Abstract
Pattern recognition receptors such as Mincle (Clec4e) play a significant role in the regulation of inflammation. Enhanced signaling of Mincle through the release of damage-associated molecular patterns during sterile inflammation has been shown to be important in the progression and manifestation of several diseases. A limitation to Mincle-targeted therapeutics is the feasibility of human-scale antibody therapy and the lack of alternative small-molecule inhibitors. Herein, we describe a highly specific neutralizing DNA aptamer targeting Mincle and demonstrate its therapeutic potential. Our data demonstrate that AptMincle selectively binds to both human and mouse Mincle with high affinity and is able to directly target and reduce Mincle activation. AptMincle can specifically reduce trehalose-6,6-dibehenate (TDB)-induced Syk and P65 phosphorylation in vitro in a manner comparable to that of the commercially available neutralizing antibody in vitro. Moreover, a bio-stable modified aptamer, AptMincleDRBL, was successful in reducing disease activity in a dextran sodium sulfate (DSS)-induced model of ulcerative colitis in a dose- and sequence-dependent manner. The results present an alternative, highly specific DNA aptamer with antagonistic function for use in the investigation of Mincle-associated diseases. The data also show the translational potential of Mincle-targeting aptamers as a new category of biologic therapy in the treatment of inflammatory bowel disease (IBD).
Keywords: MT: Oligonucleotides: Therapies and Applications, Mincle, aptamer, inflammatory bowel disease, ulcerative colitis, inflammation
Graphical abstract
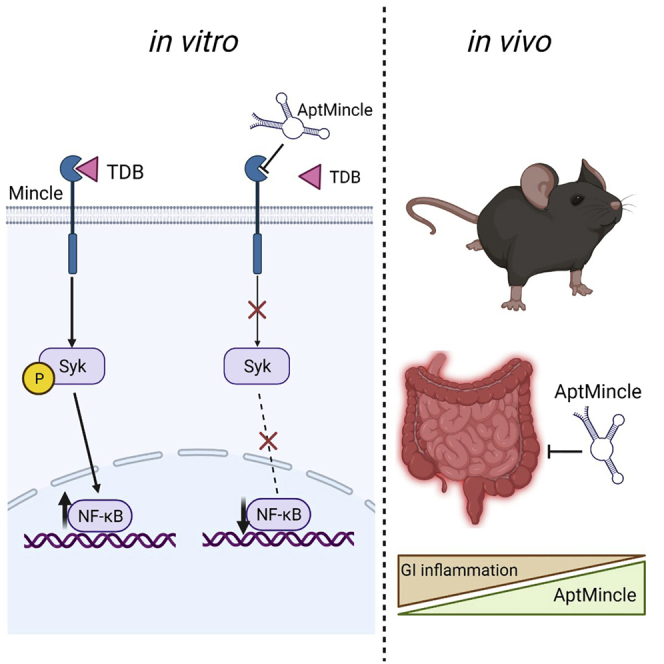
Herein, we develop and characterize an ssDNA aptamer (AptMincle) that targets the pattern recognition receptor Mincle (CLEC4E). AptMincle was tested for specificity and comparable functionality on a commercial monoclonal antibody and for its therapeutic potential in a mouse model of inflammatory bowel disease.
Introduction
An expanding field of biological therapy aims to target individual pattern recognition receptors (PRRs) due to their proposed roles in the initiation and continuation of chronic inflammatory diseases.1, 2, 3, 4 Indeed, many PRRs have been found to be dysregulated within the diseased host and described to contribute directly to mounting inflammation through the spontaneous or uncontrolled production of inflammatory mediators.4,5 A subset of these receptors, which includes inducible PRRs (iPRRs), are thought to play a distinct role in the pathogenesis of many diseases, including inflammatory bowel disease (IBD), psoriasis, multiple sclerosis, and some parasitic infections such as leishmaniasis.5, 6, 7, 8 Macrophage-inducible C-type lectin (CLEC4E/Mincle) was first identified in the late 1990s and was observed to be transcriptionally upregulated in macrophages following stimulation with a variety of inflammatory stimuli. During homeostasis, Mincle expression is low, but expression can be induced through the activation of several PRRs, including TLR2 and TLR4, or inflammatory cytokines such as IFN-γ, TNF-α, and IL-6.9 Importantly, the increase in Mincle expression is seemingly dependent on MyD88 and NF-κB (P65/RELA). Mincle expression has been shown to be limited to a subset of cell types, predominantly myeloid cells such as neutrophils, monocytes, monocyte-derived macrophages, and dendritic cells.10, 11, 12, 13
Mincle, through Syk-mediated signaling, mediates a variety of proinflammatory responses within the host that can promote or even prolong inflammation.13 Interestingly, while Mincle has been described to serve as a receptor for a variety of mycobacterial,14,15 bacterial,16 and fungal pathogens,17 it can be directly activated by several host-derived damaged-associated molecular patterns (DAMPs), suggesting an intimate involvement in sterile inflammatory insult.18 Specifically, previous studies have reported that Mincle senses aberrant levels of cholesterol sulfate7 and SAP130,19 DAMPs associated with cellular damage and necrosis, which, when bound to Mincle, trigger a proinflammatory cytokine response in the host. Furthermore, through the use of knockout mice, neutralizing Mincle antibodies, targeted siRNA, and pan-Syk inhibitors, the distinct role of Mincle in these disease models has been significantly investigated.5, 6, 7, 8,16,18
The expanding field of nucleic acid therapeutics has highlighted the potential use of aptamers as a direct substitute for commercially available, but rather expensive, monoclonal antibodies while avoiding the non-specific nature of kinase-directed pharmacological inhibitors.20 The inherent nature of aptamers with their high specificity and non-immunogenic qualities makes them desirable for the targeted blockade of inflammation as an alternative to small immune-pathway chemical inhibitors and their possible off-target effects.21 Promised for almost 20 years, aptamer-based therapeutics, such as the anti-VEGF aptamer pegaptanib (Macugen),22 have begun to spill into the market. However, with the familiarity of monoclonal antibodies, the need for aptamer biosimilars is overlooked.
IBD, subdivided into ulcerative colitis (UC) and Crohn’s disease (CD), is a rapidly expanding cohort of chronic inflammatory intestinal diseases, predominant within the developed world.23 UC is a chronic idiopathic disorder of the colon manifesting in contiguous mucosal inflammation that can extend from the distal ileum to the anus. Bimodal in its age distribution, UC is the most common form of IBD and is a significant form of morbidity worldwide.24 Patients with UC frequently experience acute cyclical episodes of disease manifestation with symptoms including bloody diarrhea, pain, fever, and weight loss. With several proinflammatory cytokines elevated during the disease flares, current biological therapies revolve almost entirely around modulating systemic TNF-α, through the prohibitively expensive use of monoclonal antibody therapies such as infliximab, adalimumab, certolizumab, or golimumab.25,26 Furthermore, the underlying cause of the overproduction of TNF-α within these patients is still debated. It is with this incomplete understanding of disease pathogenesis that the discovery and investigation of new targets for therapeutic intervention must be made. PRRs, such as Mincle, present a novel and promising target upstream of the canonical neutralization of circulating inflammatory cytokines.
While Mincle has been implicated in the progression of several experimentally induced inflammatory disease models, the lack of commercially available, Mincle-directed therapeutic agents has meant that its translational potential into the clinic is currently based only in theory. Therein, aptamers provide an attractive alternative to costly monoclonal antibody development. Highly specific, affordable, and modifiable, DNA aptamers can circumvent many issues associated with monoclonal antibody therapy. In addition to being non-immunogenic, simplistic modifications of size and structural integrity means aptamer delivery can be tailored for acute delivery and clearance from the biological system (occurring within hours) or protected from exonuclease action and renal filtration through size modification (extending half-life to several days in vivo).27 In this study, we develop, characterize, and investigate an antagonistic Mincle-targeting single-stranded DNA (ssDNA) aptamer and its protective effects within the chemically induced mouse dextran sodium sulfate (DSS) model of UC.
Results
Generation of Mincle-specific DNA aptamers
The full-length amino acid sequences of human and mouse Mincle are 65% identical (UniProt: CLC4E_Mouse Q9R0Q8, CLC4E_Human Q9ULY5). The recombinant fragment of extracellular Mincle used within this project contains 85% overall amino acid sequence identity. Given that functional studies are typically carried out in murine models, we examined whether a human Mincle-targeting ssDNA aptamer could function in multiple species while retaining immediate translational potential into human systems in the future.
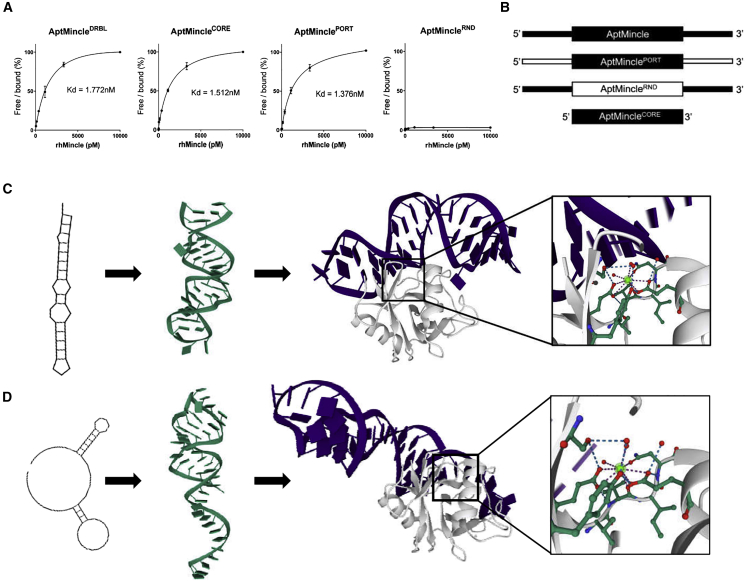
Human Mincle-targeting aptamers were identified using the nitrocellulose filter SELEX (detailed under Materials and methods). The cloned and sequenced aptamers and recurrent sequences were analyzed using Clustal Omega alignment algorithms and categorized into predicted familial groups based on their homology (Figure S1). In total, eight highly abundant sequences (72% of the sequenced community) were taken forward for in silico structural predictions and protein interaction modeling. Identification and validation of one of these sequences, herein referred to as AptMincle, was validated for specific binding to Mincle. AptMincle was synthesized with a 5′-biotin and 3′-inverted dT—this modified sequence is referred to as AptMincleDRBL—and using a modified enzyme-linked oligonucleotide assay (ELONA) protocol, its binding efficiency was quantified (KD = 1.722 nM). Through comparative analysis and replacement of key portions of the sequence, it was determined that the initially random, n = 40, region at the core of the sequence (AptMincleCORE, Kd = 1.512 nM) was responsible for its binding affinity toward Mincle. It was only by randomizing that core region (AptMincleRND, Kd N/A) that functionality was lost. Replacement of the flanking primer sequences (AptMinclePORT KD = 1.376 nM) had no discernible negative impact on binding affinity toward Mincle (Figures 1A and 1B).
Figure 1.
Characterization of the in vitro function of aptamers with Mincle affinity
(A) Binding characteristics of AptMincle, and its modified counterparts, with rhMincle as determined by ELONA. (B) Graphical depiction of the modifications made to the aptamer sequences (black, original; white, modified). (C) The predicted secondary and tertiary structures and docking simulation of AptMincleCORE (purple) with Mincle (white). Box: highlighted predicted region of interaction surrounding a calcium molecule. (D) The predicted secondary and tertiary structures and docking simulation of AptMincleRND (purple) with Mincle (white). Box: highlighted predicted region of interaction surrounding a calcium molecule. Dissociation constants were calculated on GraphPad Prism 8 using a non-linear regression binding analysis with assumed one-site target parameters.
Secondary structures, predicted in silico by Vienna Webfold, revealed that AptMincleCORE forms a long-stem stable hairpin structure. In silico 3D-structure predictions of AptMincleCORE in RNAComposer were combined to create simulations of docking to the crystal structure of human Mincle (PDB: 3WH3). Note that, as there is an absence of modeling software for the prediction of ssDNA tertiary structures, we have assumed the sequence can be modeled as ssRNA. The generated interactions suggested that the short hairpin structure of the core sequence of AptMincleCORE (40 bases in length), specifically nucleotides 18–30, potentially interacts with Mincle in regions near calcium binding domains, suggesting a possible site of interference through allosteric hindrance (amino acids Ser90–Val152), a highly conserved region between human and mouse Mincle (93.5% homology) (Figure 1C). To further support the prediction, the n = 40 scrambled oligonucleotide sequence of AptMincleRND was modeled in the same manner and was poorly predicted to bind to a nondescript region of the extracellular domain fragment (Figure 1D).
Functionality assessment of AptMincle in vitro
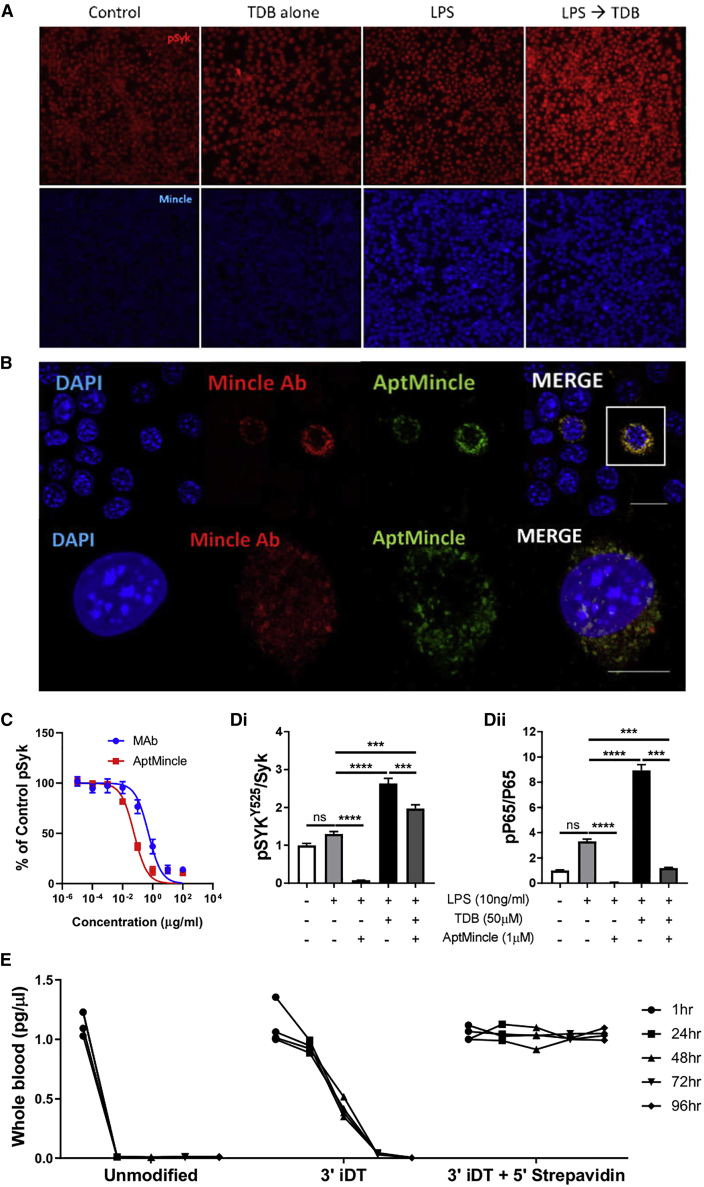
As an inducible PRR, Mincle is expressed at low levels in unstimulated macrophages (Figure 2A; control). However, this expression can be greatly increased via overnight stimulation with the TLR4 agonist lipopolysaccharide (LPS) (Figure 2A; LPS). Within LPS-primed J774.1 macrophages (Minclehigh), stimulation with a ligand of Mincle, trehalose-6,6-dibehenate (TDB), induces a robust phosphorylation of the Mincle adapter, SykY525 (Figure 2A; pSyk). This phosphorylation is not found in control cells stimulated with TDB only (Figure 2A; TDB alone). This LPS-induced increase in Mincle protein expression can be detected in the primed cells using commercially available monoclonal antibody immunostaining (Figure 1A; Mincle). Combining two suspensions of J774.1 macrophages, one primed with LPS (Minclehigh) and the other untreated (Minclelow), we formed a heterogeneous population of Mincle-expressing cells. Immunofluorescence staining revealed that both the antibody and a 5′-Cy3-conjugated AptMincle could detect and discriminate between these cell populations, staining the primed murine lymphocytes in a highly conserved pattern (Figure 2B). Determining that AptMincle specifically bound Mincle in vitro, we aimed to determine whether it could impede phosphorylation of the Mincle adapter SykY525 in an antagonistic/blocking manner as structurally predicted in Figure 1D. Phosphorylation of SykY525 in LPS-primed and TDB-stimulated J774 macrophages could be effectively neutralized in a dose-dependent manner using both a commercially available Mincle-neutralizing antibody and the AptMincle sequence, from which the IC50 of each was calculated (Figure 2C). The IC50 of AptMincle (IC50 = 0.05853 μg/mL) was shown to be almost 10 times less than that of the commercial antibody (IC50 = 0.5274 μg/mL) within this system. AptMincle displays continued downstream antagonistic activity toward murine Mincle, confirmed through analysis of both the phosphorylation of the Mincle adapter Syk and NF-κB subunit P65 phosphorylation measured in TDB-stimulated cells by whole-cell lysate direct ELISA (Figures 2Di and 2Dii). Highly abundant in exonucleases, the blood of mice, or humans, would rapidly degrade naked DNA sequences to prevent aberrant immune responses through other PRRs such as TLR9. This effect can be compounded in inflammatory settings, where serum and intracellular exonucleases can be altered in expression and function. Furthermore, the body’s natural filtration system, the nephron-based renal exclusion system, has been previously shown to exclude unmodified aptamers due their relatively low molecular weight (<25 kDa).28 In an attempt to combat these eventualities, two modifications were added to protect the sequences. The first was an inverted deoxythymidine base (iDT) added at the 3′ terminus of the sequence, preventing degradation by 3′ exonucleases while simultaneously preventing extension by DNA polymerases.29 This significantly improved stability in isolated serum; however, the viability within the host was improved to only between 24 and 48 h, as the excess was likely filtered and excreted in the urine. The latter modification, an additive 5′ biotin, allowed for the direct conjugation of a variety of streptavidin-linked moieties (Figure 2E). In this project we also wanted to increase the molecular weight of the sequence to improve its bioavailability by reducing renal filtration. This was achieved through the addition of a 5′ streptavidin linker, which increased AptMincle’s molecular weight to ∼75 kDa. In combination with the 3′ iDT, the 5′ streptavidin improved the bioavailability of AptMincle in vivo past 4 days (confirmed by quantitative analysis of AptMincle in whole blood). These data suggest that AptMincle significantly depletes endogenous TDB-induced Mincle Syk and P65 phosphorylation within macrophages (Figures 2C and 2D) and that synthesis with 3′ iDT and biotin-streptavidin 5′ modification protect it from in vitro degradation. The aptamer will herein be referred to as AptMincleDRBL and was used as the primary aptamer sequence for in vivo investigation.
Figure 2.
In vitro binding characteristics of aptamer AptMincle in comparison with antibody
(A) Immunofluorescence imaging of the relative expression of pSykY525 and Mincle in unstimulated, TDB-stimulated (50 μM), LPS-primed (10 ng/mL, 24 h), or LPS → TDB (10 ng/mL LPS + 50 μM)-stimulated J774.1 macrophages (left to right). (B) Staining of a heterogeneous population of control (Minclelow) and LPS-primed (Minclehigh) J774.1 macrophages co-stained with anti-CLEC4E antibody (InvivoGen, USA) and 5′-Cy3-conjugated AptMincle. (C) Dose-dependent inhibition of SykY525 phosphorylation in J774.1 macrophages by anti-Mincle antibody (InvivoGen) compared with AptMincle. (D) Whole-cell ELISA of (i) pSykY525/Syk and (ii) pP65/P65 relative expression in LPS-primed, TDB-treated J774.1 macrophages when challenged with AptMincle (1 μM). (E) Whole-blood assessment of AptMincleDRBL presence in mice via fully quantitative RT-PCR. IC50 was calculated on GraphPad Prism 8 using a non-linear fit: y = 100/(1 + x/IC50). One-way ANOVA with Tukey’s post hoc test was performed for multiple non-parametric data comparisons. ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Scale bar = 10μm.
AptMincleDRBL demonstrates a dose-dependent protective effect in DSS-induced colitis
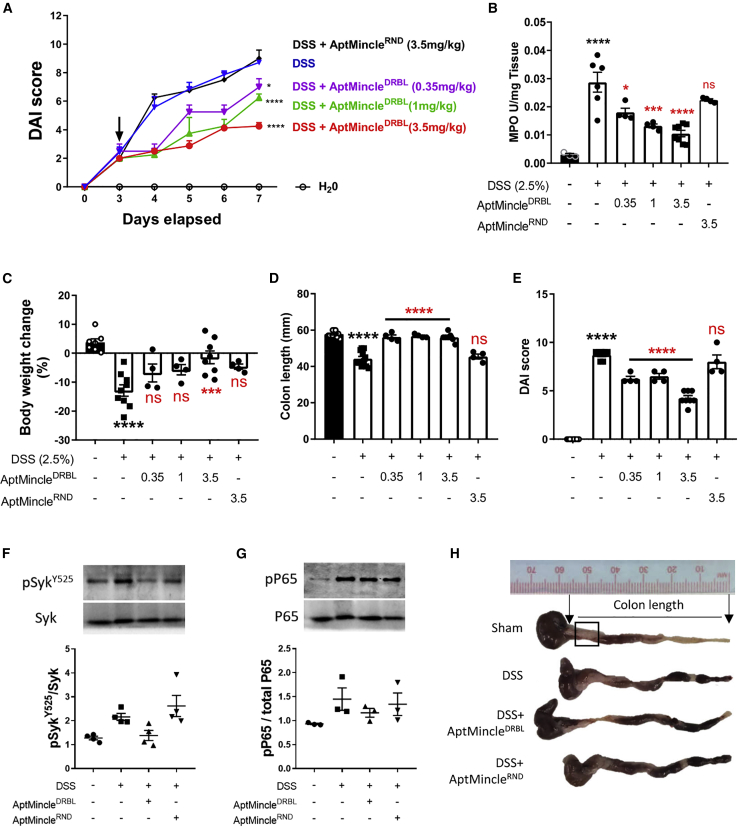
Having validated the AptMincle sequence and improved its stability in vivo, we aimed to test the therapeutic potential of the sequence in a characterized disease model previously shown to be regulated in part by Mincle.5 We used the chemically induced DSS model of UC, which results in an acute inflammation of the gastrointestinal tract, manifesting primarily within the colon and reproducing many of the pathological signs of human UC, such as weight loss, diarrhea, intestinal bleeding, colon shortening, and submucosal inflammation.4,30,31 Previously, we highlighted that the earliest DSS-induced colitis macroscopically displayed within the 7 days of DSS treatment is at day 3, whereby fecal consistency and weight loss are prominent. Therefore, at day 3, a single dose of AptMincleDRBL was injected intraperitoneally into mice (0.35, 1, 3.5 mg/kg) as a therapeutic intervention rather than prophylactically. This treatment method is in line with our previously published data of the protective effect of TLR4 inhibition in the same model.4 The data show a protective effect of AptMincleDRBL, which was achieved in a dose-dependent manner. The disease activity index (DAI) was significantly improved following the aptamer administration (day 4, Figure 3A; black arrow). At the termination of the experiment (day 7), colonic inflammation, indicated by myeloperoxidase (MPO) activity, was significantly reduced with AptMincleDRBL treatment (MPO reduced by 0.35 mg/kg = 37.2%, 1 mg/kg = 52.3%, 3.5 mg/kg = 63.5%) (Figure 3B). Loss of body weight accompanying DSS treatment was also significantly reduced at the highest dose of AptMincleDRBL (Figure 3C, 3.5 mg/kg, ANOVA, p = 0.0002) and was tending to significance at the lower doses. Interestingly, at all therapeutic concentrations of AptMincleDRBL, colonic shortening (an indicator of intestinal inflammation and fibrosis) was completely resolved (Figures 3D and 3H, p < 0.0001). All AptMincleDRBL treatment groups, regardless of dose, showed a significant resolution of day 7 DSS-associated DAI (Figure 3E, p < 0.0001), while the random-sequence oligonucleotide control, AptMincleRND, had no protective effect in any assessed metric (Figure 3E). Analysis of colon sample lysates using western blot and band densitometry analysis showed a trend toward a reduction of DSS-induced total Syk (p = 0.068) and P65 phosphorylation within the tissue in mice treated with the highest dose of AptMincleDRBL (Figures 3F and 3G). Finally, DSS-induced colon changes, as well as alterations of the luminal content of the colon, can be seen in representative examples of harvested tissue (Figure 3H). Overall, these data highlight the in vivo protective effect of AptMincleDRBL at multiple therapeutic doses, and through the concurrent assessment and use of the AptMincleRND control sequence, we show the nucleotide sequence specificity of the aptamer core region and the gross anti-inflammatory action created by the active sequence.
Figure 3.
AptMincleDRBL suppresses DSS-induced colitis through a dose-dependent inhibition of colonic inflammation
(A) Daily changes in disease activity score (DAI) over the onset of DSS-induced colitis. (B) MPO activity within colon. (C) Body weight percentage change at the end of the experiment. (D) Colon length and (E) terminal DAI measured at day 7 with relation to treatment groups with AptMincleDRBL (0.35, 1, or 3.5 mg/kg) or AptMincleRND (3.5 mg/kg). Immunoblot analysis of (F) Syk or (G) P65 phosphorylation of total colonic lysates. (H) Representative photographs of colons from mice (DSS + AptMincleDRBL is 3.5 mg/kg group only). These data are presented as the mean ± SEM and were analyzed by a repeated measures ANOVA with Tukey’s post hoc test. Black asterisks indicate significance against sham group, while red asterisks indicate significance compared with DSS group: ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Mice are a composite of 10-week-old mice, n = 4–12, with experiments performed on separate days.
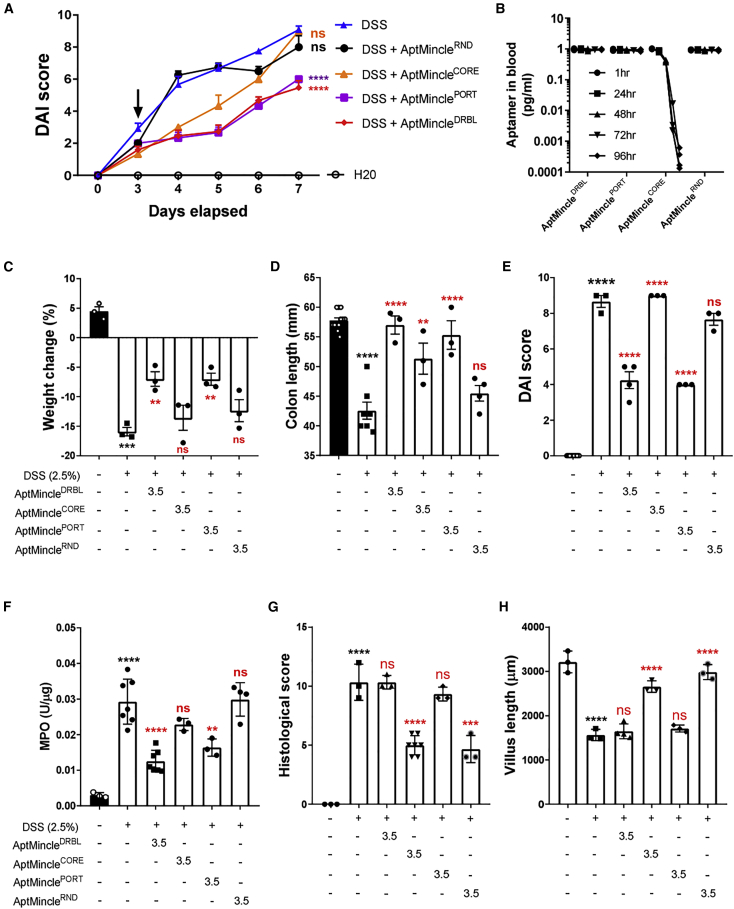
AptMincle’s bio-protective effect in DSS-induced colitis is sequence specific
As shown throughout this article, it appeared that the core oligonucleotide sequence of AptMincle is solely responsible for the aptamer’s binding function in vitro. In an attempt to confirm whether the protective effect of AptMincleDRBL in vivo was too sequence specific, we evaluated the potential of the alternate sequences: AptMinclePORT (alternative flanking primers but with conserved core n = 40 sequence),32 AptMincleCORE (removal of flanking primers, having an overall reduced molecular weight), and AptMincleRND (scrambled n = 40 core region, but with conserved flanking primers). As presented in Figure 4A, by day 7, the single “high-dose” (3.5 mg/kg) of AptMincleDRBL or AptMinclePORT had a significant effect in attenuating the DAI compared with DSS controls (ANOVA, p < 0.0001), while AptMincleRND had no discernible effect (ANOVA, p = 0.705). Interestingly, AptMincleCORE initially showed a potent preventative effect upon DAI in the mice (day 4, ANOVA, p = 0.003), but this was rapidly lost at day 6 (ANOVA, p = 0.292), 3 days after injection. This correlated with the sudden loss of detectable AptMincleCORE within the serum of the treated mice as determined by fully quantitative RT-PCR (Figure 4B). We theorize that while the 3′ iDT present on the AptMincleCORE sequence shielded it from degradation within the host, the drastic reduction in molecular weight meant it fell short of the renal filtration threshold and was likely excreted from the system in the urine. Histological assessment of the colon of mice demonstrated that during DSS treatment there was significant leukocyte infiltrate, goblet cell loss, crypt density and hyperplasia, muscle thickening, and submucosal infiltrate compared with healthy controls, with a loss of villus length, as detailed in Koelink et al.33 The histology scoring and villus length were shown to significantly improve in the DSS + AptMincleDRBL and DSS + AptMinclePORT groups compared with DSS counterparts (Figures 4G, 4H, and S2). The results compound the evidence that the size (molecular weight) and nucleotide sequence of AptMincle are important for its longevity and action, respectively, in vivo.
Figure 4.
The protective effect of AptMincleDRBL in DSS-induced colitis is sequence specific
(A) Disease activity score (DAI) during the onset of DSS-induced colitis compared with treatment groups AptMincleDRBL/RND/CORE/PORT (3.5 mg/kg) given at day 3. (B) Presence of detectable AptMincle sequences within the serum of treated mice. (C) Overall body weight percentage change during the experiments. (D) Colon length at termination. (E) DAI measured at termination at day 7. (F) MPO activity in the colon of mice at day 7. (G) The 14-point histological assessment of terminal colon of the groups and (H) the measured villus length of colon samples. These data are presented as the mean ± SEM and were analyzed by a repeated measures ANOVA with Tukey’s post hoc test. Black asterisks indicate significance compared with sham control, while red asterisks are significance against the DSS group: ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Mice are a composite of 10-week-old mice, n = 3, with experiments performed on separate days.
Discussion
In the current study, we developed, characterized, and validated a DNA aptamer targeting the PRR Mincle (CLEC4E). Termed AptMincle, this oligonucleotide aptamer demonstrates specific binding and antagonistic activity toward both human and murine Mincle and, subsequently, had a protective effect against experimental colitis in mice, a setting where Mincle has been previously demonstrated to play a crucial role in the maintenance of disease.5 We first identified eight highly enriched aptamer sequences that bound recombinant human Mincle, utilizing a modified nitrocellulose-based SELEX protocol (Figure S1). This protocol selected a community of putative Mincle-specific agonistic, neutral, or antagonistic aptamers targeting a recombinant fragment of the extracellular domain of human Mincle (accession no. Q9ULY5, amino acids 41–219). In silico structural prediction using the Vienna Webfold server suggested that AptMincle formed a tight hairpin structure specifically at its core region (n = 40). Due to the lack of tertiary structure modeling software for ssDNA we have assumed the aptamer can be modeled as RNA. RNAComposer successfully modeled AptMincleCORE (Figure 1C).34 For subsequent protein-RNA association modeling, we combined the PDB crystal structure of Mincle (PDB: 3wh3) with the predicted tertiary structure of RNA-AptMincleCORE using PatchDock.35,36 It was noted that the proposed 3D model suggested a likelihood that AptMincleCORE bound to Mincle near calcium binding regions and proposed potential antagonistic properties of the aptamer sequence. As aptamers bind to proteins based on van der Waals, hydrogen bonding, and electrostatic interactions, it was promising to note that the confirmational structure of human and mouse Mincle was highly conserved. Using the alphafold structural prediction of murine Mincle (as no crystal structure existed as of the time of writing this paper), we aligned the sequences using PyMOL 2.0. The predicted root mean square deviation (RSMD) was <1 Å (0.612). The authors highlight and acknowledge that these data are speculative and are provided for reference purposes only. Structural binding of AptMincle to Mincle must be performed in the future to support these claims.
When evaluated, the binding of AptMincle to recombinant Mincle was determined to be specific to the core 40-nucleotide (initially variable) region. This specificity was confirmed through post-SELEX truncation of the sequence removing the flanking primers (AptMincleCORE), substituting the primer sequences for alternatives (AptMinclePORT), or a scrambled control (AptMincleRND). It was noted that only the AptMincleRND lost its binding capacity to rhMincle, while the AptMincleCORE and AptMinclePORT sequences maintained function (Figure 1). As previously mentioned, one of the main project goals was to develop an antibody alternative for targeting Mincle. Comparative analysis and the use of commercially available anti-Mincle antibodies allowed us to demonstrate similar, if not better, functionality in neutralization assays and immunofluorescence imaging (Figure 2). Based on our data the IC50 of AptMincle is significantly less than that of the monoclonal antibody, suggesting a much more powerful mechanism of inhibition. However, we accredit this to the aptamer’s vastly smaller molecular weight (∼25 kDa) and therefore an ∼6× abundance of molecules within mol-mol equivalents (1 μg/mL AptMincle = 40 nM = 240 × 1014 molecules, while 1 μg/mL monoclonal antibody = 6.67 nM = 40 × 1014 molecules). As AptMincle was able to be successfully used in an ELONA and in immunofluorescence (IF) imaging, it is feasible that the sequence can be used for all techniques where the confirmational structure of Mincle is maintained (non-denaturing PAGE, immunoprecipitation, flow cytometry, etc.). These applications will require validation in the future. Serum exonucleases, combined with renal filtration and excretion, commonly result in rapid degradation and removal of oligonucleotides from the system.37,38 Thankfully, due to their highly modifiable nature, the half-life of nucleic acid aptamers within a system can be readily and easily altered. We wanted AptMincle to be stable within the host to inhibit Mincle-induced proinflammatory responses during inflammation. However, the unmodified oligonucleotide was rapidly degraded in isolated serum and undetectable after 30 min exposure at room temperature (data not shown). In an attempt to circumvent these issues, modifications to the 5′ and 3′ ends of the AptMincle sequences were made to (1) inhibit exonuclease digestion and (2) limit renal secretion. Both of these modifications were made during the sequence synthesis process and allowed for flexible modification of the sequence for specific application (Figure 2, Figure 3, Figure 4). The inclusion of an inverted dT on the 3′ end of the sequence alone improved the stability of AptMincle within the serum, but it was still noted to be depleted in vivo, likely due to size exclusion during nephron filtration within the kidneys (Figures 2 and 4). The addition of the 5′ biotin moiety and subsequent streptavidin conjugation increased the molecular weight of AptMincle to ∼50–75 kDa, far exceeding the filtration threshold.27 This modification in combination with its exonuclease resistance allowed the aptamer to persist within the system in excess of 4 days (Figure 4). It is also important to discuss that without these modifications, any aptamer’s biological function would likely be reduced to short (<1 h) bursts following administration, a strategy that could aid in preventing long-term or off-target cellular toxicities, although no such eventualities have currently been identified. Previous work has implicated Mincle in the pathogenesis of several diseases, including models of UC.5 We therefore wanted to validate the protective effect of AptMincle within the same model, murine DSS-induced colitis. The addition of the 5′ biotin-streptavidin and 3′ iDT allowed for treatment to consist of a single dose of AptMincleDRBL (0.5, 1, or 3.5 mg/kg) due to minimal clearance from the host and no additional benefit with multiple doses (Figure S3). Initial experimentation showed a clear dose-dependent therapeutic response of the aptamer with significant reduction in several disease markers (Figure 4), with the data demonstrating a significant reduction in DAI, weight loss, colonic shortening, and colonic MPO. Interestingly, disease activity scoring, which is a combination of three distinct parameters (weight loss, stool consistency, and occult rectal bleeding), indicated that a majority of the reduction was due to improvements in weight and stool consistency, but the presence of blood in the feces was not affected, suggesting a Mincle-independent role of vascular leakage and rectal bleeding. Furthermore, the significant reduction in DSS-induced colonic MPO improved histological scoring and colon length, suggesting the primary mechanism of action is through the prevention of a Mincle-driven inflammatory response. In an attempt to further support the validity of the AptMincle sequence in vivo, multiple versions of the sequence were created. Previous data had shown that the CORE nucleotide sequence was solely responsible for binding to recombinant Mincle in vitro. We therefore assessed the biological impact of these alterations by comparing the protective capacity of each sequence to the DSS colitis model. Data show that AptMincleDRBL and AptMinclePORT (alternative flanking sequences) were equally effective in reducing the severity of disease activity in treated mice. Interestingly, AptMincleCORE had a potent effect during the early stages of pathology, negating all external markers of disease activity until day 5, at which time all mice rapidly returned to similar DSS-control values. We predict that this phenomenon is driven by two intrinsic mechanisms. First, as we treated mice with 3.5 mg/kg of each modified aptamer, it is possible that the reduced size of the AptMincleCORE (MW 12,451.10 Da/28.1 μM) versus AptMincleDRBL/PORT (23,656.36 Da/14.8 μM and 24,354.87 Da/14.4 μM, respectively) increased the overall concentration of the therapy by up to 2-fold. Combining this with the previously determined half-life of the unmodified AptMincle sequence in vivo, we can hypothesize that these are the key reasons the protective effect of AptMincleCORE is transient. Importantly, the efficacy of AptMincle in a model of UC presents and supports the use of this newly developed aptamer in patients unresponsive to conventional anti-inflammatory therapeutics. The absence of toxicity and tractable modifications made to the sequence makes it an encouraging candidate for further clinical assessment in IBD and other Mincle-related diseases. While sequence homology between human and mouse Mincle is calculated at 65% (UniProt alignment codes Q9ULY5/Q9R0Q8), the evidence presented here suggests a conserved activity of AptMincle on murine Mincle despite being raised against a recombinant human protein. Furthermore, AptMincle did not bind significantly to BSA, mouse serum components, or recombinant CLEC4D (MCL) despite the latter sharing 35% sequence homology with CLEC4E (Mincle) and a common ligand, TDB, further validating the specific targeting of Mincle by AptMincle. Other aptamers against PRRs have been developed that show protective effects in disease-relevant animal models.39,40 Rather than targeting secreted, serum, or tissue-born cytokines, as is the case with conventional antibody-based biologic therapies, it is proposed that the targeting of aberrant PRR activity is a more effective way to treat the inflammation at the causative source. The aptameric targeting of inducible PRRs like Mincle, however, has never been attempted. AptMincle’s development creates a novel avenue for intervention medicine whereby the uncontrolled and sustained inflammatory response can be dampened, increasing the chance of tissue regeneration and disease resolution. AptMincle, presented here, provides a tool for the investigation and treatment of Mincle-driven diseases, including UC. Before the aptamer can be used in human settings, additional tests and validation methodologies will need to be performed in animal models to characterize biodistribution and long-term immunogenicity responses. Once fully validated, we envision that AptMincle will be assessed for translational potential into humans for the treatment of UC. Furthermore, investigation into the therapeutic benefits in mouse models of Crohn’s disease, psoriasis, and multiple sclerosis, diseases all shown to be exacerbated or driven in part by Mincle, is underway. With limited effective treatments in all of the aforementioned disease groups, the addition of a new therapeutic is greatly needed. Furthermore, the stability, cost effectiveness, and low immunogenicity associated with nucleic acid-based aptamers, compared with current antibody biologic therapies, provide an appealing alternative to both clinicians and patients.
Materials and methods
Ethics committee approval and mouse use
All mice were housed at constant temperature (22°C) on a 12:12-h light-dark cycle, with food and water ad libitum. Both male and female mice were used at 8–10 weeks of age. The animal handling and experiments were approved by the University of Calgary Animal Care and Ethics Committee and conformed to the guidelines established by the Canadian Council on Animal Care (Protocol AC17-0186).
Chemicals and oligonucleotides
All oligonucleotides were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA, USA). The sequences of all ssDNA anti-Mincle aptamers created by this project can be found in Figure 1. All chemicals were purchased from Sigma-Aldrich Canada (Oakville, ON, Canada) unless otherwise stated.
Aptamer selection protocol (SELEX)
Mincle-specific ssDNA aptamers were isolated and identified using nitrocellulose filter SELEX. The synthetic library of ssDNAs was composed of 76-nucleotide-long ssDNAs with a random region of 40 bases (RND40) flanked by conserved primer sequences: 5′-GCGGATGAAGACTGGTGT-[N]40-GCCCTAAATACGAGCAAC 3′, N = A/T/C/G.
In the first round of the SELEX procedure, 1 nmol (6.022 × 1014 individual sequences) of the RND40 population (10 μL of 100 μM stock) was added to 90 μL of SELEX buffer (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM MgCl2), denatured at 90°C for 10 min, and then cooled on ice for 10 min. These aptamers are then termed “folded.” The folded RND pool was then mixed with 1 μg (14 pmol) of rhMincle (ab152015; Abcam, Toronto, ON, Canada) (reconstituted in phenol red-free DMEM + 10% fetal calf serum [FCS]) in 90 μL of SELEX buffer (200 μL total) and incubated at 37°C for 1 h (rounds 1–3) or 30 min (rounds 4–6).
The bound aptamer-rhMincle complexes were purified using nitrocellulose adherence as previously described41 and, after being washed three times with SELEX buffer, the ssDNA-protein complexes were boiled off of the membrane in 200 μL of distilled H2O and amplified by PCR. Simply, 20 μL of the eluted aptamers from the previous round was amplified for 15–25 cycles using F′ and R′ primers (detailed below) in complete 2× PCR buffer under the conditions of 1 μM/primer, 250 μM dNTPs, in a final volume of 100 μL as above. Counterselection steps were performed before the initial round and after the third and sixth selection rounds using the reconstitution medium (DMEM +10% FCS) and the nitrocellulose membrane as the non-specific targets. All aptamers that bound to the membrane at these points were discarded and the unbound fraction (Mincle-targeted) was retained. The PCR amplifications performed at the end of each round of selection showed increased amounts of DNA product after the 15 cycles, but they did not reach saturation, consistent with the molar ratio of Mincle protein target used consistently throughout the process.
Primers used in serum analysis of aptamer
Detection of AptMincle within the blood was analyzed using fully quantitative PCR with an eight-part AptMincle standard curve. Briefly, 1 μL of collected whole blood from mice was used as a template for RT-PCR and quantified relative to the standard curve. Primers used for detection were as follows: AptMincleDRBL/RND forward, 5′-GCGGATGAAGACTGGTGT-3′, reverse, 5′-GTTGCTCGTATTTAGGGC-3′; AptMinclePORT forward, 5′-AGAAACGCTGGTGAAAGT-3′, reverse, 5′-CGCAAGCATAAAGTGTAAAG-3′; and AptMincleCORE forward, 5′-TCGGTGACGGTGGTTATT-3′, reverse, 5′CAAAACAAAAGAACTTAA-3′.
The amplification protocol consisted of an initial denaturation step at 98°C, followed by 30 cycles of 95°C for 30 s, 55°C–59°C for 15 s, 72°C for 30 s, and a final extension 72°C for 5 min. Amplicon specificity was determined by agarose gel electrophoresis.
Educated aptamer pool cloning and sequencing
The final pool of putative aptamer sequences was cloned into pJET1.2 blunt cloning vectors using the CloneJet PCR cloning kit (Thermo Fisher Scientific, Burlington, ON, Canada). Successfully transformed DH10B competent E. coli were grown on ampicillin (100 μg/mL) LB selection plates, after which colonies were picked and plasmids sequenced using pJET1.2 Forward and Reverse sequencing primers and the Sanger Sequencing Platform at the University of Calgary’s Centre for Health Genomics and Informatics Facility. Aptamer flanking primer regions were identified by hand and sequences were compiled.
Aptamer modification
AptMincle (with biotin modification at the 5′ end) was incubated in a 1:4 molar ratio with streptavidin (BioLegend, San Diego, CA, USA) in SELEX buffer for 1 h at 37°C. Successful conjugation was confirmed by aptamer band retardation assessment within a 2.5% agarose gel.
Binding affinity determination (ELONA)
Recombinant human Mincle (0, 1, 3.33, 10, 33.3, 100 nM) was bound to an ELISA capture plate before being probed with 5′biotin-AptMincle (100 nM) at 37°C for 1 h. In addition, rhMincle (100 nM) or BSA (100 nM) was incubated and probed with AptMincle to confirm the specificity of the aptamer. After binding of 5′biotin-AptMincle, the samples were washed before being incubated with streptavidin-horseradish peroxidase (HRP) for 30 min at room temperature. Samples were washed again before colorimetric assessment of binding via TMB (3,3',5,5'-Tetramethylbenzidine) substrate conversion and acid neutralization. The dissociation constant (Kd) was calculated by GraphPad Prism 8, using the equation y = Amax × x/(Kd + x). The value of AptMincle binding to rhMincle was represented as percent bound using PBS alone as the baseline reference.
Mincle-aptamer agonism/antagonism assessment in J774.1 macrophages
J774.1 murine macrophages were cultured and maintained in DMEM supplemented with 10% FCS, 1% pen/strep. To induce expression of Mincle within the resting macrophages, 1 ng/mL E. coli LPS was added to the stimulation medium for 24 h (termed J774-Mn+). Induction of Mincle expression was confirmed through immunofluorescence imaging. The prestimulated J774-Mn+ cells were then challenged with TDB (InvivoGen, San Diego, CA, USA; 50 μg/mL) or aptamer species (1 μM) for 30 min, and phosphorylation of Syk and P65 was assessed by western blot analysis as proxies of Mincle receptor activation. Agonistic effects of aptamers were assessed by incubation of J774-Mn+ cells with 1 μM individual folded aptamer species, while antagonistic effects were determined by preincubation with 1 μM aptamer species followed by stimulation with 50 μg/mL TDB (InvivoGen, cat. no. tlrl-tdb).
Protein collection and quantification
Cell culture and tissue samples were prepared in lysis buffer as previously described.42 Briefly, cells or cleaned and mechanically homogenized tissues were lysed in NP-40 lysis buffer (100 mM NaCl, 200 mM Tris-HCl [pH 8.0], 0.5% SDS, and 1 mM EDTA supplemented with: phosphatase inhibitor cocktail [cat. no. C755C25], 25 mM sodium fluoride [1 M], 1 mM sodium orthovanadate, and 1% Triton X-100). Samples were sonicated to ensure complete cellular lysis and dissociation of proteins before being clarified at 12,000g for 10 min at 4°C. Protein concentration was then confirmed and normalized using the Pierce BCA protein assay kit (Thermo Fisher Scientific, Burlington, ON, Canada).
Direct enzyme-linked immunosorbent assay
Protein (10 μg diluted in 1% BSA in TBST [0.1% Tween 20]) was loaded in triplicate per sample into wells of a 96-well plate and incubated overnight at 4°C to allow for protein binding. All wells were subsequently blocked for 1 h at room temperature (RT) in 2% BSA (TBST) before being washed three times with TBST. Sample wells were then incubated with primary antibodies for 2 h at RT on an orbital shaker. After three more washes with TBST, relevant HRP-conjugated secondary antibodies were incubated in corresponding wells for 1 h on an orbital shaker. Samples were washed for a final three times before being developed, acid-stopped (1 M HCl), and analyzed using a Victor X4 SpectraMax plate reader. Phosphorylated protein values were normalized to total protein for analysis and comparison.
Western blot
Previously collected protein lysates were combined with 5× loading buffer (20% SDS, 1 M Tris [pH 6.8], bromophenol blue, β-mercaptoethanol, glycerol, and water) and boiled for 10 min. Samples (30 μg) were loaded and run on 10% Tris-glycine SDS-PAGE gels made by hand according to standard protocols. Following resolution, the proteins were transferred to a nitrocellulose membrane for 1.5 h on ice. Ponceau staining confirmed efficient and complete transfer. Blots were washed and then blocked for 1 h in 5% BSA or milk in TBST (0.1% Tween 20) at room temperature on an orbital shaker. Blots were subsequently incubated with primary antibodies at predetermined manufacturer dilutions overnight under agitation at 4°C. Blots were washed and incubated with the appropriate HRP-conjugated secondary antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA) for 1 h at room temperature. Finally, the blots were incubated with extended-duration ECL chemiluminescent substrate (Thermo Fisher Scientific, cat. no. PIA34075) and visualized using Bio-Rad ChemiDoc MP.
Immunofluorescent staining and imaging
The protocols used for the immunofluorescent staining of sectioned tissue or cells in culture are the same. Briefly, samples were fixed in 4% paraformaldehyde (PFA) for 20–60 min at room temperature before being transferred into PBS (containing 0.1% sodium azide). Colonic tissue samples were further processed by overnight fixation in 30% sucrose solution before being embedded in OCT and sectioned into 10-μm sections on a cryotome. Slides were blocked with 2% BSA in PBST (0.1% Triton X-100) for 1 h at RT before being incubated overnight with primary antibodies at 4°C. The samples were then washed three times for 5 min in PBST before being probed with secondary antibodies or complementary conjugated primary antibodies for 2 h at RT under agitation. The samples were washed a further three times for 10 min before being sealed with Vectashield fluorescence mounting medium and a coverslip. All images were taken on an inverted Leica SP8 confocal microscope and analyzed using Leica LASX software (LEICA Microsystems Canada, Inc., Richmond Hill, ON, Canada).
Induction of colitis and administration of treatments
Six-week-old C57BL/6 mice were obtained from The Jackson Laboratory (Bar Harbor, ME, USA). Colitis was induced in these mice by administration of 2.5% (wt/vol) DSS (Affymetrix, Cleveland, OH, USA) in their drinking water for 7 days. Sham mice were given normal drinking water. Intraperitoneal injections of aptamer sequence variations of AptMincle (0.5–3.5 mg/kg) were administered at day 3 in 200 μL total volume saline; control mice received saline only. Mice were euthanized at day 7 by exposure to isoflurane (Pharmaceutical Partners of Canada, Richmond Hill, ON, Canada) and cervical dislocation.
Disease evaluation
To assess the severity of DSS-induced inflammation a multi-parameter approach was used. DAI is a combinatory metrics consisting of weight loss, fecal consistency, and fecal blood content. Calculation parameters of DAI are detailed in Table 1. Colon shortening, a common sign of inflammation-driven fibrosis, and MPO assays were used as measures of colonic inflammation.
Myeloperoxidase assay
MPO activity was measured in colonic tissue samples by quantification of the absorbance of a colored compound produced by MPO interacting with O-dianisidine and hydrogen peroxide. Absorbance readings were normalized to tissue weights.
Aptamer structure prediction and docking simulation
The 2D structure of AptMincle was predicted using the Vienna web server (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi). Due to the lack of ssDNA 3D structure prediction models, we have assumed that AptMincle could be modeled as an ssRNA aptamer. We used RNAComposer (http://rnacomposer.cs.put.poznan.pl/) for 3D-structure prediction rendering. Docking simulations for AptMincle and Mincle (PDB: 3WH3) itself were performed using the PatchDock Server (http://bioinfo3d.cs.tau.ac.il/PatchDock/) and images were taken using the PDB online viewer (https://www.rcsb.org/3d-view).
Statistics
Data are expressed as the mean ± 1 standard error of the mean (SEM). Experiments were performed as a minimum experimental triplicate with technical triplicates performed for each experiment. Statistical significance was assessed through the use of two-tailed unpaired Student’s t test for parametric data, while the Mann-Whitney test was performed for non-parametric data. GraphPad column analysis function was used to determine what previously mentioned tests were used on what dataset due to normal distribution characteristics. Multiple analyses were performed using a one-way ANOVA with Tukey’s post hoc test where indicated. A p value of 0.05 was considered significant: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Acknowledgments
We thank Jacques du Toit and Flavia Neto de Jesus for their input in the lab during the development of the project. We would also like to thank James McClellan and Robert Gowland for providing thoughtful insight into the original aptamer development protocols. This study was supported by the Lymphedema Research and Education Program, Snyder Institute for Chronic Diseases, Cumming School of Medicine, University of Calgary to P.-Y.v.d.W.
Author contribution
Conceptualization, M.S.; methodology, M.S., K.K., and S.R.; validation, S.R.; investigation, M.S., K.K., and S.R.; writing – original draft, M.S. and P.-Y.v.d.W.; writing – review & editing, M.S., K.K., S.R., S.L., and P.-Y.v.d.W.; funding acquisition, P.-Y.v.d.W.; resources, S.L. and P.-Y.v.d.W.; supervision, M.S., S.L., and P.-Y.v.d.W.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.05.026.
Supplemental information
References
- 1.Sacre S., Lo A., Gregory B., Stephens M., Chamberlain G., Stott P., Brennan F. Oligodeoxynucleotide inhibition of Toll-like receptors 3, 7, 8, and 9 suppresses cytokine production in a human rheumatoid arthritis model. Eur. J. Immunol. 2016;46:772–781. doi: 10.1002/eji.201546123. [DOI] [PubMed] [Google Scholar]
- 2.Mullen L.M., Chamberlain G., Sacre S. Pattern recognition receptors as potential therapeutic targets in inflammatory rheumatic disease. Arthritis Res. Ther. 2015/05/15 2015;17:122. doi: 10.1186/s13075-015-0645-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thwaites R., Chamberlain G., Sacre S. Emerging role of endosomal toll-like receptors in rheumatoid arthritis. Front. Immunol. 2014;5:1. doi: 10.3389/fimmu.2014.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stephens M., Liao S., von der Weid P.Y. Mesenteric lymphatic alterations observed during DSS induced intestinal inflammation are driven in a TLR4-PAMP/DAMP discriminative manner. Front. Immunol. 2019;10:557. doi: 10.3389/fimmu.2019.00557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gong W., Zheng T., Guo K., Fang M., Xie H., Li W., Tang Q., Hong Z., Ren H., Gu G., et al. Mincle/syk signalling promotes intestinal mucosal inflammation through induction of macrophage pyroptosis in crohn's disease. J Crohns Colitis. 2020;14:1734–1747. doi: 10.1093/ecco-jcc/jjaa088. [DOI] [PubMed] [Google Scholar]
- 6.Iborra S., Martínez-López M., Cueto F.J., Conde-Garrosa R., Del Fresno C., Izquierdo H.M., Abram C.L., Mori D., Campos-Martín Y., Reguera R.M., et al. Leishmania uses Mincle to target an inhibitory ITAM signaling pathway in dendritic cells that dampens adaptive immunity to infection. Immunity. 2016;45:788–801. doi: 10.1016/j.immuni.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kostarnoy A.V., Gancheva P.G., Lepenies B., Tukhvatulin A.I., Dzharullaeva A.S., Polyakov N.B., Grumov D.A., Egorova D.A., Kulibin A.Y., Bobrov M.A., et al. Receptor Mincle promotes skin allergies and is capable of recognizing cholesterol sulfate. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E2758–E2765. doi: 10.1073/pnas.1611665114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.N'diaye M., Brauner S., Flytzani S., Kular L., Warnecke A., Adzemovic M.Z., Piket E., Min J.H., Edwards W., Mela F., et al. C-type lectin receptors Mcl and Mincle control development of multiple sclerosis-like neuroinflammation. J. Clin. Invest. 2020;130:838–852. doi: 10.1172/JCI125857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsumoto M., Tanaka T., Kaisho T., Sanjo H., Copeland N.G., Gilbert D.J., Jenkins N.A., Akira S. A novel LPS-inducible C-type lectin is a transcriptional target of NF-IL6 in macrophages. J. Immunol. 1999;163:5039–5048. [PubMed] [Google Scholar]
- 10.Lee W.B., Kang J.S., Yan J.J., Lee M.S., Jeon B.Y., Cho S.N., Kim Y.J. Neutrophils promote mycobacterial trehalose dimycolate-induced lung inflammation via the Mincle pathway. PLoS Pathog. 2012;8:e1002614. doi: 10.1371/journal.ppat.1002614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Behler F., Maus R., Bohling J., Knippenberg S., Kirchhof G., Nagata M., Jonigk D., Izykowski N., Mägel L., Welte T., et al. Macrophage-inducible C-type lectin Mincle-expressing dendritic cells contribute to control of splenic Mycobacterium bovis BCG infection in mice. Infect. Immun. 2015;83:184–196. doi: 10.1128/IAI.02500-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flornes L.M., Bryceson Y.T., Spurkland A., Lorentzen J.C., Dissen E., Fossum S. Identification of lectin-like receptors expressed by antigen presenting cells and neutrophils and their mapping to a novel gene complex. Immunogenetics. 2004;56:506–517. doi: 10.1007/s00251-004-0714-x. [DOI] [PubMed] [Google Scholar]
- 13.Richardson M.B., Williams S.J. MCL and Mincle: C-type lectin receptors that sense damaged self and pathogen-associated molecular patterns. Front. Immunol. 2014;5:288. doi: 10.3389/fimmu.2014.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsunaga I., Moody D.B. Mincle is a long sought receptor for mycobacterial cord factor. J. Exp. Med. 2009;206:2865–2868. doi: 10.1084/jem.20092533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishikawa E., Ishikawa T., Morita Y.S., Toyonaga K., Yamada H., Takeuchi O., Kinoshita T., Akira S., Yoshikai Y., Yamasaki S. Direct recognition of the mycobacterial glycolipid, trehalose dimycolate, by C-type lectin Mincle. J. Exp. Med. 2009;206:2879–2888. doi: 10.1084/jem.20091750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rabes A., Zimmermann S., Reppe K., Lang R., Seeberger P.H., Suttorp N., Witzenrath M., Lepenies B., Opitz B. The C-type lectin receptor Mincle binds to Streptococcus pneumoniae but plays a limited role in the anti-pneumococcal innate immune response. PLoS One. 2015;10:e0117022. doi: 10.1371/journal.pone.0117022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamasaki S., Matsumoto M., Takeuchi O., Matsuzawa T., Ishikawa E., Sakuma M., Tateno H., Uno J., Hirabayashi J., Mikami Y., et al. C-type lectin Mincle is an activating receptor for pathogenic fungus, Malassezia. Proc. Natl. Acad. Sci. U. S. A. 2009;106:1897–1902. doi: 10.1073/pnas.0805177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiffoleau E. C-type lectin-like receptors as emerging orchestrators of sterile inflammation represent potential therapeutic targets. Front. Immunol. 2018;9:227. doi: 10.3389/fimmu.2018.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamasaki S., Ishikawa E., Sakuma M., Hara H., Ogata K., Saito T. Mincle is an ITAM-coupled activating receptor that senses damaged cells. Nat. Immunol. 2008;9:1179–1188. doi: 10.1038/ni.1651. [DOI] [PubMed] [Google Scholar]
- 20.Wynn M.L., Ventura A.C., Sepulchre J.A., García H.J., Merajver S.D. Kinase inhibitors can produce off-target effects and activate linked pathways by retroactivity. BMC Syst. Biol. 2011;5:156. doi: 10.1186/1752-0509-5-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keefe A.D., Pai S., Ellington A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng E.W.M., Shima D.T., Calias P., Cunningham E.T., Guyer D.R., Adamis A.P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006;5:123–132. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- 23.Alatab S., Sepanlou S.G., Ikuta K., Vahedi H., Bisignano C., Safiri S., Sadeghi A., Nixon M.R., Abdoli A., Abolhassani H., et al. The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol. 2020;5:17–30. doi: 10.1016/S2468-1253(19)30333-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mak W.Y., Zhao M., Ng S.C., Burisch J. The epidemiology of inflammatory bowel disease: east meets west. J Gastroenterol Hepatol. Mar. 2020;35:380–389. doi: 10.1111/jgh.14872. [DOI] [PubMed] [Google Scholar]
- 25.Arora Z., Shen B. Biological therapy for ulcerative colitis. Gastroenterol Rep (Oxf) 2015;3:103–109. doi: 10.1093/gastro/gou070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chao Y.S., Loshak H. Canadian Agency for Drugs and Technologies in Health; 2019. CADTH rapid response reports. Biologics versus immunomodulators for the treatment of ulcerative colitis: a Review of comparative clinical effectiveness and cost-effectiveness. [PubMed] [Google Scholar]
- 27.Bouchard P.R., Hutabarat R.M., Thompson K.M. Discovery and development of therapeutic aptamers. Annu. Rev. Pharmacol. Toxicol. 2010;50:237–257. doi: 10.1146/annurev.pharmtox.010909.105547. [DOI] [PubMed] [Google Scholar]
- 28.Haraldsson B., Nyström J., Deen W.M. Properties of the glomerular barrier and mechanisms of proteinuria. Physiol. Rev. 2008;88:451–487. doi: 10.1152/physrev.00055.2006. [DOI] [PubMed] [Google Scholar]
- 29.Kratschmer C., Levy M. Effect of chemical modifications on aptamer stability in serum. Nucleic Acid Ther. Dec. 2017;27:335–344. doi: 10.1089/nat.2017.0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chassaing B., Aitken J.D., Malleshappa M., Vijay-Kumar M. Dextran sulfate sodium (DSS)-induced colitis in mice. Curr. Protoc. Im. 2014;104:5.25.1–15.25.14. doi: 10.1002/0471142735.im1525s104. Unit-15.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laroui H., Ingersoll S.A., Liu H.C., Baker M.T., Ayyadurai S., Charania M.A., Laroui F., Yan Y., Sitaraman S.V., Merlin D. Dextran sodium sulfate (DSS) induces colitis in mice by forming nano-lipocomplexes with medium-chain-length fatty acids in the colon. PLoS One. 2012;7:e32084. doi: 10.1371/journal.pone.0032084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang Z., Lao R., Wang J., Liu Y., Wang L., Huang Q., Song S., Li G., Fan C. Solubilization of single-walled carbon nanotubes with single-stranded DNA generated from asymmetric PCR. Int. J. Mol. Sci. 2007;8:705–713. doi: 10.3390/i8070705. [DOI] [Google Scholar]
- 33.Koelink P.J., Wildenberg M.E., Stitt L.W., Feagan B.G., Koldijk M., van 't Wout A.B., Atreya R., Vieth M., Brandse J.F., Duijst S., et al. Development of reliable, valid and responsive scoring systems for endoscopy and histology in animal models for inflammatory bowel disease. Journal of Crohn's & colitis. 2018;12:794–803. doi: 10.1093/ecco-jcc/jjy035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Antczak M., Popenda M., Zok T., Sarzynska J., Ratajczak T., Tomczyk K., Adamiak R.W., Szachniuk M. New functionality of RNAComposer: application to shape the axis of miR160 precursor structure. Acta Biochim. Pol. 2017;63:737–744. doi: 10.18388/abp.2016_1329. [DOI] [PubMed] [Google Scholar]
- 35.Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H.J. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:W363–W367. doi: 10.1093/nar/gki481. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duhovny D., Nussinov R., Wolfson H.J. Springer Berlin Heidelberg; 2002. Efficient Unbound Docking of Rigid Molecules; pp. 185–200. [Google Scholar]
- 37.Kratschmer C., Levy M. Effect of chemical modifications on aptamer stability in serum. Nucleic Acid Therapeut. 2017;27:335–344. doi: 10.1089/nat.2017.0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Healy J.M., Lewis S.D., Kurz M., Boomer R.M., Thompson K.M., Wilson C., McCauley T.G. Pharmacokinetics and biodistribution of novel aptamer compositions. Pharmaceut. Res. 2004;21:2234–2246. doi: 10.1007/s11095-004-7676-4. [DOI] [PubMed] [Google Scholar]
- 39.Fernández G., Moraga A., Cuartero M.I., García-Culebras A., Peña-Martínez C., Pradillo J.M., Hernández-Jiménez M., Sacristán S., Ayuso M.I., Gonzalo-Gobernado R., et al. TLR4-Binding DNA aptamers show a protective effect against acute stroke in animal models. Mol. Ther. : the journal of the American Society of Gene Therapy. 2018;26:2047–2059. doi: 10.1016/j.ymthe.2018.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang Y.-C., Kao W.-C., Wang W.-Y., Wang W.-Y., Yang R.-B., Peck K. Identification and characterization of oligonucleotides that inhibit Toll-like receptor 2-associated immune responses. Faseb. J. 2009;23:3078–3088. doi: 10.1096/fj.09-129312. [DOI] [PubMed] [Google Scholar]
- 41.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 42.Stephens M., Liao S., von der Weid P.-Y. Ultra-purification of Lipopolysaccharides reveals species-specific signalling bias of TLR4: importance in macrophage function. Sci. Rep. 2021;11:1335. doi: 10.1038/s41598-020-79145-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.