Abstract
Monoclonal antibodies (mAbs) are appealing as potential therapeutics and prophylactics for viral infections owing to characteristics such as their high specificity and their ability to enhance immune responses. Furthermore, antibody engineering can be used to strengthen effector function and prolong mAb half-life, and advances in structural biology have enabled the selection and optimization of potent neutralizing mAbs through identification of vulnerable regions in viral proteins, which can also be relevant for vaccine design. The COVID-19 pandemic has stimulated extensive efforts to develop neutralizing mAbs against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), with several mAbs now having received authorization for emergency use, providing not just an important component of strategies to combat COVID-19 but also a boost to efforts to harness mAbs in therapeutic and preventive settings for other infectious diseases. Here, we describe advances in antibody discovery and engineering that have led to the development of mAbs for use against infections caused by viruses including SARS-CoV-2, respiratory syncytial virus (RSV), Ebola virus (EBOV), human cytomegalovirus (HCMV) and influenza. We also discuss the rationale for moving from empirical to structure-guided strategies in vaccine development, based on identifying optimal candidate antigens and vulnerable regions within them that can be targeted by antibodies to result in a strong protective immune response.
Subject terms: Infection, Antibodies, Viral infection
Monoclonal antibodies (mAbs) are appealing as potential therapeutics and prophylactics for viral infections. This Review describes advances in antibody discovery and engineering that have led to the development of mAbs that target viruses such as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), respiratory syncytial virus and Ebola virus, and also considers the implications for vaccine development.
Introduction
Since the development of the hybridoma technology enabling the generation of monoclonal antibodies (mAbs) by Köhler and Milstein in the 1970s1, mAbs have become a key class of drugs for cancer and immune disorders, with the 100th antibody-based therapeutic recently gaining US Food and Drug Administration (FDA) approval2. However, although harnessing antibodies to combat infectious diseases has a history stretching back more than a century to the applications of serum conferring protection against diphtheria toxin3, only a small number of mAb drugs are used to treat or prevent infectious diseases. At the time of writing in 2022, six mAbs targeting pathogens have so far been granted full approval by the FDA (Table 1), for indications including prevention of respiratory syncytial virus (RSV) infection, prevention and treatment of anthrax infection, prevention of recurrence of Clostridioides difficile infection and the treatment of Ebola virus (EBOV) infection.
Table 1.
FDA-approved mAbs for infectious disease indications
| Drug (brand name; company) | Target | Format | Technology | Indication | Year of FDA approval |
|---|---|---|---|---|---|
| Palivizumab (Synagis; MedImmune/AbbVie) | RSV | Humanized IgG1 | Hybridoma | Prevention of RSV infection | 1998 |
| Raxibacumab (ABthrax/Anthrin; GlaxoSmithKline/Human Genome Sciences) | Bacillus anthrasis PA | Human IgG1 | Human scFv phage display library | Anthrax infection | 2012 |
| Bezlotoxumab (Zinplava; Merck & Co.) | Clostridioides difficile enterotoxin B | Human IgG1 | Transgenic mice | Prevention of C. difficile infection recurrence | 2016 |
| Obiltoxaximab (Anthim; Elusys Therapeutics) | B. anthrasis PA | Chimeric IgG1 | Hybridoma | Prevention of inhalational anthrax | 2016 |
| Ibalizumaba (Trogarzo; TaiMed Biologics) | CD4 receptor (domain 2) | Humanized IgG4 | Mice | Treatment of HIV-1 infection | 2018 |
| Ansuvimab (Ebanga; MedImmune/Ridgeback Biotherapeutics) | Ebola glycoprotein | Human IgG1 | Human | Prevention and treatment of Ebola infection | 2020 |
| Atoltivimab, maftivimab and odesivimab (Inmazeb; Regeneron Pharmaceuticals) | Ebola glycoprotein | Human IgG1 | Transgenic mice | Prevention and treatment of Ebola infection | 2020 |
FDA, US Food and Drug Administration; mAb, monoclonal antibody; PA, protective antigen; RSV, respiratory syncytial virus; scFv, single-chain variable fragment. aAntibody with a host target, rather than a pathogen target.
The rapid spread of COVID-19 in 2020 led to intense efforts to develop neutralizing mAbs that target severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) for the treatment and prevention of COVID-19. As a result, more than 20 mAbs entered clinical development. So far, several of these mAbs have received emergency use authorization (EUA) from the FDA (Table 2) and other regulatory agencies worldwide, and more authorizations are anticipated. Although vaccines have been the mainstay of efforts to tackle COVID-19, mAbs can provide an important contribution for vulnerable populations before or after exposure to SARS-CoV-2, such as people who are immunocompromised or people with mild to moderate COVID-19 who are at high risk of developing severe disease. The mAbs under development providing broadly neutralizing activity against coronaviruses could also be instrumental for preparedness against future pandemics. Furthermore, the momentum built and the knowledge gained from the development of mAbs for SARS-CoV-2 may help accelerate the development of mAbs to combat other infectious diseases.
Table 2.
Selected mAbs marketed or in late-stage clinical studies for COVID-19
| Drug (brand name; company) | Origin | Engineering | Statusa | Omicron VOC neutralization | Ref. |
|---|---|---|---|---|---|
| Casirivimab and imdevimab (Ronapreve; Regeneron Pharmaceuticals) | Genetically humanized mice and B cells from a convalescent patient infected with SARS-CoV-2 | Unmodified | EUA granted by FDA for treatment and prevention of COVID-19 in 2020b | –, in vitro study322 | 118 |
| Bamlanivimab and etesevimab (NA; AbCellera/Eli Lilly) | B cells from convalescent patients infected with SARS-CoV-2 | Unmodified (bamlanivimab); LALA modification in Fc domain to extend half-life (etesevimab) | EUA granted by FDA for treatment and prevention of COVID-19 in 2021b | –, in vitro study322 | 124 |
| Sotrovimab (Xevudy; Vir Biotechnology/GlaxoSmithKline) | B cells from an individual infected with SARS-CoV | LS modification in Fc domain to extend half-life | EUA granted by FDA for the treatment of mild to moderate COVID-19 in 2021b | +/–, in vitro and clinical studies133,322–325 | 78 |
| Tixagevimab and cilgavimab (Evusheld; AstraZeneca) | B cells from convalescent patients infected with SARS-CoV-2 | YTE and TM modifications in Fc domain to extend half-life and reduce effector function, respectively | EUA granted by FDA for pre-exposure prophylaxis of COVID-19 in 2021 | +/–, in vitro study322 | 128 |
| Bebtelovimab (NA; AbCellera/Eli Lilly) | B cells from convalescent patients infected with SARS-CoV-2 | Unmodified | EUA granted by FDA for treatment of mild to moderate COVID-19 in 2022 | +++, in vitro study131 | 131 |
| Regdanvimab (Regkirona; Celltrion) | B cells from convalescent patient infected with SARS-CoV-2 | Modifications to Fc domain to reduce effector function | Approved in Republic of Korea and EU | –, in vitro study326 | 130 |
| Amubarvimab and romlusevimab (NA; Brii Biosciences) | B cells from convalescent patients infected with SARS-CoV-2 | YTE modification in Fc domain to extend half-life | Approved in China; EUA requested | +/–, in vitro and clinical studies323–325 | 327 |
| Adintrevimab (NA; Adagio Therapeutics) | B cells from convalescent patients infected with SARS-CoV-2 | LALA modification in Fc domain to extend half-life | Phase II/III | +/–, in vitro study323 | 328 |
All monoclonal antibodies (mAbs) listed are known to target the receptor-binding domain (RBD) of the spike (S) glycoprotein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Fc, crystallizable fragment; NA, not available; –, no neutralizing activity; +/–, partial neutralizing activity; +++, potent neutralizing activity. aInformation on status is with regard to the initial US Food and Drug Administration (FDA) emergency use authorizations (EUAs), unless the product was initially further advanced in other regions or countries. bEUA since withdrawn owing to likely ineffectiveness against the Omicron variant of concern (VOC).
In this Review, we first provide an overview of the technologies for the discovery and engineering of mAbs to target pathogens, with a focus on viruses. We then describe the progress in the development of mAbs against a range of viruses, including SARS-CoV-2, RSV, Ebola, cytomegalovirus (CMV) and influenza. Efforts to harness mAbs to combat bacterial infections have been reviewed elsewhere4,5 and are summarized briefly in Box 1. Finally, we also discuss how rapid mAb discovery combined with structural vaccinology can support the development of vaccines and therapeutic mAbs.
Box 1 Monoclonal antibodies as antibacterial agents.
Antimicrobial resistance is one of the top ten global public health threats facing humanity according to the World Health Organization (WHO). Although traditional small-molecule drug discovery still dominates the landscape for antibacterial solutions to antimicrobial resistance, the prophylactic or therapeutic use of monoclonal antibodies (mAbs) for bacterial infection could have an important role330. One of the main advantages of these biologics is their low toxicity, as bacterial virulence proteins are targeted instead of proteins required for survival, therefore avoiding the disruption of the microbiome and, potentially, limiting the development of resistance.
So far, three antibacterial mAbs have been approved for the treatment and prophylaxis of bacterial infections. Raxibacumab (ABthrax/Anthrin)36 and obiltoxaximab (Anthim)331 are both mAbs targeting the protective antigen (PA) component of the lethal toxin of Bacillus anthracis and are approved to treat inhalation anthrax due to B. anthracis. Bezlotoxumab is a mAb that binds to Clostridioides difficile enterotoxin B332. This product does not protect against or treat initial C. difficile infection but, rather, is used to reduce the recurrence of infection, which is often seen with C. difficile.
Antibody characteristics and engineering
Antibody structure and function
Antibodies are natural biomolecules generated by plasma cells or stimulated memory B cells after a pathogen infection or vaccination3. Structurally, they are Y-shaped heterodimers composed of two light chains of 25 kDa each and two heavy chains of at least 50 kDa, depending on the immunoglobulin isotype. The heavy and light chains are linked by multiple disulfide bridges and non-covalent interactions (Fig. 1a), with variations in the number of interactions and bridges depending on the immunoglobulin isotypes.
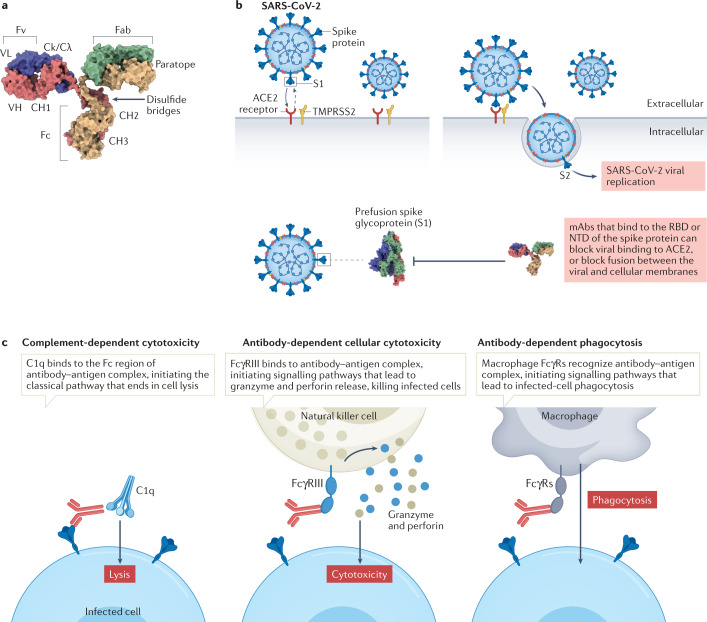
Fig. 1. Mechanism of action of monoclonal antibodies during viral infection.
a | Overview of monoclonal antibody (mAb) structure; heavy chains shown in yellow and red, light chains in blue and green. b | Binding of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) to the angiotensin-converting enzyme 2 (ACE2) receptor on host cells is mediated by the viral spike (S) protein, which comprises an S1 subunit (including a receptor-binding domain (RBD) and an amino-terminal domain (NTD)) and an S2 subunit. Priming of coronavirus spike proteins by host cell proteases such as the transmembrane serine protease TMPRSS2 through cleavage at S1/S2 and S2′ sites (see Fig. 3) is essential for viral entry. Therapeutic antibodies and antibodies elicited by vaccination that bind to the RBD or NTD can block viral binding to ACE2, or block fusion between viral and cellular membranes (see Fig. 3). c | Effector functions of antibodies. mAbs can facilitate target cell death via complement fixation and membrane attack complex (MAC) activation, which is known as complement-dependent cytotoxicity (CDC). Antibody-dependent cellular cytotoxicity (ADCC) is a mechanism of cell-mediated immune defence whereby an effector cell (natural killer cell, macrophage, neutrophil or eosinophil) of the immune system actively lyses a target cell, whose membrane has been bound by specific antibodies. Natural killer cells release cytotoxic factors (perforin and proteases known as granzymes) that cause death of the infected cell. Antibody-dependent cellular phagocytosis is the mechanism by which antibody-opsonized target cells activate Fcγ receptors (FcγRs) on the surface of macrophages to induce phagocytosis, resulting in internalization and degradation of the target cell through phagosome acidification. Fab, fragment antigen-binding domain; Fc, crystallizable fragment; Fv, variable fragment; VH, variable heavy; VL, variable light.
Antibodies can also be divided into functional components (Fig. 1b). The two fragment antigen-binding domains (Fabs) bind to and neutralize pathogens. These are linked to the crystallizable fragment (Fc) domain by a hinge region that gives the Fabs a large degree of conformation flexibility relative to the Fc domain, allowing them to strongly interact with any antigen regardless of its orientation. The glycosylated Fc domain binds to other proteins, including Fcγ receptors (FcγRs) on various immune cells and complement protein C1q, to mediate effector functions such as antibody-dependent cellular cytotoxicity (ADCC), complement-dependent cytotoxicity (CDC) and antibody-dependent cellular phagocytosis (Fig. 1c). The Fc domain also influences antibody pharmacokinetics via interaction with the neonatal Fc receptor (FcRn).
Antibodies vary in isotype depending on whether the alpha, mu, gamma, epsilon or delta gene segments recombine with the variable region. In humans, the following genes generate different subclasses of antibodies: two alpha gene segments (IgA1 and IgA2), four gamma gene segments (IgG1, IgG2, IgG3 and IgG4), one mu segment (IgM), one epsilon segment (IgE) and one delta segment (IgD). Each subclass specializes in the elimination of different types of pathogens, except for IgD, for which the function is still poorly characterized6. The IgG class is the principal isotype in the blood and extracellular fluid. An important aspect of the different isotypes is that their sequence variation determines their affinities and specificities for FcRn, FcγRs and complement protein C1q. Notably, the IgG1 isotype allows ADCC and CDC (Fig. 1c), IgG2 and IgG4 are poor CDC activators and IgG3 is a potent CDC activator. Most therapeutic mAbs in clinical use or development against infectious diseases are of the human IgG1 isotype, which has affinity for activating FcγRs but also exhibits binding to the inhibitory FcγRIIb, thereby limiting protective Fc effector activities7.
Antibodies as antivirals
For more than a century, passive immunization with monoclonal or polyclonal antibodies has been used in the treatment and prevention of infectious diseases, particularly in individuals with immunodeficiencies or individuals for whom vaccination is contraindicated.
Antibodies can combat viral infections through several mechanisms. First, antibodies can prevent viral glycoproteins of enveloped viruses or the protein shell of non-enveloped viruses from binding to the target host cells8,9. These viral proteins have two major functions in the viral life cycle: binding to cellular receptors and mediating the fusion of viral and cellular membranes (in the case of enveloped viruses) or penetration into the cytosol (in the case of non-enveloped viruses). For example, the entry of SARS-CoV-2 into host cells is mediated by the interaction between the viral spike (S) glycoprotein and the angiotensin-converting enzyme 2 (ACE2) receptor on the host cell surface. ACE2 is expressed on cells of the respiratory system, gastrointestinal tract and endothelium10 (Fig. 1b). The spike–ACE2 interaction can be blocked by antibodies targeting the spike receptor-binding domain (RBD)11, which inhibits viral infection, as discussed later in the Review (Fig. 1b).
Antibody effector functions mediated by engagement with complement protein C1q or FcγRs on leukocytes can also be involved in combating viral infections (Fig. 1c). Complement activation by antibodies leads to direct lysis of the virus and/or the infected host cell, and antibodies can also promote or induce phagocytosis, or trigger the release of toxic chemicals, such as cytokines or reactive oxygen species12. For instance, it was recently shown that Fc effector functions are required for optimal protection by mAb therapy for SARS-CoV-2; when given after infection, intact mAbs reduced the SARS-CoV-2 burden and lung disease in animals better than loss-of-function Fc variant mAbs13–15 by mitigating inflammation and improving respiratory mechanics.
Conversely, in rare cases, suboptimal binding of antibodies to virions can facilitate viral pathogenesis through a process known as antibody-dependent enhancement (ADE)16, in which recognition of virion–antibody complexes by FcγRs enhances viral entry into host immune cells. ADE was first observed with Dengue virus in the presence of sub-neutralizing antibody concentrations. For example, when the level of maternal antibodies against Dengue virus in newborns wanes, some individuals will experience an interval during which their antibody level will drop below its protective capacity, leading to severe disease following infection17. Dengue virus co-circulates as four serotypes, and the increased severity of some secondary infections is thought to be due to enhancement of viral entry by pre-existing antibodies generated following a primary infection with a different serotype that are not able to neutralize the second serotype18,19. Enhanced disease was also observed after vaccination with a formalin-inactivated RSV vaccine in infants20. The risk of ADE can be reduced by engineering the antibody Fc domain to reduce binding to FcγRs, as noted below.
The first passive immunization approaches used serum derived from animals actively immunized with an antigen such as diphtheria toxin, but such approaches come with the risk of provoking an immune response against non-human antibodies. These risks can be mitigated by using blood from people who have recovered from an illness as the source of antibodies, known as convalescent plasma therapy (CPT)21. Although CPT has been historically successful in combating infections22, it has proved to be an inconsistent tool, as antibody responses between individuals are highly variable. CPT also has safety risks, including allergic reactions and low risk of infections by other viruses such as HIV, hepatitis B and hepatitis C. Furthermore, widespread use of CPT in the context of a pandemic such as COVID-19 would depend on the availability of a sufficient number of plasma donors and facilities for appropriate processing. Overall, the role of CPT in such cases may be restricted to the early epidemic phase when therapeutic options are limited. Indeed, although plasma obtained from convalescent donors has previously been used as a therapy for coronavirus infections23 and was investigated as a potential therapeutic option for treatment of patients severely ill with COVID-19, the results of randomized controlled trials indicated that CPT has no benefit for patients with moderate, severe or critical COVID-19 infection24,25. Nevertheless, no adverse effects were reported and a potential beneficial effect in younger patients was observed26.
Given the limitations of CPT, there has been an increasing focus on the use of neutralizing mAbs for passive immunization for infectious diseases. Neutralizing mAbs with high specificity and potency can be developed and extensively characterized, and lack the risk of blood-borne disease associated with CPT. Furthermore, they can be produced at a large scale in a reasonable time frame with well-established processes.
Strategies to generate human therapeutic antibodies for viral infections
Most strategies to identify human mAbs to combat pathogens can be classified as either targeted, in which mAbs that bind to a known antigen are directly isolated, or target agnostic, in which functional assays are performed on secreted immunoglobulins obtained from the supernatant of single cell cultures.
The first efficient targeted approach for mAb identification involved panning phage display libraries constructed from the immunoglobulin variable genes of immunized or infected individuals based on binding to a target antigen27. Alternatively, random synthetic libraries were also used28. Although these methods have led to the isolation of neutralizing antibodies against multiple pathogens (for example, HIV29, SARS-CoV-2 (ref.30) and the anthrax toxin31), the obtained mAbs did not represent the natural antibody repertoire as the antibody fragments were generated from random pairings of immunoglobulin variable heavy (VH) and variable light (VL) regions (Fig. 2a). Indeed, several libraries were based on a single or limited set of V region frameworks, leading to diversification of the CDRH3 only32. Moreover, VH/VL pairing is known to be an important diversity factor, and artificial pairings can generate autoreactive molecules33, as no negative selection is present34,35. Nevertheless, a naïve human single-chain variable fragment (scFv) phage display library was used to develop a potent antitoxin mAb for anthrax, raxibacumab36, which was approved in 2012 (Box 1 and Table 1).
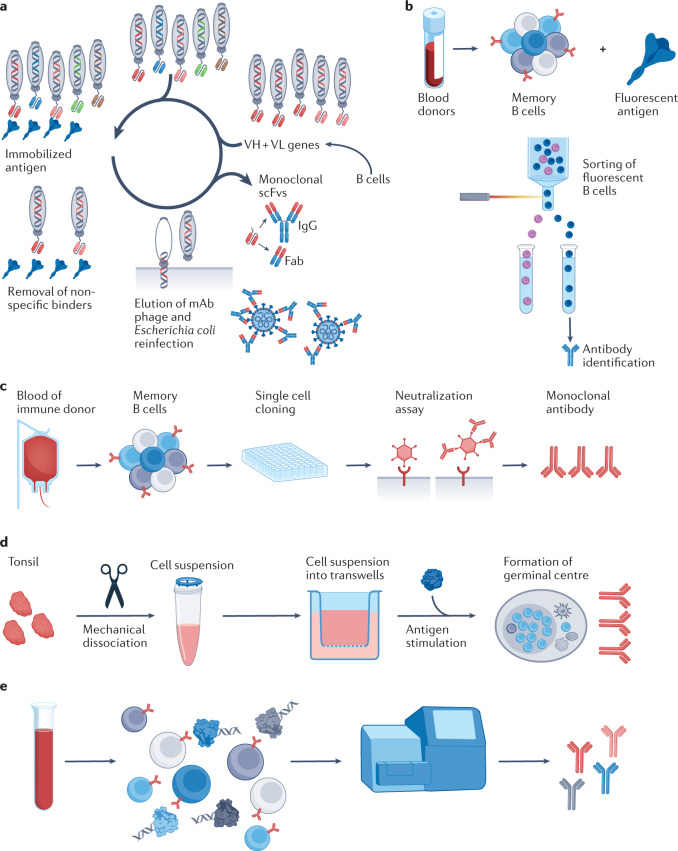
Fig. 2. Antibody discovery approaches.
a | Phage bio-panning is based on a library of phages that contain genes coding for variable heavy (VH)/variable light (VL) domains, leading to production of encoded antibodies on phage surfaces. Selection of antibodies produced by phages involves immobilization of the ligand of interest on a solid support (spike (S) protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) shown), followed by applying the phage display library to immobilized ligand to allow binding of specific variants. To eliminate adherent non-binders, multiple rounds of washing are usually performed, and remaining bound phages are eluted and re-amplified. b | Recombinant antigens conjugated to a fluorescent marker are incubated with class-switched memory B cells and sorted according to their capacity to bind the antigen of interest (such as the S protein of SARS-CoV-2) by fluorescence-activated flow cytometry, followed by identification of the antibodies they produce. c | Target-agnostic approaches using single B cell culture. Single B cells are seeded on a feeder layer in the presence of a cytokine mix and a Toll-like receptor (TLR) activator. Culture supernatant is screened for neutralization activity and clones of interest are retrieved and sequenced. d | Workflow for organoid reconstitution from human tonsils to develop an in vitro system that recapitulates key germinal centre features, including production of antigen-specific antibodies, somatic hypermutation and affinity maturation59. e | Workflow for single cell immune profiling69. B cells are encapsulated with barcoded gel beads in a single partition and undergo reverse transcription followed by PCR. Each cDNA is barcoded from its individual cell of origin and processed for next-generation sequencing. Fab, fragment antigen-binding domain; mAb, monoclonal antibody; scFv, single-chain variable fragment. Panel a is adapted from ref.329, under a Creative Commons license CC BY 3.0.
A second targeted approach developed subsequently is the direct isolation of antigen-specific memory B cells based on their capacity to bind fluorescent bait antigens, followed by identification of the mAbs they produce37. The memory B cells can originate from the plasma of convalescent patients38, or from transgenic mice carrying human immunoglobulin loci that produce fully human antibodies in response to immunization with a target antigen39. This approach has been particularly successful in the isolation of broadly neutralizing antibodies (bNAbs) targeting the CD4-binding site in the V1/V2 and V3 regions of gp120 and the membrane-proximal external region of gp41 of HIV40–45. Furthermore, mAbs against the hepatitis B virus viral S antigen (HBsAg)46 and the SARS-CoV-2 spike protein47 (Fig. 2b) have also been obtained, as discussed below.
The major limitation of targeted approaches is that target antigens must be known in advance because the selection process is based on binding affinity to the purified antigen rather than neutralization potency. Target-agnostic approaches present a viable alternative when limited information is available on the pathogen to be neutralized48. Various methods to obtain single cell cultures of memory B cells or plasma cells have been described49,50. Memory B cell immortalization using Epstein–Barr virus51,52 remains an attractive method because of its limited cost53 (Fig. 2c). A limitation of the Epstein–Barr virus approach is the suboptimal immortalization of B cells, which plateaus at approximately 35%51. However, the development of single cell cultures without the need for B cell immortalization can overcome this limitation54–56, and has been used for the identification of antibodies against pathogens such as group 1 and group 2 influenza A viruses57 and BK/JC polyomaviruses58. Recently, a functional organotypic system for antibody generation has been reported. The organoid recapitulates germinal centre features in vitro, such as the production of antigen-specific antibodies with affinity maturation and class-switch recombination from human tonsils, and this is a promising step forward for the field of mAb development59 (Fig. 2d).
In all cases, once identified, the mAb candidates must be sequenced for further recombinant expression. Cloning and expression of individual antibodies was traditionally labour-intensive, and throughput was largely limited to a few hundred clones. However, recent technological advances using nanofluidic devices have considerably increased the throughput of this approach60. In addition, advances in next-generation sequencing have enabled high-throughput screening and sequencing of paired antibody repertoires61. Currently, isolated B memory cells or plasma cells are injected into microfluidic devices (such as the 10x Genomics platform), generating droplets containing a single cell and lysis buffer with microbeads covered by barcoded primers to generate cDNA encoding VH and VL sequences60,62–65. These approaches allow the discovery of antibodies that are potentially useful as therapeutics66, in addition to the possibility of studying the human antibody repertoire at an unprecedented resolution67, and could be deployed as emergency response platforms for investigating mAbs from the blood of people who have recovered from emerging viral infections68.
Recently, the LIBRA-seq (linking B cell receptor to antigen specificity through sequencing) methodology was developed for high-throughput mapping of paired heavy-chain and light-chain B cell receptor sequences to their cognate antigen specificities69. In this approach, B cells are mixed with a panel of DNA-barcoded antigens and both the antigen barcode(s) and B cell receptor sequences are recovered via single cell next-generation sequencing. This enabled the antigen specificity of thousands of B cells from two subjects infected with HIV to be mapped, and the predicted specificities were confirmed for numerous HIV, influenza and SARS-CoV-2-specific antibodies, including known and unknown bNAbs (Fig. 2e). Indeed, large antibody data sets can be analysed computationally to infer antibody sequences and binding modality, and this has emerged as a powerful method for the identification of structurally related antibodies from sequence databases70–72. However, a limitation of the LIBRA-seq methodology, which is inherent to targeted approaches, is the need for a recombinant bait antigen that requires extensive validation73.
Antibody engineering
Several regions of mAbs can be engineered to improve their therapeutic characteristics.
In addition to the variability induced by isotype usage, several mutations in the Fc domain have been identified to increase or decrease the effectiveness of ADCC and/or CDC. For example, etesevimab, an anti-SARS-CoV-2 mAb developed by Eli Lilly that has received an EUA for COVID-19, has been engineered to lack FcγRI and FcγRII-binding activity with L234A and L235A mutations (the LALA modification)74 in the Fc domain to reduce safety concerns over the potential to exacerbate disease through ADE mechanisms75. Another example is provided by AZD7442, a cocktail of the anti-SARS-CoV-2 mAbs tixagevimab (AZD8895) and cilgavimab (AZD1061)76 developed by AstraZeneca, which has received an EUA for COVID-19. Both mAbs in the combination have engineered Fc domains including L234F/L235/P331S substitutions77 (the TM modification), resulting in little or no binding to various FcγRs or complement protein C1q, and little or no effector function in vitro76. Other mAbs against COVID-19 and their modifications are described in Table 2.
Engineering efforts have also focused on improving the mAb half-life in vivo by reducing IgG catabolism. This is regulated by mAb interaction with FcRn, which functions as a recycling or transcytosis receptor and is responsible for maintaining IgG and albumin in circulation and bidirectional transport across polarized cellular barriers. FcRn binds to IgG at the CH2–CH3 junction in a pH-dependent manner. IgG tightly binds at acidic pH (pH 6.0) but not at physiological pH (pH 7.4). Moreover, hydrophobic interactions between FcRn and Fc are stabilized by salt bridges formed between anionic residues on FcRn and protonated histidine or glutamic acid residues of the IgG Fc region in positions 117, 132 and 137 or 310, 435 and 436, respectively. Therefore, mutagenesis of Fc region residues at the FcRn–Fc interface is used to increase the half-life of IgG in the circulation.
Multiple antiviral mAbs with engineered Fc regions to extend their half-life have entered clinical development. Among the furthest advanced is sotrovimab (also known as VIR-7831 and GSK4182136), a mAb developed by Vir Biotechnology and GlaxoSmithKline against SARS-CoV-2 that received a EUA from the FDA in 2021. Sotrovimab was developed with an Fc domain that includes M428L and N434S amino acid substitutions (the LS modification) to extend antibody half-life78. Vir Biotechnology also incorporated the LS modification into VIR-3434, as well as G236A/A330L/I332E amino acid substitutions (the GAALIE modification)79 for enhanced FcγRIIIa binding. This mAb was designed to prevent chronic infections of hepatocytes by all ten hepatitis B virus genotypes and is in a phase II trial (NCT04856085).
Further antiviral mAbs that use the LS modification include VRC01LS, VRC07-523LS and elipovimab (GS-9722) for HIV80. VRC01LS, a broadly neutralizing mAb that was developed by the National Institutes of Health (NIH), shows an approximately fourfold longer serum half-life than the parent antibody with a wild-type Fc domain (VRC01) and showed similar neutralizing activity in serum to VRC01 during 48 weeks of a phase I trial81. It has also been studied in a phase I trial (NCT02256631) in combination with VRC01 and another variant of VRC01, VRC07-532LS (ref.82), that also has the LS modification. Elipovimab, developed by Gilead, is derived from the HIV-neutralizing antibody PGT121 (ref.83) and has an engineered Fab region to lower the immunogenicity and improve the stability at low pH, as well as the LS modification in the Fc domain to extend its half-life.
The triple amino acid mutation at M252Y/S254T/T256E (the YTE modification) was shown to promote a fourfold increase in serum half-life of mAbs due to increased binding to FcRn84, and was used in tixagevimab and cilgavimab on top of the TM modification (see above) to extend their half-life76. Another example using the YTE modification is nirsevimab, a mAb targeting the RSV fusion (F) glycoprotein developed by AstraZeneca and Sanofi Pasteur85 (see below). In a phase III clinical trial in healthy preterm infants, nirsevimab showed an extended half-life, offering protection from RSV for a typical 5-month season with a single intramuscular dose (50 mg)86,87.
Antibody engineering can also be used to generate novel antibody formats, such as bispecific antibodies (bsAbs) designed to recognize two different epitopes or antigens. A single bsAb can therefore bind to two different proteins or two different sites on the same protein. A wide range of bsAb formats have been developed, particularly for oncology applications88, but there have also been a few bsAbs investigated for infectious diseases. For example, a bsAb targeting both the receptor-binding site (RBS) of the Niemann-Pick C1 (NPC1) protein and a conserved surface-exposed epitope on the EBOV glycoprotein was shown to neutralize all known EBOVs by co-opting viral particles for endosomal delivery and conferred post-exposure protection against multiple EBOVs in mice89. Another example, a bsAb for HIV-1, also illustrates the potential of engineering the hinge domain to provide flexibility of orientation and rotation to the Fab region in regard to the Fc fragment. Bispecific anti-Env neutralizing antibodies with an engineered IgG3 hinge domain to increase Fab domain flexibility demonstrated improved neutralization potency and enhanced in vivo protective activity in HIV-1-infected humanized mice90.
Extending the multi-specificity concept further, trispecific antibodies engineered to interact with three independent HIV envelope determinants (the CD4-binding site, the membrane-proximal external region and the V1/V2 glycan) conferred complete immunity against a mixture of simian-human immunodeficiency viruses in non-human primates (NHPs), and showed higher potency and breadth than any previously described single broadly neutralizing mAb91. One such agent, SAR441236, has entered phase I development (NCT03705169).
Other novel engineered antibody formats that could be applied in antiviral agents include small camelid VHHs (15 kDa), known as nanobodies, that retain full antigen specificity, in contrast to mouse and human antibody-binding domains (50 kDa). Furthermore, nanobodies possess extended complementarity-determining regions, enabling binding of epitopes that are not normally accessible to conventional antibodies92, such as conserved viral domains that are often masked by glycan shields. Clinical trials demonstrated that they are safe and possess low immunogenicity93. Interestingly, transgenic mice encoding 18 alpaca, 7 dromedary and 5 Bactrian camel VHH genes were shown to generate potent neutralizing nanobodies against SARS-CoV-2 (ref.94).
Finally, antibody mimetics such as designed ankyrin repeat proteins (DARPins)95 can provide high affinity and offer multi-specificity. For instance, a multi-DARPin (ensovibep) that binds simultaneously to all three units of the SARS-CoV-2 spike RBD96 developed by Molecular Partners and Novartis97,98 is in clinical trials (NCT04828161).
Antibodies to combat viral infections
Coronaviruses, including SARS-CoV-2
Coronaviruses are enveloped positive-sense single-stranded RNA viruses belonging to the Coronaviridae family99. They can infect a wide variety of mammalian and avian species, causing respiratory and/or intestinal tract diseases. Human coronaviruses are major causes of the common cold and are responsible for up to 30% of mild respiratory tract infections and atypical pneumonia in humans100. Four different coronaviruses usually circulate in the human population: HCoV-OC43, HCoV-HKU1, HCoV-NL63 and HCoV-229E101.
In the past two decades, three coronaviruses with the potential to cause life-threatening disease in humans have emerged. Severe acute respiratory syndrome coronavirus (SARS-CoV) emerged in 2002 in China and spread, resulting in 8,100 infections and nearly 800 deaths in 37 countries102,103. Ten years later, the Middle East respiratory syndrome coronavirus (MERS-CoV) emerged in the Arabian Peninsula and spread to 21 countries, causing outbreaks in humans and infecting around 2,500 individuals, with a fatality rate of 35%104,105. In 2019, infections by a coronavirus now known as SARS-CoV-2 that can cause fever, severe respiratory illness, pneumonia, diarrhoea, dyspnoea and multiple organ failure were identified in China106,107. As of February 2022, more than 414 million cases have been confirmed, leading to at least 5.8 million deaths, according to the World Health Organization (WHO)108.
All coronaviruses enter the host cells using a trimeric spike (S) transmembrane glycoprotein109. The S protein is a type I membrane class I fusion protein110 and is organized into two functional subunits, which remain non-covalently bound in the pre-fusion conformation of the protein (Fig. 3a). The amino-terminal S1 subunit is responsible for binding to the host cell receptors, whereas the carboxy-terminal S2 subunit is responsible for fusion of the viral and cellular membranes111. The S1 subunit is further divided into an N-terminal domain (NTD) and a RBD (Fig. 3a).
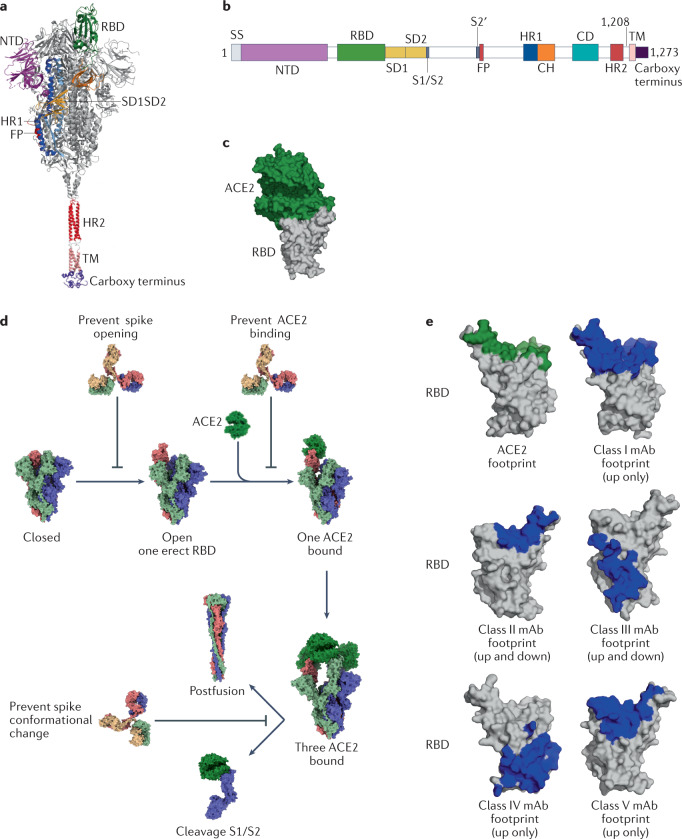
Fig. 3. The SARS-CoV-2 spike protein and neutralizing antibodies targeting the receptor-binding domain.
a | Structural representation of the spike (S) protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) with modelled carboxy-terminal part. b | SARS-CoV-2 spike protein domain architecture. c | Structure of the receptor-binding domain (RBD) (grey) bound to angiotensin-converting enzyme 2 (ACE2) (forest green) [PDB:6M0J]. d | Neutralizing antibodies can prevent opening of the S protein to block RBD erection, block the RBD to prevent binding to ACE2, block the amino-terminal domain (NTD) or block the fusion step. e | Footprints of ACE2 (green) or footprints of different ultrapotent antibody neutralizer classes (blue) shown on the RBD (grey). To generate footprints, the ‘Clashes/Contacts’ tool in UCSF Chimera was used to identify residues on the RBD that contact monoclonal antibodies (mAbs). Default contact criteria with van der Waals overlap of ≥–0.4 Å or more were used. CH, central helix; CD, connector domain; FP, fusion peptide; HR1, heptad repeat 1; HR2, heptad repeat 2; SD1, subdomain 1; SD2, subdomain 2; SS, signal sequence; TM, transmembrane region.
SARS-CoV-2 is phylogenetically closely related to SARS-CoV, sharing approximately 79.6% genomic sequence identity112, and similar to SARS-CoV uses the S1 RBD to bind to ACE2 receptors on host cell types such as pneumocytes and enterocytes113. After host cell binding, a conformational change in the S2 subunit results in virus fusion and entry into the target cell114 (Fig. 3b).
The S glycoprotein has been the primary focus of efforts to develop mAbs to target SARS-CoV-2, as it was already known to be a target of potent neutralizing mAbs against SARS-CoV115 and MERS-CoV116. More than 20 mAbs that target the S glycoprotein, originating either from the B cells of convalescent patients with COVID-19 or immunization of humanized mice, have been tested in clinical trials, and some have received an EUA from the FDA for the treatment of patients with mild to moderate COVID-19 or for pre-exposure prophylaxis (Table 2). These include sotrovimab117, the combination of casirivimab and imdevimab118–121, bamlanivimab122–124 (used as a monotherapy or in combination with etesevimab125), the combination of cilgavimab126 and tixagevimab76,127, regdanvimab128–130 and bebtelovimab131.
However, a major limitation of most mAbs evaluated so far against COVID-19 has been the rapid appearance of SARS-CoV-2 variants of concern (VOCs) that can escape both single mAbs and cocktails of mAbs132 (Table 2). VOCs such as Alpha, Beta, Gamma and Delta have 9–12 mutations in regions of the S glycoprotein, which typically have only a partial impact on the effectiveness of therapeutic mAbs. However, Omicron variants have accumulated more than 35 mutations in the S glycoprotein, of which 15 occur in the RBD, which is not only the site that binds to the host receptor ACE2 but also the key target of therapeutic mAbs, as well as neutralizing antibodies produced by the natural and vaccine-induced immune response. The emergence of Omicron VOCs (BA.1, BA.1.1 and BA.2) has rendered numerous mAbs with EUAs and/or in advanced clinical development partially or almost ineffective. These include the combination of casirivimab and imdevimab, the combination of bamlanivimab and etesevimab, and sotrovimab (Table 2). These mAbs have all been developed from patients infected with SARS-CoV-2, with the exception of sotrovimab, which was isolated from an individual infected with SARS-CoV. Nevertheless, two recently developed mAbs, bebtelovimab and P2G3, retain full activity against the Omicron VOCs131,133.
Additional, ultrapotent neutralizing antibodies binding the RBD have been identified, and some are currently in clinical development (Table 2). These mAbs are categorized into five groups based on their clustering and binding to the RBD (Fig. 3c). The group 1 mAbs bind the receptor-binding motif (RBM) similarly to ACE2 on the left side of the ridge, binding L455, F456, F486, N487 and Y489; members of this group include REGN10933 (ref.134), S2E12 (ref.135), COVA2-40 (ref.136), BD-236 (ref.134), C102 and C105 (ref.137). The group 2 mAbs also bind the RBM, but are positioned more upright and straddle the centre of the ridge, binding Y449, G485 and F486; this group includes P5C3 (ref.138), S2M11 (ref.135), COVA2-39 (ref.139) and mAb 2-4 (ref.140). Group 3 mAbs bind on the right side of the ridge opposite from group 1 and target Y449, E484 and F490; this group includes BD-368-2 (ref.134), CVO7-270 (ref.141) and P2B-2F6 (ref.142). The group 4 mAbs bind the lower half of the left side of the RBD, targeting Y369, C379, P384 and T385, and include mAbs binding the CR3022 cryptic site143; mAbs of this group are thought to act by destabilizing the pre-fusion conformation of the trimeric S protein144. Finally, group 5 mAbs bind the rear right side of the protein and include S309 (ref.145) and REGN10987 (ref.146) (Fig. 3d).
Although the RBD is immunodominant, additional regions of the S protein are immunogenic, most notably the NTD140,147,148 (Fig. 3a). Structural characterization of NTD-specific antibodies 4A8 (ref.149) and 4-8 (ref.140) showed that mAbs targeted the upper side of the protruding area of the NTD. Epitope mapping of 41 NTD-specific mAbs led to the identification of six antigenic sites, one of which is recognized by all known NTD-specific neutralizing antibodies and has been termed the ‘NTD supersite’, consisting of residues 14–20, 140–158 and 245–264 (ref.147). The mechanism of neutralization by which NTD-specific antibodies act remains to be fully determined, although it may involve the inhibition of conformational changes.
Cross-reactive conformational S2 epitopes have also been described150. Moreover, five mAbs cross-reacting with the stem helix of multiple betacoronavirus S proteins were recently identified in individuals convalescing after COVID-19 infection151. The biological significance of these different mAbs is still under investigation.
It is crucial to establish the target populations for treatment with mAbs and to define what should be the optimal timing for their use. It appears that mAbs may play a prophylactic role in individuals deemed to be at high risk of severe COVID-19, such as older people and/or individuals with polymorbidities, and immunocompromised individuals with poor or no response to vaccination. Several reports suggested that mAbs prevent COVID-19 in high-risk individuals potentially exposed to SARS-CoV-2 in nursing homes or within households152. Another question to address is how to increase the duration of action of mAbs, as they usually only allow a temporary window of protection.
Finally, although there were initial concerns about the risk of anti-SARS-CoV-2 mAbs causing ADE, there is currently no evidence to show ADE occurs with any of the mAbs tested in clinical trials. For a review of this issue, please see refs75,153.
Human respiratory syncytial virus
RSV is an enveloped negative-stranded RNA virus belonging to the Pneumoviridae family154. RSV infections are extremely common and typically result in mild respiratory symptoms. However, infection in infants and older adults accounts for a substantial hospitalization burden in both age groups.
High levels of RSV-neutralizing mAb titres correlate with protection in children and adults, including older people155,156. The first RSV intravenous immunoglobulin infusion preparation, named RespiGam, was used prophylactically from the late 1990s to the early 2000s to prevent severe RSV-associated lower respiratory tract disease in young children with bronchopulmonary dysplasia or premature birth157,158. The use of RespiGam was discontinued in 2003 and replaced by prophylaxis with palivizumab, the first neutralizing mAb developed to treat severe RSV infection in high-risk infants159, which was approved in 1998.
Of the three RSV surface proteins (F, G and SH), F-specific antibodies account for the majority of neutralizing activity in the sera of infected humans160,161, and so the F glycoprotein has been the focus of mAb development for RSV. F is a trimeric type I fusion glycoprotein responsible for merging the viral membrane with cellular membranes, and similar to many other viral fusion glycoproteins it undergoes major structural rearrangements during the transition from the pre-fusion to the post-fusion state110. Palivizumab is a humanized mAb that binds to antigenic site II of the F glycoprotein. Importantly, site II as well as sites I, III and IV are present in both pre-fusion and post-fusion conformations of the F glycoprotein (Fig. 4a). Therefore, palivizumab does not prevent triggering of conformational changes in F, and presumably blocks entry and membrane fusion by preventing the pre-hairpin to hairpin or the hairpin to post-fusion conformational change162. Motavizumab, an affinity-matured derivative of palivizumab that binds to the F glycoprotein with tenfold greater potency163, was shown to generate a relative decrease of 26% in RSV hospitalization compared with palivizumab. However, results from phase III clinical trials in 2010 showed only a marginal improvement in comparison with palivizumab164 and an increase of adverse events164, leading to termination of its development.
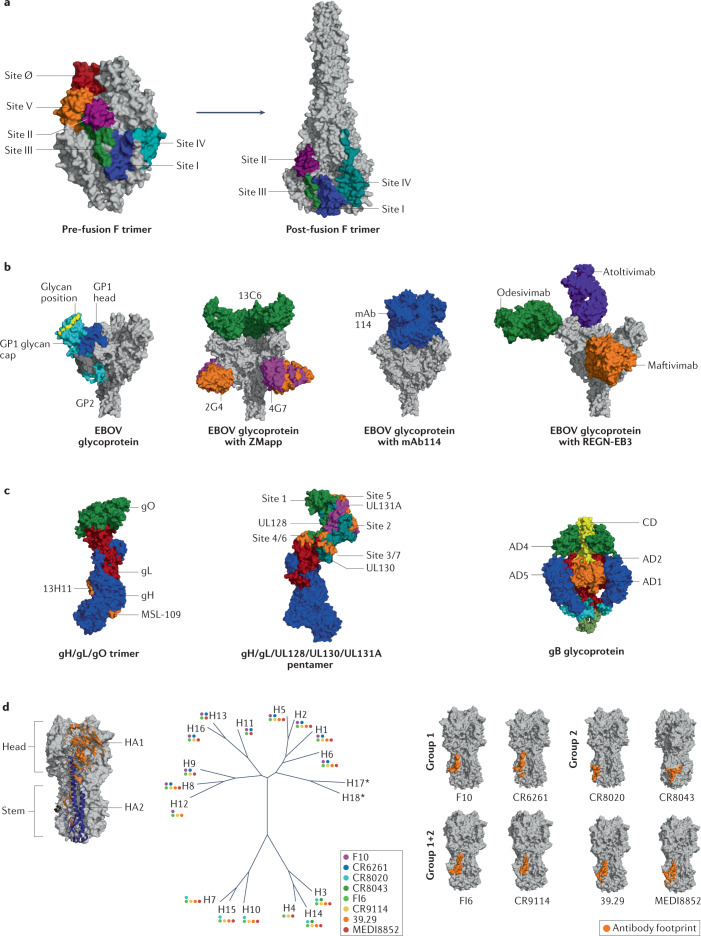
Fig. 4. Neutralizing antibodies that bind to glycoproteins from various pathogens.
a | Antigenic sites of the fusion protein (F) of respiratory syncytial virus (RSV). Locations of six antigenic sites on pre-fusion structure shown on left [PDB:4MMU], with locations on post-fusion structure shown on right [PDB:6APB]. b | Crystal structure of Zaire Ebola virus (EBOV) glycoprotein [PDB:5JQ3]: GP2 in dark grey, GP1 head in blue, GP1 glycan cap in cyan and glycans in yellow. To the right, trimeric glycoprotein shown with the ZMapp antibody cocktail [PDB:5KEN and PDB:5KEL], with mAb114 [PDB:5FHC] or with three monoclonal antibodies (mAbs) that make up REGN-EB3. c | Glycoproteins of human cytomegalovirus (HCMV) and antibody-binding sites. Trimer gH/gL/gO structure [PDB:7LBE] shown with antibody footprint (orange) for MSL-109 and 13H11 mAb on gH (left) and gH/gL/UL128/UL130/UL131A pentamer [PDB:5VOD] (centre) shown with footprint of antigenic sites present on UL. On the right, structures of gB in pre-fusion [PDB:7KDP] shown with coloured antigenic sites (AD1 in orange, AD2 in red, AD4 in green and AD5 in blue). d | Influenza haemagglutinin (HA) consists of three homotrimers built from two chains, HA1 and HA2, that are linked via a disulfide bond. HA molecules are commonly divided into immunodominant head and immunosubdominant stem. Based on sequence variations of HA, influenza is divided into two phylogenetic groups that are further subdivided into a total of 18 strains. Colour-coded dots represent observed binding of various broadly neutralizing antibodies (bNAbs) against different HAs [PDB:3S12]. Footprints of several group 1-specific and group 2-specific antibodies as well as pan-group reactive antibodies on HA stem shown to right. CD, core domain.
Novel mAbs under evaluation for RSV such as nirsevimab (MEDI8897) and suptavumab (REGN2222) target antigenic sites that are present only in the pre-fusion conformation of the F glycoprotein. Nirsevimab, which is derived from a mAb called D25 isolated by AIMM Therapeutics using a target-agnostic approach165, targets the antigenic site Ø166 and is more potent than palivizumab in vitro167. It has also been engineered for extended half-life through ‘YTE’ substitutions in the Fc region (see Antibody engineering section), which means that only one dose of nirsevimab may be required to cover a typical 5-month RSV season, rather than the five once-monthly doses that would be required for palivizumab. Indeed, nirsevimab showed an extended half-life offering protection from RSV for a typical 5-month season with a single intramuscular dose in a phase III trial86. However, suptavumab, which targets the antigenic site V, failed to meet the clinical end points in a phase III study due to the appearance of resistant RSV B strains168,169.
Ebola virus
Ebolaviruses are single-stranded RNA viruses that belong to the Filoviridae family. The genus Ebolavirus contains six species with different designations: Bundibugyo ebolavirus (Bundibugyo virus), Reston ebolavirus (Reston virus), Sudan ebolavirus (Sudan virus), Taï Forest ebolavirus (Taï Forest virus), Bombali ebolavirus (Bombali virus) and Zaire ebolavirus (Ebola virus (EBOV)). EBOV causes severe disease with a high case fatality rate of 25–90%170.
The glycoprotein of EBOV is a trimeric class I fusion protein formed by three disulfide-linked GP1–GP2 heterodimers forming a chalice-shaped trimer on the viral surface171. The GP1 subunit binds to the EBOV receptor, NPC1, allowing GP2-mediated fusion of the viral and host cell membranes172 (Fig. 4b). GP1 bears the RBS, glycan cap and mucin-like domain. GP2 contains an N-terminal peptide, internal fusion loop, stalk and transmembrane domain. Of note, the GP1 subunit contains a core domain which is shielded by a ‘glycan cap’, made by the heavily glycosylated mucin-like domain. The mucin-like domain is dispensable for viral entry, but is a decoy target for host antibody responses173.
Given the seriousness of the Ebola virus disease (EVD) and potential challenges associated with a large outbreak, there is an urgent need for therapies. The success in NHPs of ZMapp, which is a combination of three chimeric mAbs, 13C6 from MB-003 and 4G7 with 2G4 from ZMab174, illustrated the potential use of mAb therapies against EVD. The ZMapp cocktail was evaluated in humans during the 2014–2016 Ebola outbreak in West Africa, although efforts in NHPs to simplify the ZMapp regimen to contain fewer mAbs have not been successful175. Therefore, the possibility of obtaining fully human mAbs from individuals who survived EVD was investigated.
Two mAbs (mAb100 and mAb114) were isolated from an individual who survived Ebola in the Democratic Republic of Congo in 1995 (ref.176) (Fig. 4b). mAb114 binds to the GP1 head on an epitope at the physical intersection of the glycoprotein subunits177. It demonstrated neutralization and Fc-dependent cell-targeting activities in vitro, and potently activated phagocytosis and natural killer cells176. REGN-EB3 (marketed as Inmazeb, Regeneron Pharmaceuticals) is a combination of three fully human mAbs (REGN3470, REGN3479 and REGN3471) targeting the EBOV glycoprotein. These antibodies were obtained from humanized transgenic mice that had been immunized with DNA constructs encoding the EBOV glycoprotein and/or recombinant purified virus glycoprotein178. REGN3479 (now known as maftivimab) recognizes the conserved GP2 fusion loop and provides neutralizing activity, whereas REGN3471 (now known as odesivimab) recognizes the outer glycan cap and has cell-targeting functions. REGN3470 (now known as atoltivimab) binds to the GP1 head and offers both neutralization and cell-targeting activities, including FcγRIIIa and other FcγR-related functions.
During the EVD outbreak that occurred in the Democratic Republic of Congo in 2018, the triple mAb cocktail ZMapp, the monotherapy mAb114 (ansuvinmab, also known as VRC 608; developed by Ridgeback Biotherapeutics) and the triple mAb combination REGN-EB3 (maftivimab, odesivimab and atoltivimab) were evaluated in an umbrella trial. After an interim analysis, mAb114 monotherapy and REGN-EB3 were both found to be superior to ZMapp with respect to the primary outcome and patient mortality179, and were approved by the FDA for the treatment of EVD in 2020 (refs180,181).
Human cytomegalovirus
Human cytomegalovirus (HCMV) is an enveloped double-stranded DNA virus with a genome size of more than 235 kb, making it the largest known genome of human herpesviruses. HCMV is a member of the betaherpesvirus family and can establish lifelong latency in healthy individuals. Primary infection is generally asymptomatic; however, viral reactivation in immunocompromised hosts can be a life-threatening disease and vertical virus transmission during pregnancy is one of the leading causes of congenital birth defects182,183.
HCMV utilizes different glycoprotein complexes to allow cellular entry into a large variety of cells. Indeed, two gH/gL-containing complexes regulate viral tropism184,185; more specifically, the gH/gL/gO trimer binds to PDGFRα and is primarily required for infection in fibroblasts186,187, whereas the gH/gL/UL128/UL130/UL131A pentamer binds to neuropilin 2 and is required for viral entry into epithelial, endothelial and myeloid cells188–190 (Fig. 4c). In addition, the gB homotrimer catalyses membrane fusion between the virus and infected cells191–194 (Fig. 4c).
Several mAbs targeting the gH/gL or gB complexes have been isolated and have shown modest efficacy in an in vitro model of infection. To identify the most potent HCMV neutralizing antibodies, the Lanzavechia group isolated a large panel of mAbs from memory B cells in naturally infected donors and found that the pentameric complex represented the main target of neutralization against HCMV, eliciting neutralizing antibodies with a potency several orders of magnitude greater than any other HCMV complex8,48. This discovery shed light on the importance of the antigen selection to identify potent neutralizing mAbs and to design efficient vaccines.
The use of HCMV-specific mAbs for the prevention of HCMV infection and disease after allogeneic haematopoietic stem cell transplantation195 or solid organ transplant has been studied extensively196. Among the mAbs that have been evaluated in clinical trials, MSL-109, a human mAb targeting HCMV surface glycoprotein gH197, was tested as a supplementary treatment for patients with AIDS with HCMV-induced retinitis, but development was halted during phase II/III trials owing to lack of efficacy198. RG7667, a mixture of two mAbs binding the gH/gL and pentamer complexes199, could potently neutralize HCMV infections of all the cell types tested. However, when evaluated in a phase II trial for recipients of kidney transplants200, RG7667 did not meet the primary end point within 12 weeks post-transplant. Next, CSJ148, which consists of two anti-HCMV mAbs, an anti-gB mAb (LJP538) and an anti-pentamer mAb (LJP539)201, was evaluated in a phase II trial for prophylaxis of HCMV in patients undergoing haematopoietic stem cell transplantation202 and also did not achieve the primary end point. Nevertheless, in NHPs, the presence of durable and potently neutralizing antibodies at the time of primary infection was shown to prevent transmission of systemically replicating maternal rhesus CMV to the developing fetus203.
So, despite these significant development efforts, no anti-HCMV mAb has yet been FDA-approved. The only clinically available antibody-based therapy is still Cytogam, a preparation of CMV immunoglobulin used for intravenous injection; however, the neutralization capacity of this preparation is suboptimal204,205.
Influenza
Influenza, a member of the Orthomyxoviridae family, has four types — influenza A, B, C and D — all of which have a segmented, negative-sense, single-stranded RNA genome. Influenza A and B are responsible for severe infections in humans, whereas influenza C causes only mild symptoms and influenza D is not known to infect humans206. The viral genome consists of eight segments that encode at least 12 proteins: haemagglutinin (HA), neuraminidase (NA), PB2, PB1, PB1-F2, PA, PA-X, NP, M1, M2, NS1 and NS2 (ref.207).
The two glycoproteins on the viral surface are HA and NA, with HA being more abundant (~500 molecules per virion) than NA, with a ratio of 4:1 to 5:1 (ref.208). In total, 18 different HA subtypes (H1–H18) have been identified and can be separated into two phylogenetic groups according to their genetic sequences. Group 1 includes H1, H2, H5, H6, H8, H9, H11, H12, H13, H16, H17 and H18, whereas group 2 includes H3, H4, H7, H10, H14 and H15 (Fig. 4d). Eleven different NA subtypes (N1–N11) are known; group 1 includes N1, N4, N5 and N8; group 2 includes N2, N3, N6, N7 and N9; and group 3 includes N10 and N11 (ref.209). Each subtype encompasses several viral strains that represent the seasonal changes of the virus.
The HA head is the major target of the antibody response, providing only limited breadth due to its high sequence diversity and changes in glycosylation sites210. Although the structural architecture of HA is conserved overall, the sequence as well as glycosylation patterns differ among different subtypes211. Due to its high sequence variability, the elicited antibody response against the immunodominant head is strain-specific and provides only short-lived immunity212. The head contains the RBS that is responsible for viral attachment to the host cells through binding to sialic acid receptors. The RBS forms a shallow pocket and consists of four segments: the 190 helix and the 130, 150 and 220 loops213. The RBS itself is relatively conserved except for the 220 loop, whereas the remaining head is highly diverse in sequence.
Antibodies generally target five major antigenic sites, Ca1, Ca2, Cb, Sa and Sb for H1, and sites A–E for H3, which are located around the RBS206. Antibodies to the RBS site are potent as they block viral attachment or prevent receptor-mediated endocytosis, and therefore neutralize the virus, rendering it unable to infect cells214. However, these antibodies are generally strain-specific and thus not capable of providing immunity against drifted strains. Some exceptions have been observed, such as the bNAbs C05 (ref.215), S139/1 (ref.216) and F045-092 (ref.217), which target the RBS and are able to neutralize within their group. Furthermore, recently identified bNAbs target hidden epitopes at the HA trimer interfaces near the HA head domain that become accessible during a ‘breathing motion’ of the subunits218. FluA-20 has shown broad reactivity to most subtypes by recognizing a conserved epitope at the trimer interface and functions by inhibiting cell to cell spread of the virus219.
On the other hand, the HA stem is highly conserved mainly within subtypes and, to some extent, across subtypes, evolving at a much slower rate than the head domain220 (Fig. 4d). Several bNAbs against this region have recently been characterized in humans after infection or vaccination, making it a highly interesting vaccine design target221. These bNAbs generally target a hydrophobic pocket around the Trp21 HA2 residue on the HA stem in close proximity to the fusion peptide and block membrane fusion by retaining the HA in its pre-fusion state222.
Other mechanisms of action for stem-directed antibodies involve inhibition of proteolytic cleavage, reduction of viral egress by blocking NA activity through steric hindrance, ADCC and antibody-dependent cellular phagocytosis223. These antibodies tend to be less prevalent in humans and often demonstrate little or no neutralizing activity, but are protective in challenge studies221. Many of these antibodies are group-specific due to differing glycosylation patterns between group 1 and group 2 influenza HA at position N38, which is only present in the group 2 HA adjacent to the major antigenic site on the HA stem and, thus, can interfere with antibody binding. The F10 (ref.224) and CR6261 (ref.225) mAbs are specific for group 1 strains, whereas CR8020 (ref.226) and CR8043 (ref.227) mAbs are group 2-specific. CR8020 and CR8043 are encoded by VH1-18 and VH1-3 germline regions, whereas group 1 stem-specific neutralizing antibodies have been shown to generally derive from the VH1-69 germline gene228 (Fig. 4d). Recognition of both group 1 and group 2 has been shown to commonly involve VH1-18-derived, VH6-1-derived, VH3-23-derived or VH3-30-derived antibodies. Some of the broadest neutralizing antibodies, such as FI6v3 (ref.229), CR9114 (ref.230), 39.29 (ref.231) and MEDI8852 (ref.232), are able to engage HAs from both group 1 and group 2. The exact mechanisms driving differences in immunogenicity of the head versus the stem are not fully understood. However, several explanations have been proposed, including the restricted spatial availability of the stem domain to B cell receptors because of its close proximity to the viral membrane233.
MEDI8852 is a human IgG1 mAb isolated from a patient with uncomplicated influenza A infection. MEDI8852 was evaluated in phase IIa trial and is still in development by MedImmune234. Vir Biotechnology is developing VIR-2482, an influenza A neutralizing mAb that binds to the conserved region of HA and neutralizes all major strains since the Spanish flu in 1918 (H1N1)232. It is therefore tempting to speculate that this type of antibody could be used as a universal prophylactic agent, overcoming the limitations of current influenza vaccines, for which the antibody response is dependent on individual seasonal antigens. In addition, because the serum half-life of VIR-2482 has been increased by Fc domain engineering, a single dose can last the entire influenza season of around 5–6 months.
Implications for vaccine development
Antigen–antibody interactions and structural vaccinology for vaccine design
Vaccines have proven to be the most effective prophylactic strategy for infectious diseases235. However, traditional vaccine development approaches236, which rely on three categories of vaccines (live-attenuated, inactivated and dissociated pathogens), have failed for viruses such as HIV, RSV, influenza, hepatitis C virus, HCMV or EBOV.
For most vaccines, the antibody response is crucial and, thus, the identification of antibodies that can potently neutralize a pathogen is a key factor for accelerating vaccine development. Reverse vaccinology 2.0, also known as antibody-based vaccinology, aims to overcome the limitations of traditional approaches by engineering novel vaccines based on the structural characterization of antigens in complex with their cognate antibodies, with the antigen-specific antibody response acting as a correlate of protection237 (Fig. 5).
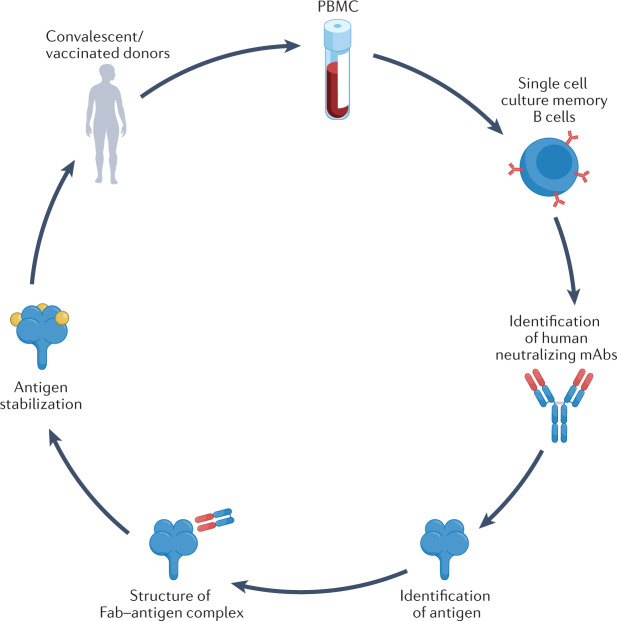
Fig. 5. Reverse vaccinology 2.0.
Single cell cultures of plasma or memory B cells from a convalescent patient or vaccinated donor are used to screen for monoclonal antibodies (mAbs) with neutralizing activity against the target pathogen. Next, recombinant mAbs are used to identify the antigen and investigate the 3D structure of the antigen–mAb complex. This structural information is used to optimize a stabilized antigen for a vaccine. Fab, fragment antigen-binding domain; PBMC, peripheral blood mononuclear cell.
Antigens are generally identified by proteomic analysis of crude homogenates of infected cells and, then, chosen based on immunogenicity and their ability to stimulate an immune response. However, this approach is time-consuming, and structural integrity or immunogenicity is not guaranteed upon expression of a newly identified protein. Experimentally driven isolation of neutralizing mAbs and identification of their target is still the best approach to identify vaccine candidates. One of the main advantages of antibody-driven vaccinology is isolation of potent neutralizing mAbs. These mAbs will be useful for passive immunization of immunocompromised patients, and/or as therapeutic agents during the acute phase of infection. An additional advantage of the antibody-driven approach is that mAbs can be instrumental in identifying the optimal vaccine antigen for which mAb binding can block virus transmission.
This identification step can be done by immunoprecipitation coupled to mass spectrometry. In addition, once identified, both the antigen and the mAb can be used for structural vaccinology. The latter approach aims at elucidating the atomic structures of the viral antigens with a neutralizing Fab (Fig. 5). Structural vaccinology is a valuable source of information to engineer antigens for stabilization purposes. The combination of antibody-driven and structure-based antigen design strategies is particularly efficient in developing rapid responses to emerging infectious disease threats.
Facilitated by the improvements in high-throughput B cell technologies, the structural insights into human mAbs have been instrumental in directing the immune response to conserved antigenic sites72,238–241. Complementary to this approach, structure-based design has been employed to completely remove domains containing sites targeted by non-neutralizing antibodies or to identify possible positions for the introduction of glycosylation sites to mask such epitopes242.
Respiratory syncytial virus
RSV provides a case study for the reverse vaccinology 2.0 concept. For RSV, neutralizing antibodies mainly target the head domain of the fusion (F) protein243 (Fig. 4a). To focus the immune response against antigenic sites in the F head, Boyington et al.244 designed truncated F immunogens containing only the head region. These immunogens were successful in eliciting neutralization titres comparable with full-length pre-fusion F protein. These proteins are especially vulnerable in their pre-fusion state, and thus have been the target of structure-guided stabilization efforts to lock antigens in their pre-fusion conformation in order to promote a neutralizing antibody response that prevents membrane fusion160. This structural information can be used to identify stabilizing mutations such as disulfide bonds or proline mutations to rigidify the protein backbone and mutations to fill cavities to impede transition from pre-fusion to post-fusion states. Additionally, structure-based design has been applied to fuse domains for multimerization or to remove potential unstable regions242.
A prime example for the successful stabilization of a pre-fusion antigen is the F protein, where the elicitation of a potent neutralizing antibody response relies on targeting the pre-fusion state160. Multiple structure-based design strategies were applied to stabilize the F protein in its pre-fusion conformation. McLellan et al. designed intra-protomer disulfide bonds coupled with additional mutations to fill cavities in the native antigen to achieve a stabilized pre-fusion antigen (Ds-Cav1)166. A different design approach by Krarup et al. focused on introducing proline residues to prevent structural rearrangements of the F protein from occurring during the adoption of the post-fusion conformation245. Immunization studies in animals confirmed the improved neutralization potential of pre-fusion F constructs over post-fusion antigens, further supported by a subsequent study by Joyce et al. which highlighted that stability improvements of the DS-Cav1 immunogen in the pre-fusion state increased the neutralizing antibody response fourfold as compared with Ds-Cav1 (ref.246).
A comparatively young field in structure-based vaccine design is epitope scaffolding, which relies on the transplantation of viral epitopes onto unrelated carrier proteins, so-called scaffolds, to focus the immune response against conserved, functional sites that are known to be targeted by neutralizing antibodies247. This strategy has first been applied to the design of novel immunogens for HIV248–251, although one limitation in antigen transplant is the stabilization of distant discontinuous epitopes252. For RSV, computational design of immunogens was first achieved by McLellan et al., who designed immunogens for antigenic site II of the F protein by grafting of the epitope of motavizumab onto an unrelated protein scaffold253. However, immunization of mice with this epitope scaffold did not elicit neutralizing antibodies, although it did elicit sera with F-binding activity253. Subsequent design efforts focusing on the same antigenic site resulted in a novel epitope scaffold that engaged site-specific antibodies with high affinity and boosted subdominant antibodies with enhanced neutralization254,255. A recent study by Sesterhenn et al. demonstrated that a cocktail formulation of three computationally designed immunogens displaying RSV F sites Ø, II and IV elicited a neutralizing antibody response and allowed for the focusing of the antibody response against specific antigenic sites upon immunization of mice and NHPs256. Using a strategy termed motif-centric design, the authors computationally designed de novo topologies around the extracted F antigenic sites to improve epitope stabilization and accurate display of the antigenic site256.
Although epitope scaffolding approaches have facilitated the design of novel immunogens that display neutralization epitopes, these designed immunogens have mostly been restricted to small, continuous epitopes248,249,254 with few exceptions256,257. In contrast, observed epitopes often consist of multiple segments258,259 that are challenging for structure-based design. As an example, Marcandalli et al. demonstrated that the use of structure-based design of a self-assembling protein nanoparticle presenting a pre-fusion-stabilized DS-Cav1 in a repetitive array on the nanoparticle exterior induced neutralizing antibody responses up to tenfold higher than trimeric DS-Cav1 alone260. The same nanoparticle was used to derive promising vaccines against SARS-CoV-2 (ref.261), influenza262 and HCMV194.
Ebola virus
Vaccines from several platforms have shown protection against EVD in human and NHPs, and two that are not based on the reverse vaccinology concept have been licensed: Ervebo (rVSV-ZEBOV) and a two-dose combination of Zabdeno/Mvabea171,177,263–265. Both vaccines are based on viral vectors, and recent analysis of human B cell responses from vaccinated humans demonstrated low levels of somatic hypermutations in mAbs induced by the vaccine, even if they were protective266. Rational design efforts for protein-based subunit vaccines are ongoing267.
Ervebo is a live, attenuated recombinant vesicular stomatitis virus-based vector expressing the envelope glycoprotein gene of Zaire EBOV instead of the VSV-G gene (Fig. 4b). NHP studies demonstrated rVSV-ZEBOV efficacy in stringent conditions, where a single inoculation at ~1 × 107 pfu was shown to protect against illness, viraemia and death after challenge with a high dose of EBOV (1,000 pfu, generally thought to represent 100–1,000 times the lethal dose (median lethal dose) in experimental animal studies)268–270. Zabdeno/Mvabea each contain a monovalent replication-incompetent virus. Zabdeno, an adenoviral vector of serotype 26 that encodes the EBOV-GP Mayinga variant (Ad26.ZEBOV), is used for the priming injection, and Mvabea, a modified vaccinia virus Ankara-Bavarian Nordic Filo-vector encoding the same glycoprotein (MVA-BN-Filo)271, is used for the booster.
Other vaccines in development include cAd3-EBO Z, an attenuated version of a chimpanzee adenovirus (cAd3) encoding the EBOV-GP glycoprotein272. Finally, a recombinant nanoparticle vaccine with the EBOV-GP Makona strain was found to induce a potent immune response273.
Human cytomegalovirus
The development of HCMV vaccines began in the early 1970s, and two attenuated virus strains were isolated for laboratory work: AD169 and Towne274,275. The AD169 attenuated strain was quickly abandoned whereas the Towne attenuated strain progressed to extensive testing in recipients of solid organ transplants and healthy volunteers276. Recipients of kidney transplants were shown to be highly protected against serious CMV disease and graft rejection. However, protection against viral infection was not statistically significant. The next development in the quest for CMV vaccines was the identification of a surface protein of CMV called glycoprotein B or gB (Fig. 4c). When combined with the MF59 oil-in-water adjuvant, the vaccine was safe and partially effective277, but the levels of neutralizing antibodies were weak in humans after three injections277–279. The subunit gB protein was also combined with the AS01 adjuvant to stimulate Toll-like receptor 4 (TLR4), which elicited higher and more prolonged levels of anti-gB antibodies in humans. Unfortunately, the adjuvanted vaccine was never tested for efficacy.
The gB antigen is a class III trimeric fusion protein and is used in the post-fusion conformation110. Therefore, it was suggested that gB in the pre-fusion conformation could generate a higher neutralizing response, but recent results have, surprisingly, indicated that this might not be the case280. The pentameric complex of proteins present on the surface of CMV consists of glycoprotein H (gH), glycoprotein L (gL) and the products of genes UL128, 130 and 131A. The complex has been shown to generate far higher titres of neutralizing antibodies than gB48,281 (Fig. 4c). This discovery has since driven much of the HCMV vaccine field and vaccine trials are ongoing. However, if neutralizing antibodies are necessary to prevent infection and spread of HCMV, a strong T cell response is also needed to suppress reactivation of the virus in patients who are seropositive. Forthcoming clinical trials using either a recombinant protein subunit (NCT05089630) or mRNA (NCT05105048) will indicate whether the pentamer can be used alone or whether it needs to be injected together with gB195,282.
Influenza
One of the major obstacles for effective influenza vaccines is the immunodominance of the highly diverse HA head that is responsible for viral attachment. Structure-based design methods have been focused on the conserved but immunorecessive HA stem region (Fig. 4d).
To shift the antibody response towards the stem domain, chimeric HA molecules have been engineered, consisting of a common HA stem paired with different HA heads283–285. Repeated immunizations of mice demonstrated that a stem-specific antibody response can be mounted, resulting in heterologous and hetero-subtypic immunity284. In ferrets, these constructs were shown to reduce viral loads after influenza virus challenge286,287. Chimeric HA has been tested in a phase I clinical trial and was found to be safe, and able to induce a broad, strong, durable and functional immune response288.
Although Kanekiyo et al. demonstrated that the isolation of RBDs from different HA strains and multimerization on ferritin nanoparticles resulted in the elicitation of a B cell response against conserved epitopes289, the most promising design strategies are based on isolated HA stem-only antigens created by removing the HA head290,291. Two such design strategies are headless HAs from Yassine et al.220 and the mini-HAs from Impagliazzo et al.233.
The headless HA immunogens were developed by removing the HA head from an H1 strain, followed by multiple rounds of structure-based design that yielded stabilized stem immunogens220. Lethal influenza challenge with a hetero-subtypic H5N1 strain showed complete protection in mice and partial protection in ferrets; however, no cross-group reactivity with group 2 viruses was observed220. Applying the same structure-based design strategy to H3 and H7 HA resulted in two group 2 headless immunogens that were able to elicit protective, homo-subtypic antibodies in mice292. Immunization of NHPs with headless H3 generated neutralizing antibodies against divergent H3N2 strains and selected H10N8 as well as H7N9 strains293.
The mini-HA molecules developed by Impagliazzo et al. follow a similar design approach. A combination of rational and library-based design approaches was applied to generate stabilized HA stem molecules that lack the immunodominant head and the transmembrane region233. The designs were based on an H1 subtype and were shown to be protective in lethal heterologous and hetero-subtypic challenge in mouse models233. In pre-exposed NHPs, the mini-HAs elicited an expanded influenza-specific humoral immune response when compared with trivalent inactivated influenza vaccine294. Together, these results demonstrate that immunogens lacking the immunodominant head domain can elicit a group-specific, hetero-subtypic immune response293,294. Recently, a ferritin nanoparticle-based vaccine incorporating the ectodomain of HA from an H2N2 pandemic strain was demonstrated to be safe and immunogenic in a phase I clinical trial, supporting its potential application in pandemic preparedness and universal influenza vaccine development295,296.
A different approach to focus the immune response against conserved antigenic sites that is structurally less demanding relies on the masking of non-neutralizing antigenic sites in the HA head through hyper-glycosylation and removal of glycans from the HA stem297,298. Eggink et al. applied this strategy to hyper-glycosylated immunodominant epitopes in the HA head, resulting in the silencing of immunodominant sites. Immunization of mice confirmed a shift of the antibody response towards the immunorecessive stem domain and improved protection of mice after viral challenge299. Beyond focusing the immune response against the HA stem, this strategy can be also used to improve the immune response against subdominant epitopes in the HA head218.
COVID-19
Acquisition of structural data for the SARS-CoV-2 S protein played an important role in the development of multiple COVID-19 vaccines, including the extraordinarily successful mRNA vaccines. Another critical aspect in the development of effective vaccines was the intensive experience gained from studies of the S protein of other coronaviruses such as HCoV-HKU1 (ref.300) and MERS-CoV301,302 led by the McLellan and Veesler laboratories. This allowed the design of stabilizing mutations in the versions of the S protein encoded in the mRNA vaccines (BNT162b2 mRNA from Pfizer-BioNTech and mRNA-1273 from Moderna) and some vectored vaccines (Ad26.COV2.S from Janssen)303, such as deletion of the polybasic cleavage site, inclusion of stabilizing mutations304 and inclusion of trimerization domains305,306.
HIV
Research on the development of an HIV vaccine has strongly benefited from the isolation of human bNAbs, reviewed in depth elsewhere307–310. The use of bNAb-instructed stabilization of HIV Env trimers is the basis for many promising immunogens311. In addition, the most promising strategies use stabilized trimers with the aims of activating particular bNAb-producing cell precursors and guiding their affinity maturation to generate mature bNAbs — a strategy called germline targeting312. Recently, 97% seroconversion was reported313 in healthy subjects vaccinated with the eOD-GT8 immunogen314,315, demonstrating the strong potential of the germline-targeting strategy. Molecular analysis of the B cells induced during the clinical trial (NCT03547245) will provide a road map to accelerate progress towards an HIV vaccine. A phase I clinical trial in which an mRNA vaccine encoding the eOD-GT8 immunogen is used for the priming injection has recently been initiated (NCT05001373).
Closing perspectives
The emergence of SARS-CoV-2 and the devastating COVID-19 pandemic have emphasized the severity of the threat of emerging infectious diseases, especially those of zoonotic origin. In response, there has been unprecedented success in the discovery and development, manufacturing and regulatory evaluation of several anti-COVID-19 vaccines and mAb therapeutics in a very short time, achieved through exceptional mobilization of public and private resources. These advances have been based on the enormous scientific progress made in immunology and vaccinology over the last few decades, and also the crucial previous knowledge on the biology of coronaviruses, such as the S protein as a target for neutralizing antibodies.
Efforts to harness antibodies to combat COVID-19 have also benefited from technological advances and expertise gained particularly in the area of human bNAbs from the HIV field. It is expected that the generation of anti-infective mAbs that are urgently needed will benefit from the recent clinical successes and case studies highlighted in this article and elsewhere316. However, mAbs still have limitations. The potential for mutations in the viral targets of antibodies to allow viruses to escape neutralization has recently been highlighted with SARS-CoV-2 Omicron VOCs, and also by the development of resistant variants when selective pressure is applied in the setting of drug treatment, which has been observed in immunocompromised patients treated with bamlanivimab and etesevimab317, sotrovimab318 or REGN-COV2 (ref.319).
With the recent advances in machine-learning algorithms, and the dramatic increases in the repertoire of available antibodies, further research should provide insight into the structural properties required for bNAbs320. Moreover, the progress made with gene-editing technology opens the possibility of engineering functionalities into human cells. Indeed, B cells engineered to carry broadly neutralizing B cell receptors are likely to represent a milestone to address pathogens with no vaccines or inefficient vaccines321. Vaccination by edited B cells able to differentiate into memory B cells, plasmablasts and long-lived plasma cells could be a valuable option for HIV and influenza prevention. Finally, given that a key current limitation of mAbs is still their limited distribution in tissues, novel formats or ways to specifically direct mAbs to the targeted tissues may render mAbs even more powerful tools in fighting viral infections.
Acknowledgements
The authors thank M. Perreau and M. Foglierini from Lausanne University Hospital (CHUV), Service of Immunology and Allergy, Switzerland for critical reading of the manuscript. The Swiss National Science Foundation (SNSF) supports work in the Perez laboratory (Grant number: 310030_204679).
Glossary
- Emergency use authorization
(EUA). A mechanism to facilitate the availability and use of medical countermeasures during a public health emergency. US Food and Drug Administration (FDA) issuance of a EUA permits the use of unapproved medical products or unapproved uses of approved medical products when no adequate alternatives are available.
- Fcγ receptors
Humans express six Fcγ receptors (FcγRs) that modulate effector cells upon binding to IgG. FcγRI, FcγRIIA, FcγRIIC, FcγRIIIA and FcγRIIIB are activating receptors, whereas FcγRIIB is inhibitory.
- Antibody-dependent cellular cytotoxicity
(ADCC). A mechanism of cell-mediated immune defence whereby an effector cell of the immune system actively lyses a target cell, whose membrane-surface antigens have been bound by specific antibodies.
- Complement-dependent cytotoxicity
(CDC). The complement system is a network of proteins that form an important part of the immune response by enhancing the opsonization of pathogens, cell lysis and inflammation. CDC is a mechanism of complement-mediated immune defence in which an antibody bound to its antigen activates the complement cascade.
- Glycoproteins
Proteins with oligosaccharide chains (glycans) covalently attached to amino acid side chains. Virus surface glycoproteins embedded in the membrane often have a role in interactions with host cells, including receptor binding, and are commonly targeted by host antibodies.
- Opsonized
A state of a pathogen in which antibodies or complement factors are bound to its surface.
Peer review
Peer review information
Nature Reviews Drug Discovery thanks Davide Corti, Bryan Jones, Jason McLellan and Rino Rappuoli for their contribution to the peer review of this work.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 2.Mullard A. FDA approves 100th monoclonal antibody product. Nat. Rev. Drug Discov. 2021;20:491–495. doi: 10.1038/d41573-021-00079-7. [DOI] [PubMed] [Google Scholar]
- 3.Behring EV, Kitasato S. Über das Zustandekommen der Diphtherie-Immunität und der Tetanus-Immunität bei Thieren. Dtsch Med. Wochenschrift. 1890;49:1113–1114. [PubMed] [Google Scholar]
- 4.Motley MP, Banerjee K, Fries BC. Monoclonal antibody-based therapies for bacterial infections. Curr. Opin. Infect. Dis. 2019;32:210–216. doi: 10.1097/QCO.0000000000000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raafat D, Otto M, Reppschläger K, Iqbal J, Holtfreter S. Fighting Staphylococcus aureus biofilms with monoclonal antibodies. Trends Microbiol. 2019;27:303–322. doi: 10.1016/j.tim.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hjelholt A, Christiansen G, Sørensen US, Birkelund S. IgG subclass profiles in normal human sera of antibodies specific to five kinds of microbial antigens. Pathog. Dis. 2013;67:206–213. doi: 10.1111/2049-632X.12034. [DOI] [PubMed] [Google Scholar]
- 7.Bournazos S, Corti D, Virgin HW, Ravetch JV. Fc-optimized antibodies elicit CD8 immunity to viral respiratory infection. Nature. 2020;588:485–490. doi: 10.1038/s41586-020-2838-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kabanova A, et al. Antibody-driven design of a human cytomegalovirus gHgLpUL128L subunit vaccine that selectively elicits potent neutralizing antibodies. Proc. Natl Acad. Sci. USA. 2014;111:17965–17970. doi: 10.1073/pnas.1415310111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng Q, et al. Viral neutralization by antibody-imposed physical disruption. Proc. Natl Acad. Sci. USA. 2019;116:26933–26940. doi: 10.1073/pnas.1916028116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hikmet F, et al. The protein expression profile of ACE2 in human tissues. Mol. Syst. Biol. 2020;16:e9610. doi: 10.15252/msb.20209610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor PC, et al. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat. Rev. Immunol. 2021;21:382–393. doi: 10.1038/s41577-021-00542-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu LL, Suscovich TJ, Fortune SM, Alter G. Beyond binding: antibody effector functions in infectious diseases. Nat. Rev. Immunol. 2018;18:46–61. doi: 10.1038/nri.2017.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winkler ES, et al. Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions for optimal therapeutic protection. Cell. 2021;184:1804–1820.e16. doi: 10.1016/j.cell.2021.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schäfer, A. et al. Antibody potency, effector function, and combinations in protection and therapy for SARS-CoV-2 infection in vivo. J. Exp. Med. 10.1084/jem.20201993 (2021). [DOI] [PMC free article] [PubMed]
- 15.Case, J. B. et al. Resilience of S309 and AZD7442 monoclonal antibody treatments against infection by SARS-CoV-2 Omicron lineage strains. Preprint at bioRxiv10.1101/2022.03.17.484787 (2022). [DOI] [PMC free article] [PubMed]
- 16.Bournazos S, Gupta A, Ravetch JV. The role of IgG Fc receptors in antibody-dependent enhancement. Nat. Rev. Immunol. 2020;20:633–643. doi: 10.1038/s41577-020-00410-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halstead SB, et al. Dengue hemorrhagic fever in infants: research opportunities ignored. Emerg. Infect. Dis. 2002;8:1474–1479. doi: 10.3201/eid0812.020170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dejnirattisai W, et al. Cross-reacting antibodies enhance dengue virus infection in humans. Science. 2010;328:745–748. doi: 10.1126/science.1185181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beltramello M, et al. The human immune response to Dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe. 2010;8:271–283. doi: 10.1016/j.chom.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu, L., Ma, Z., Li, Y., Pang, Z. & Xiao, S. in Advances in Immunology Vol. 151 (eds Frederick W. A. & Kenneth M. M.) 99–133 (Academic, 2021). [DOI] [PMC free article] [PubMed]
- 21.Mair-Jenkins J, et al. The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of severe acute respiratory infections of viral etiology: a systematic review and exploratory meta-analysis. J. Infect. Dis. 2015;211:80–90. doi: 10.1093/infdis/jiu396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Casadevall A, Dadachova E, Pirofski L-A. Passive antibody therapy for infectious diseases. Nat. Rev. Microbiol. 2004;2:695–703. doi: 10.1038/nrmicro974. [DOI] [PubMed] [Google Scholar]
- 23.Hammarström L, Marcotte H, Piralla A, Baldanti F, Pan-Hammarström Q. Antibody therapy for COVID-19. Curr. Opin. Allergy Clin. Immunol. 2021;21:553–558. doi: 10.1097/ACI.0000000000000787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simonovich VA, et al. A randomized trial of convalescent plasma in COVID-19 severe pneumonia. N. Engl. J. Med. 2021;384:619–629. doi: 10.1056/NEJMoa2031304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bégin P, et al. Convalescent plasma for hospitalized patients with COVID-19: an open-label, randomized controlled trial. Nat. Med. 2021;27:2012–2024. doi: 10.1038/s41591-021-01488-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ray Y, et al. A phase 2 single center open label randomised control trial for convalescent plasma therapy in patients with severe COVID-19. Nat. Commun. 2022;13:383. doi: 10.1038/s41467-022-28064-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCafferty J, Griffiths AD, Winter G, Chiswell DJ. Phage antibodies: filamentous phage displaying antibody variable domains. Nature. 1990;348:552–554. doi: 10.1038/348552a0. [DOI] [PubMed] [Google Scholar]
- 28.Hoogenboom HR. Overview of antibody phage-display technology and its applications. Methods Mol. Biol. 2002;178:1–37. doi: 10.1385/1-59259-240-6:001. [DOI] [PubMed] [Google Scholar]
- 29.Burton DR, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 30.Ferrara F, et al. A pandemic-enabled comparison of discovery platforms demonstrates a naïve antibody library can match the best immune-sourced antibodies. Nat. Commun. 2022;13:462. doi: 10.1038/s41467-021-27799-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maynard JA, et al. Protection against anthrax toxin by recombinant antibody fragments correlates with antigen affinity. Nat. Biotechnol. 2002;20:597–601. doi: 10.1038/nbt0602-597. [DOI] [PubMed] [Google Scholar]
- 32.Burkovitz A, Ofran Y. Understanding differences between synthetic and natural antibodies can help improve antibody engineering. mAbs. 2016;8:278–287. doi: 10.1080/19420862.2015.1123365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novobrantseva T, et al. Stochastic pairing of Ig heavy and light chains frequently generates B cell antigen receptors that are subject to editing in vivo. Int. Immunol. 2005;17:343–350. doi: 10.1093/intimm/dxh214. [DOI] [PubMed] [Google Scholar]
- 34.Tiegs SL, Russell DM, Nemazee D. Receptor editing in self-reactive bone marrow B cells. J. Exp. Med. 1993;177:1009–1020. doi: 10.1084/jem.177.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gay D, Saunders T, Camper S, Weigert M. Receptor editing: an approach by autoreactive B cells to escape tolerance. J. Exp. Med. 1993;177:999–1008. doi: 10.1084/jem.177.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Norris MH, Blackburn JK. Raxibacumab: a panacea for anthrax disease? Lancet. Infect. Dis. 2020;20:886–887. doi: 10.1016/S1473-3099(20)30164-X. [DOI] [PubMed] [Google Scholar]
- 37.Pedrioli A, Oxenius A. Single B cell technologies for monoclonal antibody discovery. Trends Immunol. 2021;42:1143–1158. doi: 10.1016/j.it.2021.10.008. [DOI] [PubMed] [Google Scholar]
- 38.Klein U, Küppers R, Rajewsky K. Variable region gene analysis of B cell subsets derived from a 4-year-old child: somatically mutated memory B cells accumulate in the peripheral blood already at young age. J. Exp. Med. 1994;180:1383–1393. doi: 10.1084/jem.180.4.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee EC, et al. Complete humanization of the mouse immunoglobulin loci enables efficient therapeutic antibody discovery. Nat. Biotechnol. 2014;32:356–363. doi: 10.1038/nbt.2825. [DOI] [PubMed] [Google Scholar]
- 40.Scheid JF, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 41.Wu X, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang J, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kong L, et al. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat. Struct. Mol. Biol. 2013;20:796–803. doi: 10.1038/nsmb.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mascola JR, et al. Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J. Virol. 1999;73:4009–4018. doi: 10.1128/JVI.73.5.4009-4018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moldt B, et al. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proc. Natl Acad. Sci. USA. 2012;109:18921–18925. doi: 10.1073/pnas.1214785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Q, et al. A combination of human broadly neutralizing antibodies against hepatitis B virus HBsAg with distinct epitopes suppresses escape mutations. Cell Host Microbe. 2020;28:335–349.e6. doi: 10.1016/j.chom.2020.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hartley GE, et al. Rapid generation of durable B cell memory to SARS-CoV-2 spike and nucleocapsid proteins in COVID-19 and convalescence. Sci. Immunol. 2020;5:eabf8891. doi: 10.1126/sciimmunol.abf8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Macagno A, et al. Isolation of human monoclonal antibodies that potently neutralize human cytomegalovirus infection by targeting different epitopes on the gH/gL/UL128-131A complex. J. Virol. 2010;84:1005–1013. doi: 10.1128/JVI.01809-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCoy LE, Burton DR. Identification and specificity of broadly neutralizing antibodies against HIV. Immunol. Rev. 2017;275:11–20. doi: 10.1111/imr.12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kwakkenbos MJ, et al. Generation of stable monoclonal antibody-producing B cell receptor-positive human memory B cells by genetic programming. Nat. Med. 2010;16:123–128. doi: 10.1038/nm.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Traggiai E, et al. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat. Med. 2004;10:871–875. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steinitz M, Klein G, Koskimies S, Makel O. EB virus-induced B lymphocyte cell lines producing specific antibody. Nature. 1977;269:420–422. doi: 10.1038/269420a0. [DOI] [PubMed] [Google Scholar]
- 53.Corti, D. & Lanzavecchia, A. Efficient methods to isolate human monoclonal antibodies from memory B cells and plasma cells. Microbiol. Spectr. 10.1128/microbiolspec.AID-0018-2014 (2014). [DOI] [PubMed]
- 54.Kuraoka M, et al. Complex antigens drive permissive clonal selection in germinal centers. Immunity. 2016;44:542–552. doi: 10.1016/j.immuni.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Su K-Y, Watanabe A, Yeh C-H, Kelsoe G, Kuraoka M. Efficient culture of human naive and memory B cells for use as APCs. J. Immunol. 2016;197:4163–4176. doi: 10.4049/jimmunol.1502193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luo XM, et al. Engineering human hematopoietic stem/progenitor cells to produce a broadly neutralizing anti-HIV antibody after in vitro maturation to human B lymphocytes. Blood. 2009;113:1422–1431. doi: 10.1182/blood-2008-09-177139. [DOI] [PubMed] [Google Scholar]
- 57.McCarthy KR, et al. Memory B cells that cross-react with group 1 and group 2 influenza A viruses are abundant in adult human repertoires. Immunity. 2018;48:174–184.e9. doi: 10.1016/j.immuni.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lindner JM, et al. Human memory B cells harbor diverse cross-neutralizing antibodies against BK and JC polyomaviruses. Immunity. 2019;50:668–676.e5. doi: 10.1016/j.immuni.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 59.Wagar LE, et al. Modeling human adaptive immune responses with tonsil organoids. Nat. Med. 2021;27:125–135. doi: 10.1038/s41591-020-01145-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tanno H, et al. A facile technology for the high-throughput sequencing of the paired VH:VL and TCRβ:TCRα repertoires. Sci. Adv. 2020;6:eaay9093. doi: 10.1126/sciadv.aay9093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.DeKosky BJ, et al. In-depth determination and analysis of the human paired heavy- and light-chain antibody repertoire. Nat. Med. 2015;21:86–91. doi: 10.1038/nm.3743. [DOI] [PubMed] [Google Scholar]
- 62.Chen D, et al. Coupled analysis of transcriptome and BCR mutations reveals role of OXPHOS in affinity maturation. Nat. Immunol. 2021;22:904–913. doi: 10.1038/s41590-021-00936-y. [DOI] [PubMed] [Google Scholar]
- 63.Thompson EA, et al. Metabolic programs define dysfunctional immune responses in severe COVID-19 patients. Cell Rep. 2021;34:108863. doi: 10.1016/j.celrep.2021.108863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Buus, T. B. et al. Improving oligo-conjugated antibody signal in multimodal single-cell analysis. eLife10.7554/eLife.61973 (2021). [DOI] [PMC free article] [PubMed]
- 65.Ramaswamy A, et al. Immune dysregulation and autoreactivity correlate with disease severity in SARS-CoV-2-associated multisystem inflammatory syndrome in children. Immunity. 2021;54:1083–1095.e7. doi: 10.1016/j.immuni.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lavinder JJ, Horton AP, Georgiou G, Ippolito GC. Next-generation sequencing and protein mass spectrometry for the comprehensive analysis of human cellular and serum antibody repertoires. Curr. Opin. Chem. Biol. 2015;24:112–120. doi: 10.1016/j.cbpa.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 67.Georgiou G, et al. The promise and challenge of high-throughput sequencing of the antibody repertoire. Nat. Biotechnol. 2014;32:158–168. doi: 10.1038/nbt.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Use of Convalescent Whole Blood or Plasma Collected from Patients Recovered from Ebola Virus Disease for Transfusion, as an Empirical Treatment during Outbreaks: Interim Guidance for National Health Authorities and Blood Transfusion Services Version 1.0 (World Health Organization, 2014).
- 69.Setliff I, et al. High-throughput mapping of B cell receptor sequences to antigen specificity. Cell. 2019;179:1636–1646.e15. doi: 10.1016/j.cell.2019.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Finn JA, et al. Identification of structurally related antibodies in antibody sequence databases using rosetta-derived position-specific scoring. Structure. 2020;28:1124–1130.e5. doi: 10.1016/j.str.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sevy AM, et al. Computationally designed cyclic peptides derived from an antibody loop increase breadth of binding for influenza variants. Structure. 2020;28:1114–1123.e4. doi: 10.1016/j.str.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Foglierini M, Pappas L, Lanzavecchia A, Corti D, Perez L. AncesTree: an interactive immunoglobulin lineage tree visualizer. PLoS Comput. Biol. 2020;16:e1007731. doi: 10.1371/journal.pcbi.1007731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kramer KJ, et al. Potent neutralization of SARS-CoV-2 variants of concern by an antibody with an uncommon genetic signature and structural mode of spike recognition. Cell Rep. 2021;37:109784. doi: 10.1016/j.celrep.2021.109784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lund J, et al. Human FcγRI and FcγRII interact with distinct but overlapping sites on human IgG. J. Immunol. 1991;147:2657–2662. [PubMed] [Google Scholar]
- 75.Lee WS, Wheatley AK, Kent SJ, DeKosky BJ. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat. Microbiol. 2020;5:1185–1191. doi: 10.1038/s41564-020-00789-5. [DOI] [PubMed] [Google Scholar]
- 76.Loo Y-M, et al. The SARS-CoV-2 monoclonal antibody combination, AZD7442, is protective in nonhuman primates and has an extended half-life in humans. Sci. Transl Med. 2022;14:eabl8124. doi: 10.1126/scitranslmed.abl8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oganesyan V, Gao C, Shirinian L, Wu H, Dall’Acqua WF. Structural characterization of a human Fc fragment engineered for lack of effector functions. Acta Crystallogr. Sect. D. 2008;64:700–704. doi: 10.1107/S0907444908007877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gupta A, et al. Early treatment for COVID-19 with SARS-CoV-2 neutralizing antibody sotrovimab. N. Engl. J. Med. 2021;385:1941–1950. doi: 10.1056/NEJMoa2107934. [DOI] [PubMed] [Google Scholar]
- 79.Weitzenfeld P, Bournazos S, Ravetch JV. Antibodies targeting sialyl Lewis A mediate tumor clearance through distinct effector pathways. J. Clin. Invest. 2019;129:3952–3962. doi: 10.1172/JCI128437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grobben M, Stuart RA, van Gils MJ. The potential of engineered antibodies for HIV-1 therapy and cure. Curr. Opin. Virol. 2019;38:70–80. doi: 10.1016/j.coviro.2019.07.007. [DOI] [PubMed] [Google Scholar]
- 81.Gaudinski MR, et al. Safety and pharmacokinetics of the Fc-modified HIV-1 human monoclonal antibody VRC01LS: a phase 1 open-label clinical trial in healthy adults. PLoS Med. 2018;15:e1002493. doi: 10.1371/journal.pmed.1002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rudicell RS, et al. Enhanced potency of a broadly neutralizing HIV-1 antibody in vitro improves protection against lentiviral infection in vivo. J. Virol. 2014;88:12669–12682. doi: 10.1128/JVI.02213-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Julien J-P, et al. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog. 2013;9:e1003342. doi: 10.1371/journal.ppat.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dall’Acqua WF, Kiener PA, Wu H. Properties of human IgG1s engineered for enhanced binding to the neonatal Fc receptor (FcRn) J. Biol. Chem. 2006;281:23514–23524. doi: 10.1074/jbc.M604292200. [DOI] [PubMed] [Google Scholar]
- 85.Griffin MP, et al. Single-dose nirsevimab for prevention of RSV in preterm infants. N. Engl. J. Med. 2020;383:415–425. doi: 10.1056/NEJMoa1913556. [DOI] [PubMed] [Google Scholar]
- 86.Hammitt LL, et al. Nirsevimab for prevention of RSV in healthy late-preterm and term infants. N. Engl. J. Med. 2022;386:837–846. doi: 10.1056/NEJMoa2110275. [DOI] [PubMed] [Google Scholar]
- 87.Domachowske JB, et al. Safety, tolerability and pharmacokinetics of MEDI8897, an extended half-life single-dose respiratory syncytial virus prefusion F-targeting monoclonal antibody administered as a single dose to healthy preterm infants. Pediatr. Infect. Dis. J. 2018;37:886–892. doi: 10.1097/INF.0000000000001916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Labrijn AF, Janmaat ML, Reichert JM, Parren PWHI. Bispecific antibodies: a mechanistic review of the pipeline. Nat. Rev. Drug Discov. 2019;18:585–608. doi: 10.1038/s41573-019-0028-1. [DOI] [PubMed] [Google Scholar]
- 89.Wec AZ, et al. A “Trojan horse” bispecific-antibody strategy for broad protection against ebolaviruses. Science. 2016;354:350–354. doi: 10.1126/science.aag3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bournazos S, Gazumyan A, Seaman MS, Nussenzweig MC, Ravetch JV. Bispecific anti-HIV-1 antibodies with enhanced breadth and potency. Cell. 2016;165:1609–1620. doi: 10.1016/j.cell.2016.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xu L, et al. Trispecific broadly neutralizing HIV antibodies mediate potent SHIV protection in macaques. Science. 2017;358:85–90. doi: 10.1126/science.aan8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Muyldermans S. Nanobodies: natural single-domain antibodies. Annu. Rev. Biochem. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 93.Scully M, et al. Caplacizumab treatment for acquired thrombotic thrombocytopenic purpura. N. Engl. J. Med. 2019;380:335–346. doi: 10.1056/NEJMoa1806311. [DOI] [PubMed] [Google Scholar]
- 94.Xu J, et al. Nanobodies from camelid mice and llamas neutralize SARS-CoV-2 variants. Nature. 2021;595:278–282. doi: 10.1038/s41586-021-03676-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Plückthun A. Designed ankyrin repeat proteins (DARPins): binding proteins for research, diagnostics, and therapy. Annu. Rev. Pharmacol. Toxicol. 2015;55:489–511. doi: 10.1146/annurev-pharmtox-010611-134654. [DOI] [PubMed] [Google Scholar]
- 96.Rothenberger, S. et al. Ensovibep, a novel trispecific DARPin candidate that protects against SARS-CoV-2 variants. Preprint at bioRxiv10.1101/2021.02.03.429164 (2022).
- 97.Walser, M. et al. Highly potent anti-SARS-CoV-2 multivalent DARPin therapeutic candidates. Preprint at bioRxiv10.1101/2020.08.25.256339 (2021).
- 98.DARPins stack up as anti-COVID-19 agents. Nat. Biotechnol.38, 1369–1369 (2020). [DOI] [PubMed]
- 99.Su S, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.V’kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Killerby ME, et al. Human coronavirus circulation in the United States 2014–2017. J. Clin. Virol. 2018;101:52–56. doi: 10.1016/j.jcv.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Olsen SJ, et al. Transmission of the severe acute respiratory syndrome on aircraft. N. Engl. J. Med. 2003;349:2416–2422. doi: 10.1056/NEJMoa031349. [DOI] [PubMed] [Google Scholar]
- 103.Yount B, et al. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl Acad. Sci. USA. 2003;100:12995–13000. doi: 10.1073/pnas.1735582100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cauchemez S, et al. Middle East respiratory syndrome coronavirus: quantification of the extent of the epidemic, surveillance biases, and transmissibility. Lancet Infect. Dis. 2014;14:50–56. doi: 10.1016/S1473-3099(13)70304-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Graham RL, Donaldson EF, Baric RS. A decade after SARS: strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 2013;11:836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhu N, et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chan JF, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Coronavirus disease (COVID-19) pandemic. World Health Organizationhttps://www.who.int/emergencies/diseases/novel-coronavirus-2019 (2021). [PubMed]
- 109.Raj VS, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Harrison SC. Viral membrane fusion. Virology. 2015;479-480:498–507. doi: 10.1016/j.virol.2015.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Park Y-J, et al. Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors. Nat. Struct. Mol. Biol. 2019;26:1151–1157. doi: 10.1038/s41594-019-0334-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhou P, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shang J, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cai Y, et al. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tripp RA, et al. Monoclonal antibodies to SARS-associated coronavirus (SARS-CoV): identification of neutralizing and antibodies reactive to S, N, M and E viral proteins. J. Virol. Meth. 2005;128:21–28. doi: 10.1016/j.jviromet.2005.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jiang L, et al. Potent neutralization of MERS-CoV by human neutralizing monoclonal antibodies to the viral spike glycoprotein. Sci. Transl. Med. 2014;6:234ra259–234ra259. doi: 10.1126/scitranslmed.3008140. [DOI] [PubMed] [Google Scholar]
- 117.[No authors listed] An EUA for sotrovimab for treatment of COVID-19. Med. Lett. Drugs Ther. 2021;63:97-xx98. [PubMed] [Google Scholar]
- 118.Baum A, et al. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science. 2020;370:1110–1115. doi: 10.1126/science.abe2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Baum A, et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hansen J, et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Deeks ED. Casirivimab/Imdevimab: first approval. Drugs. 2021;81:2047–2055. doi: 10.1007/s40265-021-01620-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jones, B. E. et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci. Transl. Med. 10.1126/scitranslmed.abf1906 (2021). [DOI] [PMC free article] [PubMed]
- 123.Shi R, et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 124.Dougan M, et al. Bamlanivimab plus etesevimab in mild or moderate COVID-19. N. Engl. J. Med. 2021;385:1382–1392. doi: 10.1056/NEJMoa2102685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gottlieb RL, et al. Effect of bamlanivimab as monotherapy or in combination with etesevimab on viral load in patients with mild to moderate COVID-19: a randomized clinical trial. JAMA. 2021;325:632–644. doi: 10.1001/jama.2021.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dong J, et al. Genetic and structural basis for SARS-CoV-2 variant neutralization by a two-antibody cocktail. Nat. Microbiol. 2021;6:1233–1244. doi: 10.1038/s41564-021-00972-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zost SJ, et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.[No authors listed] Tixagevimab and cilgavimab (Evusheld) for pre-exposure prophylaxis of COVID-19. JAMA. 2022;327:384–385. doi: 10.1001/jama.2021.24931. [DOI] [PubMed] [Google Scholar]
- 129.Li T, et al. Cross-neutralizing antibodies bind a SARS-CoV-2 cryptic site and resist circulating variants. Nat. Commun. 2021;12:5652. doi: 10.1038/s41467-021-25997-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ryu D-K, et al. Therapeutic effect of CT-P59 against SARS-CoV-2 South African variant. Biochem. Biophys. Res. Commun. 2021;566:135–140. doi: 10.1016/j.bbrc.2021.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Westendorf K, et al. LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants. Cell Rep. 2022;39:110812. doi: 10.1016/j.celrep.2022.110812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hoffmann M, et al. The Omicron variant is highly resistant against antibody-mediated neutralization: implications for control of the COVID-19 pandemic. Cell. 2022;185:447–456.e11. doi: 10.1016/j.cell.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fenwick, C. et al. SARS-CoV-2 Omicron potently neutralized by a novel antibody with unique spike binding properties. Preprint at bioRxiv10.1101/2022.03.18.484873 (2022).
- 134.Du S, et al. Structurally resolved SARS-CoV-2 antibody shows high efficacy in severely infected hamsters and provides a potent cocktail pairing strategy. Cell. 2020;183:1013–1023.e13. doi: 10.1016/j.cell.2020.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tortorici MA, et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370:950–957. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Brouwer PJM, et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Barnes CO, et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Fenwick C, et al. A highly potent antibody effective against SARS-CoV-2 variants of concern. Cell Rep. 2021;37:109814. doi: 10.1016/j.celrep.2021.109814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wu NC, et al. An alternative binding mode of IGHV3-53 antibodies to the SARS-CoV-2 receptor binding domain. Cell Rep. 2020;33:108274. doi: 10.1016/j.celrep.2020.108274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Liu L, et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 141.Kreye J, et al. A therapeutic non-self-reactive SARS-CoV-2 antibody protects from lung pathology in a COVID-19 hamster model. Cell. 2020;183:1058–1069.e19. doi: 10.1016/j.cell.2020.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ju B, et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 143.Tian X, et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhou D, et al. Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient. Nat. Struct. Mol. Biol. 2020;27:950–958. doi: 10.1038/s41594-020-0480-y. [DOI] [PubMed] [Google Scholar]
- 145.Pinto D, et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 146.Cerutti G, et al. Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe. 2021;29:819–833.e7. doi: 10.1016/j.chom.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.McCallum M, et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184:2332–2347.e16. doi: 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Suryadevara N, et al. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. Cell. 2021;184:2316–2331.e15. doi: 10.1016/j.cell.2021.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chi X, et al. A neutralizing human antibody binds to the N-terminal domain of the spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Shrock, E. et al. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science10.1126/science.abd4250 (2020). [DOI] [PMC free article] [PubMed]
- 151.Pinto D, et al. Broad betacoronavirus neutralization by a stem helix-specific human antibody. Science. 2021;373:1109–1116. doi: 10.1126/science.abj3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.O’Brien, M. P. et al. Subcutaneous REGEN-COV antibody combination to prevent COVID-19. N. Engl. J. Med. 10.1056/NEJMoa2109682 (2021). [DOI] [PMC free article] [PubMed]
- 153.Arvin AM, et al. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature. 2020;584:353–363. doi: 10.1038/s41586-020-2538-8. [DOI] [PubMed] [Google Scholar]
- 154.Battles MB, McLellan JS. Respiratory syncytial virus entry and how to block it. Nat. Rev. Microbiol. 2019;17:233–245. doi: 10.1038/s41579-019-0149-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Glezen WP, Taber LH, Frank AL, Kasel JA. Risk of primary infection and reinfection with respiratory syncytial virus. Am. J. Dis. Child. 1986;140:543–546. doi: 10.1001/archpedi.1986.02140200053026. [DOI] [PubMed] [Google Scholar]
- 156.Hall CB, Walsh EE, Long CE, Schnabel KC. Immunity to and frequency of reinfection with respiratory syncytial virus. J. Infect. Dis. 1991;163:693–698. doi: 10.1093/infdis/163.4.693. [DOI] [PubMed] [Google Scholar]
- 157.American Academy of Pediatrics Committee on Infectious Diseases, Committee on Fetus and Newborn. Respiratory syncytial virus immune globulin intravenous: indications for use. Pediatrics99, 645–650 (1997). [PubMed]
- 158.Groothuis JR, et al. Prophylactic administration of respiratory syncytial virus immune globulin to high-risk infants and young children. The Respiratory Syncytial Virus Immune Globulin Study Group. N. Engl. J. Med. 1993;329:1524–1530. doi: 10.1056/NEJM199311183292102. [DOI] [PubMed] [Google Scholar]
- 159.Group TI-RS. Palivizumab, a humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. Pediatrics. 1998;102:531–537. doi: 10.1542/peds.102.3.531. [DOI] [PubMed] [Google Scholar]
- 160.Magro M, et al. Neutralizing antibodies against the preactive form of respiratory syncytial virus fusion protein offer unique possibilities for clinical intervention. Proc. Natl Acad. Sci. USA. 2012;109:3089–3094. doi: 10.1073/pnas.1115941109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ngwuta JO, et al. Prefusion F-specific antibodies determine the magnitude of RSV neutralizing activity in human sera. Sci. Transl. Med. 2015;7:309ra162. doi: 10.1126/scitranslmed.aac4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Huang K, Incognito L, Cheng X, Ulbrandt ND, Wu H. Respiratory syncytial virus-neutralizing monoclonal antibodies motavizumab and palivizumab inhibit fusion. J. Virol. 2010;84:8132–8140. doi: 10.1128/JVI.02699-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.McLellan JS, et al. Structural basis of respiratory syncytial virus neutralization by motavizumab. Nat. Struct. Mol. Biol. 2010;17:248–250. doi: 10.1038/nsmb.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Carbonell-Estrany X, et al. Motavizumab for prophylaxis of respiratory syncytial virus in high-risk children: a noninferiority trial. Pediatrics. 2010;125:e35–e51. doi: 10.1542/peds.2008-1036. [DOI] [PubMed] [Google Scholar]
- 165.Kwakkenbos MJ, et al. Generation of stable monoclonal antibody-producing B cell receptor-positive human memory B cells by genetic programming. Nat. Med. 2010;16:123–128. doi: 10.1038/nm.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.McLellan JS, et al. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science. 2013;342:592–598. doi: 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Tian D, et al. Structural basis of respiratory syncytial virus subtype-dependent neutralization by an antibody targeting the fusion glycoprotein. Nat. Commun. 2017;8:1877. doi: 10.1038/s41467-017-01858-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sivapalasingam, S. et al. Phase 1 study evaluating safety, tolerability, pharmacokinetics and immunogenicity of REGN2222 in healthy adults: a new human monoclonal RSV-F antibody for RSV prevention. Open Forum Infect. Dis. 10.1093/ofid/ofv133.628 (2015).
- 169.Simões, E. A. F. et al. Suptavumab for the prevention of medically attended respiratory syncytial virus infection in preterm infants. Clin. Infect. Dis. 10.1093/cid/ciaa951 (2020). [DOI] [PMC free article] [PubMed]
- 170.Malvy D, McElroy AK, de Clerck H, Günther S, van Griensven J. Ebola virus disease. Lancet. 2019;393:936–948. doi: 10.1016/S0140-6736(18)33132-5. [DOI] [PubMed] [Google Scholar]
- 171.Lee JE, et al. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Côté M, et al. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature. 2011;477:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Olal D, et al. Structure of an antibody in complex with its mucin domain linear epitope that is protective against Ebola virus. J. Virol. 2012;86:2809–2816. doi: 10.1128/JVI.05549-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wilson JA, et al. Epitopes involved in antibody-mediated protection from Ebola virus. Science. 2000;287:1664–1666. doi: 10.1126/science.287.5458.1664. [DOI] [PubMed] [Google Scholar]
- 175.Qiu X, et al. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature. 2014;514:47–53. doi: 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Corti D, et al. Protective monotherapy against lethal Ebola virus infection by a potently neutralizing antibody. Science. 2016;351:1339–1342. doi: 10.1126/science.aad5224. [DOI] [PubMed] [Google Scholar]
- 177.Misasi J, et al. Structural and molecular basis for Ebola virus neutralization by protective human antibodies. Science. 2016;351:1343–1346. doi: 10.1126/science.aad6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Pascal KE, et al. Development of clinical-stage human monoclonal antibodies that treat advanced Ebola virus disease in nonhuman primates. J. Infect. Dis. 2018;218:S612–s626. doi: 10.1093/infdis/jiy285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Mulangu S, et al. A randomized, controlled trial of Ebola virus disease therapeutics. N. Engl. J. Med. 2019;381:2293–2303. doi: 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Markham A. REGN-EB3: first approval. Drugs. 2021;81:175–178. doi: 10.1007/s40265-020-01452-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lee A. Ansuvimab: first approval. Drugs. 2021;81:595–598. doi: 10.1007/s40265-021-01483-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Griffiths, P. & Reeves, M. Pathogenesis of human cytomegalovirus in the immunocompromised host. Nat. Rev. Microbiol. 10.1038/s41579-021-00582-z (2021). [DOI] [PMC free article] [PubMed]
- 183.Schleiss MR. Congenital cytomegalovirus: impact on child health. Contemp. Pediatr. 2018;35:16–24. [PMC free article] [PubMed] [Google Scholar]
- 184.Ciferri C, et al. Structural and biochemical studies of HCMV gH/gL/gO and Pentamer reveal mutually exclusive cell entry complexes. Proc. Natl Acad. Sci. USA. 2015;112:1767–1772. doi: 10.1073/pnas.1424818112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Foglierini M, Marcandalli J, Perez L. HCMV envelope glycoprotein diversity demystified. Front. Microbiol. 2019;10:1005. doi: 10.3389/fmicb.2019.01005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kabanova A, et al. Platelet-derived growth factor-α receptor is the cellular receptor for human cytomegalovirus gHgLgO trimer. Nat. Microbiol. 2016;1:16082. doi: 10.1038/nmicrobiol.2016.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kschonsak M, et al. Structures of HCMV Trimer reveal the basis for receptor recognition and cell entry. Cell. 2021;184:1232–1244.e16. doi: 10.1016/j.cell.2021.01.036. [DOI] [PubMed] [Google Scholar]
- 188.Martinez-Martin, N. et al. An unbiased screen for human cytomegalovirus identifies Neuropilin-2 as a central viral receptor. Cell10.1016/j.cell.2018.06.028 (2018). [DOI] [PubMed]
- 189.Kschonsak M, et al. Structural basis for HCMV Pentamer receptor recognition and antibody neutralization. Sci. Adv. 2022;8:eabm2536. doi: 10.1126/sciadv.abm2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wrapp D, et al. Structural basis for HCMV Pentamer recognition by neuropilin 2 and neutralizing antibodies. Sci. Adv. 2022;8:eabm2546. doi: 10.1126/sciadv.abm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wille PT, Wisner TW, Ryckman B, Johnson DC. Human cytomegalovirus (HCMV) glycoprotein gB promotes virus entry in trans acting as the viral fusion protein rather than as a receptor-binding protein. MBio. 2013;4:e00332–00313. doi: 10.1128/mBio.00332-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Burke HG, Heldwein EE. Crystal structure of the human cytomegalovirus glycoprotein B. PLoS Pathog. 2015;11:e1005227. doi: 10.1371/journal.ppat.1005227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Chandramouli S, et al. Structure of HCMV glycoprotein B in the postfusion conformation bound to a neutralizing human antibody. Nat. Commun. 2015;6:8176. doi: 10.1038/ncomms9176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Perotti M, Marcandalli J, Demurtas D, Sallusto F, Perez L. Rationally designed human cytomegalovirus gB nanoparticle vaccine with improved immunogenicity. PLoS Pathog. 2020;16:e1009169. doi: 10.1371/journal.ppat.1009169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Gabanti E, et al. Early T cell reconstitution and cytokine profile may help to guide a personalized management of human cytomegalovirus infection after allogeneic hematopoietic stem cell transplantation. J. Clin. Virol. 2021;135:104734. doi: 10.1016/j.jcv.2021.104734. [DOI] [PubMed] [Google Scholar]
- 196.Baldanti F, Lilleri D, Gerna G. Monitoring human cytomegalovirus infection in transplant recipients. J. Clin. Virol. 2008;41:237–241. doi: 10.1016/j.jcv.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 197.Fouts AE, et al. Mechanism for neutralizing activity by the anti-CMV gH/gL monoclonal antibody MSL-109. Proc. Natl Acad. Sci. USA. 2014;111:8209–8214. doi: 10.1073/pnas.1404653111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Jabs DA, et al. HIV and cytomegalovirus viral load and clinical outcomes in AIDS and cytomegalovirus retinitis patients: monoclonal antibody cytomegalovirus retinitis trial. AIDS. 2002;16:877–887. doi: 10.1097/00002030-200204120-00007. [DOI] [PubMed] [Google Scholar]
- 199.Ishida JH, et al. Phase 1 randomized, double-blind, placebo-controlled study of RG7667, an anticytomegalovirus combination monoclonal antibody therapy, in healthy adults. Antimicrob. Agents Chemother. 2015;59:4919–4929. doi: 10.1128/AAC.00523-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ishida, J. H. et al. Phase 2 randomized, double-blind, placebo-controlled trial of RG7667, a combination monoclonal antibody, for prevention of cytomegalovirus infection in high-risk kidney transplant recipients. Antimicrob. Agents Chemother. 10.1128/aac.01794-16 (2017). [DOI] [PMC free article] [PubMed]
- 201.Patel HD, et al. In vitro characterization of human cytomegalovirus-targeting therapeutic monoclonal antibodies LJP538 and LJP539. Antimicrob. Agents Chemother. 2016;60:4961–4971. doi: 10.1128/AAC.00382-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Maertens, J. et al. Phase 2 study of anti-human cytomegalovirus monoclonal antibodies for prophylaxis in hematopoietic cell transplantation. Antimicrob. Agents Chemother. 10.1128/aac.02467-19 (2020). [DOI] [PMC free article] [PubMed]
- 203.Nelson, C. S. et al. Preexisting antibodies can protect against congenital cytomegalovirus infection in monkeys. JCI Insight10.1172/jci.insight.94002 (2017). [DOI] [PMC free article] [PubMed]
- 204.Revello MG, et al. A randomized trial of hyperimmune globulin to prevent congenital cytomegalovirus. N. Engl. J. Med. 2014;370:1316–1326. doi: 10.1056/NEJMoa1310214. [DOI] [PubMed] [Google Scholar]
- 205.Spinillo A, Gerna G. Hyperimmune globulin to prevent congenital CMV infection. N. Engl. J. Med. 2014;370:2544–2545. doi: 10.1056/NEJMc1405377. [DOI] [PubMed] [Google Scholar]
- 206.Yamayoshi S, Kawaoka Y. Current and future influenza vaccines. Nat. Med. 2019;25:212–220. doi: 10.1038/s41591-018-0340-z. [DOI] [PubMed] [Google Scholar]
- 207.Jones, J. E. et al. Parallel evolution between genomic segments of seasonal human influenza viruses reveals RNA-RNA relationships. eLife10.7554/eLife.66525 (2021). [DOI] [PMC free article] [PubMed]
- 208.Zost SJ, Wu NC, Hensley SE, Wilson IA. Immunodominance and antigenic variation of influenza virus hemagglutinin: implications for design of universal vaccine immunogens. J. Infect. Dis. 2019;219:S38–S45. doi: 10.1093/infdis/jiy696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Virk RK, et al. Divergent evolutionary trajectories of influenza B viruses underlie their contemporaneous epidemic activity. Proc. Natl Acad. Sci. USA. 2020;117:619–628. doi: 10.1073/pnas.1916585116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Wang C-C, et al. Glycans on influenza hemagglutinin affect receptor binding and immune response. Proc. Natl Acad. Sci. USA. 2009;106:18137–18142. doi: 10.1073/pnas.0909696106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Wu NC, Wilson IA. Structural insights into the design of novel anti-influenza therapies. Nat. Struct. Mol. Biol. 2018;25:115–121. doi: 10.1038/s41594-018-0025-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Krammer F. The human antibody response to influenza A virus infection and vaccination. Nat. Rev. Immunol. 2019;19:383–397. doi: 10.1038/s41577-019-0143-6. [DOI] [PubMed] [Google Scholar]
- 213.Tzarum N, et al. Structure and receptor binding of the hemagglutinin from a human H6N1 influenza virus. Cell Host Microbe. 2015;17:369–376. doi: 10.1016/j.chom.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Cheung CS, et al. Identification and structure of a multidonor class of head-directed influenza-neutralizing antibodies reveal the mechanism for its recurrent elicitation. Cell Rep. 2020;32:108088. doi: 10.1016/j.celrep.2020.108088. [DOI] [PubMed] [Google Scholar]
- 215.Ekiert DC, et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature. 2012;489:526–532. doi: 10.1038/nature11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Lee PS, et al. Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity. Proc. Natl Acad. Sci. USA. 2012;109:17040–17045. doi: 10.1073/pnas.1212371109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Lee PS, et al. Receptor mimicry by antibody F045–092 facilitates universal binding to the H3 subtype of influenza virus. Nat. Commun. 2014;5:3614. doi: 10.1038/ncomms4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Bajic G, et al. Influenza antigen engineering focuses immune responses to a subdominant but broadly protective viral epitope. Cell Host Microbe. 2019;25:827–835.e6. doi: 10.1016/j.chom.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Bangaru S, et al. A site of vulnerability on the influenza virus hemagglutinin head domain trimer interface. Cell. 2019;177:1136–1152.e18. doi: 10.1016/j.cell.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Yassine HM, et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat. Med. 2015;21:1065–1070. doi: 10.1038/nm.3927. [DOI] [PubMed] [Google Scholar]
- 221.Wei CJ, et al. Next-generation influenza vaccines: opportunities and challenges. Nat. Rev. Drug Discov. 2020;19:239–252. doi: 10.1038/s41573-019-0056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Harshbarger WD, et al. Unique structural solution from a V(H)3-30 antibody targeting the hemagglutinin stem of influenza A viruses. Nat. Commun. 2021;12:559. doi: 10.1038/s41467-020-20879-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.He W, et al. Epitope specificity plays a critical role in regulating antibody-dependent cell-mediated cytotoxicity against influenza A virus. Proc. Natl Acad. Sci. USA. 2016;113:11931–11936. doi: 10.1073/pnas.1609316113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Sui J, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Throsby M, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS ONE. 2008;3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Ekiert DC, et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science. 2011;333:843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Friesen RH, et al. A common solution to group 2 influenza virus neutralization. Proc. Natl Acad. Sci. USA. 2014;111:445–450. doi: 10.1073/pnas.1319058110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Giang E, et al. Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc. Natl Acad. Sci. USA. 2012;109:6205–6210. doi: 10.1073/pnas.1114927109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Corti D, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 230.Dreyfus C, et al. Highly conserved protective epitopes on influenza B viruses. Science. 2012;337:1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Nakamura G, et al. An in vivo human-plasmablast enrichment technique allows rapid identification of therapeutic influenza A antibodies. Cell Host Microbe. 2013;14:93–103. doi: 10.1016/j.chom.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 232.Kallewaard NL, et al. Structure and function analysis of an antibody recognizing all influenza A subtypes. Cell. 2016;166:596–608. doi: 10.1016/j.cell.2016.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Impagliazzo A, et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science. 2015;349:1301–1306. doi: 10.1126/science.aac7263. [DOI] [PubMed] [Google Scholar]
- 234.Ali, S. O. et al. Evaluation of MEDI8852, an anti-influenza A monoclonal antibody, in treating acute uncomplicated influenza. Antimicrob. Agents Chemother.10.1128/aac.00694-18 (2018). [DOI] [PMC free article] [PubMed]
- 235.Stern AM, Markel H. The history of vaccines and immunization: familiar patterns, new challenges. Health Aff. 2005;24:611–621. doi: 10.1377/hlthaff.24.3.611. [DOI] [PubMed] [Google Scholar]
- 236.Burton, D. R. What are the most powerful immunogen design vaccine strategies? Reverse vaccinology 2.0 shows great promise. Cold Spring Harb. Perspect. Biol. 10.1101/cshperspect.a030262 (2017). [DOI] [PMC free article] [PubMed]
- 237.Plotkin SA. Correlates of protection induced by vaccination. Clin. Vaccin. Immunol. 2010;17:1055–1065. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Malito E, Carfi A, Bottomley MJ. Protein crystallography in vaccine research and development. Int. J. Mol. Sci. 2015;16:13106–13140. doi: 10.3390/ijms160613106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Lanzavecchia A, Frühwirth A, Perez L, Corti D. Antibody-guided vaccine design: identification of protective epitopes. Curr. Opin. Immunol. 2016;41:62–67. doi: 10.1016/j.coi.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 240.Wilson PC, Andrews SF. Tools to therapeutically harness the human antibody response. Nat. Rev. Immunol. 2012;12:709–719. doi: 10.1038/nri3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Pantophlet R, Burton DR. Immunofocusing: antigen engineering to promote the induction of HIV-neutralizing antibodies. Trends Mol. Med. 2003;9:468–473. doi: 10.1016/j.molmed.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 242.Sesterhenn F, Bonet J, Correia BE. Structure-based immunogen design-leading the way to the new age of precision vaccines. Curr. Opin. Struct. Biol. 2018;51:163–169. doi: 10.1016/j.sbi.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Gilman, M. S. et al. Rapid profiling of RSV antibody repertoires from the memory B cells of naturally infected adult donors. Sci. Immunol. 10.1126/sciimmunol.aaj1879 (2016). [DOI] [PMC free article] [PubMed]
- 244.Boyington JC, et al. Structure-based design of head-only fusion glycoprotein immunogens for respiratory syncytial virus. PLOS ONE. 2016;11:e0159709. doi: 10.1371/journal.pone.0159709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Krarup A, et al. A highly stable prefusion RSV F vaccine derived from structural analysis of the fusion mechanism. Nat. Commun. 2015;6:8143. doi: 10.1038/ncomms9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Joyce MG, et al. Iterative structure-based improvement of a fusion-glycoprotein vaccine against RSV. Nat. Struct. Mol. Biol. 2016;23:811–820. doi: 10.1038/nsmb.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Liljeroos L, Malito E, Ferlenghi I, Bottomley MJ. Structural and computational biology in the design of immunogenic vaccine antigens. J. Immunol. Res. 2015;2015:156241. doi: 10.1155/2015/156241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Correia BE, et al. Computational design of epitope-scaffolds allows induction of antibodies specific for a poorly immunogenic HIV vaccine epitope. Structure. 2010;18:1116–1126. doi: 10.1016/j.str.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 249.Ofek G, et al. Elicitation of structure-specific antibodies by epitope scaffolds. Proc. Natl Acad. Sci. USA. 2010;107:17880–17887. doi: 10.1073/pnas.1004728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Guenaga J, et al. Heterologous epitope-scaffold prime:boosting immuno-focuses B cell responses to the HIV-1 gp41 2F5 neutralization determinant. PLoS ONE. 2011;6:e16074. doi: 10.1371/journal.pone.0016074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Azoitei ML, et al. Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope. J. Mol. Biol. 2012;415:175–192. doi: 10.1016/j.jmb.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Kuhlman B, Bradley P. Advances in protein structure prediction and design. Nat. Rev. Mol. Cell Biol. 2019;20:681–697. doi: 10.1038/s41580-019-0163-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.McLellan JS, et al. Design and characterization of epitope-scaffold immunogens that present the motavizumab epitope from respiratory syncytial virus. J. Mol. Biol. 2011;409:853–866. doi: 10.1016/j.jmb.2011.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Correia BE, et al. Proof of principle for epitope-focused vaccine design. Nature. 2014;507:201–206. doi: 10.1038/nature12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Sesterhenn F, et al. Boosting subdominant neutralizing antibody responses with a computationally designed epitope-focused immunogen. PLoS Biol. 2019;17:e3000164. doi: 10.1371/journal.pbio.3000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Sesterhenn, F. et al. De novo protein design enables the precise induction of RSV-neutralizing antibodies. Science10.1126/science.aay5051 (2020). [DOI] [PMC free article] [PubMed]
- 257.Azoitei ML, et al. Computation-guided backbone grafting of a discontinuous motif onto a protein scaffold. Science. 2011;334:373–376. doi: 10.1126/science.1209368. [DOI] [PubMed] [Google Scholar]
- 258.Rubinstein ND, et al. Computational characterization of B-cell epitopes. Mol. Immunol. 2008;45:3477–3489. doi: 10.1016/j.molimm.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 259.Jin L, Fendly BM, Wells JA. High resolution functional analysis of antibody-antigen interactions. J. Mol. Biol. 1992;226:851–865. doi: 10.1016/0022-2836(92)90636-X. [DOI] [PubMed] [Google Scholar]
- 260.Marcandalli J, et al. Induction of potent neutralizing antibody responses by a designed protein nanoparticle vaccine for respiratory syncytial virus. Cell. 2019;176:1420–1431.e17. doi: 10.1016/j.cell.2019.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Brouwer PJM, et al. Two-component spike nanoparticle vaccine protects macaques from SARS-CoV-2 infection. Cell. 2021;184:1188–1200.e19. doi: 10.1016/j.cell.2021.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Boyoglu-Barnum S, et al. Quadrivalent influenza nanoparticle vaccines induce broad protection. Nature. 2021;592:623–628. doi: 10.1038/s41586-021-03365-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Maruyama T, et al. Recombinant human monoclonal antibodies to Ebola virus. J. Infect. Dis. 1999;179:S235–S239. doi: 10.1086/514280. [DOI] [PubMed] [Google Scholar]
- 264.Pettitt J, et al. Therapeutic intervention of Ebola virus infection in rhesus macaques with the MB-003 monoclonal antibody cocktail. Sci. Transl. Med. 2013;5:199ra113–199ra113. doi: 10.1126/scitranslmed.3006608. [DOI] [PubMed] [Google Scholar]
- 265.Cagigi A, et al. Vaccine generation of protective ebola antibodies and identification of conserved B-cell signatures. J. Infect. Dis. 2018;218:S528–s536. doi: 10.1093/infdis/jiy333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Rijal P, et al. Therapeutic monoclonal antibodies for Ebola virus infection derived from vaccinated humans. Cell Rep. 2019;27:172–186.e7. doi: 10.1016/j.celrep.2019.03.020. [DOI] [PubMed] [Google Scholar]
- 267.He L, et al. Single-component multilayered self-assembling nanoparticles presenting rationally designed glycoprotein trimers as Ebola virus vaccines. Nat. Commun. 2021;12:2633. doi: 10.1038/s41467-021-22867-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Jones SM, et al. Live attenuated recombinant vaccine protects nonhuman primates against Ebola and Marburg viruses. Nat. Med. 2005;11:786–790. doi: 10.1038/nm1258. [DOI] [PubMed] [Google Scholar]
- 269.Marzi A, et al. Antibodies are necessary for rVSV/ZEBOV-GP-mediated protection against lethal Ebola virus challenge in nonhuman primates. Proc. Natl Acad. Sci. USA. 2013;110:1893–1898. doi: 10.1073/pnas.1209591110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Henao-Restrepo AM, et al. Efficacy and effectiveness of an rVSV-vectored vaccine in preventing Ebola virus disease: final results from the Guinea ring vaccination, open-label, cluster-randomised trial (Ebola Ça Suffit!) Lancet. 2017;389:505–518. doi: 10.1016/S0140-6736(16)32621-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Mutua G, et al. Safety and immunogenicity of a 2-dose heterologous vaccine regimen with Ad26.ZEBOV and MVA-BN-Filo Ebola vaccines: 12-month data from a phase 1 randomized clinical trial in Nairobi, Kenya. J. Infect. Dis. 2019;220:57–67. doi: 10.1093/infdis/jiz071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Ledgerwood JE, et al. Chimpanzee adenovirus vector Ebola vaccine. N. Engl. J. Med. 2017;376:928–938. doi: 10.1056/NEJMoa1410863. [DOI] [PubMed] [Google Scholar]
- 273.Thi EP, et al. Lipid nanoparticle siRNA treatment of Ebola-virus-Makona-infected nonhuman primates. Nature. 2015;521:362–365. doi: 10.1038/nature14442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Elek S, Stern H. Development of a vaccine against mental retardation caused by cytomegalovirus infection in utero. Lancet. 1974;303:1–5. doi: 10.1016/S0140-6736(74)92997-3. [DOI] [PubMed] [Google Scholar]
- 275.Plotkin SA, Furukawa T, Zygraich N, Huygelen C. Candidate cytomegalovirus strain for human vaccination. Infect. Immun. 1975;12:521–527. doi: 10.1128/iai.12.3.521-527.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Plotkin SA, et al. Multicenter trial of Towne strain attenuated virus vaccine in seronegative renal transplant recipients. Transplantation. 1994;58:1176–1178. [PubMed] [Google Scholar]
- 277.Nelson Cody S, et al. HCMV glycoprotein B subunit vaccine efficacy mediated by nonneutralizing antibody effector functions. Proc. Natl Acad. Sci. USA. 2018;115:6267–6272. doi: 10.1073/pnas.1800177115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Pass RF, et al. Vaccine prevention of maternal cytomegalovirus infection. N. Engl. J. Med. 2009;360:1191–1199. doi: 10.1056/NEJMoa0804749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Griffiths PD, et al. Cytomegalovirus glycoprotein-B vaccine with MF59 adjuvant in transplant recipients: a phase 2 randomised placebo-controlled trial. Lancet. 2011;377:1256–1263. doi: 10.1016/S0140-6736(11)60136-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Liu Y, et al. Prefusion structure of human cytomegalovirus glycoprotein B and structural basis for membrane fusion. Sci. Adv. 2021;7:eabf3178. doi: 10.1126/sciadv.abf3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Wang D, Shenk T. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc. Natl Acad. Sci. USA. 2005;102:18153–18158. doi: 10.1073/pnas.0509201102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Lilleri D, et al. Human cytomegalovirus (HCMV)-specific T cell but not neutralizing or IgG binding antibody responses to glycoprotein complexes gB, gHgLgO, and pUL128L correlate with protection against high HCMV viral load reactivation in solid-organ transplant recipients. J. Med. Virol. 2018;90:1620–1628. doi: 10.1002/jmv.25225. [DOI] [PubMed] [Google Scholar]
- 283.Hai R, et al. Influenza viruses expressing chimeric hemagglutinins: globular head and stalk domains derived from different subtypes. J. Virol. 2012;86:5774–5781. doi: 10.1128/JVI.00137-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Krammer F, Pica N, Hai R, Margine I, Palese P. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J. Virol. 2013;87:6542–6550. doi: 10.1128/JVI.00641-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Margine I, et al. Hemagglutinin stalk-based universal vaccine constructs protect against group 2 influenza A viruses. J. Virol. 2013;87:10435–10446. doi: 10.1128/JVI.01715-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Krammer F, et al. Assessment of influenza virus hemagglutinin stalk-based immunity in ferrets. J. Virol. 2014;88:3432–3442. doi: 10.1128/JVI.03004-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Nachbagauer R, et al. Hemagglutinin stalk immunity reduces influenza virus replication and transmission in ferrets. J. Virol. 2015;90:3268–3273. doi: 10.1128/JVI.02481-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Nachbagauer R, et al. A chimeric hemagglutinin-based universal influenza virus vaccine approach induces broad and long-lasting immunity in a randomized, placebo-controlled phase I trial. Nat. Med. 2021;27:106–114. doi: 10.1038/s41591-020-1118-7. [DOI] [PubMed] [Google Scholar]
- 289.Kanekiyo M, et al. Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses. Nat. Immunol. 2019;20:362–372. doi: 10.1038/s41590-018-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Lu Y, Welsh JP, Swartz JR. Production and stabilization of the trimeric influenza hemagglutinin stem domain for potentially broadly protective influenza vaccines. Proc. Natl Acad. Sci. USA. 2014;111:125–130. doi: 10.1073/pnas.1308701110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Bommakanti G, et al. Design of an HA2-based Escherichia coli expressed influenza immunogen that protects mice from pathogenic challenge. Proc. Natl Acad. Sci. USA. 2010;107:13701–13706. doi: 10.1073/pnas.1007465107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Corbett, K. S. et al. Design of nanoparticulate group 2 influenza virus hemagglutinin stem antigens that activate unmutated ancestor B cell receptors of broadly neutralizing antibody lineages. mBio10.1128/mBio.02810-18 (2019). [DOI] [PMC free article] [PubMed]
- 293.Darricarrère, N. et al. Broad neutralization of H1 and H3 viruses by adjuvanted influenza HA stem vaccines in nonhuman primates. Sci. Transl. Med. 10.1126/scitranslmed.abe5449 (2021). [DOI] [PubMed]
- 294.van der Lubbe JEM, et al. Mini-hemagglutinin vaccination induces cross-reactive antibodies in pre-exposed NHP that protect mice against lethal influenza challenge. NPJ Vaccin. 2018;3:25. doi: 10.1038/s41541-018-0063-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Andrews SF, et al. A single residue in influenza virus H2 hemagglutinin enhances the breadth of the B cell response elicited by H2 vaccination. Nat. Med. 2022;28:373–382. doi: 10.1038/s41591-021-01636-8. [DOI] [PubMed] [Google Scholar]
- 296.Houser KV, et al. Safety and immunogenicity of a ferritin nanoparticle H2 influenza vaccine in healthy adults: a phase 1 trial. Nat. Med. 2022;28:383–391. doi: 10.1038/s41591-021-01660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Tseng YC, et al. Egg-based influenza split virus vaccine with monoglycosylation induces cross-strain protection against influenza virus infections. Proc. Natl Acad. Sci. USA. 2019;116:4200–4205. doi: 10.1073/pnas.1819197116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Weidenbacher PA, Kim PS. Protect, modify, deprotect (PMD): a strategy for creating vaccines to elicit antibodies targeting a specific epitope. Proc. Natl Acad. Sci. USA. 2019;116:9947–9952. doi: 10.1073/pnas.1822062116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Eggink D, Goff PH, Palese P. Guiding the immune response against influenza virus hemagglutinin toward the conserved stalk domain by hyperglycosylation of the globular head domain. J. Virol. 2014;88:699–704. doi: 10.1128/JVI.02608-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Kirchdoerfer RN, et al. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Pallesen J, et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl Acad. Sci. USA. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Walls AC, et al. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016;531:114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Sadarangani M, Marchant A, Kollmann TR. Immunological mechanisms of vaccine-induced protection against COVID-19 in humans. Nat. Rev. Immunol. 2021;21:475–484. doi: 10.1038/s41577-021-00578-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Amanat F, et al. Introduction of two prolines and removal of the polybasic cleavage site lead to higher efficacy of a recombinant spike-based SARS-CoV-2 vaccine in the mouse model. mBio. 2021;12:e02648–02620. doi: 10.1128/mBio.02648-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Wrapp D, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Walls AC, et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e286. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Haynes, B. F., Burton, D. R. & Mascola, J. R. Multiple roles for HIV broadly neutralizing antibodies. Sci. Transl. Med. 10.1126/scitranslmed.aaz2686 (2019). [DOI] [PMC free article] [PubMed]
- 308.Sok D, Burton DR. Recent progress in broadly neutralizing antibodies to HIV. Nat. Immunol. 2018;19:1179–1188. doi: 10.1038/s41590-018-0235-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Caskey M, Klein F, Nussenzweig MC. Broadly neutralizing anti-HIV-1 monoclonal antibodies in the clinic. Nat. Med. 2019;25:547–553. doi: 10.1038/s41591-019-0412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Ferrari G, et al. Envelope-specific antibodies and antibody-derived molecules for treating and curing HIV infection. Nat. Rev. Drug Discov. 2016;15:823–834. doi: 10.1038/nrd.2016.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Derking R, Sanders RW. Structure-guided envelope trimer design in HIV-1 vaccine development: a narrative review. J. Int. AIDS Soc. 2021;24:e25797. doi: 10.1002/jia2.25797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.McGuire AT. Targeting broadly neutralizing antibody precursors: a naïve approach to vaccine design. Curr. Opin. HIV AIDS. 2019;14:294–301. doi: 10.1097/COH.0000000000000548. [DOI] [PubMed] [Google Scholar]
- 313.Venkatesen P. Preliminary phase 1 results from an HIV vaccine candidate trial. Lancet Microbe. 2021;2:E95. doi: 10.1016/S2666-5247(21)00042-2. [DOI] [PubMed] [Google Scholar]
- 314.Jardine JG, et al. HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science. 2016;351:1458–1463. doi: 10.1126/science.aad9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Sok D, et al. Priming HIV-1 broadly neutralizing antibody precursors in human Ig loci transgenic mice. Science. 2016;353:1557–1560. doi: 10.1126/science.aah3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Crowe, J. E. Jr Human antibodies for viral infections. Annu. Rev. Immunol. 10.1146/annurev-immunol-042718-041309 (2022). [DOI] [PubMed]
- 317.Vellas C, et al. Influence of treatment with neutralizing monoclonal antibodies on the SARS-CoV-2 nasopharyngeal load and quasispecies. Clin. Microbiol. Infect. 2022;28:139.e135–139.e8. doi: 10.1016/j.cmi.2021.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Rockett, R. et al. Resistance mutations in SARS-CoV-2 Delta variant after sotrovimab use. N. Engl. J. Med. 10.1056/NEJMc2120219 (2022). [DOI] [PMC free article] [PubMed]
- 319.Choi B, et al. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N. Engl. J. Med. 2020;383:2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Vajda S, Porter KA, Kozakov D. Progress toward improved understanding of antibody maturation. Curr. Opin. Struct. Biol. 2021;67:226–231. doi: 10.1016/j.sbi.2020.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Nguyen DN, et al. Polymer-stabilized Cas9 nanoparticles and modified repair templates increase genome editing efficiency. Nat. Biotechnol. 2020;38:44–49. doi: 10.1038/s41587-019-0325-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Bruel, T. et al. Serum neutralization of SARS-CoV-2 Omicron sublineages BA.1 and BA.2 in patients receiving monoclonal antibodies. Nat. Med. 10.1038/s41591-022-01792-5 (2022). [DOI] [PubMed]
- 323.Liu L, et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2022;602:676–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- 324.Iketani S, et al. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature. 2022;604:553–556. doi: 10.1038/s41586-022-04594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.ACTIV-3/Therapeutics for Inpatients with COVID-19 (TICO) Study Group. Efficacy and safety of two neutralising monoclonal antibody therapies, sotrovimab and BRII-196 plus BRII-198, for adults hospitalised with COVID-19 (TICO): a randomised controlled trial. Lancet Infect. Dis. 22, 622–635 (2022). [DOI] [PMC free article] [PubMed]
- 326.VanBlargan, L. A. et al. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat. Med. 10.1038/s41591-021-01678-y (2022). [DOI] [PMC free article] [PubMed]
- 327.Self, W. H. et al. Efficacy and safety of two neutralising monoclonal antibody therapies, sotrovimab and BRII-196 plus BRII-198, for adults hospitalised with COVID-19 (TICO): a randomised controlled trial. Lancet Infect. Dis. 10.1016/S1473-3099(21)00751-9 (2021). [DOI] [PMC free article] [PubMed]
- 328.Sakharkar M, et al. Prolonged evolution of the human B cell response to SARS-CoV-2 infection. Sci. Immunol. 2021;6:eabg6916. doi: 10.1126/sciimmunol.abg6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Schirrmann T, Meyer T, Schütte M, Frenzel A, Hust M. Phage display for the generation of antibodies for proteome research, diagnostics and therapy. Molecules. 2011;16:412–426. doi: 10.3390/molecules16010412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Zurawski, D. V. & McLendon, M. K. Monoclonal antibodies as an antibacterial approach against bacterial pathogens. Antibiotics10.3390/antibiotics9040155 (2020). [DOI] [PMC free article] [PubMed]
- 331.Mohamed N, et al. A high-affinity monoclonal antibody to anthrax protective antigen passively protects rabbits before and after aerosolized Bacillus anthracis spore challenge. Infect. Immun. 2005;73:795–802. doi: 10.1128/IAI.73.2.795-802.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Wilcox MH, et al. Bezlotoxumab for prevention of recurrent Clostridium difficile infection. N. Engl. J. Med. 2017;376:305–317. doi: 10.1056/NEJMoa1602615. [DOI] [PubMed] [Google Scholar]