo A, et al. Bird satellite tracking revealed critical protection gaps in East Asian–Australasian Flyway. Int J Environ Res Public Health. 2019;16(7):1147. doi: 10.3390/ijerph16071147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozuki E, Arima Y, Matsui T, et al. Human psittacosis in Japan: notification trends and differences in infection source and age distribution by gender, 2007 to 2016. Ann Epidemiol. 2020;44:60–63. doi: 10.1016/j.annepidem.2020.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Marrie TJ, Peeling RW, Reid T, De Carolis E. Chlamydia species as a cause of community-acquired pneumonia in Canada. Eur Respir J. 2003;21(5):779–784. doi: 10.1183/09031936.03.00095403. [DOI] [PubMed] [Google Scholar]
- 26.Knittler MR, Sachse K. Chlamydia psittaci: update on an underestimated zoonotic agent. Pathog Dis. 2015;73(1):1–15. doi: 10.1093/femspd/ftu007. [DOI] [PubMed] [Google Scholar]
- 27.Butaye P, Ducatelle R, De Backer P, Vermeersch H, Remon JP, Haesebrouck F. In vitro activities of doxycycline and enrofloxacin against European Chlamydia psittaci strains from turkeys. Antimicrob Agents Chemother. 1997;41(12):2800–2801. doi: 10.1128/aac.41.12.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Unterweger C, Schwarz L, Jelocnik M, et al. Isolation of tetracycline-resistant Chlamydia suis from a Pig Herd affected by reproductive disorders and conjunctivitis. Antibiotics (Basel) 2020 doi: 10.3390/antibiotics9040187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wahdan A, Rohner L, Marti H, et al. Prevalence of chlamydiaceae and tetracycline resistance genes in wild boars of central Europe. J Wildl Dis. 2020;56(3):512–522. doi: 10.7589/2019-11-275. [DOI] [PubMed] [Google Scholar]
- 30.Speciale A, Musumeci R, Blandino G, Milazzo I, Caccamo F, Nicoletti G. Minimal inhibitory concentrations and time-kill determination of moxifloxacin against aerobic and anaerobic isolates. Int J Antimicrob Agents. 2002;19(2):111–118. doi: 10.1016/s0924-8579(01)00486-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.