Abstract
Background:
Presumed serious infection (PSI) is defined as a blood culture drawn and new antibiotic course of at least 4 days among pediatric patients with Central Venous Lines (CVLs). Early PSI prediction and use of medical interventions can prevent adverse outcomes and improve the quality of care.
Methods:
Clinical features including demographics, laboratory results, vital signs, characteristics of the CVLs and medications used were extracted retrospectively from electronic medical records. Data were aggregated across all hospitals within a single pediatric health system and used to train machine learning models (XGBoost and ElasticNet) to predict the occurrence of PSI 8 hours prior to clinical suspicion. Prediction for PSI was benchmarked against PRISM-III.
Results:
Our model achieved an area under the receiver operating characteristic curve of 0.84 (95% CI = [0.82, 0.85]), sensitivity of 0.73 [0.69, 0.74], and positive predictive value (PPV) of 0.36 [0.34, 0.36]. The PRISM-III conversely achieved a lower sensitivity of 0.19 [0.16, 0.22] and PPV of 0.30 [0.26, 0.34] at a cut-off of >= 10. The features with the most impact on the PSI prediction were maximum diastolic blood pressure prior to PSI prediction (mean SHAP = 3.4), height (mean SHAP = 3.2), and maximum temperature prior to PSI prediction (mean SHAP = 2.6).
Conclusion:
A machine learning model using common features in the electronic medical records can predict the onset of serious infections in children with central venous lines at least 8 hours prior to when a clinical team drew a blood culture.
Keywords: Machine learning, infection, CLABSI, predictive model, sepsis
Introduction
Children with central venous lines (CVLs) are at high risk of morbidity and mortality from hospital acquired infections (HAI), including central-line associated bloodstream infections (CLABSIs) and sepsis. While specific definitions for these entities exist in pediatrics, they often have inadequate sensitivity for clinically important infections and may be difficult to generalize across electronic medical record (EMR) platforms [1–2]. The presumed serious infection (PSI) case definition was developed initially by adult sepsis epidemiologists to allow for retrospective surveillance of infection and organ dysfunction that could be applied across diverse EMRs [3–5]. It is defined as at least one blood culture draw followed by at least four consecutive days (or fewer if the patient dies or is transferred out) of antimicrobial agents that were not administered in the week prior to the blood culture draw. The definitions for PSI, as well as organ dysfunction, were adapted for pediatrics by Hsu et al. [6] and have been validated [7]. Successful prediction of PSI, or sepsis in general, among hospitalized children or the adult population could lead to decreased costs while improving the quality of care [8–10].
Many serious infections and adverse outcomes in hospitalized children are likely preventable through interventions that prevent PSIs or identify them early to initiate antimicrobial therapy. On the other hand, excessive use of antimicrobials can lead to adverse events and worsening antimicrobial resistance. In this setting, predictive models to identify patients at the highest risk for serious infections can help clinicians better achieve the balance between early intervention and antimicrobial overuse.
Machine learning models have been used to address clinical problems [11–13]. Most of these models have employed biomarkers or clinical risk predictions to predict the onset of events [14–15]. The deployment of machine learning models and early recognition of the adverse events decreased the mortality and morbidity among patients [4,16]. However, there are still open challenges about developing machine learning models for low prevalence outcomes [17]. Within the pediatric domain, a recent study reported the incidence of CLABSI in pediatric cardiac ICUs to be 0.32% among children aged between 1–18 years [18], representing tremendous challenge for classical machine learning techniques which assume a balanced distribution of cases and controls. These limitations suggest that there is a need to develop robust machine learning algorithms that can adequately predict low prevalence conditions, particularly for pediatric cohorts.
Class-imbalanced data classification is a challenge of training predictive models in many fields such as the medical domain. Studies have been done in this area, and different algorithms have been proposed [19–21]. The proposed solutions mainly include sampling techniques [22–25], such as oversampling the minority class or undersampling the majority class, and employing ensemble learning approaches [26].
This paper introduces our proposed framework named pBoost which was developed based on our adapted study design and contributes both clinically and technically to the literature. From the clinical perspective, we incorporated a novel case definition to identify the pediatric patient with CVL who were at a higher risk of experiencing a serious infection episode during their hospitalization time. From the technical point of view, we proposed a framework that can perform a series of tasks such as data engineering, data preprocessing and feature transformation to model hyperparameter tuning, providing multiple performance metrics and the most influential features. We further highlight clinical features which contributed the PSI prediction using data from the EMR. This study was conducted according to Emory University protocol number 19–012.
Methods
We proposed pBoost framework which performs multiple tasks; feature engineering, data preprocessing (e.g., feature transformation, missing values imputation, removing multicollinearity, etc.), optimize the classifier setting with Bayesian optimization technique, provide performance metrics and the features with the most significant effect on the model’s decision-making process. The GitHub repository for pBoost framework is publicly available.
We performed a retrospective cohort study of all hospitalized patients with a CVL at a single pediatric health system. Patients were included in the cohort if they were admitted to one of three freestanding children’s hospitals between January 1st, 2013 and December 31st, 2018 and had a central line documented in the system before or at the time of admission or received at least one CVL during the hospitalization. We extracted data routinely available across electronic health records systems including demographics, vital signs, laboratory values, prior diagnoses, medication administrations, microbiology results, respiratory support, CVL properties, and CVL care documentation. The full set of data collected are listed in Appendix A.
PSI T0 time is defined as the first specimen collection time associated with a blood culture after a central line is inserted, followed by four days of new antibiotic administration. For the patients who already had a CVL at the time of admission, we considered all the blood cultures on their records. For patients who had a CVL placed during the hospitalization, we only included blood cultures after their first central line was inserted (Figure 1-A and 1-B). A patient may develop multiple PSIs during the stay at the hospital. We disregarded the first and last 12 hours of the hospitalization then selected a random event time in the intermediate period to limit selection bias from specific admission/discharge workups.
Figure 1. Summary of the methods.
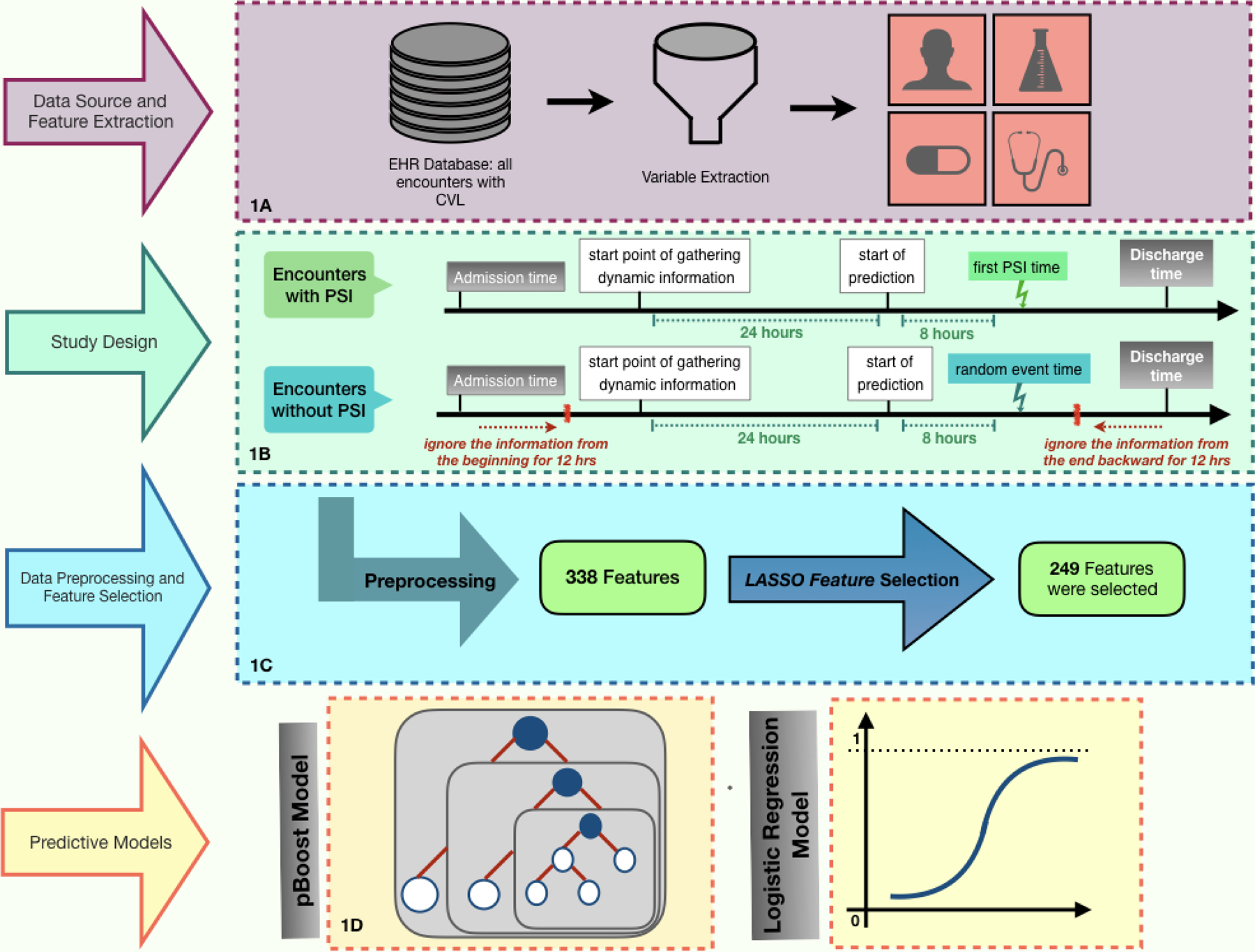
(1-A) A cohort of patients with CVL and demographic, clinical, and laboratory characteristics were extracted from the EMR database. (1-B) If an encounter has a PSI, the time of PSI (tPSI) is marked as the event time and the prediction time is set at 8 hours prior (tPSI- 8 hours). Minimum and maximum of the time-variant variables, such as laboratory results and vital signs, are considered from 24 hours prior to the prediction time. If a patient encounter does not have a PSI, the first and last 12 hours of patient information is ignored. Then a random point in time is selected as the hypothetical event time, and the prediction time is set at 8 hours prior. (1-C) There were 338 features extracted in the study. To eliminate the features without a significant effect on the outcome, we applied LASSO feature selection, which led to a 249-dimensional feature space. (1-D) XGBoost and ElasticNet models were employed to predict if a patient will develop PSI during the next 8 hours from the prediction time using the selected 249 features. Cross-validation and Bayesian optimization were applied in the training process of both models in order to find the best settings of the predictive models and avoid overfitting.
Data was preprocessed, and machine learning features developed as described in Supplemental Appendix A and Figure 1-C. Model development was performed using XGBoost [27] and logistic regression with a L1L2 regularization also known as ElasticNet [28] (Figure 1-D). ElasticNet has the ability to identify more important features and penalize the less informative ones, and XGBoost is a scalable tree-based boosting technique that is popular for supervised machine learning involving highly imbalanced (i.e. low prevalence) and missing data. Additional details on the machine learning algorithms are described in Supplement Appendix C.
We calculated Area Under the Receiver Operating Characteristic curve (AUC) and Area Under the Precision-Recall Curve (AUCpr) performance metrics. AUC is a commonly used measure to present binary classification results. In the presence of a class-imbalanced dataset, AUCpr can provide a more informative insight into the classifier’s performance relevant to clinician decision-making. We then calculated the true positive and true negative classification rates for each classification model.
We evaluated the importance of variables in the machine learning method by using SHaply Additive exPlanations (SHAP) [29–30] values for pBoost which is a validated interpretability tool for machine learning models that provides the average marginal contribution of each feature to the prediction. Equation 1 demonstrates the SHAP calculations in which f is the classification model, F is the complete set of features, S is a subset of F, and feature i is the feature that we want to calculate its contribution to the outcome of the classification model. fSU{i}(xSU{i}) is the classification outcome using S and feature i while fS(xS) is the outcome of the model withholding feature i. Therefore, the classification results with all possible combinations of feature in F, with and without feature i, is calculated and weighted by . Then, ɸi will be the marginal contribution of feature i on the outcome of the classification model f and represents the SHAP value of feature i.
| (Equation 1) |
We compared the performance of the pBoost model to PRISM-lll [31]. We calculated PRISM-III during the 24-hour interval prior to the PSI prediction start time. As demonstrated in Figure 1-B, the 24-hour time period before the start of the prediction is the same interval that the dynamic features for the ElasticNet and pBoost models are collected.
To assess the performance of PRISM-III in predicting PSI during the next 8 hours, we calculated the performance metrics of PRISM-lll to predict PSI using different cut-off values on the training dataset. We then applied the optimal cut-off values on the testing dataset for performance comparisons.
This manuscript was prepared using the guidelines provided by Leisman et al. [32] for reporting of prediction models.
Results
We initially screened a total of 97,424 patient encounters associated with 15,704 patients, of which 65,766 encounters were excluded due to having length-of-stay less than 24 hours. After applying the exclusion criteria mentioned in section 3, a total of 27,137 unique encounters were thus eligible to be included in the analysis. A total of 2,749 neonates, 4,076 infants, 5,580 toddlers and preschoolers, 6,500 children, and 8,232 adolescents met eligibility criteria. Figure A-1 in Supplement Appendix A presents the CONSORT diagram for this study.
We observed a statistically significant difference between the median of age, weight, height, length of stay (LOS), and race in PSI and non-PSI groups, while there was no statistical difference in patients’ sex between PSI and non-PSI groups (Table 1). The median age of patients in the PSI group was 2.9 years (IQR = [0.18, 11.8]), whereas in the non-PSI group, the median age was 6.5 years (IQR = [1.3, 13.6]). The median LOS for a patient with PSI was 30 days (IQR = [18.4, 54.2]), compared to 4.8 days (IQR = [3.1, 9.3]) for non-PSI.
Table 1.
Cohort characteristics
| PSI | non-PSI | p-value | |
|---|---|---|---|
| Age (years) | |||
| (Median [25th, 75th]) | 2.9 [0.2, 11.8]* | 6.5 [1.3, 13.6] | < 0.001 |
|
| |||
| Weight (Kg) | |||
| (Median [25th, 75th]) | 13.1 [3.8, 37.6]* | 20.9 [9.5, 46.2] | < 0.001 |
|
| |||
| Height (cm) | |||
| (Median [25th, 75th]) | 89.9 [51.9, 142.3]* | 115 [72.5, 153.5] | < 0.001 |
|
| |||
| Length of Stay (LOS) | |||
| (Median [25th, 75th]) | 30 [18.4, 54.2]* | 4.8 [3.1, 9.3] | < 0.001 |
|
| |||
| Gender | |||
| Male (%) | 45.9% | 45.4% | 0.52 |
|
| |||
| Race | |||
| Asian (%) | 3.4% | 4% | 0.07 |
| Caucasian (%) | 49.7% * | 54.9% | < 0.001 |
| African American (%) | 41% * | 35.3% | < 0.001 |
| American Indian or Alaska Native (%) | 0.3% | 0.2% | 0.22 |
| Native Hawaiian or Pacific Islander (%) | 0.1% | 0.2% | 0.21 |
| Other (%) | 5.5% | 5.4% | 0.79 |
|
| |||
| Insurance Status | |||
| Commercial (%) | 33.7% * | 40.2% | < 0.001 |
| Public - Medicaid (%) | 62.9% * | 55.8% | < 0.001 |
| Public - non-Medicaid (%) | 2.8% | 3.1% | 0.47 |
| Self-pay (%) | 0.6% * | 0.9% | 0.02 |
|
| |||
| ICU Admission (%) | 70.5% * | 41.1% | < 0.001 |
|
| |||
| Placed on Extracorporeal Membrane Oxygenation (%) | 8.3% * | 1.3% | < 0.001 |
|
| |||
| Mortality (%) | 0.08% | 0.05% | 0.53 |
Statistically significant difference between PSI positive and negative groups.
The results of the classifiers are presented in Figure 2. pBoost performed best in the testing dataset, with an average AUC of 0.84 (95% CI = [0.82, 0.85]) and AUCpr of 0.52 [0.51, 0.53] compared to the AUC of 0.79 [0.78, 0.79] And AUCpr of 0.38 [0.36, 0.39] for ElasticNet (p < 0.001). When fixing the specificity of both models at 0.80, the sensitivity of pBoost was 0.73 [0.69, 0.74] compared to sensitivity of 0.64 [0.62, 0.66] for ElasticNet (p < 0.001). The PPV of the pBoost model, 0.36 [0.34, 0.36], was slightly higher that the ElasticNet 0.33 [0.32, 0.34] (p < 0.001). However, the NPVs were nearly the same (pBoost 0.94 [0.94, 0.95], ElasticNet 0.93 [0.93, 0.94], p < 0.001).
Figure 2. The predictive models’ performance.
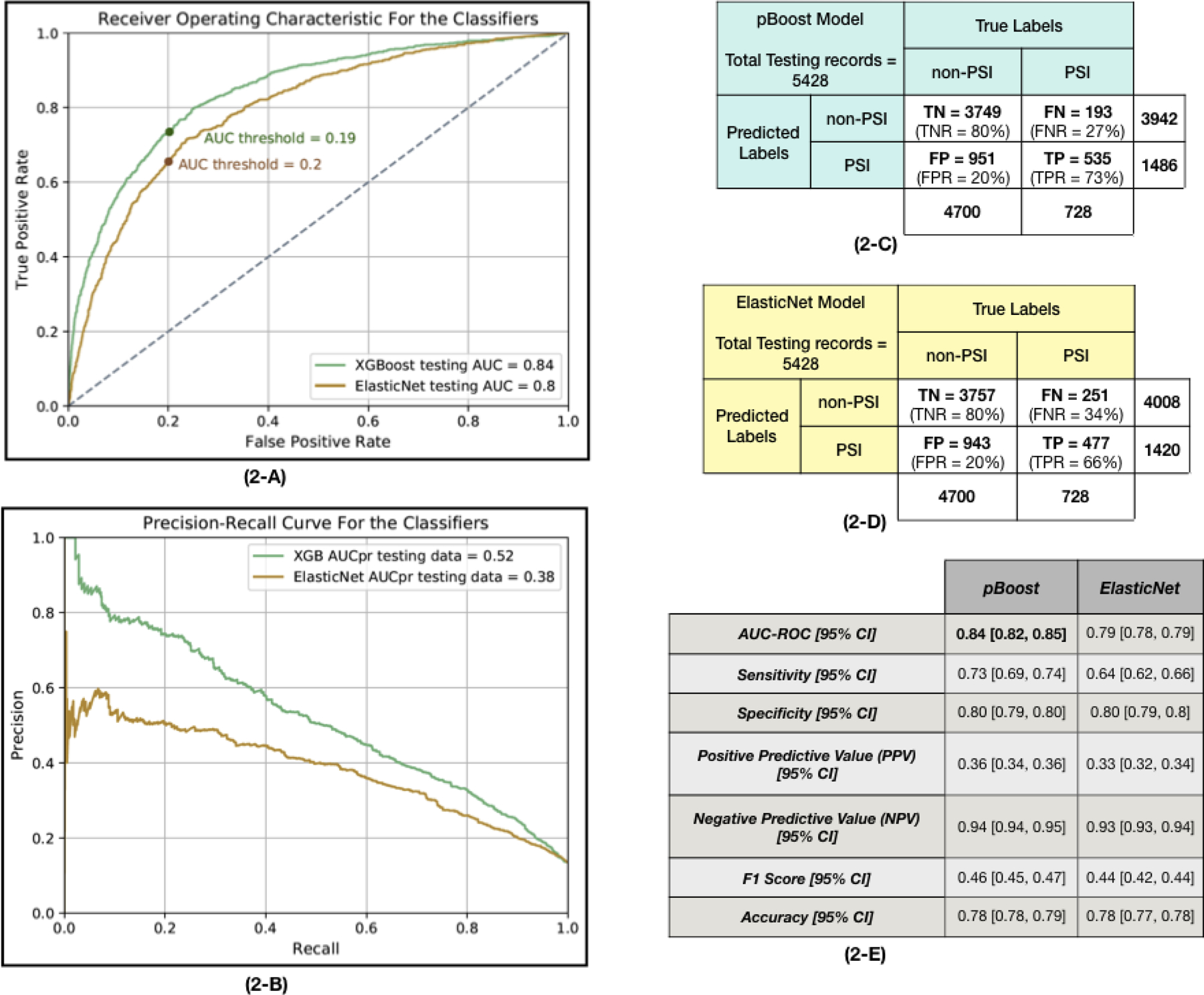
(2-A) The Receiver Operating Characteristic (ROC) plot for pBoost and ElasticNet models applied to the testing dataset. This plot demonstrates the trade-off between the sensitivity and specificity of the classifiers. To find the optimum AUC threshold, the specificity is fixed at 0.80 and the rest of the metrics are calculated. The selected AUC threshold is marked on each curve. (2-B) Precision-Recall curve (PRC) for pBoost and ElasticNet models applied to the testing dataset. A PRC plots the positive predictive value (precision or PPV in the y-axis) against the true positive rate (recall or sensitivity in the x-axis). In class-imbalanced data classification, it is more informative to look at both ROC and PRC to consider the trade-off between PPV and sensitivity. (2-C) The confusion matrix of applying the pBoost model on the testing dataset. This table presents the True Positive (TP), True Negative (TN), False Positive (FP) and False Negative (FN) values that are calculated based on the optimal AUC threshold marked on the ROC plots in part 2-A of this figure. The total number of PSI and non-PSI records and the total number of predicted labels of each class are mentioned in the confusion matrix. (2-D) The confusion matrix associated with the ElasticNet classifier. (2-E) Two predictive models, ElasticNet and pBoost, are employed to predict if a given patient encounter will develop PSI during the next 8 hours of hospital stay. The AUC values are reported for testing subsets of the data. To make a better comparison among the results, the specificity level is fixed at 0.80 and the rest of the performance measurements are calculated subsequently. The mean and 95% confidence interval of each metric are reported.
Comparing the confusion matrices of pBoost and ElasticNet models in Figure 2, true positive cases increased while false positive and false negative cases decreased when employing pBoost instead of ElasticNet model.
Explanability:
We identified which features contributed most to the prediction of PSI (Figure 3). The maximum value of the diastolic blood pressure in the 24 hours prior to the prediction period was on average the leading predictor of PSI risk. The next most important features were height, maximum temperature, minimum Hemoglobin and minimum pulse oximetry. Multiple Complete Blood Count (CBC) components were also important for PSI prediction.
Figure 3. Feature importance from pBoost model.
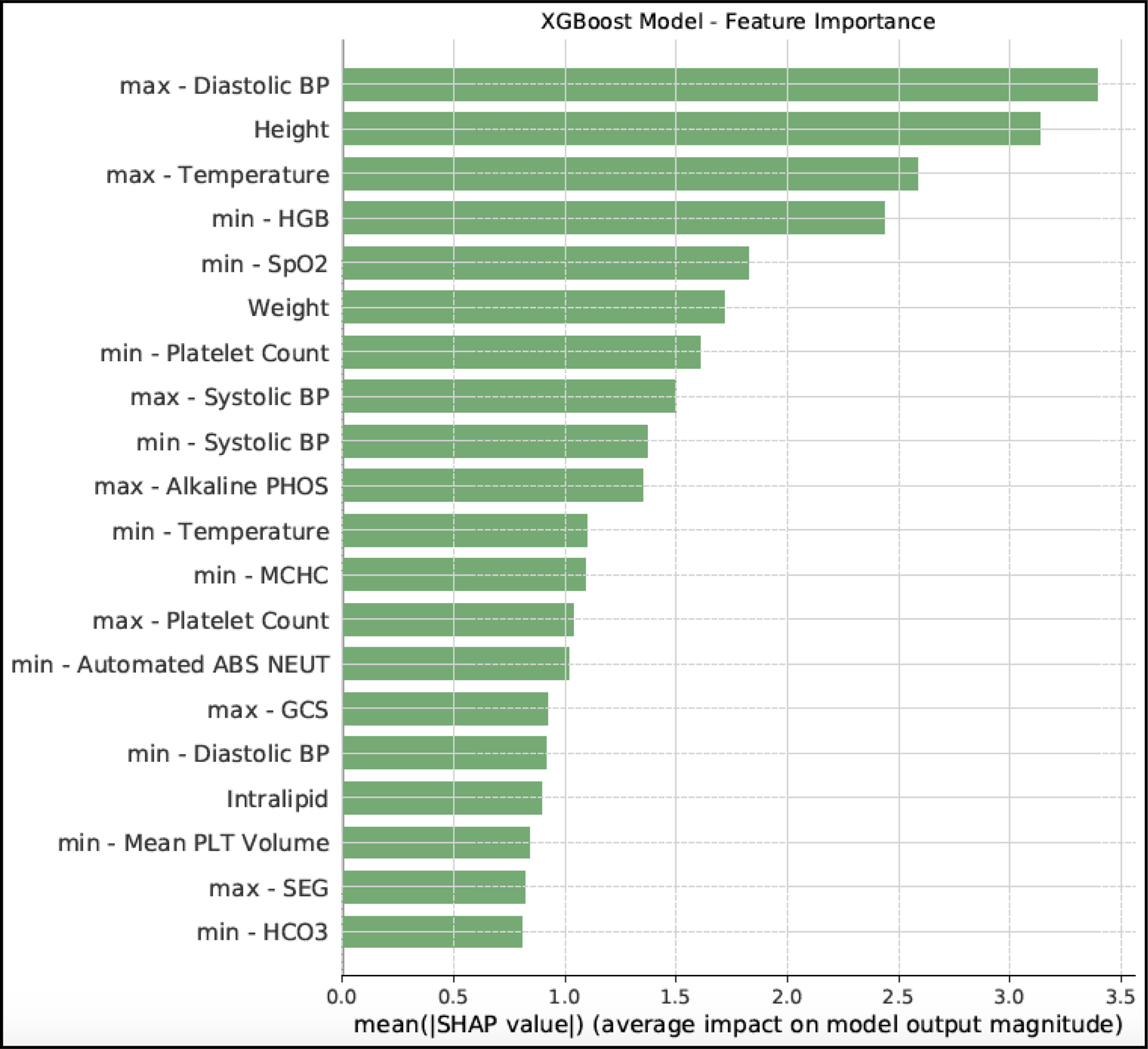
This figure presents the top 20 features according to the pBoost model using SHAP values. In tree-based models, there are different criteria to sort features based on their effect on the outcome. Employing SHAP values in tree-based models, such as pBoost, is the most reliable and consistent way to calculate feature importance. SHAP values do not provide the direction of the effect, so there is no information regarding if an important feature positively or negatively affects the outcome. The min or max mentioned in the feature names, refers to the minimum or maximum of the time-variant variables, such as laboratory results or vital signs, during the 24 hours prior to the PSI prediction time.
Comparison to PRISM-III:
Table 2 demonstrates the performance of PRISM-III in predicting PSI. At a PRISM-III score of >= 10, the sensitivity was 0.19 [0.16, 0.22], a drop of 54% compared to pBoost, the PPV (0.30 [0.26, 0.34]) was reduced by 6%, and the NPV (0.88 [0.87, 0.90]) was 6% worse for the same comparison. Reducing the cut-off to >= 5 improved the sensitivity (0.48 [0.44, 0.51]); however, there was an 13% drop in PPV (0.17 [0.15, 0.19]) and 1% improvement in NPV (0.89 [0.88, 0.90]).
Table 2.
Performance of PRISM-III score as the clinical benchmark to predict PSI in the next 8 hours of hospitalization
| cut-off value = 5 |
cut-off value = 6 |
cut-off value = 7 |
cut-off value = 8 |
cut-off value = 9 |
cut-off value = 10 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Train | Test | Train | Test | Train | Test | Train | Test | Train | Test | Train | Test | |
|
Sensitivity Mean [95% CI] |
0.50 [0.48, 0.52] | 0.48 [0.44, 0.51] | 0.45 [0.43, 0.47] | 0.44 [0.40, 0.47] | 0.35 [0.34, 0.37] | 0.34 [0.30, 0.38] | 0.29 [0.27, 0.31] | 0.28 [0.24, 0.31] | 0.23 [0.22, 0.25] | 0.22 [0.19, 0.25] | 0.19 [0.17, 0.20] | 0.19 [0.16, 0.22] |
|
Specificity Mean [95% CI] |
0.64 [0.63, 0.64] | 0.64 [0.62, 0.65] | 0.68 [0.67, 0.69] | 0.68 [0.67, 0.70] | 0.82 [0.81, 0.82] | 0.82 [0.80, 0.83] | 0.86 [0.86, 0.87] | 0.87 [0.86, 0.88] | 0.90 [0.90, 0.91] | 0.91 [0.90, 0.91] | 0.93 [0.93, 0.94] | 0.93 [0.92, 0.94] |
|
Positive Predictive Value (PPV) Mean [95% CI] |
0.18 [0.17, 0.18] | 0.17 [0.15, 0.19] | 0.18 [0.17, 0.19] | 0.18 [0.16, 0.19] | 0.23 [0.22, 0.24] | 0.22 [0.20, 0.25] | 0.25 [0.23, 0.26] | 0.24 [0.21, 0.27] | 0.27 [0.25, 0.29] | 0.27 [0.23, 0.30] | 0.29 [0.27, 0.31] | 0.30 [0.26, 0.34] |
|
Negative Predictive Value (NPV) Mean [95% CI] |
0.89 [0.89, 0.90] | 0.89 [0.88, 0.90] | 0.89 [0.88, 0.89] | 0.89 [0.88, 0.90] | 0.89 [0.89, 0.90] | 0.89 [0.88, 0.90] | 0.89 [0.88, 0.89] | 0.89 [0.88, 0.89] | 0.88 [0.88, 0.89] | 0.88 [0.87, 0.89] | 0.88 [0.88, 0.89] | 0.88 [0.87, 0.90] |
|
F1 Score Mean [95% CI] |
0.26 [0.25, 0.27] | 0.25 [0.23, 0.27] | 0.26 [0.24, 0.27] | 0.26 [0.23, 0.27] | 0.28 [0.27, 0.29] | 0.27 [0.24, 0.30] | 0.27 [0.25, 0.28] | 0.26 [0.23, 0.29] | 0.25 [0.23, 0.26] | 0.24 [0.21, 0.27] | 0.23 [0.21, 0.25] | 0.23 [0.20, 0.27] |
On the training dataset, different cut-off points are examined for the PRISM-III threshold such that if a PRISM-III score of a patient is equal or greater than that threshold, then the predicted value will be that patient will develop PSI in the next 8 hours of hospital stay. We selected the PRISM-III cut-off values that lead to better sensitivity and specificity values. Then, check the selected threshold performances on the testing dataset. The mean and 95% confidence interval for each metric is presented in this table.
Sensitivity analysis of age groups:
We compared PSI prevalence and model performance across all age groups. The highest and lowest PSI prevalence, 25.5% and 9.7%, were observed in neonates and children, respectively. We investigated the performance of pBoost model on each age group broken down by male and female (Tables F-1 to F-6 in Supplemental Digital Content). The pBoost model performed best on adolescents with average AUC of 0.84 [0.83, 0.85] with slight drop in performance in other age groups; an average AUC of 0.81 [0.80, 0.83] for children, 0.80 [0.78, 0.82] for toddlers and infants, and 0.74 [0.72, 0.76] for neonates.
Discussion
In this study, we developed a novel algorithm to predict the risk of PSI among hospitalized children using readily available features from the EMR. The pBoost model has a high NPV and a PPV of nearly three times the baseline PSI prevalence. We found that diastolic blood pressure, height, temperature, and CBC components were the most useful clinical variables for predicting PSI, consistent with other measures of patient acuity. The pBoost model is based on the XGBoost method, which enhances prediction of low prevalence events through boosting, where misclassified cases from one round of model training are weighted more heavily in subsequent rounds. This approach outperformed the ElasticNet model which does not have any boosting structure. Both models had statistically superior performance in predicting PSI compared to PRISM-lll alone. While PRISM-III score is a well-established clinical benchmark, we included more features in the pBoost framework that may help with boosting the prediction performance of the machine learning model. Moreover, a machine learning model does not have the limitation of the clinical benchmark and can learn the patterns for PSI positive and negative patient conditions regardless of their PRISM-III score. The superior performance of the pBoost framework demonstrated the potential of machine learning techniques to inform infection prevention and management.
PSI is a relatively new definition of serious infection compared to sepsis and CLABSI. However, its simplicity and ease of implementation is advantageous particularly in pediatrics. In our study, we found that patients with PSI had a longer LOS (median of 30 days vs. 4.8 days, p-value < 0.001), were more likely to be admitted to the ICU (70.5% vs. 41.1%, p-value < 0.001), more likely to be placed on extracorporeal membrane oxygenation (8.3% vs. 1.3%, p-value < 0.001), and a higher mortality rate (0.08% vs. 0.05%, p-value = 0.53). We also found that PSI was more common in African Americans (41% vs. 35.3%, p-value < 0.001) and those with Medicaid insurance (0.62.9% vs. 55.8%, p-value < 0.001), consistent with health disparities seen in adult patients with sepsis [33]. Thus, efforts aimed at predicting, preventing and managing PSI are likely to improve outcomes for children.
Diastolic blood pressure had the largest contribution to the PSI prediction, consistent with prior studies of early predictors of pediatric sepsis [34]. Other studies in pediatric or adult sepsis prediction identified body temperature [35–36], hemoglobin [37], SpO2 [36], platelet count [37] and systolic blood pressure [38] as the features with significant impacts on sepsis prediction, which were aligned with our findings from investigating important features by mean SHAP values.
The predictive performance of pBoost as measured by AUC was superior to other models of pediatric sepsis relying on EMR data alone. Le et al. [36] and Desautels et al. [39] achieved an AUC of 0.73 to predict pediatric severe sepsis four hours prior to the onset using boosted ensembles of decision trees. Several factors may contribute to higher AUC in our model; first, PSI has higher prevalence than severe sepsis and may be easier to predict. Second, we restricted our population to patients with CVL which increases the prevalence of the serious infection and may facilitate prediction. Finally, in our study we used a much broader range of features which may provide additional predictive power compared to vital signs and a small set of laboratory values.
Limitations:
Our study has several important limitations. First, even though our data was derived from a large multi-hospital tertiary pediatric health system, it represents a single instance of the EMR and may reflect workflows specific to this setting. Therefore, any practices and procedures that are unique to the system may bias the outcomes of the model if applied to an external site. Second, we investigated only two methods of predictive modeling specifically to handle imbalanced (i.e. low prevalence) data. Deep learning approaches may improve on the performance achieved with XGBoost and ElasticNet. We also did not consider unstructured data, which may contain further information about the patient’s acuity and infection risk. This represents a future opportunity to improve on the performance of our model. Third, our patient cohort represents a highly diverse population that is unique to the South, U.S.A, and therefore may limit the generalizability to other clinical contexts. Finally, our approach used a lookback method in which we compared performance of the model 8 hours prior to known PSI events with 8 hours randomly selected from encounters without PSI. This “lookback” method is a standard practice in the development and validation of many predictive models, but it nonetheless increases the prevalence of the event in our sample compared to what would be observed clinically moving forward in time. Thus, the PPVs of our models may be higher than what might be seen in prospective validation where predictions may be assessed at regular time intervals throughout a hospitalization.
Recent studies have suggested that machine learning algorithms can outperform clinical rules-based criteria [40]. In our study, the pBoost achieved a sensitivity and PPV (0.73 [0.69, 0.74] and 0.36 [0.34, 0.36]) higher than the benchmark using PRISM-lll (0.19 [0.16, 0.22] and 0.30 [0.26, 0.34], p-values < 0.001) at a cut-off of >= 10. This indicates that the improved performance of pBoost can meaningfully improve clinical decision making by alerting clinicians earlier, thereby allowing for more rapid intervention and by that potentially improving outcomes.
Conclusions
pBoost, a novel machine learning model using routinely available information in the EMR, predicted the occurrence of PSI in the next 8 hours for hospitalized patients with CVL with reasonable sensitivity, specificity, and PPV, outperforming PRISM-III. Diastolic blood pressure, height, temperature, other hemodynamic variables, and CBC components were the most useful features for predicting PSI. Integrating natural language processing features and/or blood-based biomarkers used in pediatric sepsis may improve the predictive performance [41–42]. Future studies evaluating this and related models prospectively [43] could improve outcomes for patients with CVLs through targeted efforts to prevent and treat infections as early as possible while minimizing adverse events.
Supplementary Material
Acknowledgments
We would like to thank Children’s Healthcare of Atlanta and The Pediatric Technology Center at Georgia Tech, in conjunction with the Health Analytics Council for the generous grant, which supported this work. GC is also supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under Award Number UL1TR002378. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Larsen EN, Gavin N, Marsh N, et al. A systematic review of central-line–associated bloodstream infection (CLABSI) diagnostic reliability and error. Infection Control & Hospital Epidemiology 2019. Oct;40(10):1100–6. [DOI] [PubMed] [Google Scholar]
- [2].Bagchi S, Watkins J, Pollock DA, et al. State health department validations of central line–associated bloodstream infection events reported via the National Healthcare Safety Network. American Journal of Infection Control 2018. Nov 1;46(11):1290–5. [DOI] [PubMed] [Google Scholar]
- [3].Rhee C, Dantes R, Epstein L, et al. Incidence and trends of sepsis in US hospitals using clinical vs claims data, 2009–2014. JAMA 2017. Oct 3;318(13):1241–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Rhee C, Kadri S, Huang SS, et al. Objective sepsis surveillance using electronic clinical data. Infection Control & Hospital Epidemiology 2016. Feb;37(2):163–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Rhee C, Dantes RB, Epstein L, et al. Using objective clinical data to track progress on preventing and treating sepsis: CDC’s new ‘Adult Sepsis Event’surveillance strategy. BMJ Quality & Safety 2019. Apr 1;28(4):305–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hsu HE, Abanyie F, Agus MS, et al. A National Approach to Pediatric Sepsis Surveillance. Pediatrics 2019. Dec 1;144(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Weiss SL, Balamuth F, Chilutti M, et al. Identification of Pediatric Sepsis for Epidemiologic Surveillance Using Electronic Clinical Data. Pediatr Crit Care Med 2020. Feb 1;21(2):113–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Miller MR, Griswold M, Harris JM, et al. Decreasing PICU catheter-associated bloodstream infections: NACHRI’s quality transformation efforts. Pediatrics 2010. Feb 1;125(2):206–13. [DOI] [PubMed] [Google Scholar]
- [9].Reyna MA, Josef C, Seyedi S, et al. Early prediction of sepsis from clinical data: the PhysioNet/Computing in Cardiology Challenge 2019. Comput Cardiol (2019) 2019. Sep 8 (pp. Page-1). IEEE. [Google Scholar]
- [10].Nemati S, Holder A, Razmi F, et al. An interpretable machine learning model for accurate prediction of sepsis in the ICU. Crit Care Med 2018. Apr;46(4):547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Le S, Hoffman J, Barton C, et al. Pediatric severe sepsis prediction using machine learning. Front Pediatr 2019. Oct 11;7:413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Desautels T, Calvert J, Hoffman J, et al. Prediction of sepsis in the intensive care unit with minimal electronic health record data: a machine learning approach. JMIR Med Inform 2016;4(3):e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Biassoni R, Di Marco E, Squillario M, et al. Gut Microbiota in T1DM-Onset Pediatric Patients: Machine-Learning Algorithms to Classify Microorganisms as Disease Linked. Int J Clin Endocrinol Metab 2020. Sep;105(9):dgaa407. [DOI] [PubMed] [Google Scholar]
- [14].Shin EK, Mahajan R, Akbilgic O, et al. Sociomarkers and biomarkers: predictive modeling in identifying pediatric asthma patients at risk of hospital revisits. NPJ Digit Med 2018. Oct 2;1(1):1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Iwasawa K, Suda W, Tsunoda T, et al. Dysbiosis of the salivary microbiota in pediatric-onset primary sclerosing cholangitis and its potential as a biomarker. Scientific Reports 2018. Apr 3;8(1):1–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Raita Y, Camargo CA, Macias CG, et al. Machine learning-based prediction of acute severity in infants hospitalized for bronchiolitis: a multicenter prospective study. Scientific Reports 2020. Jul 3;10(1):1–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Schaefer J, Lehne M, Schepers J, et al. The use of machine learning in rare diseases: a scoping review. Orphanet J Rare Dis 2020. Dec;15(1):1–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Alten JA, Rahman AF, Zaccagni HJ, et al. The epidemiology of health-care associated infections in pediatric cardiac intensive care units. Pediatr Infect Dis J 2018. Aug;37(8):768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Li C, Liu S. A comparative study of the class imbalance problem in Twitter spam detection. Concurrency and Computation: Practice and Experience 2018. Mar 10;30(5):e4281. [Google Scholar]
- [20].Hassan AK, Abraham A. Modeling insurance fraud detection using imbalanced data classification. In Advances in nature and biologically inspired computing 2016. (pp. 117–127). Springer, Cham. [Google Scholar]
- [21].Fotouhi S, Asadi S, Kattan MW. A comprehensive data level analysis for cancer diagnosis on imbalanced data. Journal of biomedical informatics 2019. Feb 1;90:103089. [DOI] [PubMed] [Google Scholar]
- [22].Kubat M, Matwin S. Addressing the curse of imbalanced training sets: one-sided selection. InIcml 1997. Jul 8 (Vol. 97, pp. 179–186). [Google Scholar]
- [23].Japkowicz N The class imbalance problem: Significance and strategies. InProc. of the Int’l Conf. on Artificial Intelligence 2000. Jun 26 (Vol. 56). [Google Scholar]
- [24].Lewis DD, Catlett J. Heterogeneous uncertainty sampling for supervised learning. InMachine learning proceedings 1994 1994. Jan 1 (pp. 148–156). Morgan Kaufmann. [Google Scholar]
- [25].Ling CX, Li C. Data mining for direct marketing: Problems and solutions. InKdd 1998. Aug 27 (Vol. 98, pp. 73–79). [Google Scholar]
- [26].Breiman L Random forests. Machine learning 2001. Oct 1;45(1):5–32. [Google Scholar]
- [27].Chen T, Guestrin C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 2016 Aug 13 (pp. 785–794). [Google Scholar]
- [28].Zou H, Hastie T. Regularization and variable selection via the elastic net. J R Stat Soc Series B Stat Methodol 2005. Apr 1;67(2):301–20. [Google Scholar]
- [29].Lundberg SM, Lee SI. A unified approach to interpreting model predictions. Adv Neural Inf Process Syst 2017. (pp. 4765–4774). [Google Scholar]
- [30].Lundberg SM, Erion G, Chen H, et al. From local explanations to global understanding with explainable AI for trees. Nat Mach Intell 2020;2(1):2522–5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Pollack MM, Patel KM, Ruttimann UE. PRISM III: an updated Pediatric Risk of Mortality score. Crit Care Med 1996. May 1;24(5):743–52. [DOI] [PubMed] [Google Scholar]
- [32].Leisman DE, Harhay MO, Lederer DJ, et al. Development and reporting of prediction models: guidance for authors from editors of respiratory, sleep, and critical care journals. Crit Care Med 2020. May;48(5):623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Chaudhary NS, Donnelly JP, Wang HE. Racial Differences in Sepsis Mortality at United States Academic Medical Center-Affiliated Hospitals. Crit Care Med 2018. Jun;46(6):878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kamaleswaran R, Akbilgic O, Hallman MA, et al. Applying artificial intelligence to identify physiomarkers predicting severe sepsis in the PICU. Pediatr Crit Care Med 2018. Oct 1;19(10):e495–503. [DOI] [PubMed] [Google Scholar]
- [35].Khojandi A, Tansakul V, Li X, et al. Prediction of sepsis and in-hospital mortality using electronic health records. Methods Inf Med 2018. Sep 1;57(04):185–93. [DOI] [PubMed] [Google Scholar]
- [36].Le S, Hoffman J, Barton C, et al. Pediatric severe sepsis prediction using machine learning. Front Pediatr 2019. Oct 11;7:413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Saqib M, Sha Y, Wang MD. Early prediction of sepsis in EMR records using traditional ML techniques and deep learning LSTM networks. 40th Annu Int Conf IEEE Eng Med Biol Soc (2018) Jul 18 (pp. 4038–4041). IEEE. [DOI] [PubMed] [Google Scholar]
- [38].Mao Q, Jay M, Hoffman JL, et al. Multicentre validation of a sepsis prediction algorithm using only vital sign data in the emergency department, general ward and ICU. BMJ open 2018. Jan 1;8(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Desautels T, Hoffman J, Barton C, et al. Pediatric severe sepsis prediction using machine learning. bioRxiv 2017. Jan 1:223289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zhang G, Xu J, Yu M, et al. A machine learning approach for mortality prediction only using non-invasive parameters. Med Biol Eng Comput 2020. Jul 20:1–44. [DOI] [PubMed] [Google Scholar]
- [41].Mohammed A, Cui Y, Mas VR, et al. Differential gene expression analysis reveals novel genes and pathways in pediatric septic shock patients. Scientific Reports 2019. Aug 2;9(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Banerjee S, Mohammed A, Wong HR, et al. Machine Learning Identifies Complicated Sepsis Trajectory and Subsequent Mortality Based on 20 Genes in Peripheral Blood Immune Cells at 24 Hours post ICU admission. bioRxiv 2020. Jan 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Sutton JR, Mahajan R, Akbilgic O, et al. PhysOnline: an open source machine learning pipeline for real-time analysis of streaming physiological waveform. IEEE J Biomed Health Inform 2018. May 2;23(1):59–65. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


