Abstract
Biological processes underlying cerebral small vessel disease (cSVD) are largely unknown. We hypothesized that identification of clusters of inter-related bood-based biomarkers that are associated with the burden of cSVD provides leads on underlying biological processes. In 494 participants (mean age 67.6 ± 8.7 years; 36% female; 75% cardiovascular diseases; 25% reference participants) we assessed the relation between 92 blood-based biomarkers from the OLINK cardiovascular III panel and cSVD, using cluster-based analyses. We focused particularly on white matter hyperintensities (WMH). Nineteen biomarkers individually correlated with WMH ratio (r range: 0.16–0.27, Bonferroni corrected p-values <0.05), of which sixteen biomarkers formed one biomarker cluster. Pathway analysis showed that this biomarker cluster predominantly reflected coagulation processes. This cluster related also significantly to other cSVD manifestations (lacunar infarcts, microbleeds, and enlarged perivascular spaces), which supports generalizability beyond WMHs. To study possible causal effects of biological processes reflected by the cluster we performed a mediation analysis that showed a mediation effect of the cluster on the relation between age and WMH ratio (proportion mediated 17%), and hypertension and WMH-volume (proportion mediated 21%). In conclusion, we identified a cluster of blood-based biomarkers reflecting coagulation, that is related to manifestations of cSVD, corroborating involvement of coagulation abnormalities in the etiology of cSVD.
Keywords: Biomarkers, cerebral small vessel diseases, coagulation, proteins, unsupervised cluster analysis
Introduction
Cerebral small vessel disease (cSVD) is an important cause of cognitive decline, dementia, and stroke. 1 Although the clinical consequences of cSVD are evident, the exact pathogenesis of cSVD is still unclear. Clinical and experimental studies in both humans and animals have identified pathophysiological mechanisms of cSVD, including dysfunction of the cerebrovascular endothelium, changes in the blood-brain barrier components (pericytes, astrocytic end-feet, and extracellular matrix), impaired vasodilation, vessel stiffening and dysfunctional blood flow. 2 These microvessel abnormalities may ultimately contribute to injury in the cerebral white and deep grey matter, including white matter hyperintensities (WMH), ischemia, microbleeds and enlarged perivascular spaces. 2 Nonetheless, it is still unknown which biological processes underlie these abnormalities and how dysfunction of the small vessel eventually leads to brain injury.
Blood-based biomarkers may reflect biological processes underlying cSVD. Previous studies observed relations between cSVD and individual biomarkers, such as C-Reactive Protein (CRP) and interleukin-6 as markers for inflammation, 3 and Von Willebrand factor as a marker for coagulation. 4 It is important to consider, however, that most biological processes work in concert under upstream regulators and may therefore be associated with changes in multiple blood-based biomarkers. Vice versa, the levels of individual biomarkers may be affected by more than one biological process. Hence, exploring shared and differential relations of multiple biomarkers with cSVD at once may provide a more comprehensive perspective on possible disease mechanisms.
We hypothesized that clustering of blood-based biomarkers in relation to cSVD burden may provide leads on possible underlying biological processes involved in cSVD. We explored such clustering in individuals with a variable burden of cSVD using an existing panel of cardiovascular protein biomarkers, in relation to white matter hyperintensity (WMH) volume, but also lacunar infarcts, microbleeds and enlarged perivascular spaces.
Material and methods
Study population
We tested our hypothesis in a cohort of individuals with different manifestations of cardiovascular diseases who participated in the Heart Brain Connection (HBC) study. Participants had a variable burden of cSVD. The rationale and design of the HBC study have been described elsewhere. 5 In brief, the HBC study is a multicenter, observational study investigating hemodynamic and cardiovascular contributions to the pathophysiology of cognitive impairment in patients with manifest cardiovascular disease, including vascular cognitive impairment (VCI), carotid occlusive disease (COD), and heart failure (HF), and a reference group. All patients were 50 years or older and independent in daily living. For the current study, we included participants from the HBC study with an available cardiovascular protein biomarker panel and brain MRI.
Inclusion criteria for patients with VCI were cognitive complaints in the absence of moderate and severe dementia (Clinical Dementia Rating score ≤1 and a Mini-Mental State Examination ≥20), combined with at least moderate vascular brain injury (operationalized moderate to severe WMH (Fazekas ≥2) and/or (lacunar) infarct(s) and/or intracerebral (micro)hemorrhage(s)) or mild vascular brain injury (mild WMHs (Fazekas = 1)) and the presence of at least two vascular risk factors. Patients with COD had a stenosis >80% or occlusion of an internal carotid artery on MR angiography and were not scheduled for surgical intervention. Patients with HF were diagnosed according to the European Society of Cardiology guidelines 6 and were clinically stable for at least 6 months. Patients were recruited from cardiology, memory, and neurology outpatient clinics in four university medical centers in The Netherlands. Reference participants were recruited via advertising leaflets and among spouses of patients. The most important exclusion criteria were the inability to complete the study due to a life expectancy shorter than three years or plans to move out of the area of investigation or the presence of other psychiatric or neurological disorders (including neurodegenerative disease other than VCI or Alzheimer’s’ dementia) that could affect cognitive performance.
All participants provided written informed consent before research-related procedures. The Medical Ethics Review Committee of the Leiden University Medical Center performed central approval. Local medical ethical committees of all sites approved the local performance of the study. The Heart-Brain Study is performed in accordance with the declaration of Helsinki (version 2013) and the Medical Research Involving Human Subjects
Act (WMO).
Clinical characteristics
Clinical characteristics of all participants were registered by trained physicians or research nurses using a standardized interview and physical examination. Hypertension was defined as presence in medical history, use of antihypertensive drugs, or newly diagnosed hypertension defined as a systolic blood pressure ≥140 mm Hg or a diastolic blood pressure ≥90 mm Hg (systolic and diastolic blood pressure were measured on the left and right arm with an automatic blood pressure monitor, the mean of these two readings was used for analyses). Diabetes was defined as the presence in medical history with or without the use of anti-diabetic drugs. Obesity was defined as a body mass index of ≥30. Ischemic heart disease was defined as a history of myocardial infarction or coronary revascularization (percutaneous coronary intervention or coronary artery bypass graft). History of stroke was defined as presence of an ischemic or hemorrhagic stroke in medical history. Participants underwent blood tests to determine LDL-cholesterol in mg/dL.
Assessment of blood-based cardiovascular biomarkers
Participants provided blood samples which were collected into ethylenediaminetetraacetic acid (EDTA) plasma vacutainer tubes. Cardiovascular protein biomarkers were determined with a multiplex immunoassay using the OLINK® Proteomics Cardiovascular III panel (OLINK® Proteomics, Uppsala, Sweden). 7 This panel comprises 92 cardiovascular disease related biomarker proteins which were selected in collaboration with experts from the cardiovascular field. Raw biomarker values were converted to OLINK®'s arbitrary unit, Normalized Protein eXpression (NPX). NPX is a relative unit on a log2-scale, with higher values corresponding to higher protein concentrations in the sample. A list of the 92 blood-based cardiovascular protein biomarkers that are included in the OLINK® Cardiovascular III panel can be found in Supplemental Table S1. The blood samples that were used for the cardiovascular biomarker assessment were collected at the same day as the clinical characteristics.
MRI protocol and analysis
Brain MRI was acquired on Philips Ingenia, Achieva and Gemini 3 TMRI scanners. The brain protocol included a 3D T1-weighted image (resolution= 1.0 × 1.0 × 1.0 mm3; TR = 8.2 ms; TE = 4.5 ms; inversion delay 990 ms), a fluid-attenuated inversion recovery (FLAIR) image (resolution = 1.11 × 1.11 × 1.11 mm3; TR = 4.8 ms; TE = 313 ms; TI= 1.65 ms and a 3 D susceptibility-weighted image (SWI) (resolution 0.8 × 0.8 × 0.8 mm3; TR = 45 ms; TE = 31 ms.
Brain volumes, including WMH volume, were calculated with an automated pipeline (Quantib brain, Rotterdam, the Netherlands) after manual segmentation of infarcts and other focal pathologies. 8 WMH volumes were expressed as a percentage of the total brain volume (TBV) (WMH ratio) to correct for differences in head size and degree of atrophy. Presence of (defined as ≥1 lesion) lacunar infarcts (small 3–15 mm lesions), microbleeds (small homogeneous round foci at any anatomical location), and enlarged perivascular spaces (small fluid filled spaces, following the course of penetrating vessels, in the basal ganglia) were visually rated by an experienced neuroradiologist (JB) according to the STRIVE-criteria. 9 These manifestations of cSVD were entered in a dichotomous fashion in the analyses.
Statistical analysis
To study the relationship between the panel of 92 cardiovascular protein biomarkers and cSVD (particularly WMHs), we performed a 4-step analysis.
Step 1: Identification of biomarker cluster(s) related to WMH ratio
To test which of the 92 biomarkers individually correlated with WMH ratio, we performed Pearson correlation analyses with Bonferroni corrections (p-value threshold: 0.05/92 = 0.00054). Within the selection of biomarkers that individually significantly (Bonferroni corrected p <0.05) correlated with WMH ratio we identified clusters based on prior biological knowledge by using the STRING database, 10 and in an unsupervised data-driven way based on the OLINK NPX values. We used the affinity propagation 11 algorithm to identify the optimal number of clusters, after which we performed agglomerative clustering based on the Pearson’s correlation matrix. Thereby, we set the numbers of clusters to numbers identified with affinity propagation. Furthermore, we determined whether the data-driven clustering method suggested additional relationships, next to the relations identified in the STRING database. If a biomarker was clustered together with biomarkers involved in the STRING cluster(s) in 500 bootstrapped subsample replicates of the data, we added this biomarker to the cluster(s). Subsequently, to create a variable that captures the values of the biomarker cluster(s) in one score per cluster, we calculated a Biomarker Compound Score (BCS). This variable initially contained average Z-scores of the biomarker values of the cluster(s) and was optimized by minimizing the Euclidean distance between the biomarkers values of the cluster(s) and the average Z-score. 11 We performed a Pearson correlation analysis to test the correlation between the BCS and WMH ratio across and within the participants groups (VCI, COD, HF and reference participants), and adjusted for antithrombotic use.
Of note, several other biomedical factors (such as age and sex) might be associated with both the burden of cSVD and blood-based biomarkers. Simply adding these biomedical factors to the analyses as potential confounders might lead to overadjustment, if a biomarker would be in a causal path between a factor and the outcome WMH. Hence, in this first step we used unadjusted analyses, except for antithrombotic use. In step 4 we describe how we investigated the possible interplay between certain biomedical factors, the identified biomarker cluster(s) and WMH ratio.
Step 2: Biological interpretations of the identified biomarker cluster(s)
To identify involved biological processes within the identified biomarker cluster(s), we used the Reactome pathway analysis tool. 12 This tool identifies biological pathways that are enriched within our biomarker cluster(s) more than would be expected by chance. Reactome uses Fisher’s exact test, to identify enriched pathways within the cluster(s). 12 Pathways with p-values <0.05 and false discovery rates (FDR) <0.05 were considered statistically significant enriched and were selected for biological interpretation of the cluster(s). Since the OLINK panel we analyzed is a preselected cardiovascular biomarker panel, we first verified which pathways were overall significantly enriched in the entire panel of 92 biomarkers. We compared these findings with the enriched pathways within our identified cluster(s) 12 and selected the same number of significantly enriched pathways as in the previous step.
Step 3: Relation of the identified biomarker cluster(s) with other cSVD manifestations
To investigate the generalizability of the identified biomarker cluster(s) to other cSVD manifestations we assessed the relation between the BCS and the presence of lacunar infarcts, microbleeds, and enlarged perivascular spaces by performing t-tests of BCS values between the group with and the group without the presence of each manifestation.
Step 4: Causal mediation analysis
To evaluate possible causal effects of biological processes reflected by the identified biomarker cluster(s) we performed a causal mediation analysis. In this analysis we studied the potential mediating role of biological processes reflected by the identified biomarker cluster(s) in the relation between known vascular risk factors and WMH. Known vascular risk factors (age, hypertension and sex) were used as exposure variables, the BCS as potential mediator variable and WMH ratio as outcome variable.
We used the natural logarithm of the WMH ratio in all correlation and linear regression analyses because of the right-skewed distribution of the WMH ratio. For 11 participants, WMH ratios lower than 0.1% were set at 0.1% to approximate the normal distribution and to avoid log-transformation of 0. We used R v3.6.3 13 with mediation 14 to perform mediation analyses and Python v3.9 15 with scikit-learn 16 to perform data-driven cluster analyses.
Results
A total of 494 participants were included in this study. Table 1 shows the detailed characteristics of the study population. Participants were on average 67.6 ± 8.7 years old, 36% were female, 29% of the participants had a diagnosis of heart failure, 28% of VCI, 18% of COD and 25% were from the reference group. Median WMH ratio was 0.15% (IQR 0.04; 0.56). Patients with VCI had the highest WMH ratios (median 0.64%, IQR 0.19; 1.70) and participants from the reference group the lowest (median 0.05%, IQR 0.02; 0.18). Lacunar infarcts were present in 35%, microbleeds in 25% and enlarged perivascular spaces in the basal ganglia in 28% of the participants.
Table 1.
Baseline characteristics.
| Participants(n = 494) | |
|---|---|
| Sociodemographics | |
| Age (years), mean (sd) | 67.6 (8.7) |
| Female sex, n (%) | 177 (36) |
| Years of education, mean (sd) | 13.5 (4.3) |
| Vascular risk factors | |
| Hypertension, n (%) | 386 (78) |
| LDL cholesterol (mg/dL), mean (sd) | 105.2 (36.7) |
| Diabetes Mellitus, n (%) | 69 (14) |
| Current smoker, n (%) | 82 (17) |
| Obesity, n (%) | 103 (21) |
| History of ischemic heart disease, n (%) | 146 (30) |
| History of TIA, n (%) | 119 (24) |
| History of stroke, n (%) | 113 (23) |
| Participant group | |
| Vascular Cognitive Impairment, n (%) | 140 (28) |
| Carotid Occlusive Disease, n (%) | 90 (18) |
| Heart failure, n (%) | 141 (29) |
| Reference group, n (%) | 123 (25) |
| Neuro-imaging markers for cSVD | |
| WMH ratio (as % of TBV), median (IQR) | 0.15 (0.04; 0.56) |
| TBV (ml), mean (sd) | 1096.4 (111.4) |
| Lacunar infarcts, n (%) | 174 (35) |
| Microbleeds (lobar or non-lobar), n (%) | 125 (25) |
| Enlarged perivascular spaces, n (%) | 140 (28) |
IQR: interquartile range (25th to 75th percentile); LDL: low-density lipoprotein; sd: standard deviation; TBV: total brain volume: WMH: white matter hyperintensity.
Data are expressed as mean (sd), median (IQR), or number (percentage).
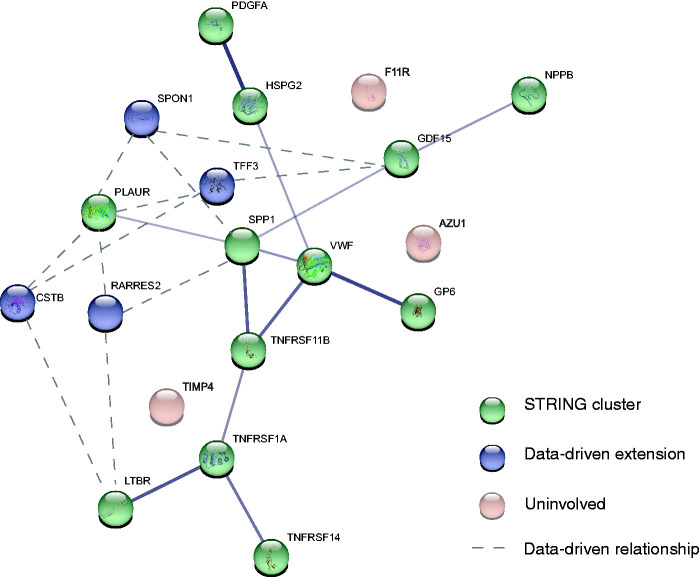
Cluster analyses point towards a 16-component biomarker cluster of potential interest
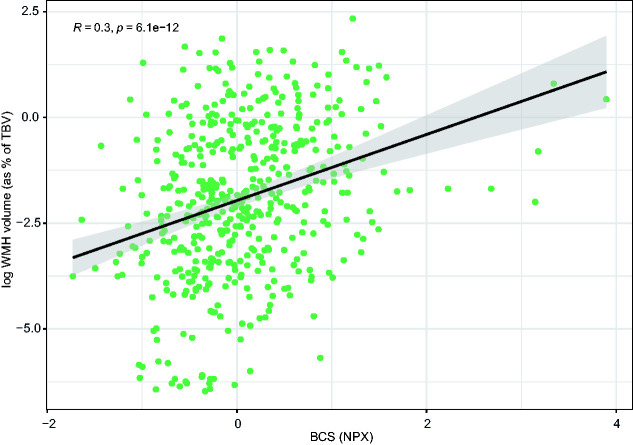
To identify cluster(s) of biomarkers related to WMHs within the measured panel, we first performed a Pearson's correlation analysis to establish associations between individual biomarkers and WMH ratio. In total, 19 of the 92 biomarkers (21%) correlated positively with WMH ratio (Pearson's r ranging from 0.16 to 0.27, Bonferroni corrected p-values <0.05) (Table 2, Figure 1). Twelve of these 19 biomarkers were found to be interrelated according to prior knowledge from the STRING database (protein-protein interaction (PPI) p-value <0.001). Data-driven cluster analyses identified 4 additional biomarkers: Cystatin B (CSTB), Retinoic acid receptor responder 2 (RARRES2), Spondin 1 (SPON1), and Trefoil factor 3 (TFF3) (Figure 1), with strong positive correlation (r > 0.8) with the other 12 biomarkers identified through STRING (Supplemental Figure 1). The combined set of 16 biomarkers (i.e. 12 from STRING, 4 data-driven) formed a cluster with a central role for the Von Willebrand Factor (VWF), Secreted phosphoprotein 1 (SPP1), TNF receptor superfamily member 11 b (TNFRSF11B), and Growth differentiation factor 15 (GDF15). The calculated BCS based on the identified cluster showed a significant positive correlation with WMH ratio (Pearson's r 0.3, p <0.001) (Figure 2). We found no confounding effect of Platelet inhibitor (p = 0.656) and Vitamin K antagonist (p = 0.396) use, as also supported by analyses stratified by medication use (Supplemental Figure 2). After stratification by participant group, we found a significant positive correlation between the BCS and WMH ratio within each of the patient groups (VCI, COD, and HF). This correlation was non-significant in reference participants (p = 0.16) (Supplemental Figure 3).
Table 2.
Overview of the 19 significantly correlating biomarkers with white matter hyperintensity volume.a
| Biomarker (abbreviation) | Pearson's r | Bonferroni correctedp-value |
|---|---|---|
| Growth differentiation factor 15 (GDF15) | 0.270 | <0.001 |
| Cystatin B (CSTB) | 0.242 | <0.001 |
| Secreted phosphoprotein 1 (SPP1) | 0.233 | <0.001 |
| TNF receptor superfamily member 14 (TNFRSF14) | 0.219 | <0.001 |
| TNF receptor superfamily member 11 b (TNFRSF11B) | 0.215 | <0.001 |
| TIMP metallopeptidase inhibitor 4 (TIMP4) | 0.203 | <0.001 |
| Trefoil factor 3 (TFF3) | 0.202 | <0.001 |
| Plasminogen activator, urokinase receptor (PLAUR) | 0.198 | <0.001 |
| Spondin 1 (SPON1) | 0.180 | 0.005 |
| F11 receptor (F11R) | 0.180 | 0.006 |
| TNF receptor superfamily member 1 A (TNFRSF1A) | 0.179 | 0.006 |
| Azurocidin 1 (AZU1) | 0.175 | 0.008 |
| Lymphotoxin beta receptor (LTBR) | 0.173 | 0.010 |
| Natriuretic peptide B (NPPB) | 0.172 | 0.010 |
| Retinoic acid receptor responder 2 (RARRES2) | 0.172 | 0.012 |
| Glycoprotein VI platelet (GP6) | 0.170 | 0.016 |
| Heparan sulfate proteoglycan 2 (HSPG2) | 0.169 | 0.014 |
| Platelet derived growth factor subunit A (PDGFA) | 0.160 | 0.030 |
| Von Willebrand factor (VWF) | 0.156 | 0.045 |
Data are expressed as correlation coefficients (Pearson’s r) with corresponding Bonferroni corrected p-values.
aWe entered the natural logarithm of white matter hyperintensity volume (expressed as % of total brain volume) in the Pearson correlation analyses.
Figure 1.
Interrelations of the 19 identified protein biomarkers that were found to correlate significantly with white matter hyperintensity (WMH) ratio. 16 out of 19 biomarkers that correlated significantly to WMH ratio were found to form one cluster. Biomarkers that were found to be involved based on information from the STRING database are shown in green. 4 biomarkers that were found to extend this cluster based on data-driven cluster analysis are shown in blue. 3 biomarkers that were not found to be involved in the cluster based on both information from the STRING database and the data-driven cluster analysis are shown in pink. Line width reflects the strength of data support. For the blue, data-driven extensions, the three strongest correlations are shown as dashed lines (r > 0.8).
Figure 2.
Correlation between the Biomarker Compound Score and white matter hyperintensity volume. The Biomarker Compound Score (BCS) was expressed as Normalized Protein eXpression (NPX) and the natural log of white matter hyperintensity (WMH) volume was expressed as percentage of total brain volume (TBV). Correlation is unadjusted (see the method section).
Pathway analysis shows that the identified biomarker cluster predominantly reflects coagulation processes
Through pathway analysis of the identified biomarker cluster, we identified 14 statistically significant enriched pathways (p-values <0.05, FDR <0.05), pointing mainly towards coagulation and to a lesser extent towards extracellular matrix organization, inflammation and angiogenesis processes (Table 3). In the top-14 statistically significant enriched pathways present in the entire panel of 92 biomarkers, we predominantly found processes involved in inflammation and to a lesser extent extracellular matrix organization and cell organization processes (Supplemental Table S2). These results indicate that the identified biomarker cluster related to WMH ratio predominantly reflects coagulation and to a lesser extent extracellular matrix organization, inflammation and angiogenesis processes, whereas the entire 92 protein cardiovascular biomarker panel that was tested predominantly reflected inflammation.
Table 3.
Statistically enriched biological pathways reflected by the identified 16-component biomarker cluster.
| Pathway | No. of biomarkers found/Total no. of biomarkers inpathway (Reactome) | p-value | FDR | Biomarkers |
|---|---|---|---|---|
| Coagulation pathways | ||||
| Platelet Adhesion to exposed collagen | 2/25 | <0.001 | <0.001 | GP6;VWF |
| Platelet activation, signaling and aggregation | 4/264 | <0.001 | 0.012 | GP6;VWF;RARRES2;PDGFA |
| Platelet degranulation | 3/128 | 0.001 | 0.012 | VWF;RARRES2;PDGFA |
| Response to elevated plateletcytosolic Ca2+ | 3/133 | 0.001 | 0.012 | VWF;RARRES2;PDGFA |
| Defective F8 binding to vonWillebrand factor | 1/2 | 0.003 | 0.031 | VWF |
| Hemostasis | 5/726 | 0.004 | 0.035 | PLAUR;GP6;VWF;RARRES2;PDGFA |
| Defective F8 cleavage by thrombin | 1/3 | 0.004 | 0.038 | VWF |
| Inflammation pathways | ||||
| TNFs bind their physiological receptors | 3/30 | <0.001 | <0.001 | TNFRSF1A;TNFRSF14;TNFRSF11B |
| TNFR2 non-canonical NF-kB pathway | 4/102 | <0.001 | <0.001 | TNFRSF1A;TNFRSF14;LTBR;TNFRSF11B |
| Angiogenesis pathways | ||||
| Signaling by PDGF | 2/60 | <0.001 | <0.001 | SPP1;PDGFA |
| Downstream signal transduction | 1/31 | 0.001 | 0.012 | PDGFA |
| ECM organization pathways | ||||
| Integrin cell surface interactions | 3/85 | <0.001 | 0.007 | VWF;SPP1;HSPG2 |
| ECM organization | 4/301 | 0.001 | 0.012 | PDGFA;VWF;SPP1;HSPG2 |
| Non-integrin membrane-ECM interactions | 2/59 | 0.003 | 0.035 | PDGFA;HSPG2 |
FDR: false discovery rate; TNF: tumor necrosis factor; TNFR2: tumor necrosis factor receptor 2; No.: number; NF-kB: nuclear factor-kB; PDGF: Platelet-derived Growth Factor; ECM: extracellular matrix.
Data are presented as the number of biomarkers found per pathway (expressed as a fraction of the total number of biomarkers known to be involved per pathway based on the Reactome database) with corresponding p-values and FDRs. A statistically significant p-value implies that the number of biomarkers identified within a pathway is higher than would be expected by chance.
Generalizability of the identified biomarker cluster to other cSVD manifestations
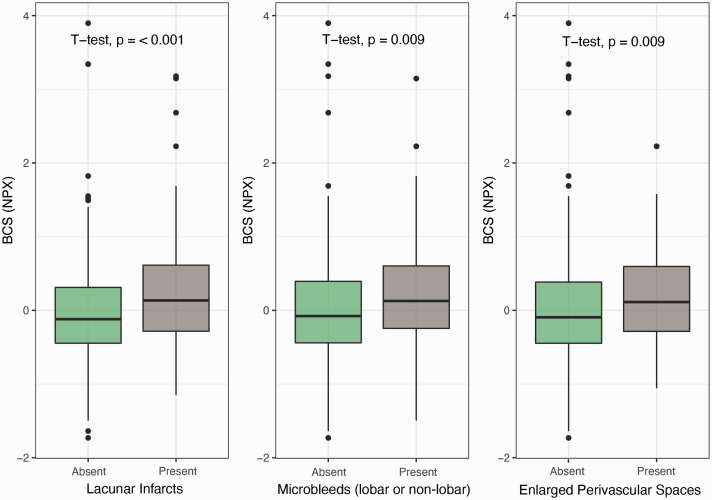
The BCS also related significantly to the presence of lacunar infarcts (p <0.05), microbleeds (p <0.01) and enlarged perivascular spaces (p <0.05) (Figure 3). These findings confirm generalizability of the found effects of the clusters underlying cSVD in different manifestations of cSVD.
Figure 3.
The BCS and other cerebral small vessel manifestations. Boxplots of the Biomarker Compound Score (BCS) expressed as Normalized Protein eXpression (NPX) according to presence of lacunar infarcts, microbleeds, and enlarged perivascular spaces.
Causal effects of the biological processes reflected by the biomarker cluster
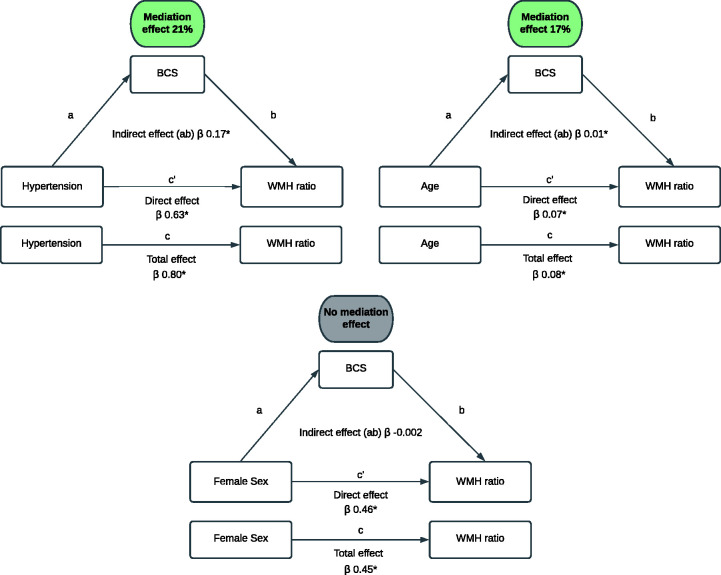
To study the possible causal effects of the biological processes reflected by the found cluster, we performed causal mediation analysis with known vascular risk factors. Figure 4 shows the potential mediating role of the identified biomarker cluster on the relation between age, hypertension and sex and WMH ratio. Age (β = 0.03, p <0.001) and hypertension (β = 0.24, p <0.01) were significantly associated with the BCS. Causal mediation analysis showed a significant mediation effect of the BCS in the relation between age and WMH ratio (proportion mediated 17%, p <0.001) and the relation between hypertension and WMH ratio (proportion mediated 21%, p <0.001) (Figure 4). We found no association between sex and the BCS (β = −0.003, p = 0.96) and the BCS did therefore not mediate the relation between sex and WMH ratio.
Figure 4.
Results of the causal mediation analysis. The total effect (c) of known vascular risk factors (age, hypertension and sex) on white matter hyperintensity (WMH) ratio is composed of an indirect effect (ab) and direct effect (c′) and all are presented in β with corresponding p-values. The mediation effect is the indirect effect expressed as a percentage of the total effect, that is, the proportion of the relation between vascular risk factors (age, hypertension and sex) and WMH ratio attributable to mediation of the identified biomarker cluster (BCS).
*p < 0.05.
Discussion
In patients with various manifestations of cardiovascular disease, we identified a cluster of 16 blood-based proteins related to WMH ratio, based on both known protein-protein interactions and data-driven unsupervised clustering. This biomarker cluster points towards coagulation processes as a unifying process linking the identified proteins to WMH burden. The identified biomarker cluster was also significantly associated with manifestations of cSVD other than WMH. The cluster mediated the relation between known WMH risk factors, age and hypertension, and WMH-volume, further highlighting biological relevance.
To the best of our knowledge, this study is the first that assessed the relation between a large panel of blood-based biomarkers and manifestations of cSVD. Because identification of patterns in a large biomarker panel can be challenging, we pre-defined a comprehensive cluster analysis, to identify a cluster of protein biomarkers related to the burden of cSVD. Such an approach, in which prior biological knowledge and data-driven cluster analyses are combined, has already shown to be an effective method for data-reduction and pathway identification in several other diseases, such as heart failure,17,18 diabetes 19 and cancer.20,21 The biomarker cluster we identified with data-driven cluster analysis was largely consistent with information from the STRING-database, 10 which demonstrates the biological coherence of our findings. Previous studies have related several single blood-based biomarkers or small biomarkers panels to the appearance of MRI findings in cSVD, such as WMHs and lacunar infarcts.3,4,22 Most of these studies focused on biomarkers that reflect inflammation, endothelial dysfunction and coagulation processes. Two protein biomarkers, VWF and GDF15, that have a central role in our identified biomarker cluster, have been previously related to cSVD.23–26 The other 14 biomarkers of our cluster have not been related to cSVD before.
Pathway analysis showed that our identified biomarker cluster predominantly reflects coagulation processes and to a lesser extent extracellular matrix organization, inflammation and angiogenesis processes. All of these biological processes are considered to play a role in the function, maintenance, and repair of the vascular endothelium.4,27 These findings are in line with previous clinical and experimental studies that identified a prominent role for endothelial dysfunction in the etiology of cSVD. 2 Several rodent models point towards dysfunctional endothelial cells in the pathophysiology of cSVD.2,28 Also in humans considerable evidence from studies including advanced neuro-imaging models support that endothelial dysfunction is important in the etiology of cSVD. 2 These studies demonstrated diffuse blood-brain barrier dysfunction 29 and impaired cerebrovascular reactivity in cSVD. 30 Also previous studies that assessed single or small panel blood-based biomarkers identified molecules related to endothelial dysfunction. 4 Thus, together with evidence from previous studies, our findings support that endothelial dysfunction plays a prominent role in the etiology of cSVD. Moreover, our study extends previous findings by indicating that particularly coagulation abnormalities appear to underlie dysfunction of the cerebrovascular endothelium.
The observed effects of the identified biomarker cluster hold true for patients with VCI, COD and HF. In reference participants, who as expected had the lowest burden of WMHs, the cluster did not significantly relate to WMH ratio. Additionally, also related to cSVD manifestations beyond WMH, including lacunar infarcts, microbleeds and enlarged perivascular spaces. Thus, despite that the etiology of cSVD is thought to include different mechanisms 2 that are likely to depend on both underlying cardiovascular diseases and the manifestations of cSVD, our findings indicate that coagulation abnormalities play a prominent role in different patient subtypes with cSVD, irrespective of their underlying cardiovascular diseases and manifestations of cSVD. In addition to the demonstrated generalizability of our biomarker cluster, we demonstrated the potential etiological relevance of the cluster by means of a causal mediation analysis. This analysis revealed a significant mediation effect of the identified cluster and WMHs, indicating that biological processes (particularly coagulation) reflected by the biomarker cluster appear to mediate the relation between advanced age and hypertension and the burden of WMHs. These findings may support current evidence that advanced age 31 and hypertension 32 lead to the activation of among other, coagulation pathways, which, in turn, may lead to WMHs.
The implications of this study are twofold. First, we demonstrated that identification of biomarker clusters with prior-knowledge based and data-driven cluster analyses can help to gain more insight in etiological processes involved in cSVD. Future studies could use the same approach with other, even larger biomarker panels to identify involvement of hitherto unknown processes in the etiology of cSVD. Second, with this approach we identified a biomarker cluster reflecting specific processes involved in endothelial dysfunction, particularly concerning coagulation processes and to a lesser extent extracellular matrix organization, inflammation and angiogenesis processes. These results may represent important contributions in better understanding of involved biological processes in cSVD. If validated in future studies, our findings could support considering these biological processes as targets for prevention and therapy of cSVD.
Strengths of this study are the comprehensive cluster analyses and standardized work-up including optimal imaging modalities for vascular brain injury assessment. However, also some limitations should be considered. First, the cross-sectional design precludes us to study relations between the biomarker cluster and progression of WMHs and, clearly, cross-sectional observational data cannot be used to actually prove causality. Moreover, as explained in the methods section, in our primary analytical steps we did not adjust for biomedical factors like age, that may be related to both the biomarkers and the outcome. We addressed this interplay between biomedical factors, biomarkers, and WMH with the mediation analysis, but these results should also be interpreted with caution in a cross-sectional study because temporal ordering of our variables could be reversed and studied pathways bidirectional. Yet, our findings concur with prior knowledge on this topic.31,32 Furthermore, we studied a heterogenous study population including participants with different prototypical cardiovascular conditions with a variable burden of cSVD. However, stratified analysis per participant group showed that our findings hold through for different manifestations of cardiovascular diseases including VCI, COD and HF. Importantly, the manifestations of cSVD on MRI we have studied may not only be due to cSVD but also to large vessel disease or even other disease processes. It is therefore of interest to further explore the biomarker cluster also in relation to other possible manifestations of SVD, such as MR measures of microvascular function, in future studies. One could also argue that the non-directed approach involves multiple testing with risk of spurious findings. However, since we have demonstrated biological plausibility of our identified biomarker cluster in multiple ways, and we have (at least partially) mitigated fortuitous correlations by applying a Bonferroni correction, we consider it unlikely that our results are coincidental. Furthermore, although we used a non-directed analytical approach, the biomarker panel we used is not entirely non-directed: it was designed to include only a subset of all proteins in the circulation, selected based on their known involvement in cardiovascular diseases. 7 On the other hand, pathway analysis of the entire biomarker panel revealed other biological processes (no coagulation) than our identified biomarker cluster, which makes it unlikely that our results are biased by the use of a pre-selected protein panel. Nevertheless, future studies with alternative and larger biomarker panels should be performed to validate and extend our findings. Lastly, we observed relatively weaker correlations between PDGFA, GP6, NPPB and VWF and other biomarkers involved in the cluster. In our current study, we tried to find direct associations within a subset of biomarkers. We assessed direct interaction of the BCS proteins with each other based on the information from the STRING database without including further proteins from the respective biological pathway, as these were not included in the targeted proteomics approach. Furthermore, the associations found in STRING are not only based on linear direct associations, but also on proven co-expression, associations in curated databases, high-throughput lab experiments, and/or automated text-mining. Hence, we are not able to provide direct evidence for causality and thus can only speculated about the underlying reasons for the observed weaker correlations for the mentioned markers.
Conclusion
With comprehensive cluster analyses in patients with various manifestations of cardiovascular disease, we identified a biomarker cluster, predominantly reflecting coagulation abnormalities, that relates to the burden of cSVD. This study also underscores the potential of non-directed cluster analyses in proteomics approaches to gain insight in biologic processes involved in cSVD.
Supplemental Material
Supplemental material, sj-pdf-1-jcb-10.1177_0271678X221077339 for A cluster of blood-based protein biomarkers reflecting coagulation relates to the burden of cerebral small vessel disease by Sanne Kuipers, L Malin Overmars, Bram van Es, Jeroen de Bresser, Esther E Bron, Imo E Hoefer, L Jaap Kappelle, Charlotte E Teunissen, Geert Jan Biessels and Saskia Haitjema in Journal of Cerebral Blood Flow & Metabolism
Acknowledgements
We gratefully acknowledge the contribution of researchers and participants of the HBC (Heart-Brain Connection) Consortium.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The HBC (Heart-Brain Connection) Consortium is supported by the Netherlands CardioVascular Research Initiative: the Dutch Heart Foundation (CVON 2018-28 & 2012-06 Heart Brain Connection), Dutch Federation of University Medical Centers, the Netherlands Organization for Health Research and Development, and the Royal Netherlands Academy of Sciences. J. de Bresser is supported by Alzheimer Nederland (WE.03-2019-08). E.E. Bron is supported by Dutch Heart Foundation (PPP Allowance, 2018B011). GJ Biessels is supported by ZonMw, The Netherlands, Organization for Health Research and Development Vici Grant 918.16.616).
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Authors’ contributions: SK, MO, GB and SH substantially contributed to the conception and design of this manuscript. SK, JB, EB, IH, JK, CT and GB participated in the data acquisition. SK, MO and BE analyzed the data. SK, MO, GB and SH drafted the manuscript and all authors revised it critically for important intellectual content. All authors read and approved the final draft of the manuscript.
ORCID iDs: Sanne Kuipers https://orcid.org/0000-0002-3839-4311
L Malin Overmars https://orcid.org/0000-0001-7086-0864
Supplemental material: Supplemental material for this article is available online.
References
- 1.Rensma SP, van Sloten TT, Launer LJ, et al. Cerebral small vessel disease and risk of incident stroke, dementia and depression, and all-cause mortality: a systematic review and Meta-analysis. Neurosci Biobehav Rev 2018; 90: 164–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wardlaw JM, Smith C, Dichgans M. Small vessel disease: mechanisms and clinical implications. Lancet Neurol 2019; 18: 684–696. [DOI] [PubMed] [Google Scholar]
- 3.Low A, Mak E, Rowe JB, et al. Inflammation and cerebral small vessel disease: a systematic review. Ageing Res Rev 2019; 53: 100916. [DOI] [PubMed] [Google Scholar]
- 4.Poggesi A, Pasi M, Pescini F, et al. Circulating biologic markers of endothelial dysfunction in cerebral small vessel disease: a review. J Cereb Blood Flow Metab 2016; 36: 72–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hooghiemstra AM, Bertens AS, Leeuwis AE, et al. The missing link in the pathophysiology of vascular cognitive impairment: design of the heart-brain study. Cerebrovasc Dis Extra 2017; 7: 140–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ponikowski P, Voors AA, Anker SD, et al. 2016 ESC guidelines for the diagnosis and treatment of acute and chronic heart failure: the task force for the diagnosis and treatment of acute and chronic heart failure of the European society of cardiology (ESC). Developed with the special contribution of the heart failure association (HFA) of the ESC. Eur J Heart Fail 2016; 18: 891–975. [DOI] [PubMed] [Google Scholar]
- 7.Olink proteomics. Biomarkers in Olink Cardiovascular III 2019, www.olink.com/products/cvd‐iii‐panel (2019, accessed 21 April 2021).
- 8.de Boer R, Vrooman HA, van der Lijn F, et al. White matter lesion extension to automatic brain tissue segmentation on MRI. Neuroimage 2009; 45: 1151–1161. [DOI] [PubMed] [Google Scholar]
- 9.Wardlaw JM, Smith EE, Biessels GJ, et al. Neuroimaging standards for research into small vessel disease and its contribution to ageing and neurodegeneration. Lancet Neurol 2013; 12: 822–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019; 47: D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frey BJ, Dueck D. Clustering by passing messages between data points. Science 2007; 315: 972–976. [DOI] [PubMed] [Google Scholar]
- 12.Jassal B, Matthews L, Viteri G, et al. The reactome pathway knowledgebase. Nucleic Acids Res 2020; 48: D498–D503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.R Core Team. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing, 2020. [Google Scholar]
- 14.Tingley D, Yamamoto T, Hirose K, et al. Mediation: R package for causal mediation analysis. UCLA Statistics/American Statistical Association, https://dspace.mit.edu/handle/1721.1/91154 (2014, accessed 28 April 2021).
- 15.Van Rossum G, Drake FL. Python 3 reference manual. Scotts Valley, CA: CreateSpace, 2009. [Google Scholar]
- 16.Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in python. J Mach Learn Res 2011; 12: 2825–2830. [Google Scholar]
- 17.Sanders-van Wijk S, Tromp J, Beussink-Nelson L, et al. Proteomic evaluation of the comorbidity-Inflammation paradigm in heart failure with preserved ejection fraction: results from the PROMIS-HFpEF study. Circulation 2020; 142: 2029–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sama IE, Woolley RJ, Nauta JF, et al. A network analysis to identify pathophysiological pathways distinguishing ischaemic from non-ischaemic heart failure. Eur J Heart Fail 2020; 22: 821–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, Qiao Z, Zheng W, et al. Network cluster analysis of protein-protein interaction network-identified biomarker for type 2 diabetes. Diabetes Technol Ther 2015; 17: 475–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo T, Wu S, Shen X, et al. Network cluster analysis of protein-protein interaction network identified biomarker for early onset colorectal cancer. Mol Biol Rep 2013; 40: 6561–6568. [DOI] [PubMed] [Google Scholar]
- 21.Hu K, Chen F. Identification of significant pathways in gastric cancer based on protein-protein interaction networks and cluster analysis. Genet Mol Biol 2012; 35: 701–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vilar-Bergua A, Riba-Llena I, Nafría C, et al. Blood and CSF biomarkers in brain subcortical ischemic vascular disease: involved pathways and clinical applicability. J Cereb Blood Flow Metab 2016; 36: 55–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kario K, Matsuo T, Kobayashi H, et al. Hyperinsulinemia and hemostatic abnormalities are associated with silent lacunar cerebral infarcts in elderly hypertensive subjects. J Am Coll Cardiol 2001; 37: 871–877. [DOI] [PubMed] [Google Scholar]
- 24.Altendahl M, Maillard P, Harvey D, et al. An IL-18-centered inflammatory network as a biomarker for cerebral white matter injury. PLoS One 2020; 15: e0227835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chai YL, Hilal S, Chong JPC, et al. Growth differentiation factor-15 and white matter hyperintensities in cognitive impairment and dementia. Medicine (Baltimore) 2016; 95: e4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gottesman RF, Cummiskey C, Chambless L, et al. Hemostatic factors and subclinical brain infarction in a community-based sample: the ARIC study. Cerebrovasc Dis 2009; 28: 589–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arroyo AG, Iruela-Arispe ML. Extracellular matrix, inflammation, and the angiogenic response. Cardiovasc Res 2010; 86: 226–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajani RM, Quick S, Ruigrok SR, et al. Reversal of endothelial dysfunction reduces white matter vulnerability in cerebral small vessel disease in rats. Sci Transl Med July 2018; 10: 4. [DOI] [PubMed] [Google Scholar]
- 29.Wardlaw JM, Makin SJ, Valdés Hernández MC, et al. Blood-brain barrier failure as a core mechanism in cerebral small vessel disease and dementia: evidence from a cohort study. Alzheimer's Dementia 2017; 13: 634–643. [Google Scholar]
- 30.Sam K, Crawley AP, Conklin J, et al. Development of white matter hyperintensity is preceded by reduced cerebrovascular reactivity. Ann Neurol 2016; 80: 277–285. [DOI] [PubMed] [Google Scholar]
- 31.Kurachi K, Zhang K, Ameri A, et al. Genetic and molecular mechanisms of age regulation (homeostasis) of blood coagulation. IUBMB Life 2000; 49: 189–196. [DOI] [PubMed] [Google Scholar]
- 32.Lip GY. Hypertension and the prothrombotic state. J Hum Hypertens 2000; 14: 687–690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-jcb-10.1177_0271678X221077339 for A cluster of blood-based protein biomarkers reflecting coagulation relates to the burden of cerebral small vessel disease by Sanne Kuipers, L Malin Overmars, Bram van Es, Jeroen de Bresser, Esther E Bron, Imo E Hoefer, L Jaap Kappelle, Charlotte E Teunissen, Geert Jan Biessels and Saskia Haitjema in Journal of Cerebral Blood Flow & Metabolism