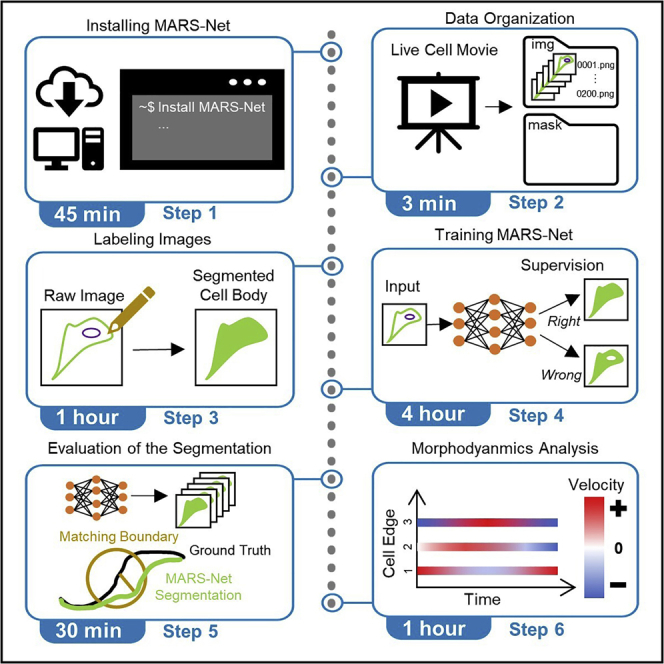
Summary
Quantitative studies of cellular morphodynamics rely on accurate cell segmentation in live cell images. However, fluorescence and phase contrast imaging hinder accurate edge localization. To address this challenge, we developed MARS-Net, a deep learning model integrating ImageNet-pretrained VGG19 encoder and U-Net decoder trained on the datasets from multiple types of microscopy images. Here, we provide the protocol for installing MARS-Net, labeling images, training MARS-Net for edge localization, evaluating the trained models’ performance, and performing the quantitative profiling of cellular morphodynamics.
For complete details on the use and execution of this protocol, please refer to Jang et al. (2021).
Subject areas: Bioinformatics, Cell Biology, Microscopy, Computer sciences
Graphical abstract
Highlights
-
•
Deep learning-based segmentation pipeline for live cell movies
-
•
Semi-automatic labeling tool for ground truth masks
-
•
Train segmentation models and evaluate their segmentation accuracy
-
•
Quantification of cellular morphodynamics from detected cell edges
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Quantitative studies of cellular morphodynamics rely on accurate cell segmentation in live cell images. However, fluorescence and phase contrast imaging hinder accurate edge localization. To address this challenge, we developed MARS-Net, a deep learning model integrating ImageNet-pretrained VGG19 encoder and U-Net decoder trained on the datasets from multiple types of microscopy images. Here, we provide the protocol for installing MARS-Net, labeling images, training MARS-Net for edge localization, evaluating the trained models’ performance, and performing the quantitative profiling of cellular morphodynamics.
Before you begin
Data collection
MARS-Net was developed to leverage the datasets from multiple types of microscopy images for the most accurate segmentation results. The protocol below describes the specific steps for training on our multiple-microscopy-type datasets from phase contrast, spinning disk confocal, and total internal reflection fluorescence microscopes. However, this protocol can be used for single-microscopy datasets.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Software and algorithms | ||
| MARS-Net | This paper | https://github.com/kleelab-bch/MARS-Net |
| ImageJ | Schneider et al. (2012) | https://imagej.nih.gov/ij/ |
| MATLAB 2019b–2021b | MathWorks | https://www.mathworks.com/products/matlab.html |
| Python 3.6.8 | Python Software Foundation | https://www.python.org/ |
| Anaconda v4.5.11 | Anaconda | https://www.anaconda.com/ |
| CUDA v10.1 | NVIDIA | https://developer.nvidia.com/cuda-toolkit |
| cuDNN v7.6.5 | NVIDIA | https://developer.nvidia.com/rdp/cudnn-archive |
| Extended Berkeley Segmentation Benchmark | David Stutz | https://github.com/davidstutz/extended-berkeley-segmentation-benchmark |
| Windowing and Protrusion Package | Gaudenz Danuser Lab | https://github.com/DanuserLab/Windowing-Protrusion |
| npy-matlab | Kwik Team | https://github.com/kwikteam/npy-matlab |
| TensorFlow v2.3 | TensorFlow | https://www.tensorflow.org/ |
| Matplotlib v3.3.4 | Matplotlib | https://matplotlib.org/ |
| NumPy v1.18.5 | NumPy | https://numpy.org/ |
| Absl-py (v0.12.0) | Abseil Python Common Libraries | https://github.com/abseil/abseil-py |
| Grpcio (v1.37.0) | The gRPC Authors | https://pypi.org/project/grpcio/ |
| H5py (v2.10.0) | Andrew Collette and contributors | https://www.h5py.org/ |
| Imageio (v2.9.0) | Imageio | https://imageio.readthedocs.io/en/stable/ |
| Importlib-metadata (v3.10.0) | Python | https://docs.python.org/3/library/importlib.metadata.html |
| Joblib (v1.0.1) | Joblib | https://joblib.readthedocs.io/en/latest/ |
| Keras-preprocessing (v1.1.2) | TensorFlow | https://www.tensorflow.org/api_docs/python/tf/keras/preprocessing |
| Numba (v0.53.1) | Numba | https://numba.pydata.org/ |
| OpenCV-contrib-python (v4.5.1.48) | OpenCV | https://docs.opencv.org/4.x/d6/d00/tutorial_py_root.html |
| OpenCV-python (v4.5.1.48) | OpenCV | https://docs.opencv.org/4.x/d6/d00/tutorial_py_root.html |
| Pillow (v8.2.0) | Tidelift | https://github.com/python-pillow/Pillow |
| Psutil (v5.8.0) | Giampaolo Rodola | https://github.com/giampaolo/psutil |
| Scikit-image (v0.17.2) | scikit-image | https://scikit-image.org/ |
| Scikit-learn (v0.24.1) | scikit-learn | https://scikit-learn.org/stable/ |
| Scipy (v1.4.1) | SciPy | https://scipy.org/ |
| Tqdm (v4.60.0) | Casper da Costa-Luis | https://github.com/tqdm/tqdm |
| Umap-learn (v0.5.1) | Leland McInnes | https://umap-learn.readthedocs.io/en/latest/ |
| Other | ||
| CPU (1 required) | any core | N/A |
| RAM Memory | N/A | 16 GB or greater recommended |
| GPU | NVIDIA GTX 1080Ti, 2080Ti or Titan RTX | N/A |
| OS | Ubuntu v16.04, v18.04, or Windows 10 | N/A |
Materials and equipment
-
•
Microscopy images from either phase contrast, spinning disk confocal or total internal reflection fluorescence microscopes. Other microscopy images have not been tested.
Python Software and required packages. The required packages will be automatically installed when users run the installation using Anaconda with environment.yml, which is explained later. While different versions of the Python software and associated packages may work correctly with MARS-Net, the authors use Python v3.6.8 and the following packages at the indicated versions when writing this protocol. When new versions become available, our software will be updated accordingly. The aforementioned software and packages are listed above in the key resources table.
Step-by-step method details
Part 1. Installing MARS-Net
Timing: 45 min
Full installation of MARS-Net includes downloading the MARS-Net package from GitHub and installing its software requirements.
-
1.Setup the Anaconda environment by running the following commands.
-
a.In the command prompt in Windows 10/11:>conda env create --name marsnet –file environment_windows.yml
-
b.In the Linux Terminal in Ubuntu:>conda env create –name marsnet –file environment_linux.yml>conda activate marsnet
-
a.
-
2.
To download the MARS-Net pipeline from GitHub Repository, click the link and navigate to the green “Code” button on the page. Click this, and then select “Download ZIP”. Be sure to also install the requirements listed in the key resources table above (also located on the MARS-Net pipeline GitHub page).
-
3.
Unzip the downloaded zip file from the GitHub and open the unzipped folder.
-
4.
Open “UserParams.py” file with a text editor or IDE such as PyCharm.
Note: UserParams.py is a file that contains all the configurations necessary for running Mars-Net.
-
5.Edit the following parameters inside __init__() function within UserParams class. These specify the type of the deep learning model to use, location of datasets, and training configurations. The parameters for training step and prediction step are independent and must be set separately. For example, if statement block from line 62 to 81 specify parameters for training the ‘multi_micro’ model and the if statement block from line 422 to 441 specify parameters for segmenting live cell movies using the trained ‘multi_micro’ model.
-
a.“strategy_type”.
-
i.Set as the type of deep learning model to use (ex. VGG19_dropout, unet).Note: We recommend users to always use the VGG19_dropout model since we experimentally showed that the VGG19_dropout model segmented all live cell movies in our dataset with the highest accuracy. Other options are U-Net and plain VGG19 which are the baseline models.
-
i.
-
b.“dataset_folders”.
-
i.Set as the location where your images and masks are stored (ex. [“..\assets\”, “..\assets\second_dataset\”].Note: The sample images and labels are in the “..\assets\040119_PtK1_S01_01_phase_ROI2\” directory so that users can practice the written procedures.
-
i.
-
c.“img_type”.
-
i.Set as the type of file your image is (ex. “.png").
-
i.
-
d.“img_folders”.
-
i.Set as a list of folders where images are located (ex. ["/img/","/img/"]).
-
i.
-
e.“mask_folders”.
-
i.Set as a list of folders where masks are located (ex. ["/mask/"," mask"]).Note: These are the folders for ground truth masks of the images.
-
i.
-
f.“frame_list”.
-
i.Set as a list of the number of images to train the model (ex. [1,2,6,10]).Note: [1,2,6,10] means that the first model will be trained on one of the randomly selected images in the dataset folder. Then the second model will be trained on two images, and so on. Model can train on as few as just one image and its corresponding ground truth mask or as many frames as possible from the live cell movie but more training frames require more labeling efforts.
-
i.
-
g.“dataset_names”.
-
i.Set as a list of the dataset names, which will be parent folders containing img and mask folders. (ex. ["dataset1"," dataset2"]).Note: For training, more than one dataset name should be specified in the list because our pipeline performs a leave-one-movie-out cross validation. For instance, given the list of datasets m1, m2, m3 and m4, first three datasets (m1, m2 and m3) will be used for training and m4 will be used as a test set for evaluating the segmentation accuracy. Then, other combination of three datasets such as (m1, m3, m4) will be used for training and m2 will be used as a test set for evaluating the segmentation accuracy. For prediction, only one dataset can be specified in the list, which would segment one live cell movie using a trained model. See part 4. Training MARS-Net and segmenting movies for more details.
-
i.
-
h.“model_names”.
-
i.Set as a list of the model names, these names can be any distinguishing characters (ex. ["A"," B"]).Note: As described in the note in step g, each model name in the list represents a different model trained during the leave-one-movie-out cross validation. These models use the same deep learning architecture specified in the “strategy_type” but are trained on different combinations of datasets.
-
i.
-
i.“REPEAT_MAX”.
-
i.Set as the number of times to repeat cross validation (ex. 1 or 5).Note: Repeating the cross validation is for evaluating the deep learning model robustly. The same deep learning model trained with different “random seed” can yield slightly different performances. Therefore, in each repetition, we vary the random seed and train the same model repetitively to see the variance of our models’ performance. 5 is preferred for robust evaluation of the model but it also takes five times longer to train and evaluate. So, in practice, it is set to 1.Note: Make sure the list length is equal for “model_names”, “img_folders”, “mask_folders”, “dataset_folders”, and “dataset_names”.
-
i.
-
a.
Part 2. Data organization
Timing: 3 min per movie
Organizing the data with the correct labels, in the correct folders, etc. is a key component in insuring MARS-Net is run effectively.
-
6.
Create a folder with a unique name, and inside that folder, create subfolders with names “img” and “mask”.
-
7.If the dataset is a movie, separate the movie into images (we used PNG format) with ImageJ.
-
a.Open ImageJ, load in the movie, click “File”, “Save As” then “Image Sequence”.
-
a.
-
8.Make sure that the images are generated in order with filenames having the four-digit frame id with leading zeros at the end, starting with ‘0001’.
-
a.For example, given a movie with 200 frames, the first frame’s filename is cell_0001.png, the second frame’s filename is cell_0002.png, and the last frame’s filename is cell_0200.png.
-
a.
-
9.
Put images into the “img” folder and leave the mask folder empty. The mask folder will be filled with labeled images later.
-
10.
Repeat previous steps for each additional movie.
Part 3. Labeling images
Timing: 3 min per image
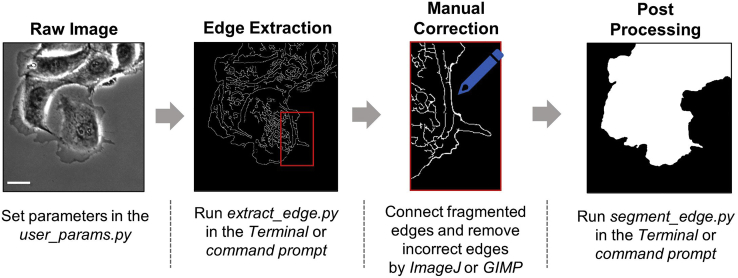
The labeling tool (Figure 1) facilitates labeling raw images semi-automatically for training MARS-Net. Gaussian, bilateral, and guided blurring operations denoise the raw images, followed by the Canny edge detector (Canny, 1986) extracting edges from three blurred images. And then, they are combined into one preliminary edge image. Human annotators should fix any errors of the preliminary edges manually (Figure 2). All files for labeling tools are found in the “label_tool” folder. The timing is from labeling the image of size 392 × 474 by an expert.
Note: This Part can be skipped if no movie needs to be labeled for training and evaluation and a user just wants to use our pretrained model to segment live cell movies. If a user chooses to label a dataset, we recommend labeling about 1% of the data for training. For a robust evaluation of the trained model, we recommend labeling about 10% of the data. Throughout this Part, it is recommended to use a similar naming scheme for folders and images as the example in MARS-Net Github.
-
11.Specify the location of image files to the label in the “user_params.py”:
-
a.Set variable “a_dataset” to the name of the folder with original images.
-
b.Set variable “img_root_path” to the path where original images are located.
-
c.Save “user_params.py”.
-
a.
Note: “user_params.py” is not the same as “UserParams.py” used in previous steps. The “user_params.py” file is specifically for the labeling tool.
-
12.Determine the optimal hysteresis thresholding for canny detector and kernel size for blurring (parameters located in “user_params.py”):
-
a.Run the following code:>python explore_edge_extraction_user_params.py
-
b.When “explore_edge_extraction_user_params.py” completes all edge extractions, results will be located in the “generated_explore_edge” folder.
-
i.Select each generated image and determine which parameter values are best for optimizing hysteresis thresholding.
-
i.
-
c.Set the selected best thresholding values in “user_params.py” for “canny_std_multiplier” and “denoise_kernel_size” parameters located at around line 11 and 12.
-
d.Save “user_params.py”.
-
a.
-
13.Extract edges with optimized parameters:
-
a.Run the following code:>python extract_edge.py
-
b.When “extract_edge.py” completes all generated edge images, results will be located in the “generated_edge” folder.
-
a.
-
14.Manually fix the generated edge images:
-
a.The image edges will not always be correctly connected, even with the optimized parameters. Hence, it is important to manually connect any fragmented edges and remove the wrong edges in the image. Download ImageJ or GIMP to manually fix after overlaying the edge on the original image∗
-
b.Replace any generated edge images in the “generated_edge” folder with manually fixed edge images.
-
a.
Note: ∗When using ImageJ for manual edge fixing, use the freehand tool to obtain the true edges of the cells. When using GIMP, overlay the image with a mask that is 50% transparent. From here, draw the true edges of the cell with the paintbrush tool.
-
15.Post-process the edge images to fill the edge images:
-
a.Run the following code:>python segment_edge.py
-
i.The “segment_edge.py” will ask for how many backgrounds to fill in your image, and one pair of (x, y) coordinates in each background. The total number of backgrounds will be determined based on the image of the cell. Once you locate all backgrounds in the image, provide any (x, y) coordinates in each respective background.
-
i.
-
b.When “segment_edge.py” completes all segmentations, results will be located in the “generated_segmentation” folder.
-
c.Move these labeled images to the “assets” folder to begin training the model.
-
a.
-
16.
Repeat previous steps in this Part on all cell movies before moving to the next Part. All movies are labeled for training multiple models for cross-validation in the evaluation Part. Labeling all live cell movies is only necessary for training multiple models and evaluating them by leave-one-movie-out cross validation.
Figure 1.
Labeling procedure
Simplified diagram of four steps involved in semi-automatically labeling live cell movie. In the first step, prepare the raw image and set the parameters of the label tool. In the second step, extract the edge from the raw image. It is inaccurate since edge extraction is performed by a traditional algorithm. Therefore, in the third step, a user needs to fix extracted edge image by connecting fragmented edges or removing incorrect edges. A manually corrected image is shown as an example. Then in the fourth step, a simple flood fill algorithm can segment the cell body region in the fully connected edge image. Scale Bar: 32.5 μm.
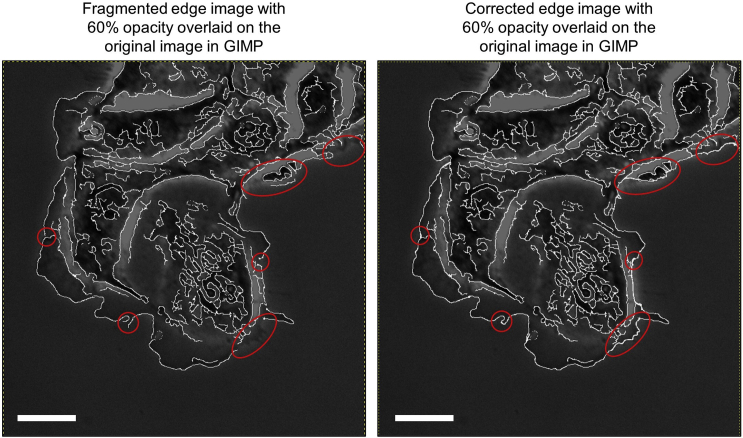
Figure 2.
Manual correction of labels
The image on the left contains fragmented edges or isolated edges that are extracted due to noise. The red circles are used to indicate some of those regions. The image on the right shows the result after all fragmented or isolated edges that are corrected manually in GIMP. Scale bars: 32.5 μm.
Part 4. Training MARS-Net and segmenting movies
Timing: 4 h
MARS-Net (Jang et al., 2021) takes a transfer learning approach (Bertasius et al., 2015; Iglovikov et al., 2018; Kim et al., 2018; Long et al., 2015; Vaidyanathan et al., 2021) by integrating ImageNet-pretrained VGG19 encoder and U-Net decoder with additional dropout layers (Deng et al., 2009; Ronneberger et al., 2015; Simonyan and Zisserman, 2015; Srivastavanitish et al., 2014), trained on the datasets from multiple types of microscopy. MARS-Net accepts the segmented images generated in Part 3 of this protocol. The code described in the following steps crops each image of the dataset into 200 of 128 × 128 patches, trains the deep learning model, and predicts the segmentation of the images in the test set, which is defined in UserParams.py in Part 1. The cropping code samples 60% of the patches from the cell edges, 20% of the patches from the cell interior and the other 20% from the background outside the cell. Titan RTX took about 4 h, but training time can vary based on which GPU is used.
-
17.
In the terminal, navigate to the “crop” folder and run the “crop_augment_split.py” file to begin cropping patches for training:
>cd “location of crop folder”
>python crop_augment_split.py
-
18.
When “crop_augment_split.py” finishes cropping the images, navigate to the “models” folder in the terminal and run the following code to begin training MARS-Net:
>cd “location of model folder”
>python train_mars.py
-
19.
During training which might take a few hours, the terminal will show the training loss and dice coefficient on the training set and the validation loss and dice coefficient on the validation set for each epoch. For metrics on both training and validation sets, usually high dice coefficient at around 0.98 and low loss at around 0.01 indicates the successful training.
-
20.
While the model is training, set up parameters for prediction in UserParams.py as described in the step 5 in Part 1 to specify which live cell movie to segment.
-
21.
When “train_mars.py” finishes training the model, navigate to the “models” folder and run the following code to segment the live cell movies:
>python predict.py
-
22.
The segmented live cell movie will be generated in the “models/results/predict_wholeframe_round1_∗“directory where ∗ represents the model name specified in the “strategy_type” parameter.
Note: Please remember this directory of segmented result since it will be used in Part 6 for quantification of morphodynamics.
CRITICAL: MARS-Net will over-write previous cropped files, trained models and predicted results saved with the same “strategy_type”, so please use a unique “strategy_type” in UserParams.py before cropping, training and prediction.
Note: A user can predict segmentation of our sample live cell movie with pretrained U-Net and VGG19D models without going through the cropping and training steps above. Follow the instructions below if you want to use our pretrained model without cropping and training. Also, follow the instructions below if you don’t want to perform leave-one-movie-out cross validation but segment a live cell movie using one of the trained models. The instructions below are a more user-friendly way to use our pipeline that only requires four parameters from the user, without the need to specify any parameters in UserParams.py:
-
23.
To use our pretrained U-Net or VGG19D models, download them from the following google drive as a zip file: https://drive.google.com/drive/folders/1FLP0D-Y9-DHQmhC-LBZChdUSe6W5zyPw?usp=sharing.
Note: The sample images and labels are in the “..\assets\040119_PtK1_S01_01_phase_ROI2\” directory so that users can practice the written procedures.
-
24.
Create the new folder named “results” under the “models” folder.
-
25.
Move the downloaded zip file from the google drive into the “results” folder and unzip it. Then, there should be “model_round1_Multi_VGG19D” folder “model_round1_Single_Unet” folder inside the “results” folder.
-
26.
Navigate to the “models” folder in the terminal and run the following code to segment the live cell movie. Please specify trained_model_path, live_cell_images_path, save_path, and img_format as input arguments in the command line as follows:
> python predict_simple.py --trained_model_path ./results/model_round1_Multi_VGG19D/model_frame2_D_repeat0.hdf5 --live_cell_images_path ../assets/040119_PtK1_S01_01_phase_2_DMSO_nd_02/img_all/ --save_path ./results/ --img_format .png
-
27.
To use one of the trained models from leave-one-movie-out cross validation scheme above, navigate to the folder in the directory “models/results/model_round1_∗/” where ∗ stands for the name of the model specified in the “strategy_type” parameter. Inside that folder, there should be one or more trained models with the name “model_∗_repeat0.hdf5”. Choose one of those models for prediction in step 26 based on the evaluation of each cross validation in Part 5.
Part 5. Evaluation of the segmentation results
Timing: 30 min
Now that MARS-Net model training and prediction are complete, we can evaluate and visualize the segmentation results. Segmentation results are evaluated by Precision, Recall, and F1 score (Arbelaez et al., 2010). To briefly explain these criteria; precision measures how well the model identified only the real cell boundaries compared to non-cell boundaries, recall measures if the model found all the real cell boundaries and F1 measures a harmonic mean of precision and recall. For comparison of the model against other models, the evaluated results from multiple models are shown together in a bar graph, edge evolution diagram, violin plot, line graph, and bubble plot. All evaluations follow the leave-one-movie-out cross-validation scheme, which means that given ‘n’ number of movies, ‘n-1’ movies are used for training the model, and one movie is segmented, and segmentation results are evaluated. Cross validation repeats such training and evaluation until each movie is used for validation once. The visualization codes in this Part show the cross-validation results by averaging the evaluation results from the segmentation of each movie. For more details on what each plot describes, refer to the MARS-Net (Jang et al., 2021).
CRITICAL: Correspondence Algorithm(Arbelaez et al., 2010) necessary for calculating F1, precision and recall is only supported in Linux. Windows 10 user still can perform steps 35, 36 and 40 in this Part or go to the Part 6.
-
28.
Navigate to the “evaluation” folder in the MARS-Net folder and open the “GlobalConfig.m” file in MATLAB.
-
29.Edit the following parameters that specify the location of the segmentation results, name of the deep learning architecture to evaluate, the number of training frames used to train the model, the location to save the evaluated results and the location of the input images.:
-
a.“prediction_path_list”.
-
i.Set as folder path to a newly made file in “results” folder called “predict_wholeframe_round{#}_{project name}”.
-
i.
-
b.“display_names”.
-
i.Set as the project name, or a name that is to be displayed on results.
-
i.
-
c.“frame_list”.
-
i.Set as the number of frames that was used to train the model.Note: This is the list of the number of training frames used in the training dataset. Since we train multiple models with different number of training frames, we specify which models to evaluate here. This is useful to see the how the segmentation accuracy changes as the size of training dataset increases.
-
i.
-
d.“root_path”.
-
i.Set as a folder path to the “evaluation” folder.Note: This is a location to save the evaluation results.
-
i.
-
e.“img_root_path”.
-
i.Set as a folder path to the “assets” folder.Note: This is a location of input images which are necessary for overlaying segmentation results on top of input images.Note: Below is the example parameters for comparing the segmentation results from Single_Unet and Multi_VGG19D models in the location described in step 22 in Part 4. User can copy parameters below and paste to line 244 in GlobalConfig.m to set the parameters for evaluation. Instead of using the example parameters below as is, please change the part ‘C:\Users\JunbongJang\PycharmProjects’ to a folder path where MARS-Net is installed in the user’s computer.prediction_path_list = {'C:\Users\JunbongJang\PycharmProjects\MARS-Net\models\results\predict_wholeframe_round1_ Single_Unet\'; 'C:\Users\JunbongJang\PycharmProjects\MARS-Net\models\results\predict_wholeframe_round1_ Multi_VGG19D\'};display_names = {‘Single Unet’;’Multi VGG19D’};frame_list = [2;2];root_path = 'C:\Users\JunbongJang\PycharmProjects\MARS-Net\evaluation\';img_root_path = 'C:\Users\JunbongJang\PycharmProjects\MARS-Net\assets\';
-
i.
-
a.
-
30.Optionally in GlobalConfig.m, update_config function can be edited to process a new result with the user’s project name. Currently, update_config function handles results from phase contrast, spinning disk confocal, total internal reflection fluorescence (TIRF), single-microscopy, and multi-microscopy models.
-
a.For instance, add another else if statement at around line 308 in GlobalConfig.m and specify parameters such as: img_root_path, mask_type, img_type, dataset_list, and fold_name_list. They are similar to the parameters specified in UserParams.py in Part 1.
-
a.
-
31.
Download NPC Reader and Correspondence Algorithm (Arbelaez et al., 2010). Add the NPC Reader folder to the “evaluation” folder, and add the Correspondence Algorithm folder to the “evaluation_f1” folder located in the “evaluation” folder.
-
32.Calculate F1, precision and recall from the segmented movies by running the following code in the Terminal after going to the “evalutation_f1” folder:>cd “location of evaluation_f1 folder”>matlab -nodisplay -nosplash -nodesktop -r “run(‘run_overlap_mask_prediction.m’);exit;”
-
a.Alternatively, this can be run in the MATLAB command window by typing “run_overlap_mask_prediction”.
-
a.
-
33.
Open the generated MATLAB data files with the following name “Recall_Precision_F_score_frame#.mat” located in the “results” folder. # in the name can be any number.
-
34.Draw the violin plot of F1, precision, and recall of the opened MATLAB data file by running the following code in the terminal:>cd “evalutation_f1 folder”>matlab -nodisplay -nosplash -nodesktop -r “run(‘visualize_a_model.m’);exit;”
-
a.Alternatively, this can be run in MATLAB by typing “visualize_a_model”.
 Pause point: Instructions below are for comparing the multiple models’ segmentation results against the ground truth segmentation labeled in Part 3, so make sure to train and predict segmentation of movies and run 'run_overlap_mask_prediction.m' for each model before comparing them. Some of the models can be our VGG19_dropout (VGG19D) or U-Net for instance and other new models that users developed.
Pause point: Instructions below are for comparing the multiple models’ segmentation results against the ground truth segmentation labeled in Part 3, so make sure to train and predict segmentation of movies and run 'run_overlap_mask_prediction.m' for each model before comparing them. Some of the models can be our VGG19_dropout (VGG19D) or U-Net for instance and other new models that users developed.
-
a.
-
35.
Draw the learning curves by running the following code located in the “evaluation” folder:
>cd “location of evaluation folder”
>python draw_learning_curve.py
-
36.
Draw bubble plots by running the following code located in the “evaluation” folder:
>cd “location of evaluation folder”
>python bubble_training_curve.ipynb
-
37.
Draw bar graphs and line graphs across different training frames by running the following code in the terminal located in the “evalutation_f1” folder:
>cd “location of evaluation_f1 folder”
>matlab -nodisplay -nosplash -nodesktop -r “run(‘visualize_across_frames_datasets.m’);exit;”
-
38.
Draw edge evolution by running the following code in the terminal located in the “evalutation_f1” folder:
>cd “location of evaluation_f1 folder”
>matlab -nodisplay -nosplash -nodesktop -r “run(‘run_overlap_compare.m’);exit;”
-
39.
Draw violin plots by running the following code in the terminal located in the “evaluation” folder:
>cd “location of evaluation folder”
>matlab -nodisplay -nosplash -nodesktop -r “run(‘run_violin_compare.m‘);exit;”
-
40.
Independent to above MATLAB evaluation codes for segmentation accuracy, trained models’ weights can be visualized by drawing activation maps on each live cell image using SEG-GRAD-CAM (Vinogradova et al., 2020). Visualizing trained model’s weights is helpful for understanding how each layer in the model uses the information of the cell images to segment the cell in the image. Run the following code in the terminal:
>python SegGradCAM/main.py
-
41.
View evaluation results in the “results” or “generated” folder located in the directory where the corresponding code was run.
Part 6. Quantification of morphodynamics of the single cell
Timing: 1 h
This Part can be performed right after Part 4, but we recommend first evaluating the segmentation results in Part 5. This Part uses the segmentation results in the location described in step 22 in Part 4. By verifying the accuracy of the segmentation results, quantification of morphodynamics is more reliable. Window and Protrusion package is developed by Danuser Lab at UT Southwestern (Machacek and Danuser, 2006; Machacek et al., 2009) to quantify morphodynamics of the single cell (Lee et al., 2015; Machacek et al., 2009; Wang et al., 2018). The quantified morphodynamics can be further processed to discover morphodynamics phenotypes (Choi et al., 2021; Ma et al., 2018; Wang et al., 2018, 2021) and causal relationships without molecular perturbations (Noh et al., 2021). For more details on Window and Protrusion package, refer to its GitHub page.
-
42.
For single cell cropping the segmented movie, navigate to the “img_proc” folder and run the following code in the terminal:
>cd “location of img_proc folder”
>python rotate_crop_img.py
-
43.
Download the Window and Protrusion package and in MATLAB, add the downloaded package’s folder as a directory, including its subdirectories.
-
44.
Type the following code in the command window in MATLAB:
>movieSelectorGUI
-
45.When the movieSelectorGUI pops up, click the button “New” to add segmented images as a movie and specify the dataset’s details.
-
a.We specified details of our live cell movies as follows: pixel size is 72 mm, time interval is 5 s, numerical aperture is 1.4, and camera bit depth is 16.
-
a.
-
46.
After clicking “Save” button to save the movie’s information, select the radio button next to Windowing and click “continue”.
-
47.Run the following operations in order: Thresholding, Mask Refinement, Protrusion, Windowing, Protrusion Sampling, Window Sampling.
-
a.Thresholding: choose “External Segmentation” among segmentation methods and specify the location of the folder described in step 22 in Part 4 to load the segmented images.
-
b.Mask Refinement: Use the default settings which are 10 pixels for minimum size, 3 pixels for closure radius, 1 object number, and check marks for fill holes and fill boundary holes.
-
c.Protrusion: Choose Mask Refinement for mask process, 50 for down sampling parameter and 30 for spline tolerance value.
-
d.Windowing: Use the default settings, which are 10 for minimum size of objects to window, 7 pixels for window size parallel and perpendicular to the mask edge, constant number for the number of windows, and 2 for StartContour number.
-
e.Protrusion Sampling: There are no parameters to set.
-
f.Window Sampling: Choose raw images with 1 channel for images to sample.
-
a.
Expected outcomes
Segmented images
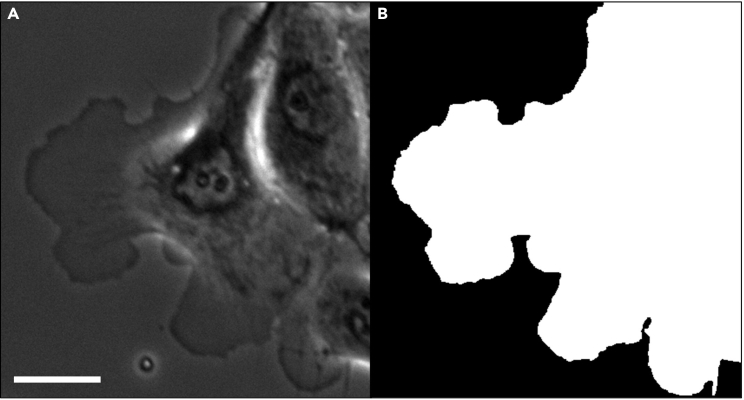
MARS-Net is designed to have segmented images inputted for training and prediction. Figure 3 shows an example of what a single segmented image of a cell should look like after following the Part 3, “Labeling Images”. Note the cell is the white part of the image, and the background is the black part of the image.
Figure 3.
Segmented image example
(A) Original phase contrast image from the dataset being analyzed.
(B) Segmented phase contrast image from the dataset being analyzed. Scale Bar: 32.5 μm.
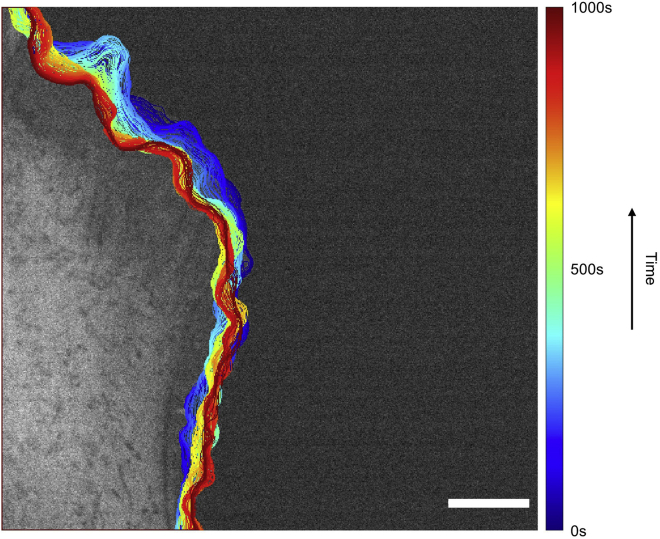
Edge progression
The example output from running “run_overlap_compare.m” in step 38 in Part 5 is shown in Figure 4. This figure shows the progression of cell edges segmented by the deep learning model overlaid on the first frame of the movie. The legend was made separately and is not part of the output.
Figure 4.
Edge progression image example
Edge progression overlaid on the original phase contrast image. Note that the dark blue color indicates initial progression at t=0 s while the dark red indicates the end of the progression at t=1000 s. Scale Bar: 32.5 μm.
Evaluation of segmentation
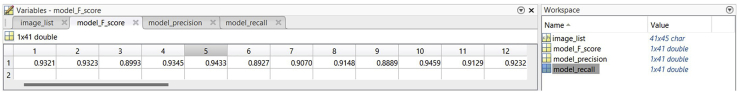
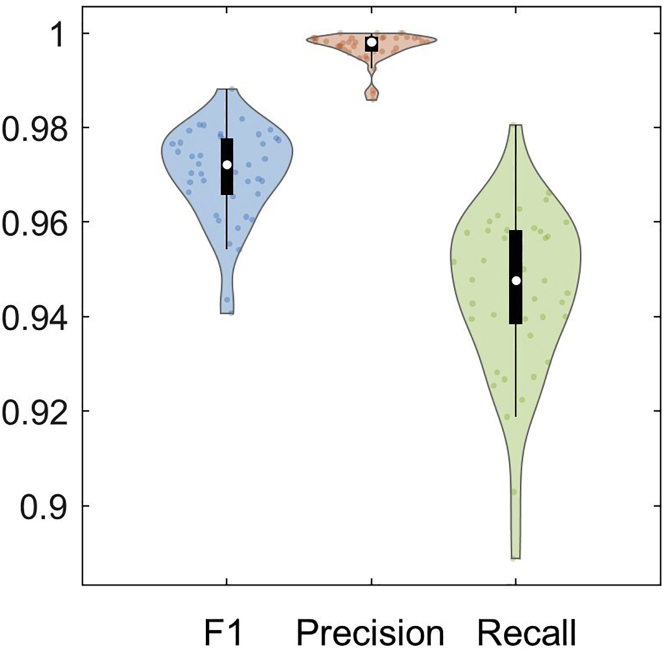
The evaluation results in Recall_Precision_F_score_frame#.mat files contain four variables: image_list, model_F_score, model_precision, and model_recall (Figure 5). Image_list is a list of input image names, and each row of image_list corresponds to the evaluated image name. model_F_score, model_precision, and model_recall contain F1-score, Precision, and Recall values, respectively, from comparing ground truth mask and predicted segmentation mask of the live cell movie. F1-score, precision and recall at around 0.92 on average yields a good enough segmentation but higher is preferred. The F1 score, precision, and recall have the number of columns equal the number of evaluated segmentation masks. The evaluation results are visualized as shown in Figure 6 by running “visualize_a_model.m”.
Figure 5.
Evaluation results viewed in MATLAB
The main window on the left shows the list of values of one variable opened by the user’s double click. There are only 12 values shown in the window, but the user can scroll right to view more values. The workspace on the right shows four variables stored in the .mat file and the size of each variable. The row index of image_list corresponds to the column index of model_F_score, model_precision, and model_recall. There are 41 evaluated images in this example.
Figure 6.
Violin plot of evaluation results
The distribution of F1 score, precision, and recall are shown in the violin plot. There is a boxplot in black within the violin plot with a median indicated by the white circle. Individual evaluated results from 41 frames are plotted as small dots within the violin plot. This model has high precision and low recall, but the performance is good overall, with a median F1-score higher than 0.97.
Morphodynamical quantification
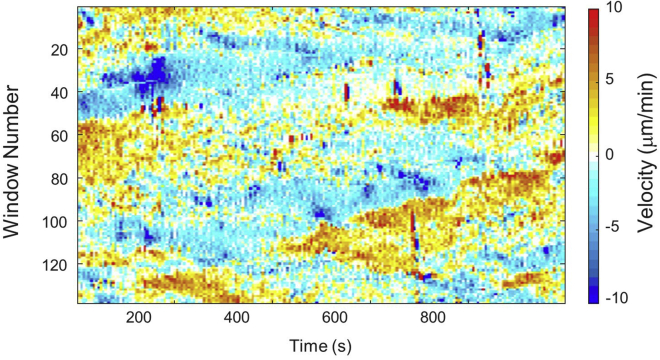
Successful Quantification of Morphodynamics should yield a result similar to Figure 7. The maximum time on the x-axis, the maximum number of windows on the y-axis, and the range of velocity in the heatmap can be different based on the input movie. If you want the colormap or range of velocity in the heatmap, you can change its colormap or scale velocity and threshold outliers in MATLAB.
Figure 7.
Protrusion velocity map example
The heatmap is colored to represent the cell edge velocity ranging from -10 μm/min to +10 μm/min. The red-colored region indicates a portion of the protruding cell edge, and the blue colored region indicates a portion of the retracting cell edge. In cellular dynamics of morphology, protrusion means the cell edge is moving away from the cell body and retraction means the cell edge is moving toward the cell body. The horizontal axis represents the duration of the movie for which the cell was observed, and the vertical axis represents window id or fragments of cell edge. Given a cell edge with two endpoints, the window id 1 represents one endpoint, and the window id 140 represents the other endpoint.
Limitations
MARS-Net segments the entire cluster of cells together as one body and was not tested to identify cell-to-cell boundaries. The quantification of the morphodynamics of a single cell is possible only when the cell does not move outside the cropping box throughout the movie.
Troubleshooting
Problem 1
MARS-Net cannot find input data located in the folder paths specified in UserParams.py. (In step 5 in part 1. Installing MARS-Net).
Potential solution
Double check that your folders and folder paths are written correctly. File separators in the path are different based on the user’s operating system (\ for Windows, / for Linux). Also, check to see what type of image file your images are, and make sure the “img_format” in the “UserParams.py” file is the same. MARS-Net works with .png, .jpg and .tiff.
Problem 2
Failure in running crop.py script for cropping input dataset. (In step 17 in part 4. Training MARS-Net and segmenting movies).
Potential solution
Make sure that the assets folder contains the same number of masks and images. For instance, the error occurs when the image folder has 100 images, and the mask folder has 101 masks.
Also, try converting images and masks to 8-bit grayscale.
Problem 3
CUDA & cuDNN are not installed or incorrect versions are installed in the user’s computer. (In step 18 in part 4. Training MARS-Net and segmenting movies).
When running train.py or predict.py in the command line, the following warning message is shown:
W tensorflow/stream_executor/platform/default/dso_loader.cc:59] Could not load dynamic library ‘cudart64_101.dll’; dlerror: cudart64_101.dll not found.
I tensorflow/stream_executor/cuda/cudart_stub.cc:29] Ignore above cudart dlerror if you do not have a GPU set up on your machine.
Or the following error message is shown:
tensorflow.python.framework.errors_impl.InvalidArgumentError: Default MaxPoolingOp only supports NHWC on device type CPU.
[[node functional_1/block1_pool/MaxPool (defined at train_mars.py:209) ]] [Op:__inference_train_function_3474].
Potential solution
Make sure that the correct CUDA version is installed in a user’s computer for the corresponding TensorFlow version. For instance, TensorFlow v2.3 requires CUDA 10.1 and TensorFlow v1.15 requires CUDA 10.0. The authors downloaded CUDA 10.1 and cuDNN v7.6.5 from the following websites: (https://developer.nvidia.com/cuda-10.1-download-archive-base), (https://developer.nvidia.com/rdp/cudnn-archive).
Installation of CUDA is just as simple as running the .exe file in Windows 10. To install cuDNN, please refer to its installation guides for more details (https://docs.nvidia.com/deeplearning/cudnn/install-guide/index.html).
Problem 4
Not enough GPU memory to train the model. (In step 18 in part 4. Training MARS-Net and segmenting movies).
When running train_mars.py, the following error message is shown:
tensorflow.python.framework.errors_impl.ResourceExhaustedError: OOM when allocating tensor with shape[64,128,128,128] and type float on /job:localhost/replica:0/task:0/device:GPU:0 by allocator GPU_0_bfc.
Potential solution
Ideally, use a GPU with more memory or train with multiple GPUs.
Otherwise, reduce the batch size in UserParams.py. For instance, set train_batch_size = 8 in the get_args(self) function at around line 793. Originally, the authors used 64 as the batch size. Please note that decreasing the batch size might affect the segmentation accuracy since smaller batch size increase the variance during stochastic gradient descent.
Problem 5
Error when running the MATLAB code in the terminal: "-bash: syntax error near unexpected token `('" (In step 32 in part 5. Evaluation of the segmentation results).
Potential solution
This error most commonly occurs when you copy and paste the code to run any MATLAB file. To potentially overcome this error, delete and replace each double quotation mark (i.e., “”) and each single quotation mark (i.e.,”) in the code. Alternatively, type the command without copying and pasting.
Problem 6
Failure in running MATLAB evaluation codes. (In step 29 in part 5. Evaluation of the segmentation results).
Potential solution
Verify that the paths specified in GlobalConfig.m are correct.
Also, “prediction_path_list”, “display_names”, “frame_list” are matrices that should have the same number of rows. In MATLAB’s matrix, semi-colon indicates a new row, and comma indicates a new column.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Kwonmoo Lee (kwonmoo.lee@childrens.harvard.edu).
Materials availability
The authors did not generate any unique reagents in this study.
Acknowledgments
This work was supported by NIH, United States (grant R15GM122012 and R35GM133725).
Author contributions
Writing, J.J., C.H., K.L.; development, J.J.; funding acquisition, K.L.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Junbong Jang, Email: junbongjang@kaist.ac.kr.
Kwonmoo Lee, Email: kwonmoo.lee@childrens.harvard.edu.
Data and code availability
The sample dataset and the code for our deep learning-based segmentation pipeline are available at GitHub (https://github.com/kleelab-bch/MARS-Net) and Zenodo (https://doi.org/10.5281/zenodo.6558761).
References
- Arbelaez P., Maire M., Fowlkes C., Malik J. Contour detection and hierarchical image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2010;33:898–916. doi: 10.1109/TPAMI.2010.161. [DOI] [PubMed] [Google Scholar]
- Bertasius G., Shi J., Torresani L. Deepedge: a multi-scale bifurcated deep network for top-down contour detection. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2015 doi: 10.1109/CVPR.2015.7299067. [DOI] [Google Scholar]
- Canny J. A computational approach to edge detection. IEEE Trans. Pattern Anal. Mach. Intell. 1986;8:679–698. doi: 10.1109/tpami.1986.4767851. [DOI] [PubMed] [Google Scholar]
- Choi H.J., Wang C., Pan X., Jang J., Cao M., Brazzo J.A., 3rd, Bae Y., Lee K. Emerging machine learning approaches to phenotyping cellular motility and morphodynamics. Phys. Biol. 2021;18:041001. doi: 10.1088/1478-3975/abffbe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J., Dong W., Socher R., Li L.-J., Kai L., Li F.-F. IEEE; 2009. ImageNet: A Large-Scale Hierarchical Image Database. [Google Scholar]
- Iglovikov V., Seferbekov S.S., Buslaev A., Shvets A. TernausNetV2: fully convolutional network for instance segmentation. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops. 2018 doi: 10.1109/CVPRW.2018.00042. [DOI] [Google Scholar]
- Jang J., Wang C., Zhang X., Choi H.J., Pan X., Lin B., Yu Y., Whittle C., Ryan M., Chen Y., Lee K. A deep learning-based segmentation pipeline for profiling cellular morphodynamics using multiple types of live cell microscopy. Cell Rep. Methods. 2021;1:100105. doi: 10.1016/j.crmeth.2021.100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.J., Wang C., Zhao B., Im H., Min J., Choi H.J., Tadros J., Choi N.R., Castro C.M., Weissleder R., et al. Deep transfer learning-based hologram classification for molecular diagnostics. Sci. Rep. 2018;8:17003. doi: 10.1038/s41598-018-35274-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Elliott H.L., Oak Y., Zee C.T., Groisman A., Tytell J.D., Danuser G. Functional hierarchy of redundant actin assembly factors revealed by fine-grained registration of intrinsic image fluctuations. Cell Syst. 2015;1:37–50. doi: 10.1016/j.cels.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J., Shelhamer E., Darrell T. Fully convolutional networks for semantic segmentation. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2015 doi: 10.1109/CVPR.2015.7298965. [DOI] [PubMed] [Google Scholar]
- Ma X., Dagliyan O., Hahn K.M., Danuser G. Profiling cellular morphodynamics by spatiotemporal spectrum decomposition. PLoS Comput. Biol. 2018;14:e1006321. doi: 10.1371/journal.pcbi.1006321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machacek M., Danuser G. Morphodynamic profiling of protrusion phenotypes. Biophys. J. 2006;90:1439–1452. doi: 10.1529/biophysj.105.070383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machacek M., Hodgson L., Welch C., Elliott H., Pertz O., Nalbant P., Abell A., Johnson G.L., Hahn K.M., Danuser G. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh J., Isogai T., Chi J., Bhatt K., Danuser G. Inference of Granger-causal relations in molecular systems — a case study of the functional hierarchy among actin regulators in lamellipodia. bioRxiv. 2021 doi: 10.1101/2021.05.21.445144. Preprint at. [DOI] [Google Scholar]
- Ronneberger O., Fischer P., Brox T. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Navab N., Hornegger J., Wells W., Frangi A., editors. Springer, Cham; 2015. U-net: convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- Schneider C., Rasband W., Eliceiri K. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonyan K., Zisserman A. Very deep convolutional networks for large-scale image recognition. Preprint at arXiv. 2015 doi: 10.48550/arXiv.1409.1556. [DOI] [Google Scholar]
- Srivastavanitish N., Hinton G., Krizhevskykriz A., Sutskeverilya I., Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014;15:1929–1958. doi: 10.5555/2627435.2670313. [DOI] [Google Scholar]
- Vaidyanathan K., Wang C., Krajnik A., Yu Y., Choi M., Lin B., Jang J., Heo S.J., Kolega J., Lee K., Bae Y. A machine learning pipeline revealing heterogeneous responses to drug perturbations on vascular smooth muscle cell spheroid morphology and formation. Sci. Rep. 2021;11:23285. doi: 10.1038/s41598-021-02683-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradova K., Dibrov A., Myers G. Towards interpretable semantic segmentation via gradient-weighted class activation mapping (student abstract) Proc. AAAI Conf. Artif. Intell. 2020;34:13943–13944. doi: 10.1609/aaai.v34i10.7244. [DOI] [Google Scholar]
- Wang C., Choi H.J., Kim S.J., Desai A., Lee N., Kim D., Bae Y., Lee K. Deconvolution of subcellular protrusion heterogeneity and the underlying actin regulator dynamics from live cell imaging. Nat. Commun. 2018;9:1688. doi: 10.1038/s41467-018-04030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Choi H.J., Woodbury L., Lee K. Deep learning-based subcellular phenotyping of protrusion dynamics reveals fine differential drug responses at subcellular and single-cell levels. bioRxiv. 2021 doi: 10.1101/2021.05.25.445699. Preprint at. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sample dataset and the code for our deep learning-based segmentation pipeline are available at GitHub (https://github.com/kleelab-bch/MARS-Net) and Zenodo (https://doi.org/10.5281/zenodo.6558761).