Abstract
For young ruminants, starter feeding can effectively facilitate the growth and development of rumen in ruminants, but the development of rumen is an important physiological challenge as it remains unclear for the mechanism of starter feeding stimulating. In this study, we performed an analysis of ruminal microbiota and their metabolites in yak calves to explore how the ruminal microbiota and their metabolites stimulate the ruminal function. This study associated 16S rRNA sequencing with liquid chromatography-mass spectrometry (LC-MS)-based metabolomics to evaluate the effects of starter feeding on ruminal microbiota diversity and metabolites in yak calves. We designed the experiment using 20 yak calves that were assigned equally into 2 groups, based on feeding milk replacer; the control (RA) group was fed with alfalfa hay while the treatment (RAS) group was fed with alfalfa hay and starter. After the experiment, we investigated the ruminal microbiota and metabolites through 16S rRNA sequencing and LC-MS-based metabolomics. During the preweaning period, the RAS group significantly promoted the growth performance and ruminal development in yak calves, including increases in body weight, chest girth, and development of rumen (P < 0.05). The RAS group increased the relative abundance of Bacteroidota, Proteobacteria, Chloroflexi, Synergistota, and Spirochaetota and decreased the abundance of Firmicutes, Desulfobacterota, Actinobacteriota, and Actinobacteriota at the phylum level (P < 0.05). At the genus level, the ruminal content of the RAS group was significantly enriched for Rikenellaceae_RC9_gut_group and Ruminococcus, while depleted for Prevotella, Christensenellaceae_R-7_group, and NK4A214_group (P < 0.05). A total of 37 metabolites were identified between the RA group and the RAS group, of which 15 metabolites were upregulated and 22 metabolites were downregulated compared with the RA group. Metabolic pathway analyses indicated that upregulated the metabolites of the RAS group yak calves were related to carbohydrate metabolism, ubiquinone, and other terpenoid-quinone biosynthesis, while the downregulated metabolic pathway was relevant to xenobiotic biodegradation, metabolism, and nucleotide metabolism. In summary, starter feeding before weaning significantly increased the dry matter intake and body weight of yak calves, changed the diversity and abundance of ruminal microbiota, and positively regulated the good development of ruminal morphology and function, providing an important basis for high-quality cultivation and the nutritional level of nutrition of yak calves in the Qinghai Tibet plateau. This study is based on the availability of 16S rRNA sequencing and LC-MS-based metabolomics in clarifying the function of starter feeding in the yak calves.
Keywords: feeding strategies, ruminal development, ruminal microbiota, ruminal metabolomics, yak calves
Introduction
Yak calves are the foundation of the yak industry in the Qinghai-Tibetan Plateau, and the quality of yak calf rearing directly contributes to the performance of adults (Long et al., 2008). In the early stage of raising, the suckling period has a long-term impact on various biological functions (Baldwin et al., 2004; Soberon and Van Amburgh, 2013). Traditionally, yak calves are weaned naturally or artificially at 1.5–2 years of age under a wide range of conditions (Ding et al., 2013). Premature weaning of yaks is being attempted to improve ruminal growth and function (Mohr et al., 2002). However, under natural grazing conditions, there is a high mortality rate of calves due to the lack of nutrition and poor environmental conditions in the Qinghai-Tibetan Plateau (Liu et al., 2018). Taking this into account, Khan et al. (2016) proposed that a mixture of roughage and grain could be used for early weaning under the condition of house feeding. Diet, as one of the most important factors, affects the digestive systems, especially the structure and function of the ruminal microbiota, which can promote ruminal development (Berends et al., 2015; Latham et al., 2018; Ogunade et al., 2019). Coincidentally, a supplement of alfalfa stimulated the ruminal development consistent with the changes in ruminal microbiota and animal performance before and after weaning (Yang et al., 2018). Meanwhile, starter feeding decreased mRNA expression of cytokines, namely, TNF-α and IFN-γ in the colonic tissue and also in the digestive tract (Liu et al., 2017).
The digestive tract mainly develops in the early growth stage of calves, which effectively influences their long-term performance (Davis Rincker et al., 2011). As the fermentation pot of the ruminant animal, rumen fermentation produces volatile fatty acids (VFAs) that can directly stimulate the proliferation and development of the ruminal epithelium (Górka et al., 2018). Besides, the ruminal microbial composition is an effective way to resist external stimulation and maintain ruminal environmental stability (Shen et al., 2017). Therefore, the ruminal microbiota plays a central role in the efficiency of digestion in ruminants (Morgavi et al., 2013).
Although it had already been proved in dairy calves that alfalfa hay could promote ruminal epithelial and muscular development (Yang et al., 2015), but the inferior quality of alfalfa hay to be fed to calves could not provide efficient ruminal VFA required for ruminal papilla development (Mirzaei et al., 2015; Hosseini et al., 2016). Fortunately, the previous study found that the supplement of milk replacer and starter feeding can relieve the stress of weaning and raise the level of ruminal development (Sweeney et al., 2010). Therefore, in this study, 16S rRNA sequencing and metabolomics technology were used to investigate starter feeding based on alfalfa hay for weaning yak calves, providing an important reference for the research of milk replacer breeding technology after weaning in yak calves.
In recent years, next-generation high-throughput sequencing (via 16S rRNA sequencing) has been used to assess the influences of dietary on the ruminal microbial community (Pinloche et al., 2013; Kim et al., 2014). The application of metabolomics analysis provides an opportunity to measure large numbers of small molecule metabolites in cells, tissues, and biofluids (Goldansaz et al., 2017). Recent studies have applied metabolomics to predict feed efficiency and residual feed intake (Karisa et al., 2014; Artegoitia et al., 2017), examine disease conditions (Hailemariam et al., 2014), evaluate dietary responses to different feeds (Saleem et al., 2012), and assess milk quality of ruminants (Abarghuei et al., 2014). Nevertheless, variations in the types of metabolites produced as a result of starter feeding in yak calves have not been completely described. Therefore, a better comprehension of the relationship between starter feeding and ruminal factors (i.e., fermentation, morphology, microbiota community, and their metabolites) in yak calves will facilitate more accurate estimations of the starter supplement and demand under current feeding patterns.
Materials and Methods
Ethical Approval Statement
All yak calves and experimental protocols in this study were conducted following the recommendations of the Administration of Affairs Concerning Experimental Animals (Ministry of Science and Technology, China, revised in 2004).
Animals and Experimental Design
All the yak calves were maternally nursed and grazed in the Datong Yak Breeding Farm of Qinghai Province before the trail. The experiment was conducted from July to November 2020, with 20 male yak calves at the age of 30 days [body weight (BW) of 33.67 ± 3.52 kg, mean ± standard deviation (SD)] with similar body conditions randomly recruited and assigned into two groups, with ten calves per group, nursed at the Haibei Tibetan Autonomous Prefecture Plateau Ecological Animal Husbandry Science Park Management Committee. All the calves were supplied with the same milk replacer (Beijing Precision Animal Nutrition Research Center, Q/HDJZA0007-2019); while the control (RA) group received only alfalfa hay, the treatment (RAS) group was fed with alfalfa hay and starter (Beijing Precision Animal Nutrition Research Center). The ten calves in each group were individually fed in ten different pens. We fed all the yak calves twice a day, at 08:00 and 16:30 with 100–700 g of milk replacer powder dissolved in 42°C water five times. The supplementation of milk replacer increased along with the increase of body weight, approximately 80 g per week until 3 months of age, and then gradually decreased by the same rate until the end of the study. Freshwater was supplied freely to the yak calves.
In brief, during the experimental period, the alfalfa hay and starter offered were adjusted daily to ensure at least 10% orts, while daily feed supplied was recorded at 3-day intervals, and the orts were gathered as well and then pooled and weighed at 3-day intervals for the calculation of the averaged dry matter intake (DMI) over 3 days until the average daily DMI achieved 1 kg each of the yak calves. At the end of the experiment, this resulted in the numbers of the feed intakes for each calf, and the means of those intakes were used as individual replicates for the statistical analysis of the difference in feed intake between the two treatments.
Samples of the starter feed, alfalfa hay, and milk replacer were measured (AOAC International, 2000) for dry matter (oven method 930.15), sugar (colorimetric method), starch (α-amylase method), crude protein (Kjeldahl method 988.05), ether extract (alkaline treatment with Röse-Gottlieb method 932.06 for milk replacer; diethyl ether extraction method 2003.05 for starter and alfalfa hay), NDF with ash without sodium sulfite or α-amylase, ADF with ash, calcium (Ca), and phosphorus (P) (dry ashing, acid digestion, and analysis by inductively coupled plasma, method 985.01), and the nutrient compositions of the milk replacer, alfalfa hay, and starter are given in Table 1.
TABLE 1.
Nutrient composition of the milk replacer, alfalfa hay, starter used in the present study.
| Items (% of dry matter) | Milk replacer1 | Alfalfa hay | Starter feed |
| Dry matter (% as fed) | 95.00 | 93.70 | 87.80 |
| Sugar | − | − | 6.50 |
| Starch | − | − | 40.50 |
| Crude protein | 26.24 | 12.51 | 20.01 |
| Ether extract | 27.79 | 0.89 | 4.70 |
| Neutral detergent fiber | − | 56.45 | 10.90 |
| Acid detergent fiber | − | 40.40 | 4.10 |
| Calcium | 2.50 | 0.99 | 0.79 |
| Phosphorus | 1.40 | 0.16 | 0.46 |
| Lysine | 2.20 | 0.84 | 1.05 |
| Methionine | 1.00 | 0.16 | 0.34 |
1The milk replacer was stored as a powder and contained whole milk powder, whey powder, protein concentrate, vitamin A, vitamin D3, vitamin E, nicotinic acid, pantothenic acid, lysine, methionine, threonine, sodium chloride, copper, zinc manganese, and iron.
Sample Collection
When the average daily DMI achieved 1 kg each of the yak calves, yak calves were fasted for 24 h, and then, the body weight, height, length, and the chest girth of all the yak calves were recorded. Five yak calves were selected randomly from each group, killed by exsanguination, and then dissected at once.
The rumen was separated, and the content within the rumen was collected for sampling. We collected 5 ml of homogenized ruminal content samples in triplicates from the ventral sac of the rumen and stored them at -80°C for microbial DNA extraction and untargeted metabolomics. Ruminal fluid samples were collected from individual yak calves and strained through 4 layers of sterile cheesecloth, and the pH was measured immediately using a portable pH meter (HI 9024C; HANNA Instruments, Woonsocket, RI, United States). Meanwhile, another 5 ml of the ruminal fluid was collected and stored at −20°C for VFA and NH3-N analyses. Specifically, a solute with metaphosphoric acid and crotonic acid was added to 2 ml of these 5 ml ruminal fluid samples before further analyses of the VFA concentrations in gas chromatography (GC-14B, Shimadzu, Japan) (Wang et al., 2017).
Subsequently, three segments of the tissue sample (2 × 2 cm) from the ventral sac of the rumen were collected, immediately washed with saline solution, then fixed in 10% buffered formalin, and stored at 4°C until papilla length and width, and the thickness of ruminal base was measured (Ishii et al., 2005). All the tissue samples were taken from the same location in each animal. Additionally, all the other collected samples were first stored in liquid nitrogen for 24 h, unless noted otherwise, and then stored at −80°C before analyses.
Determination of the Ruminal Morphology
By using the routine method of the wax section, the development of rumen was studied. After fixing in 10% buffered formalin for 24 h, the ventral sac of the ruminal tissue samples was gradually dehydrated at different concentrations (60, 70, 80, 90, and 100%) of ethanol and cleaned. Then, the ruminal samples were trimmed into small pieces and inserted into cassettes, which were embedded in liquid paraffin. Notably, 5-μm paraffin sections were sliced using the microtome and stained with hematoxylin-eosin. Using the phase-contrast microscope (Nikon NiE200, Tokyo, Japan) the papillae length and width of the ruminal tissue and the thickness of the ruminal base were measured (Wu et al., 2018).
Determination of Volatile Fatty Acid and NH3-N Concentrations in Ruminal Fluid
The ruminal fluid samples were centrifuged at 13,000 × g for 10 min at 4°C before the VFA and NH3-N concentration measurement. Using the Agilent 6850 gas chromatograph (Agilent Technologies Inc., Santa Clara, CA, United States) equipped with a polar capillary column (HP-FFAP, 30 m × 0.25 mm × 0.25 μm) and a flame ionization detector to analyze the supernatant ruminal fluid samples is previously described (Xue et al., 2017). The NH3-N concentration in each supernatant sample was measured using a continuous-flow analyzer (SKALAR San, Skalar Co., Breda, Netherlands).
Microbial DNA Extraction, 16S rRNA Gene Amplification of the V3 + V4, and Bioinformatics Analysis
Total genome DNA from the 10 ruminal content samples of yak calves from two different treatments was extracted using the cetyltrimethylammonium bromide (CTAB) method in accordance with Henderson et al. (2013). Meanwhile, DNA extraction was assessed through the QIAamp DNA Stool Mini Kit (Qiagen, Dusseldorf, Germany). DNA concentration was monitored on 1% agarose gels. The purity was assessed from the 260: 280 nm ratio (>1.8) using a NanoDrop ND2000 spectrophotometer (Thermo Scientific, Waltham, MA, United States), and the DNA was stored at −80°C until it was used in sequencing analysis.
16S rRNA genes of 16S V3-V4 regions were amplified using specific primer set 515F (5′-GTGCCAGCMGCCGCGG-3′) and 806R (5′-GGACTACNNGGGTATCTAAT-3′) with barcodes (Yu et al., 2005; Sundberg et al., 2013). All PCR reactions were carried out with 15 μl of Phusion® High-Fidelity PCR Master Mix (New England Biolabs, Beijing, China), 2 μM of forward and reverse primers, and approximately 10 ng template DNA. Thermal cycling consisted of initial denaturation at 98°C for 1 min, followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 50°C for 30 s, and elongation at 72°C for 30 s, and finally, 5 min at 72°C. PCR product quantification and qualification: the same volume of 1 × loading buffer (contained SYB green) was mixed with PCR products and electrophoresis on 2% agarose gel was performed for detection. PCR products were mixed in equidensity ratios. Then, the mixture of PCR products was purified using the Qiagen Gel Extraction Kit (Qiagen, Dusseldorf, Germany). Sequencing libraries were generated using the TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina, San Diego, CA, United States) following the manufacturer’s recommendations and adding index codes. The library quality was assessed using the Qubit@ 2.0 Fluorometer (Thermo Scientific, Waltham, MA, United States) and Agilent Bioanalyzer 2100 system. Finally, the library was sequenced on an Illumina NovaSeq platform, and 250 bp paired-end reads were generated.
Quality filtering on the raw tags was performed under specific filtering conditions to obtain the high-quality clean tags (Bokulich et al., 2013), according to the QIIME 2 (Bolyen et al., 2019). The tags were compared with the reference database (Silva database) using the UCHIME algorithm (UCHIME) (Edgar et al., 2011) to detect chimera sequences, then the chimera sequences were removed (Haas et al., 2011), and effective tags were finally obtained.
Sequence analyses were performed using Uparse software (Uparse version 7.0.1001) (Edgar, 2013). Sequences with ≥97% similarity were assigned to the same OTUs. A representative sequence for each OTU was screened for further annotation. For each representative sequence, the Silva database (Edgar et al., 2011) was used based on the Mothur algorithm to annotate taxonomic information. To study the phylogenetic relationship of different OTUs and the difference of the dominant species in different groups, multiple sequence alignments were conducted using the MUSCLE software (Version 3.8.31) (Edgar, 2004). OTU abundance information was normalized using a standard sequence number corresponding to the sample with the least sequences. Subsequent analyses of alpha diversity and beta diversity were all performed based on this output normalized data.
The taxon abundance for each sample was determined according to phylum, class, order, family, and genus. The microbiota were compared for beta diversity using the distance matrices generated from weighted UniFrac analysis, principal coordinated analysis (PCoA), and analysis of similarities (ANOMIS). The P-value was set as <0.05, and the threshold of the linear discriminant analysis (LDA) score was set at a default value of 2.0.
Untargeted Metabolomics
The ruminal content samples (1 ml) were freeze-dried and resuspended with prechilled 80% methanol and 0.1% formic acid using a good vortex. Then, the samples were incubated on ice for 5 min and centrifuged at 15,000 g, 4°C for 15 min. Some supernatant was diluted to the final concentration containing 53% methanol by LC-MS grade water. The samples were subsequently transferred to a fresh Eppendorf tube and then centrifuged at 15,000 g, 4°C for 15 min. Finally, the supernatant was injected into the LC-MS system analysis.
Discoverer 3.1 (CD3.1, ThermoFisher, Waltham, MA, United States) was used to perform peak alignment, peak picking, and quantitation for each metabolite. Later, peak intensities were normalized to the total spectral intensity. The normalized data were used to predict the molecular formula based on additive ions, molecular ion peaks, and fragment ions. Then, peaks were matched using the mzClou, mzVault, and MassList database to obtain accurate qualitative and relative quantitative results.
Statistical analyses were performed using the statistical software R (R version R-3.4.3), Python (Python 2.7.6 version), and CentOS (CentOS release 6.6). When data were not normally distributed, normal transformations were attempted using area normalization.
These metabolites were annotated using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Principal component analysis (PCA) was performed at metaX (Wen et al., 2017) (a flexible and comprehensive software for processing metabolomics data). We applied univariate analysis (t-test) to calculate the statistical significance (P-value). The metabolites with VIP > 1, P-value < 0.05, and fold change (FC) ≥ 2 or FC ≤ 0.5 were considered to be differential metabolites. Volcano plots were used to filter metabolites of interest-based on log2(FC) and −log10(P-value) of metabolites using ggplot2 in R language.
The KEGG pathway enrichment of differential metabolites was performed, the ratio was satisfied by x/n > y/N, and the metabolic pathway was considered an enrichment, while when the P-value of the metabolic pathway < 0.05, the metabolic pathway was considered a statistically significant enrichment.
Statistical Analysis
In this study, all data were tested and presented as a normal distribution. At the beginning of the experiment, the initial body weight, height, length, and chest girth were tested using the t-test, no significant differences were identified, and the initial indices were almost constant. Therefore, for other indices except for the DMI data, analysis was performed using the one-way ANOVA procedure using SPSS 22.0 software (SPSS, Inc., Chicago, IL, United States) with replicates as experiment units, and a value of P < 0.05 was regarded as statistically significant. The differences in DMI between two groups of yak calves were further analyzed using the following model: Yij = μ + Di + Tj + εij + DT, using the MIXED procedure of SAS (SAS Institute Inc., Cary, NC, United States, 2007), which considered the significant differences of these indices induced by the time effect in the same treatment. Yij is the response variable, in particular, μ is the overall mean, Di is the fixed effect of treatment (i = maternal grazing or barn feeding), Tj is the fixed effect of time (10 days as a unit) of the experiment, DT is the interaction of dietary and time, and εij is the residual error. If a significant diet and time effect were observed, the significance between the treatment and time differences was separately identified using the Tukey’s test multiple comparison test. All data are expressed as the means with the standard error.
The statistical evaluation of 16S rRNA sequencing and the untargeted metabolomics results were analyzed using the bioinformatics methods described above. Spearman’s correlation test was used to examine relationships between the RA group and the RAS group. The correlation coefficient rho between the relative abundance of differential metabolites and the total abundance of differential bacteria was calculated using the Spearman statistical method (rho ≥ 0, P ≤ 0.05).
Results
The Effect of Starter Feeding on Growth Performance, Dry Matter Intake, and Development of the Rumen
Significant differences in daily DMI were found between the two groups of the yak calves over the whole experimental period (P = 0.033), where the higher intake was found for yak calves in the RAS group. Compared with the RA group, the RAS group had a significantly greater body weight (P < 0.001), height (P = 0.008), and chest girth (P < 0.001) (Table 2). Meanwhile, a significant increase in organ development was found in the RAS group, which included the weight of the rumen (P = 0.036) (Table 2).
TABLE 2.
Effects of the starter feeding supplementation in the preweaning period on the growth performance and development of rumen in yak calves.
| Treatment1 |
SEM | P-value | |||
| Items | RA | RAS | |||
| Growth performance | Body weight (kg) | 72.07 ± 2.47 | 80.58 ± 1.26 | 1.38 | < 0.001 |
| Body length (cm) | 90.80 ± 3.63 | 93.80 ± 1.92 | 1.34 | 0.141 | |
| Body height (cm) | 76.33 ± 2.08 | 81.25 ± 0.96 | 1.20 | 0.008 | |
| Chest girth (cm) | 104.80 ± 2.68 | 116.40 ± 2.61 | 2.22 | < 0.001 | |
| DMI (g) | 608.36 ± 60.68 | 678.71 ± 6.53 | 17.04 | 0.033 | |
| Rumen weight | Rumen (g) | 1.07 ± 0.07 | 1.17 ± 0.04 | 0.23 | 0.036 |
| Rumen index, expressed as kg of organ/kg of BW | Rumen (×10–2) | 1.47 ± 0.08 | 1.56 ± 0.13 | 0.17 | 0.221 |
1The alfalfa (RA) was fed with the milk replacer and alfalfa hay, the alfalfa hay and starter (RAS) group was fed with milk replacer, alfalfa hay and the starter.
Development of Ruminal Morphology and Ruminal Fermentation Profiles
Over the whole experimental period, when compared with the RA group, the RAS group had significantly greater papilla length (P = 0.049). The RAS group could significantly increase the ruminal fluid NH3-N concentration compared with the RA group (P = 0.001). Besides, the RA group had a significantly higher acetate concentration than the yak calves in the RAS group (P = 0.034), while the valerate concentration was significantly greater in the RAS group (P = 0.021) (Table 3).
TABLE 3.
Effects of starter feeding on the ruminal fermentation parameters and ruminal epithelium development in yak calves.
| Treatment |
SEM | P-value | |||
| Items | RA | RAS | |||
| Ruminal epithelium | Papilla width (μm) | 952.69 ± 105.45 | 824.07 ± 191.11 | 25.15 | 0.283 |
| Papilla length (μm) | 569.82 ± 100.37 | 748.72 ± 128.53 | 33.29 | 0.049 | |
| Muscle thickness (μm) | 1915.07 ± 439.49 | 2031.12 ± 643.69 | 105.68 | 0.757 | |
| Ruminal fermentation characteristics | pH | 7.35 ± 0.12 | 7.35 ± 0.116 | 0.03 | 0.978 |
| Ammonia nitrogen, NH3-N (mg/dL) | 2.46 ± 0.28 | 3.29 ± 0.23 | 0.16 | 0.001 | |
| Total VFA (mmol/L) | 57.40 ± 5.27 | 50.23 ± 5.25 | 2.07 | 0.081 | |
| Acetate (mmol/L) | 35.78 ± 3.51 | 29.60 ± 1.05 | 0.23 | 0.034 | |
| Propionate (mmol/L) | 9.39 ± 1.29 | 8.28 ± 1.07 | 0.44 | 0.232 | |
| Butyrate (mmol/L) | 5.41 ± 1.55 | 4.62 ± 1.18 | 0.43 | 0.389 | |
| Isobutyrate (mmol/L) | 1.53 ± 0.084 | 1.54 ± 0.25 | 0.06 | 0.936 | |
| Valerate (mmol/L) | 3.58 ± 0.43 | 4.23 ± 0.05 | 0.15 | 0.021 | |
| Isovalerate (mmol/L) | 1.20 ± 0.14 | 1.27 ± 0.17 | 0.05 | 0.592 | |
Ruminal Microbiota
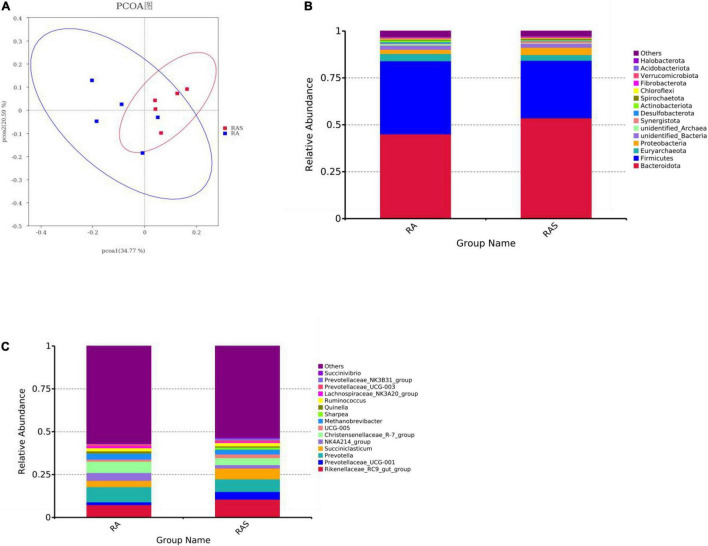
According to further analysis of ruminal microbiota, we found significantly decreased the OTUs in the RAS group (Table 4, P = 0.046), whereas the two groups showed no significant difference in Chao 1 value, Shannon indices, and Simpson index (Table 4, P = 0.468). Moreover, beta diversity analyses showed that the compositions of the gastrointestinal prokaryotic community of the yak calves in different feeding groups were different in the rumen (Figure 1A).
TABLE 4.
Effects of the starter feeding on richness and diversity index of ruminal microbiota in yak calves.
| Items | Treatment |
SEM | P-value | |
| RA | RAS | |||
| OTUs | 1457.25 ± 94.49 | 1265.00 ± 120.69 | 50.78 | 0.046 |
| Good’s coverage | 0.99 ± 0.001 | 0.99 ± 0.001 | 0.01 | 0.557 |
| ACE value | 1619.81 ± 162.03 | 1404.46 ± 132.99 | 63.33 | 0.086 |
| Chao 1 value | 1596.47 ± 143.09 | 1385.30 ± 136.44 | 60.72 | 0.077 |
| Shannon indices | 7.50 ± 0.29 | 7.18 ± 0.89 | 0.21 | 0.468 |
| Simpson indices | 0.97 ± 0.01 | 0.96 ± 0.03 | 0.01 | 0.468 |
FIGURE 1.
(A) Diversity analyses based on the ruminal microbiota; phylum (B) and genus level (C) composition of the ruminal bacteria (top 15).
Rumen whose phylum-level relative abundance was in the top 15 of all the microbiota communities is shown in Figure 1B. The genus of the top 15 is given in Figure 1C. Differential ruminal microbiota are identified between two different feeding groups. Among these, Firmicutes (P < 0.001), Desulfobacterota (P = 0.002), and Actinobacteriota (P = 0.002) were significantly greater in the RA group, while Bacteroidota (P = 0.001), Proteobacteria (P = 0.011), Chloroflexi (P = 0.004), Synergistota (P = 0.005), and Spirochaetota (P = 0.017) were significantly higher in the RAS group (Table 5). At the genus level, Prevotella (P = 0.018), Christensenellaceae_R-7_group (P = 0.013), and NK4A214_group (P = 0.018) were significantly greater in the RA group. Rikenellaceae_RC9_gut_group (P = 0.004) and Ruminococcus (P = 0.041) were identified significantly greater genera in the RAS group. Furthermore, similar results that contained similar but less differential bacteria were also identified using the Mann–Whitney U test (Table 5).
TABLE 5.
Ruminal microbiota community difference between the two groups by using the Mann–Whitney U test.
| Items | Groups |
SEM | P-value | |
| RA | RAS | |||
| Phylum | ||||
| Bacteroidota | 38.38 ± 4.96 | 53.61 ± 1.41 | 2.963 | 0.001 |
| Firmicutes | 43.98 ± 2.77 | 30.70 ± 2.41 | 2.567 | < 0.001 |
| Euryarchaeota | 3.16 ± 1.16 | 2.20 ± 0.92 | 0.387 | 0.240 |
| Proteobacteria | 2.68 ± 0.38 | 3.87 ± 0.59 | 0.263 | 0.011 |
| Verrucomicrobiota | 0.38 ± 0.13 | 0.43 ± 0.15 | 0.046 | 0.577 |
| Chloroflexi | 0.13 ± 0.04 | 0.32 ± 0.07 | 0.040 | 0.004 |
| Synergistota | 0.27 ± 0.05 | 0.90 ± 0.27 | 0.140 | 0.005 |
| Desulfobacterota | 0.68 ± 0.11 | 0.34 ± 0.08 | 0.069 | 0.002 |
| Actinobacteriota | 1.06 ± 0.28 | 0.28 ± 0.02 | 0.169 | 0.002 |
| Spirochaetota | 0.46 ± 0.19 | 1.00 ± 0.22 | 0.130 | 0.017 |
| Genus | ||||
| Prevotella | 11.04 ± 0.10 | 8.10 ± 1.43 | 0.707 | 0.018 |
| Methanobrevibacter | 2.67 ± 0.74 | 1.20 ± 0.85 | 0.290 | 0.274 |
| Quinella | 0.56 ± 0.08 | 0.56 ± 0.35 | 0.080 | 0.993 |
| Rikenellaceae_RC9_gut_group | 6.73 ± 1.74 | 12.33 ± 0.79 | 1.240 | 0.004 |
| Prevotellaceae_UCG-001 | 1.56 ± 0.31 | 1.39 ± 0.54 | 0.164 | 0.659 |
| Succiniclasticum | 3.66 ± 0.92 | 4.57 ± 0.50 | 0.337 | 0.203 |
| NK4A214_group | 2.72 ± 0.71 | 1.43 ± 0.25 | 0.311 | 0.018 |
| Christensenellaceae_R-7_group | 6.60 ± 1.11 | 4.07 ± 1.40 | 0.565 | 0.013 |
| UCG-005 | 1.37 ± 0.46 | 1.08 ± 0.11 | 0.139 | 0.351 |
| Ruminococcus | 1.59 ± 0.41 | 3.93 ± 0.23 | 0.660 | 0.041 |
Microbiological Metabolism of the Rumen
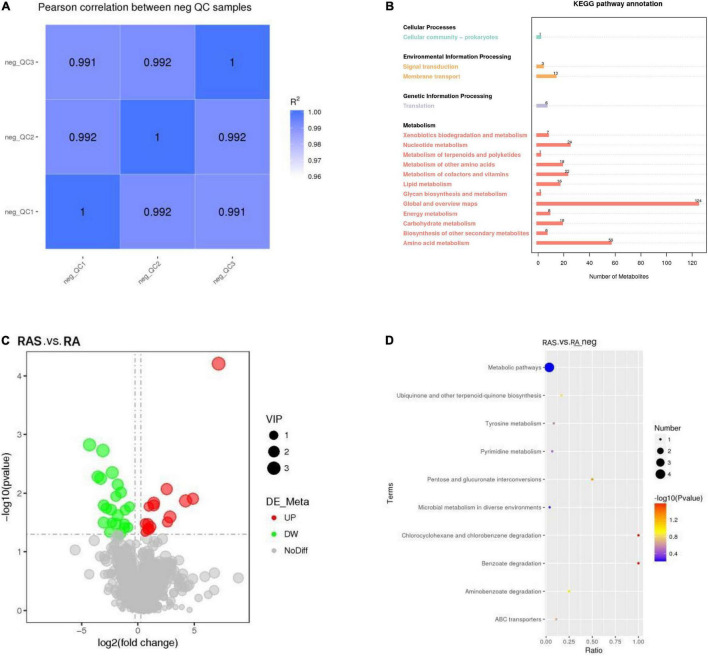
Based on the relative quantitative value of the metabolites, the Spearman correlation coefficient between the QC samples is calculated (Figure 2A). The correlation between the QC samples was closer to 1 (R2 < 1), indicating that the test process was stable and the quality of data high (Rao et al., 2016).
FIGURE 2.
(A) QC samples correlation analysis; (B) The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotation; (C) The volcano plot of difference metabolites; (D) enrichment of differential metabolites in KEGG pathways. Rich factor, ratio of the proportion of differential metabolites to the proportion of all metabolites in the pathway; the size of the dots in the graph represents the number of distinct metabolites enriched in the corresponding pathways.
In contrast, metabolism mainly participated in amino acid metabolism and nucleotide metabolism in ruminal content (Figure 2B). We detected the metabolites in the rumen by metabolomics. In our research, the relative concentrations of 37 of the total 1,057 identified metabolites were altered by starter feeding (Figure 2C), of which 15 metabolites were upregulated and 22 metabolites were downregulated in yak calves in the RAS group compared with those in the RA group (Table 6).
TABLE 6.
Differential metabolites in the rumen of yak calves in the RA and RAS groups.
| Metabolites | mzmed | rtmed | log2FC | P-value | VIP | Regulated |
| Saccharin | 181.992 | 6.042 | 7.160 | 0.000 | 3.387 | Up |
| 6-Keto-prostaglandin f1alpha | 369.228 | 12.182 | 2.556 | 0.009 | 2.381 | Up |
| delta-Tocopherol | 401.343 | 16.697 | 4.883 | 0.012 | 2.348 | Up |
| Pyridoxine O-Glucoside | 330.120 | 2.068 | 4.228 | 0.014 | 2.896 | Up |
| 2′-O-Methyluridine | 257.079 | 5.528 | 1.411 | 0.015 | 2.638 | Up |
| 15(S)-HpETE | 353.234 | 12.929 | 1.405 | 0.016 | 2.137 | Up |
| FAHFA (16:1/18:3) | 529.424 | 14.506 | 0.964 | 0.017 | 1.374 | Up |
| Isopentenyladenine | 202.110 | 10.799 | 2.845 | 0.025 | 2.731 | Up |
| N1-isopropyl-2-(1H-2-pyrrolylcarbonyl)-1-hydrazinecarboxamide | 419.212 | 9.289 | 2.629 | 0.031 | 1.717 | Up |
| N7-Methylguanosine | 298.115 | 1.361 | 0.638 | 0.033 | 1.566 | Up |
| Pepstatin | 684.455 | 15.539 | 0.884 | 0.033 | 2.031 | Up |
| (+/-)8(9)-DiHET | 337.239 | 13.317 | 1.122 | 0.038 | 2.039 | Up |
| Xylitol | 151.061 | 1.426 | 0.862 | 0.041 | 1.692 | Up |
| (+/-)9,10-dihydroxy-12Z-octadecenoic acid | 313.239 | 12.924 | 0.962 | 0.041 | 2.172 | Up |
| Cinnamoylglycine | 204.067 | 8.755 | 0.639 | 0.046 | 1.025 | Up |
| 3,5-Dihydroxybenzoic acid | 153.020 | 7.550 | –4.292 | 0.001 | 2.951 | Down |
| (3R)-8-hydroxy-3-(4-methoxyphenyl)-3,4-dihydro-1H-2-benzopyran-1-one | 269.083 | 9.621 | –3.107 | 0.002 | 3.140 | Down |
| 2-Hydroxyhippuric acid | 194.046 | 7.134 | –2.277 | 0.004 | 2.801 | Down |
| FAHFA (2:0/23:0) | 411.349 | 16.143 | –3.557 | 0.005 | 2.496 | Down |
| Hydroquinone | 109.030 | 7.551 | –3.315 | 0.006 | 2.674 | Down |
| D-Glucosyl-beta-1,1-N-palmitoyl-D-erythro-sphingosine | 698.560 | 16.209 | –1.790 | 0.007 | 2.303 | Down |
| 3-(methylsulfanyl)-5H-[1,2,4]triazino[5,6-b]indole | 215.039 | 10.572 | –1.497 | 0.010 | 2.102 | Down |
| PC (15:1/18:2) | 800.546 | 16.225 | –1.955 | 0.011 | 1.749 | Down |
| Isorhapontigenin | 257.082 | 10.283 | –3.028 | 0.016 | 2.153 | Down |
| FAHFA (2:0/18:1) | 357.265 | 13.102 | –0.751 | 0.017 | 1.655 | Down |
| 1-(2,4-dihydroxyphenyl)-2-(3,5-dimethoxyphenyl)propan-1-one | 301.108 | 7.583 | –2.793 | 0.018 | 1.856 | Down |
| 2-Furoylglycine | 168.031 | 5.992 | –2.379 | 0.019 | 2.147 | Down |
| PE (16:0/16:0) | 690.509 | 14.288 | –1.134 | 0.019 | 1.724 | Down |
| D-Glucuronic acid | 193.036 | 12.638 | –1.792 | 0.024 | 2.331 | Down |
| PG (20:0/20:4) | 825.567 | 15.518 | –3.036 | 0.032 | 2.263 | Down |
| PG (20:0/20:3) | 827.584 | 16.042 | –2.299 | 0.032 | 1.490 | Down |
| 13,14-dihydro Prostaglandin F1α | 393.243 | 14.550 | –1.955 | 0.033 | 2.025 | Down |
| PG (18:1/22:4) | 823.552 | 15.392 | –1.183 | 0.033 | 1.623 | Down |
| 3-methyl-4-[2-(2-methylphenyl)hydrazono]-4,5-dihydro-1H-pyrazol-5-one | 215.093 | 1.915 | –0.860 | 0.038 | 1.362 | Down |
| UMP | 323.029 | 1.460 | –1.210 | 0.039 | 2.018 | Down |
| N1-[4-(cyanomethyl)phenyl]-4-chlorobenzamide | 269.046 | 10.375 | –2.514 | 0.046 | 1.759 | Down |
| Catechin | 289.072 | 7.111 | –1.567 | 0.049 | 2.133 | Down |
FC, fold change; mzmed, mass-to-charge ratio of metabolites; rtmed, retention time of metabolites; VIP, variable importance in the projection.
Regulated “up” represents a higher abundance in calves in the RAS group, “down” represent a higher abundance in calves in the RA group.
As shown in Figure 2D, UMP, hydroquinone, xylitol, and delta-tocopherol were enriched in the KEGG pathway “metabolic pathways.” Hydroquinone was enriched in the KEGG pathway “Microbial metabolism in diverse environments,” “Tyrosine metabolism,” “Aminobenzoate degradation,” “Benzoate degradation,” and “Chlorocyclohexane and chlorobenzene degradation.” Xylitol was enriched in the KEGG pathway “ABC transporters” and “Pentose and glucuronate interconversions.” UMP and delta-tocopherol may be related to “Pyrimidine metabolism” and “Ubiquinone and other terpenoid-quinone biosynthesis,” respectively.
Correlation Between Ruminal Microbiota and Fermentation Profiles
There is an interaction between ruminal microbiota and growth performance of yak calves and ruminal fermentation profiles, which affects the feed efficiency and tissue development of ruminants. The correlation between them is illustrated in Figure 3.
FIGURE 3.
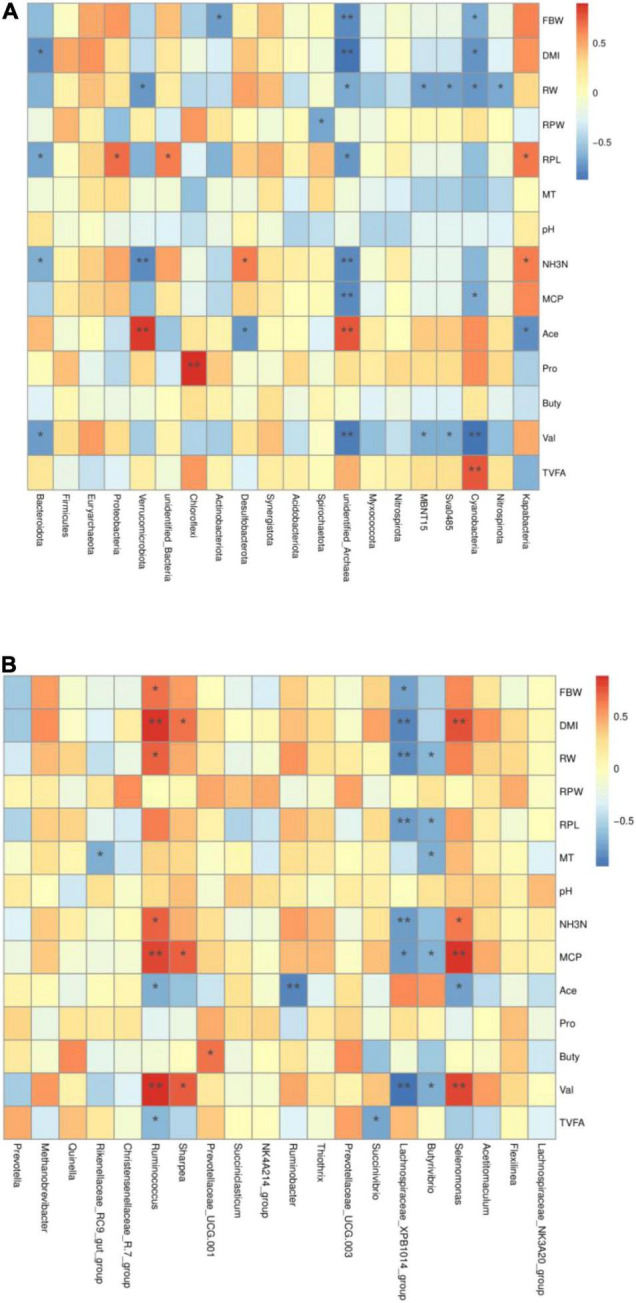
Heatmap of the correlation between microbiota [(A) phylum and (B) genus] and fermentation parameters in the rumen of yak calves. RBW, finally body weight; DMI, dry matter intake; RW, ruminal weight; RPW, ruminal papilla width; RPL, ruminal papilla length; MT, muscle thickness; NH3N, NH3-N; MCP, microbiota crude protein; Ace, acetate; Pro, propionate; But, butyrate; Ibu, isobutyrate; Val, valerate; Iva, isovalerate.
At the phylum level (Figure 3A), finally, body weight of yak calves was significantly positively correlated with Actinobacteriota, unidentified-Archaea, and Cyanobacteria. The DMI of yak calves was significantly negatively correlated with Bacteroidota, unidentified-Archaea, and Cyanobacteria. The ruminal weight of yak calves was significantly negatively correlated with Verrucomicrobiota, unidentified-Archaea, MBNT15, Sva0485, Cyanobacteria, and Nitrospinota. The width of ruminal papilla was significantly negatively correlated with Spirochaetota. The length of ruminal papilla was significantly positively correlated with Proteobacteria, unidentified-Bacteria, and Kapabacteria and significantly negatively correlated with Bacteroidota and unidentified-Archaea. The NH3-N concentration in the ruminal fluid was significantly negatively correlated with the Bacteroidota, unidentified-Archaea, and Verrucomicrobiota and was significantly positively correlated with Desulfobacterota and Kapabacteria. The MCP concentration in the ruminal fluid was significantly negatively correlated with the relative abundance of unidentified-Archaea and Cyanobacteria. Acetate concentration was significantly positively correlated with Verrucomicrobiota and unidentified-Archaea and significantly negatively correlated with Kapabacteria. Propionate concentration was significantly positively correlated with Chloroflexi. The concentration of valerate was significantly negatively correlated with Bacteroidota, unidentified-Archaea, MBNT15, Sva0485, and Cyanobacteria. TVFA concentration was significantly positively correlated with Cyanobacteria.
At the genus level (Figure 3B), there were significant positive correlations between the final body weight, DMI, ruminal weight, NH3-N, MCP concentration, and valerate concentration and the relative abundance of Ruminococcus, significant negative correlations between acetate and TVFA concentration and the relative abundance of Ruminococcus. MCP and valerate concentrations were significantly positively correlated with Sharpea. Butyrate concentration was significantly positively correlated with the Prevotaceae-UCG-001. The concentration of acetate was significantly negatively correlated with Ruminobacter. The final body weight, DMI, ruminal weight, ruminal papilla length, NH3-N, and MCP concentration and valerate concentration of yak calves were significantly negatively correlated with Lachnospiraceae-XPB1014-group.
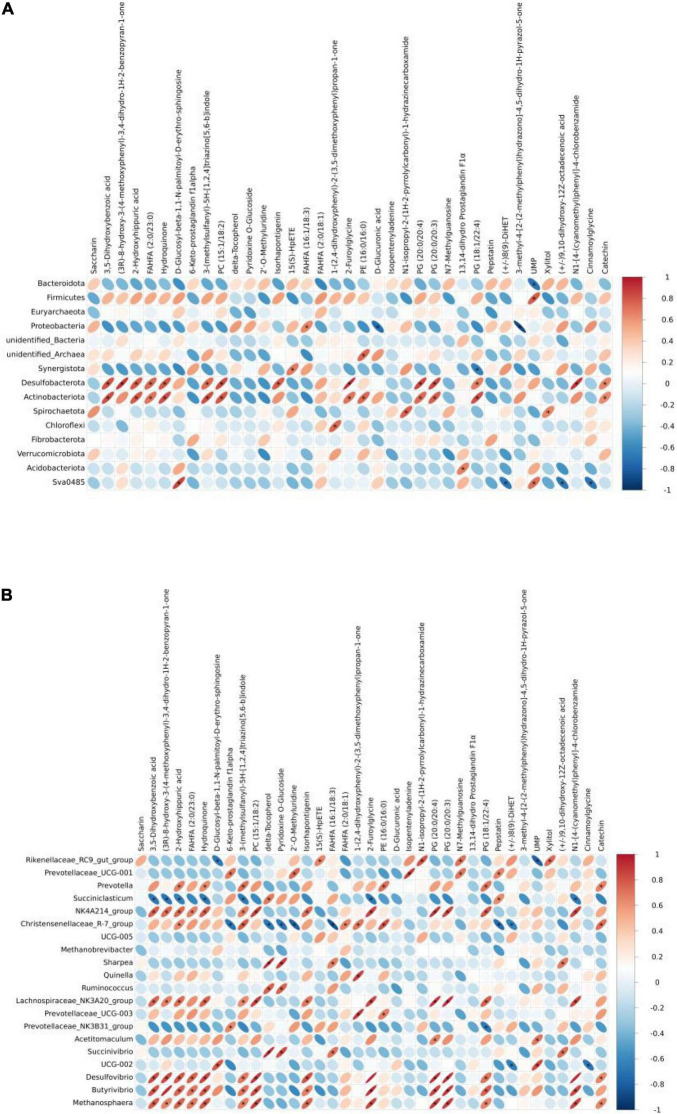
Correlation Between Ruminal Microbiota and Metabolites
In this study, the correlation analysis results of metabolites and 16S rRNA are shown. As shown in Figure 4A, a total of 44 correlations significantly differ at the phylum level, with 36 significantly positive correlations and 8 significantly negative correlations. Hydroquinone was significantly positive with Desulfobacterota and Actinobacteriota. UPM was significantly positive with Firmicutes and Sva0485, while significantly negative with Bacteroidota. Xylitol was significantly positive with Spirochaetota.
FIGURE 4.
Heatmap of the correlation between ruminal microbiota [(A) phylum and (B) genus] and metabolites of yak calves.
There were 104 significantly positive and 19 significantly negative correlations between ruminal microbiota and their metabolites at the genus level (Figure 4B), with Rikenellaceae_RC9_gut_group significantly positive with UPM and significantly positive with xylitol. Prevotella, NK4A214_group, Lachnospiraceae_NK3A20_group, Desulfovibrio, Butyrivibrio, and Methanosphaera were significantly positive with Hydroquinone, while Succiniclasticum was significantly negative with Hydroquinone. Delta-tocopherol was significantly positive with Succiniclasticum, Sharpea, Ruminococcus, and UCG-002, while it was significantly negative with Christensenellaceae_R-7_group. UMP was significantly positive with Acetitomaculum and UCG-002.
Discussion
Due to the slow development of rumen, the growth performance of newborn calves was related to the digestion of wrinkles and the intestine. The ruminal epithelium is responsible for nutrient absorption and transportation in the rumen. This study shows that starter feeding with alfalfa hay significantly increased DMI, body weight, and chest girth in yak calves, which presumably stimulated both the development of ruminal epithelia and performance (Baldwin et al., 2004; Norouzian et al., 2011; Yang et al., 2015; Lin et al., 2019), in line with the results of the previous studies in calves’ and lambs’ early life (Saro et al., 2018; Cui et al., 2020). Starter feeding also significantly promoted ruminal development in yak calves. These improvements probably attributed to the improved dry matter intake and ruminal fermentation. With the growth of the age and the solid feed intake, the ruminal epithelium can influence the net use of digestion and utilization of diet, serving as a barrier to the ruminal epithelium’s contents (Sander et al., 1959; Anderson et al., 1987). Similar to the previous study, the starter feeding contributed to the ruminal papilla length, which is also consistent with Xie et al. (2013) research on Holstein calves.
Due to various in-nutrient supplementation, significant changes occurred in the ruminal fermentation, which represented the increased carbohydrate fermentation in the rumen (Raghuvansi et al., 2007; Xue et al., 2017; Yang et al., 2018). The starter feeding contains grains, thus providing easily fermentable carbohydrates for microbial fermentation. When supplied with a starter, the RAS group produced a bigger NH3-N concentration compared with the RA group, which fed with only alfalfa hay. NH3-N concentration in the rumen is in a dynamic balance, which can provide raw materials for generating the microbial protein (Firkins et al., 2007). Increased NH3-N concentration in RAS group yak calves, this is in line with this study (Agle et al., 2010). As suggested in previous studies (Stams et al., 1984; Poudel et al., 2018; Gao et al., 2019), the alfalfa hay supplementation could increase the ruminal acetate-producing bacteria, further influencing the ruminal acetate concentration. Likewise, an increased acetate concentration level in the RA group calves that fed with the alfalfa hay reflected increased fiber digestibility because the acetate is the primary product of cellulolytic bacteria (Lu et al., 2005). The significantly increased valerate concentration happened in the starter feeding group, which demonstrated that more nutrients could be provided to increase growth performance and organ development (Orskov, 2012).
Both dietary and additives could influence the abundance of ruminal microbiota (Abecia et al., 2014), while the abundance of the microbiota also affects the use of the host’s utilization of the diet (Carberry et al., 2014). Bacteroidetes and Firmicutes are the most dominant phylum in yak calves’ rumen, which are consistent with the previous studies on ruminants (Shen et al., 2017; Wu et al., 2021). The depressed ratio of Firmicutes vs. Bacteroidetes in the rumen resulted in increasing lignocellulose digestion (Mu et al., 2019). Similar to other studies (Fernando et al., 2010; Iqbal et al., 2017), the dominant phylum was identified in this study, including Euryarchaeota and Proteobacteria. Through the correlation analysis, Proteobacteria and Synergistota were significantly positive with the NH3-N concentration, and these three indicators were also significantly higher in the starter feeding group, which indicated that starter feeding can promote ruminal microbiota to produce available NH3-N and strengthen the absorption and utilization of nutrients. Proteobacteria have the function of degrading soluble carbohydrates; the crude protein and ether extract were much higher in the feeding of the RAS group, along with the significantly increased abundance of Proteobacteria in the RAS group. Therefore, we predicted that the abundance of Proteobacteria was positive with the crude protein and ether extract content in the dairy feed.
Prevotella was the dominant genus of abundance, which had the ability to degrade fiber sources (Emerson and Weimer, 2017). Members of the genus Prevotella in the ruminal microbial community have been reported to play an important role in the utilization of dietary nutrition (Latham et al., 2018). In this study, the abundance of Prevotella was significantly negative with isobutyrate, and the abundance of Prevotella was significantly increased in the RA group, due to the higher neutral detergent fiber and acid detergent fiber level in the alfalfa hay. Part strains of Butyrivibrio degraded cellulose in the rumen, which also shows that the abundance of Butyrivibrio increased in the RA group. The abundance of Rikenellaceae_RC9_gut_group was significantly enriched in the RAS group and positive with the NH3-N and valerate concentration, which was consistent with the result of rumen fermentation. The relative abundance of UCG-002 was negative with the propionate, butyrate, valerate, and isovalerate concentration, which could be resistant to the growth performance when propionate was absorbed and converted to glucose, amino acids, and lipids (Orskov, 2012). Very few studies referred to the significant valerate concentration decrease in the RAS group, with completely unknown and unexplored functions in ruminal physiology.
In brief, the colonization of ruminal microbiota during preweaning could further influence the subsequent ruminal microbiota of adult ruminants (Ben Salem et al., 2005; Kim et al., 2016). Specifically, the effect of differentially supplementing carbohydrates in the early life of ruminants has been demonstrated (Khan et al., 2016; Meale et al., 2016). Therefore, starter feeding yak calves in early life was beneficial to the digestion and absorption function of yaks both during preweaning and in the subsequent adult period, which could be helpful for the growth of yaks.
The alteration in the ruminal metabolites we discovered as a result of feeding starter was not surprising since alteration of the ruminal microbial community generally affects the types of compounds produced by the rumen microbiota. We identified 37 different metabolites using the currently available metabolite databases.
Ruminal metabolites in pathway analysis expressed the difference in yak calves, and based on the t-test and FC analysis, nine pathways were revealed: chlorocyclohexane and chlorobenzene degradation, benzoate degradation, pentose and glucuronate interconversions, aminobenzoate degradation, ubiquinone, and other terpenoid-quinone biosynthesis, ABC transporters, tyrosine metabolism, pyrimidine metabolism, and microbial metabolism in diverse environment pathways. All relevant pathways were significantly upregulated in the starter feeding group due to increased concentrations of the associated metabolites xylitol and delta-tocopherol and decreased UMP and hydroquinone. This study has found that ruminants can meet up to 70% of their energy requirements and 50–70% of their protein requirements through rumen microbial metabolic activities (Wu et al., 2021). Xylitol, as an intermediate of sugar metabolism, is mainly involved in carbohydrate metabolism. In the case of the decreased insulin level in the body, xylitol is transported across the membrane to participate in the generation of liver glycogen and provide energy for the body cells (Di Rienzi and Britton Robert, 2020; Meyer-Gerspach et al., 2021). In the metabolic process of the body, xylitol is utilized by microorganisms and converted into xylulose, which enters the pentose phosphate pathway and generates short-chain fatty acids (propionate) to ensure the smooth metabolic pathway of pentose and glucuronic acid conversion (Uebanso et al., 2017). In this study, xylitol, a differential metabolite of yak calves, was significantly upregulated after supplying with starter feeding, indicating that starter feeding is helpful with propionate metabolism and thus improves the growth and development of yak calves. Hydroquinone was significantly downregulated, resulting in significant degradation of cyclochloroethane and chlorobenzene as well as benzoic acid degradation pathway, indicating that the metabolic pathway of xenobiotic degradation was significantly changed after starter feeding supplementation. UMP is a short uridine acid, consisting of uracil base, phosphate group, and ribose, it can promote the biosynthesis of glucuronic acid, uridine acid, and its derivatives play a key role in host immune response and metabolic regulation (Zhang et al., 2014). In feed production, the use of the internucleotide combination, including UMP, can promote the growth and development of animals and improve the immune function of the body (Grimble and Westwood, 2001). In this study, the metabolites of yak calves were significantly changed after the early intake of concentrate, which also caused changes in related metabolic pathways, further suggesting that early supplementation of concentrate can improve the growth performance of calves by changing rumen metabolites.
The relative abundance of Bacteroidota significantly decreased with the increase of DMI and showed a positive correlation with Proteobacteria. Bacteroidota, as fiber-degrading bacteria (Zapata and Quagliarello, 2015), were significantly higher in the rumen of yak calves fed only on alfalfa hay, while the nutritional monoculture and poor palatability of herbage resulted in a decrease in DMI of yak calves. Proteobacteria can use polysaccharides as a nutrient source for ruminants (Zimmermann, 1990), ruminal papilla length was significantly positively correlated with Proteobacteria, which may be due to the increased Proteobacteria abundance after starter feeding supplementation, which improves nutrient utilization efficiency of yak calves, promotes ruminal papilla development of yak calves, and thus improves production performance. Studies have shown that Ruminococcus, as a producer of carbohydrate-active enzymes (Rosewarne Carly et al., 2012), can decompose dietary fiber and improve the decomposition of hemicellulose and dietary fiber (Wang L. et al., 2019). The final body weight, DMI, and concentrations of NH3-N, MCP, and valerate in the rumen were proportional to the relative abundance of Ruminococcus, suggesting that starter feeding may improve the relative abundance of Ruminococcus and lead to the improvement of body growth performance.
Ruminal microbial interaction can decompose carbohydrate carbohydrates such as starch and fructose in the diet that cannot be directly absorbed by the digestive tract into monosaccharides or disaccharides. After microbial fermentation, glycogen is resynthesized and then transported to the intestine for decomposition and utilization in the small intestine, which is one of the important pathways for body function (Zhao et al., 2018). In ruminants, there are a large number of ciliates and other fiber-degrading bacteria in the rumen, and a large amount of cellulose in herbages is decomposed into short-chain fatty acids under the action of ruminal microbial fermentation, providing carbon skeleton for the synthesis of microbial proteins in the rumen and participating in the body circulation pathway (Gill and King, 2002; Zhang et al., 2017). Ruminal microbial community response changes with the change of dietary nutrient composition, which affects the types of small molecule metabolites in the digestive tract, and even threatens the immunity and health of the body (Yang and Duan, 2018). Therefore, exploring the relationship between ruminal microbiota and metabolomics is helpful to reasonably improve the diet structure and early prevention of diseases. In the correlation analysis, the metabolite UMP was positively correlated with microbial Bacteroidota in the rumen, and negatively correlated with Firmicutes. In the 16s rRNA sequencing, the relative abundance of Bacteroidota in yak calves fed with starter feeding significantly decreased, and the difference metabolite UMP significantly decreased, and the trend was consistent. In the functional analysis of differential metabolites, UMP was involved in the metabolism of pyrimidine in the nucleotide metabolic pathway. At the genus level, delta-tocopherol was significantly positively correlated with Ruminococcus and Sharpea, while the Ruminococcus was significantly increased after the yak calves were fed with starter feeding, and the metabolite delta-tocopherol was also significantly upregulated, which promoted the metabolism of coenzyme factors and vitamins. Therefore, UMP and delta-tocopherol can be used as potential markers for exploring the dominant ruminal microbiota. Studies have shown that the correlation between ruminal microbiota and metabolomics plays an important role in the treatment of ruminant ruminal acidosis and other diseases, and there is a direct or indirect correlation between microbiota candidate metabolomics (Mao et al., 2016; Xue et al., 2020). Wang changed the diet structure of dairy calves and found that the changes in the ruminal environment were caused by microbial community structure and metabolites (Wang B. et al., 2019). Pickard used high-throughput sequencing to detect the activity of intestinal microbiota and suggested that when metabolomics was associated with the abundance of microbiota, in-depth exploration of the correlation mechanism between the two had important advantages for monitoring the physiological state of the body and preventing diseases (Pickard Joseph and Chervonsky Alexander, 2015). How ruminal microbiota and metabolomics affect and associate with each other remains to be further explored.
The untargeted metabolomics approach depends on comparing peak intensity to evaluate differences in the relative abundance of metabolites with the disadvantage of a lack of accuracy and precision (Veenstra Timothy, 2012). Furthermore, identifying metabolites accurately is a tough challenge due to the complexity and chemical diversity of the metabolome (Aretz and Meierhofer, 2016). Another limitation is the small number of yak calves in similar conditions. Even with these limitations, our study enhances the understanding of the effects of starter feeding and confirms the usefulness of 16S rRNA sequencing and untargeted metabolomics analyses in ruminant nutrition studies, especially yak calves.
Conclusion
In summary, a supplement to the starter promoted organ development and increased the abundance of some amylolytic bacteria. Ruminal microbiota have positive associations with metabolites involved in carbohydrate metabolism, nucleotide metabolism, xenobiotic biodegradation, and metabolism, demonstrating that the starter feeding can improve the nutritional status of the yak calves.
Data Availability Statement
The original contributions presented in the study are publicly available. These data can be found here: https://dataview.ncbi.nlm.nih.gov/object/PRJNA808822, BioProject: PRJNA808822.
Ethics Statement
The animal study was reviewed and approved by the Animal Ethics Committee of College of Agriculture and Animal Husbandry in Qinghai University. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
YW, SL, and ZC conceived and designed the experiments. YW, ZC, SL, HX, QY, and DY mainly performed the experiments. YW and ZC analyzed the data. ZC and SL contributed the reagents, materials, and analysis tools, had primary responsibility for final content. YW and ZC wrote the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Funding
This study was funded by the Program for Qinghai Science and Technology (2020-ZJ-911 to ZC and 2021-NK-A5 to QY) and the award fund program of Yak Engineering and Technology Research Center in Qinghai Province (SL).
References
- Abarghuei M. J., Rouzbehan Y., Salem A. Z. M., Zamiri M. J. (2014). Nitrogen balance, blood metabolites and milk fatty acid composition of dairy cows fed pomegranate-peel extract. Livestock Sci. 164 72–80. 10.1016/j.livsci.2014.03.021 [DOI] [Google Scholar]
- Abecia L., Waddams K. E., Martínez-Fernandez G., Martín-García A. I., Ramos-Morales E., Newbold C. J., et al. (2014). An antimethanogenic nutritional intervention in early life of ruminants modifies ruminal colonization by Archaea. Archaea 2014:841463. 10.1155/2014/841463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agle M., Hristov A. N., Zaman S., Schneider C., Ndegwa P. M., Vaddella V. K. (2010). Effect of dietary concentrate on rumen fermentation, digestibility, and nitrogen losses in dairy cows. J. Dairy Sci. 93 4211–4222. 10.3168/jds.2009-2977 [DOI] [PubMed] [Google Scholar]
- Anderson K. L., Nagaraja T. G., Morrill J. L., Avery T. B., Galitzer S. J., Boyer J. E. (1987). Ruminal microbial development in conventionally or early-weaned calves. J. Anim. Sci. 64 1215–1226. 10.2527/jas1987.6441215x [DOI] [PubMed] [Google Scholar]
- AOAC International (2000). Official Methods of Analysis, 17th edn. Arlington, VA: AOAC International. [Google Scholar]
- Aretz I., Meierhofer D. (2016). Advantages and pitfalls of mass spectrometry based metabolome profiling in systems biology. Int. J. Mol. Sci. 17:632. 10.3390/ijms17050632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artegoitia V. M., Foote A. P., Lewis R. M., Freetly H. C. (2017). Rumen fluid metabolomics analysis associated with feed efficiency on crossbred steers. Sci. Rep. 7:2864. 10.1038/s41598-017-02856-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin R. L., McLeod K. R., Klotz J. L., Heitmann R. N. (2004). Rumen development, intestinal growth and hepatic metabolism in the pre-and postweaning ruminant. J. Dairy Sci. 87 E55–E65. 10.3168/jds.s0022-0302(04)70061-2 [DOI] [Google Scholar]
- Ben Salem H., Nefzaoui A., Makkar H. P. S., Hochlef H., Ben Salem I., Ben Salem L. (2005). Effect of early experience and adaptation period on voluntary intake, digestion, and growth in Barbarine lambs given tannin-containing (Acacia cyanophylla Lindl. foliage) or tannin-free (Oaten Hay) diets. Anim. Feed Sci. Technol. 122 59–77. 10.1016/j.anifeedsci.2005.04.014 [DOI] [Google Scholar]
- Berends H., van den Borne J. J., Røjen B. A., Hendriks W. H., Gerrits W. J. (2015). Effect of protein provision via milk replacer or solid feed on protein metabolism in veal calves. J. Dairy Sci. 98 1119–1126. 10.3168/jds.2014-8375 [DOI] [PubMed] [Google Scholar]
- Bokulich N. A., Subramanian S., Faith J. J., Gevers D., Gordon J. I., Knight R., et al. (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10 57–59. 10.1038/nmeth.2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E., Rideout Jai R., Dillon Matthew R., Caporaso J. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 8 852–857. 10.1038/s41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carberry C. A., Waters S. M., Kenny D. A., Creevey C. J. (2014). Rumen methanogenic genotypes differ in abundance according to host residual feed intake phenotype and diet type. Appl. Environ. Microbiol. 80 586–594. 10.1128/AEM.03131-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z., Wu S., Li J., Yang Q. E., Chai S., Wang L., et al. (2020). Effect of alfalfa hay and starter feeding intervention on gastrointestinal microbial community, growth and immune performance of yak calves. Front. Microbiol. 11:994. 10.3389/fmicb.2020.00994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis Rincker L., Vandehaar M., Wolf C., Liesman J., Chapin L., Weber Nielsen M. (2011). Effect of intensified feeding of heifer calves on growth, pubertal age, calving age, milk yield, and economics. J. Dairy Sci. 94 3554–3567. 10.3168/jds.2010-3923 [DOI] [PubMed] [Google Scholar]
- Di Rienzi S. C., Britton Robert A. (2020). Adaptation of the gut microbiota to modern dietary sugars and sweeteners. Adv. Nutr. 3 616–629. 10.1093/advances/nmz118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Wang Y., Kreuzer M., Guo X., Mi J., Gou Y., et al. (2013). Seasonal variations in the fatty acid profile of milk from yaks grazing on the Qinghai-Tibetan plateau. J. Dairy Res. 80 410–417. 10.1017/S0022029913000496 [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10 996–998. 10.1038/nmeth.2604 [DOI] [PubMed] [Google Scholar]
- Edgar R. C., Haas B. J., Clemente J. C., Quince C., Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27 2194–2200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson E. L., Weimer P. J. (2017). Fermentation of model hemicelluloses by prevotella strains and butyrivibrio fibrisolvens in pure culture and in ruminal enrichment cultures. Appl. Microbiol. Biotechnol. 101 4269–4278. 10.1007/s00253-017-8150-7 [DOI] [PubMed] [Google Scholar]
- Fernando S. C., Purvis C. T. II, Najar F. Z., Sukharnikov L. O., Krehbiel C. R., Nagaraja T. G., et al. (2010). Rumen microbial population dynamics during adaptation to a high-grain diet. Appl. Environ. Microbiol. 76 7482–7490. 10.1128/AEM.00388-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firkins J. L., Yu Z., Morrison M. (2007). Ruminal nitrogen metabolism: perspectives for integration of microbiology and nutrition for dairy. J. Dairy Sci. 90(Suppl. 1) E1–E16. 10.3168/jds.2006-518 [DOI] [PubMed] [Google Scholar]
- Gao X., Zhang Q., Zhu H. (2019). High rejection rate of polysaccharides by microfiltration benefits Christensenella minuta and acetic acid production in an anaerobic membrane bioreactor for sludge fermentation. Bioresour. Technol. 282 197–201. 10.1016/j.biortech.2019.03.015 [DOI] [PubMed] [Google Scholar]
- Gill J. W., King K. W. (2002). Rumen microbiology, characteristics of free rumen cellulases. J. Agric. Food Chem. 5 363–367. 10.1021/jf60075a006 [DOI] [Google Scholar]
- Goldansaz S. A., Guo A. C., Sajed T., Steele M. A., Plastow G. S., Wishart D. S. (2017). Livestock metabolomics and the livestock metabolome: a systematic review. PLoS One 12:e0177675. 10.1371/journal.pone.0177675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Górka P., Kowalski Z. M., Zabielski R., Guilloteau P. (2018). Invited review: use of butyrate to promote gastrointestinal tract development in calves. J. Dairy Sci. 101 4785–4800. 10.3168/jds.2017-14086 [DOI] [PubMed] [Google Scholar]
- Grimble G. K., Westwood O. M. (2001). Nucleotides as immunomodulators in clinical nutrition. Curr. Opin. Clin. Nutr. Metab. Care 4 57–64. 10.1097/00075197-200101000-00011 [DOI] [PubMed] [Google Scholar]
- Haas B. J., Gevers D., Earl A. M., Feldgarden M., Ward D. V., Giannoukos G., et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21 494–504. 10.1101/gr.112730.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailemariam D., Mandal R., Saleem F., Dunn S. M., Wishart D. S., Ametaj B. N. (2014). Identification of predictive biomarkers of disease state in transition dairy cows. J. Dairy Sci. 97 2680–2693. 10.3168/jds.2013-6803 [DOI] [PubMed] [Google Scholar]
- Henderson G., Cox F., Kittelmann S., Miri V. H., Zethof M., Noel S. J., et al. (2013). Effect of DNA extraction methods and sampling techniques on the apparent structure of cow and sheep rumen microbial communities. PLoS One 8:e74787. 10.1371/journal.pone.0074787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini S. M., Ghorbani G. R., Rezamand P., Khorvash M. (2016). Determining optimum age of Holstein dairy calves when adding chopped alfalfa hay to meal starter diets based on measures of growth and performance. Animal 10 607–615. 10.1017/S1751731115002499 [DOI] [PubMed] [Google Scholar]
- Iqbal M. W., Zhang Q., Yang Y., Li L., Zou C., Huang C., et al. (2017). Comparative study of rumen fermentation and microbial community differences between water buffalo and Jersey cows under similar feeding conditions. J. Appl. Anim. Res. 46 740–748. 10.1080/09712119.2017.1394859 [DOI] [Google Scholar]
- Ishii S., Kosaka T., Hori K., Hotta Y., Watanabe K. (2005). Coaggregation facilitates interspecies hydrogen transfer between Pelotomaculum thermopropionicum and Methanothermobacter thermautotrophicus. Appl. Environ. Microbiol. 71 7838–7845. 10.1128/AEM.71.12.7838-7845.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karisa B. K., Thomson J., Wang Z., Li C., Montanholi Y. R., Miller S. P., et al. (2014). Plasma metabolites associated with residual feed intake and other productivity performance traits in beef cattle. Livest. Sci. 2014 200–211. 10.1016/j.livsci.2014.03.002 [DOI] [Google Scholar]
- Khan M. A., Bach A., Weary D. M., von Keyserlingk M. A. G. (2016). Invited review: transitioning from milk to solid feed in dairy heifers. J. Dairy Sci. 99 885–902. 10.3168/jds.2015-9975 [DOI] [PubMed] [Google Scholar]
- Kim M., Eastridge M. L., Yu Z. (2014). Investigation of ruminal bacterial diversity in dairy cattle fed supplementary monensin alone and in combination with fat, using pyrosequencing analysis. Can. J. Microbiol. 60 65–71. 10.1139/cjm-2013-0746 [DOI] [PubMed] [Google Scholar]
- Kim Y. H., Nagata R., Ohtani N., Ichijo T., Ikuta K., Sato S. (2016). Effects of dietary forage and calf starter diet on ruminal pH and bacteria in Holstein calves during weaning transition. Front. Microbiol. 7:1575. 10.3389/fmicb.2016.01575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham E., Weldon K., Wickersham T., Coverdale J., Pinchak W. (2018). Responses in the rumen microbiome of Bos taurus and indicus steers fed a low-quality rice straw diet and supplemented protein. J. Anim. Sci. 96 1032–1044. 10.1093/jas/sky023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Xie F., Sun D., Liu J., Zhu W., Mao S. (2019). Ruminal microbiome-host crosstalk stimulates the development of the ruminal epithelium in a lamb model. Microbiome 7:83. 10.1186/s40168-019-0701-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Bian G., Sun D., Zhu W., Mao S. (2017). Starter feeding supplementation alters colonic mucosal bacterial communities and modulates mucosal immune homeostasis in newborn lambs. Front. Microbiol. 8:429. 10.3389/fmicb.2017.00429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P. P., Liu S. J., Degen A. A., Qiu Q., Dong Q. M., Jing X. P., et al. (2018). Effect of weaning strategy on behavior, blood parameters and performance of yak calves (Poephagus grunniens). Rangel. J. 40 263–270. 10.1071/RJ17112 [DOI] [Google Scholar]
- Long R. J., Ding L. M., Shang Z. H., Guo X. H. (2008). The yak grazing system on the Qinghai-Tibetan plateau and its status. Rangel. J. 30 241–246. 10.1071/RJ08012 [DOI] [Google Scholar]
- Lu C. D., Kawas J., Mahgoub O. (2005). Fibre digestion and utilization in goats. Small Rumin. Res. 60 45–52. 10.1016/j.smallrumres.2005.06.035 [DOI] [Google Scholar]
- Mao S. Y., Huo W. J., Zhu W. Y. (2016). Microbiome-metabolome analysis reveals unhealthy alterations in the composition and metabolism of ruminal microbiota with increasing dietary grain in a goat model. Environ. Microbiol. 18 525–541. 10.1111/1462-2920.12724 [DOI] [PubMed] [Google Scholar]
- Meale S. J., Li S., Azevedo P., Derakhshani H., Plaizier J. C., Khafipour E. (2016). Development of ruminal and fecal microbiomes are affected by weaning but not weaning strategy in dairy calves. Front. Microbiol. 7:582. 10.3389/fmicb.2016.00582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Gerspach A. C., Drewe J., Verbeure W., Roux C. W. L., Dellatorre-Teixeira L., Rehfeld J. F. (2021). Effect of the natural sweetener xylitol on gut hormone secretion and gastric emptying in humans: a pilot dose-ranging study. Nutrients 13:174. 10.3390/NU13010174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaei M., Khorvash M., Ghorbani G. R., Kazemi-Bonchenari M., Riasi A., Nabipour A., et al. (2015). Effects of supplementation level and particle size of alfalfa hay on growth characteristics and rumen development in dairy calves. J. Anim. Physiol. Anim. Nutr. 99 553–564. 10.1111/jpn.12229 [DOI] [PubMed] [Google Scholar]
- Mohr E., Langbein J., Nürnberg G. (2002). Heart rate variability: a noninvasive approach to measure stress in calves and cows. Physiol. Behav. 75 251–259. 10.1016/s0031-9384(01)00651-5 [DOI] [PubMed] [Google Scholar]
- Morgavi D. P., Kelly W. J., Janssen P. H., Attwood G. T. (2013). Rumen microbial (meta)genomics and its application to ruminant production. Animal 7(Suppl. 1) 184–201. 10.1017/S1751731112000419 [DOI] [PubMed] [Google Scholar]
- Mu Y., Lin X., Wang Z., Hou Q., Wang Y., Hu Z. (2019). High-production dairy cattle exhibit different rumen and fecal bacterial community and rumen metabolite profile than low-production cattle. Microbiologyopen 8:e00673. 10.1002/mbo3.673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norouzian M. A., Valizadeh R., Vahmani P. (2011). Rumen development and growth of Balouchi lambs offered alfalfa hay pre-and post-weaning. Trop. Anim. Health Prod. 43 1169–1174. 10.1007/s11250-011-9819-z [DOI] [PubMed] [Google Scholar]
- Ogunade I., Schweickart H., Mccoun M., Cannon K., Mcmanus C. (2019). Integrating 16S rRNA sequencing and LC-MS-based metabolomics to evaluate the effects of live yeast on rumen function in beef cattle. Animals 9:28. 10.3390/ani9010028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orskov E. R. (2012). Energy Nutrition in Ruminants. Berlin: Springer. [Google Scholar]
- Pickard Joseph M., Chervonsky Alexander V. (2015). Intestinal fucose as a mediator of host-microbe symbiosis. J. Immunol. 194 5588–5593. 10.4049/jimmunol.1500395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinloche E., McEwan N., Marden J. P., Bayourthe C., Auclair E., Newbold C. J. (2013). The effects of a probiotic yeast on the bacterial diversity and population structure in the rumen of cattle. PLoS One 8:e67824. 10.1371/journal.pone.0067824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poudel P., Froehlich K., Casper D. P., St-Pierre B. (2018). 438 Feeding an essential oils blend to Neonatal Holstein dairy calves increased rumen propionate concentration and resulted in higher representation of a previously uncharacterized strain of Prevotella ruminicola. J. Anim. Sci. 96(Suppl._2) 235–236. 10.1093/jas/sky073.435 32704858 [DOI] [Google Scholar]
- Raghuvansi S. K. S., Prasad R., Tripathi M. K., Mishra A. S., Chaturvedi O., Misra A. K., et al. (2007). Effect of complete feed blocks or grazing and supplementation of lambs on performance, nutrient utilisation, rumen fermentation and rumen microbial enzymes. Animal 1 221–226. 10.1017/S1751731107284058 [DOI] [PubMed] [Google Scholar]
- Rao G., Sui J., Zhang J. (2016). Metabolomics reveals significant variations in metabolites and correlations regarding the maturation of walnuts (Juglans regia L.). Biol. Open 5 829–836. 10.1242/bio.017863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosewarne Carly P., Cheung Jane L., Smith Wendy J. M., Morrison M. (2012). Draft genome sequence of Treponema sp. strain JC4, a novel spirochete isolated from the bovine rumen. J. Bacteriol. 194:4130. 10.1128/JB.00754-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem F., Ametaj B. N., Bouatra S., Mandal R., Zebeli Q., Dunn S. M., et al. (2012). A metabolomics approach to uncover the effects of grain diets on rumen health in dairy cows. J. Dairy Sci. 95 6606–6623. 10.3168/jds.2012-5403 [DOI] [PubMed] [Google Scholar]
- Sander E. G., Warner R. G., Harrison H. N. (1959). The stimulatory effect of sodium butyrate and sodium propionate on the development of rumen mucosa in the young calf. J. Dairy Sci. 42 1600–1605. 10.3168/jds.s0022-0302(59)90772-6 [DOI] [Google Scholar]
- Saro C., Hohenester U. M., Bernard M., Lagrée M., Martin C., Doreau M., et al. (2018). Effectiveness of interventions to modulate the rumen microbiota composition and function in pre-ruminant and ruminant lambs. Front. Microbiol. 9:1273. 10.3389/fmicb.2018.01273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Chen Z., Shen Z., Lu Z. (2017). Maintaining stability of the rumen ecosystem is associated with changes of microbial composition and epithelial TLR signaling. Microbiologyopen 6:e00436. 10.1002/mbo3.436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soberon F., Van Amburgh M. E. (2013). Lactation biology symposium: the effect of nutrient intake from milk or milk replacer of pre-weaned dairy calves on lactation milk yield as adults: a meta-analysis of current data. J. Anim. Sci. 91 706–712. 10.2527/jas.2012-5834 [DOI] [PubMed] [Google Scholar]
- Stams A. J., Kremer D., Nicolay K., Weenk G. H., Hansen T. A. (1984). Pathway of propionate formation in Desulfobulbus propionicus. Arch. Microbiol. 139 167–173. 10.1007/BF00401994 [DOI] [Google Scholar]
- Sundberg C., Al-Soud W., Larsson M., Alm E., Yekta S. S., Svensson B. H., et al. (2013). 454 Pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiol. Ecol. 85 612–626. 10.1111/1574-6941.12148 [DOI] [PubMed] [Google Scholar]
- Sweeney B. C., Rushen J., Weary D. M., de Passillé A. M. (2010). Duration of weaning, starter intake, and weight gain of dairy calves fed large amounts of milk. J. Dairy Sci. 93 148–152. 10.3168/jds [DOI] [PubMed] [Google Scholar]
- Uebanso T., Kano S., Yoshimoto A., Naito C., Shimohata T., Mawatari K., et al. (2017). Effects of consuming xylitol on gut microbiota and lipid metabolism in mice. Nutrients 9:756. 10.3390/nu9070756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra Timothy D. (2012). Metabolomics: the final frontier? Genome Med. 4:40. 10.1186/gm339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Ma M. P., Diao Q. Y., Tu Y. (2019). Saponin-Induced shifts in the rumen microbiome and metabolome of young cattle. Front. Microbiol. 10:356. 10.3389/fmicb.2019.00356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Zhang G., Xu H., Xin H., Zhang Y. (2019). Metagenomic analyses of microbial and carbohydrate-active enzymes in the rumen of holstein cows fed different forage-to-concentrate ratios. Front. Microbiol. 10:649. 10.3389/fmicb.2019.00649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Xu L., Liu J., Zhu W., Mao S. (2017). A high grain diet dynamically shifted the composition of mucosa-associated microbiota and induced mucosal injuries in the colon of sheep. Front. Microbiol. 8:2080. 10.3389/fmicb.2017.02080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen B., Mei Z., Zeng C., Liu S. (2017). MetaX: a flexible and comprehensive software for processing metabolomics data. BMC Bioinformatics 18:183. 10.1186/s12859-017-1579-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Cui Z., Chen X., Zheng L., Ren H., Wang D., et al. (2021). Diet-ruminal microbiome-host crosstalk contributes to differential effects of calf starter and alfalfa hay on rumen epithelial development and pancreatic α-amylase activity in yak calves. J. Dairy Sci. 104 4326–4340. 10.3168/jds.2020-18736 [DOI] [PubMed] [Google Scholar]
- Wu S., Liu Y., Duan Y., Wang F., Guo F., Yan F., et al. (2018). Intestinal toxicity of deoxynivalenol is limited by supplementation with Lactobacillus plantarum JM113 and consequentially altered gut microbiota in broiler chickens. J. Anim. Sci. Biotechnol. 9:74. 10.1186/s40104-018-0286-285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X. X., Meng Q. X., Liu P., Wu H., Li S. R., Ren L. P., et al. (2013). Effects of a mixture of steam-flaked corn and extruded soybeans on performance, ruminal development, ruminal fermentation, and intestinal absorptive capability in veal calves. J. Anim. Sci. 91 4315–4321. 10.2527/jas.2012-5731 [DOI] [PubMed] [Google Scholar]
- Xue D., Chen H., Zhao X., Xu S., Hu L., Xu T., et al. (2017). Rumen prokaryotic communities of ruminants under different feeding paradigms on the QinghaiTibetan Plateau. Syst. Appl. Microbiol. 40 227–236. 10.1016/j.syapm.2017.03.006 [DOI] [PubMed] [Google Scholar]
- Xue M. Y., Sun H. Z., Wu X. H., Liu J. X., Guan L. L. (2020). Multi-omics reveals that the rumen microbiome and its metabolome together with the host metabolome contribute to individualized dairy cow performance. Microbiome 8:64. 10.1186/s40168-020-00819-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., He B., Wang S. S., Liu J. X., Wang J. K. (2015). Early supplementation of starter pellets with alfalfa improves the performance of pre- and postweaning Hu lambs. J. Anim. Sci. 93 4984–4994. 10.2527/jas.2015-9266 [DOI] [PubMed] [Google Scholar]
- Yang B., Le J., Wu P., Liu J., Guan L. L., Wang J. (2018). alfalfa intervention alters rumen microbial community development in hu lambs during early life. Front. Microbiol. 9:574. 10.3389/fmicb.2018.00574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Duan Z. (2018). The local defender and functional mediator: gut microbiome. Digestion 97 137–145. 10.1159/000484687 [DOI] [PubMed] [Google Scholar]
- Yu Y., Lee C., Kim J., Hwang S. (2005). Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol. Bioeng. 89 670–679. 10.1002/bit.20347 [DOI] [PubMed] [Google Scholar]
- Zapata H. J., Quagliarello V. J. (2015). The microbiota and microbiome in aging: potential implications in health and age-related diseases. J. Am. Geriatr. Soc. 63 776–781. 10.1111/jgs.13310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Ye H., Liu J., Mao S. (2017). High-grain diets altered rumen fermentation and epithelial bacterial community and resulted in rumen epithelial injuries of goats. Appl. Microbiol. Biotechnol. 101 6981–6992. 10.1007/s00253-017-8427-x [DOI] [PubMed] [Google Scholar]
- Zhang X. M., Liu Y. G., Zhu Z. Y. (2014). Total polyphenols, flavonoids and antioxidant activities in mulberry (Morus alba) leaves and fruit at different maturities. J. Fruit Sci. 31 660–666. 10.13925/j.cnki.gsxb.20130473 [DOI] [Google Scholar]
- Zhao F., Ren W., Zhang A., Jiang N., Liu W., Wang F. (2018). Effects of different amylose to amylopectin ratios on rumen fermentation and development in fattening lambs. Asian Austr. J. Anim. Sci. 31 1611–1618. 10.5713/ajas.17.0833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann W. (1990). Degradation of lignin by bacteria. J. Biotechnol. 13 119–130. 10.1016/0168-1656(90)90098-V [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are publicly available. These data can be found here: https://dataview.ncbi.nlm.nih.gov/object/PRJNA808822, BioProject: PRJNA808822.