Abstract
Cell cycle dysregulation is a characteristic hallmark of malignancies, which results in uncontrolled cell proliferation and eventual tumor formation. Cyclin-dependent kinase 1 (CDK1) is a member of the family of cell cycle regulatory proteins involved in cell cycle maintenance. Given that overexpression of CDK1 has been associated with cancer, CDK1 inhibitors may restore equilibrium to the skewed cell cycle system and operate as an effective therapeutic drug. This study aimed to identify and classify inhibitors having a higher affinity for CDK1 and also evaluate the expression pattern and prognostic relevance of CDK1 in a wide range of cancers. We investigated therapeutic molecules structurally similar to dinaciclib for their ability to inhibit CDK1 selectively. To assess the therapeutic potential of screened Dinaciclib analogs, we used drug likeliness analysis, molecular docking, and simulation analysis. CDK1 was found to be highly upregulated across several malignancies and is associated with poor overall and relapse-free survival. Molecular docking and dynamics evaluation identified two novel dinaciclib analogs as potent CDK1 inhibitors with high binding affinity and stability compared to dinaciclib. The results indicate that increased CDK1 expression is associated with decreased OS and RFS. Additionally, dinaciclib analogs are prospective replacements for dinaciclib since they exhibit increased binding affinity, consistent with MDS findings, and have acceptable ADMET qualities. The discovery of new compounds may pave the road for their future application in cancer prevention through basic, preclinical, and clinical research.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12032-022-01748-2.
Keywords: CDK1, Dinaciclib, Molecular dynamic simulation, Breast cancer, Cell cycle
Introduction
Presently, cancer is one of the leading causes of mortalities and health concern throughout the world [1]. According to GLOBOCAN 2020, an estimated 19.3 million new cancer cases (18.1 million excluding nonmelanoma skin cancer) were diagnosed worldwide, with around 10.0 million cancer deaths (9.9 million excluding nonmelanoma skin cancer). Despite advances in diagnostics and therapeutics, the cancer incidence and mortalities are still alarming [2]. Therefore, there is an urgent need to explore new diagnostic and therapeutic strategies for effective management of this lethal disease [3–5]. Cell cycle disruption is a defining characteristic of cancer [6, 7]. The cell cycle is a highly conserved and tightly controlled cell process regulated by checkpoints, cyclin-dependent kinases (CDKs), and cyclins [8, 9]. Alterations in the regulatory mechanisms result in loss of the cell cycle control system and subsequently uncontrolled proliferation of cells, which ultimately leads to tumor formation [10–12]. Moreover, cell proliferation being the requisite process for the development of tumors, targeting cell cycle regulatory proteins is a promising therapeutic strategy [13–15]. Cyclin-dependent kinase 1 (CDK1), a member of a family of cell cycle kinases, is central to cell cycle progression [16, 17]. Numerous investigations have demonstrated that CDK1 dysregulation results in aggressive tumor development, chromosomal instability, and enhanced proliferation of cells [18]. The dysregulation of CDK1 in cancer is more significant as compared to other kinases because it is the universal master kinase that is being conserved from yeast to humans. Additionally, CDK1 overexpression has been found in various cancer types, such as esophageal adenocarcinoma, gastric cancer, ovarian cancer, oral squamous cell carcinoma, colorectal cancer, liver cancer, and breast cancer, thus designing and development of therapeutics will have pan-can application across several malignancies [19].
Numerous studies have demonstrated that inhibiting CDK1 can be a highly effective anticancer strategy for the treatment of cancer [16, 18]. Several studies have evaluated potent CDK1 inhibitors and its effect on tumor cell growth [16]. Dinaciclib is one of the most potent CDK1 inhibitors, with significant pharmacokinetic properties and safety profile. In addition to CDK1, dinaciclib (SCH727965) also inhibits CDK2, CDK5, and CDK9 at nanomolar doses [20, 21]. Studies have shown that dinaciclib reduces tumor growth in preclinical models and is efficacious against a broad spectrum of human cancer cell lines [22, 23]. However, due to the orphan nature of dinaciclib, screening and evaluating dinaciclib-related compounds with high effectiveness is needed. Computer-aided drug designing (CADD) has vastly enhanced and eased the drug development process [24–26]. CADD has gained academic appeal since it requires less time and produces findings more rapidly than traditional experimental approaches [27]. The recognition of sildenafil and thalidomide drugs are examples of CADD [27]. Additionally, the in-silico and CADD techniques can identify novel targets for already discovered drugs and predict the presence of adverse effects.
Herein the present study, we evaluated the expression profiles and prognostic significance of CDK1 across several malignancies. We also screened therapeutic molecules with structural similarity to dinaciclib for selective inhibition of CDK1. Drug likeliness evaluation, molecular docking, and simulation analysis were performed to analyze the therapeutic potential of screened dinaciclib-related compounds. The study identified two novel compounds with high binding affinity to CDK1 and may be potential dinaciclib replacements in treating malignancies overexpressing CDK1.
Materials and methods
Evaluation of the expression pattern of CDK1 in pan-cancer
GEPIA2 is a comprehensive online tool (http://gepia2.cancer-pku.cn/#index) that uses a standard processing pipeline to analyze the RNA sequencing expression data of 9736 tumors and 8587 normal samples from the TCGA and GTEx projects. Tumor/normal differential expression analysis, profiling according to cancer types or pathological stages, patient survival analysis, similar gene recognition, correlation analysis, and dimensionality reduction analysis are among the services that can be customized. GEPIA2 utilized RNA-Seq datasets from the UCSC Xena project (http://xena.ucsc.edu), generated using a standard procedure [28]. The CDK1 expression in diverse malignancies was assessed using the Gepia2 portal. We also created a heat map of CDK1 expression patterns across all TCGA tumors using the TIMER 2.0 database. TIMER 2.0 is an online resource for analyzing immune infiltrates and expression analysis in various cancer types [29].
KM plotter
The Kaplan–Meier plotter (https://kmplot.com/) is a web resource that can evaluate the relationship between gene expression (mRNA, miRNA, protein) and survival in 25k + samples from 21 tumor types including breast, ovarian, lung, and gastric cancer. GEO, EGA, and TCGA are some of the databases sources. The tool’s primary goal is to develop and validate survival biomarkers using meta-analysis [30, 31]. The overall survival (OS) and Relapse Free Survival (RFS) of CDK1 were evaluated across pan-cancer. Patients were grouped into two cohorts viz low expression group and high expression group, based on the median expression of CDK1. The association of CDK1 expression with overall survival (OS) and relapse-free survival (RFS) was evaluated along with the hazard ratio.
Screening of dinaciclib analogs
Dinaciclib was used as a parent compound in the study, and we retrieved 100 compounds structurally related to dinaciclib for evaluation in our study. The compounds were obtained from the PubChem database with information regarding structural properties, chemical and physical characteristics of the compounds, and pharmacological and biochemical information [32]. The compounds were downloaded in .SDF format and subjected to the Discovery Studio v18 for structure analysis.
Drug-like dataset preparation
The retrieved dinaciclib analogs from PubChem were evaluated for their drug likeliness potential. SwissADME, an online portal, assessed Lipinski’s rule of five [33, 34]. Next, SwissADME was further utilized to analyze the absorption, distribution, metabolism, excretion, and toxicity (ADMET analysis) of the selected dinaciclib analogs. The following parameters were selected for the process: absorption was set to 0 (excellent) and 1 (moderate), solubility was set to 2 (poor solubility), 3 (moderate solubility), and 4 (soluble). The blood–brain barrier (BBB) was maintained at levels 1 (high), 2 (middle), and 3 (extreme) (low).
Selection of target and its preparation
The target protein viz CDK1 was obtained from the protein data bank with PDB ID 6GU7 in the PDB format [35]. CDK1 was subject to the ‘Prepare protein’ module in DS, which standardizes atom names, eliminates alternate conformations (disorder), inserts missing main- and side-chain atoms, and minimizes the structure. Additionally, water molecules and heteroatoms were eliminated [35].
Molecular docking
The screened compounds after the ADMET analysis were subject to molecular docking to provide insight into the binding propensities of the ligands with CDK1. Auto dock v 4.2.6 was used to perform docking investigations of the screened compounds with CDK1. The predetermined co-crystallized X-ray structure from the RCSB PDB was used to calculate the binding cavity of proteins. The co-crystallized ligand was used to compute the residue locations within the 4 Å radius. As part of the cavity selection process, Chimera (https://www.cgl.ucsf.edu/chimera/) was used to remove co-crystallized ligands, and energy was reduced by energy using the steepest descent and conjugate gradient algorithms. Both target and receptor molecules were saved in pdbqt format after combining non-polar hydrogens. Molecular docking was performed within a grid box dimension 14 × 14 × 13 Å. It was necessary to design grid boxes with particular dimensions and 0.3 Å spacing. Docking studies of the protein–ligand complex were carried out following the Lamarckian Genetic Algorithm (LGA). There were three replicates of molecular docking investigations, each of which included 50 solutions, a population size of 500, 2,500,000 evaluations, a maximum generational number of 27, and all other parameters left at their default values. Once the docking was complete, the RMSD clustering maps were generated by re-clustering with the clustering tolerances of 0.25, 0.50, and 1 to find the best cluster with the lowest energy score and the most populations.
Molecular dynamics simulation (MD)
The Desmond 2020.1 from Schrödinger, LLC was used to run MD simulations on dock complexes for CDK1 with ligands having PubChem IDs 46916588 and 67133456. SPC water molecules and the OPLS-2005 force field were utilized in this system [36] in a period boundary salvation box of 10 Å × 10 Å × 10 Å dimensions. Na+ ions were supplied to the system to neutralize the charge, and 0.15 M of NaCl solutions were added to replicate the physiological environment. When retraining with the complexes CDK1 + 46916588 and CDK1 + 67133456, the system was first equilibrated using an NVT ensemble for 100 ns. After the preceding phase, a 12-ns NPT ensemble run was used to perform a quick equilibration and reduction. The NPT ensemble was set up using the Nose–Hoover chain coupling approach [37] and run at 27 °C for 1.0 ps while under a pressure of 1 bar throughout the whole study. A time step of 2 fs was employed in this experiment. With a relaxation duration of 2 ps, the Martyna–Tuckerman–Klein barostat method was utilized for pressure control. Ewald’s particle mesh approach was utilized to calculate long-range electrostatic interactions; the radius for coulomb interactions was fixed at 9 nm. The bonded forces were calculated using the RESPA integrator with a time step of 2 fs for each trajectory. Calculations were made to track the stability of MD simulations using parameters such as the root mean square deviation (RMSD), gyroradius, root mean square fluctuation (RMSF), number of hydrogen atoms (H-bonds), and solvent accessible surface area (SASA).
Results
CDK1 is highly upregulated across pan-cancers
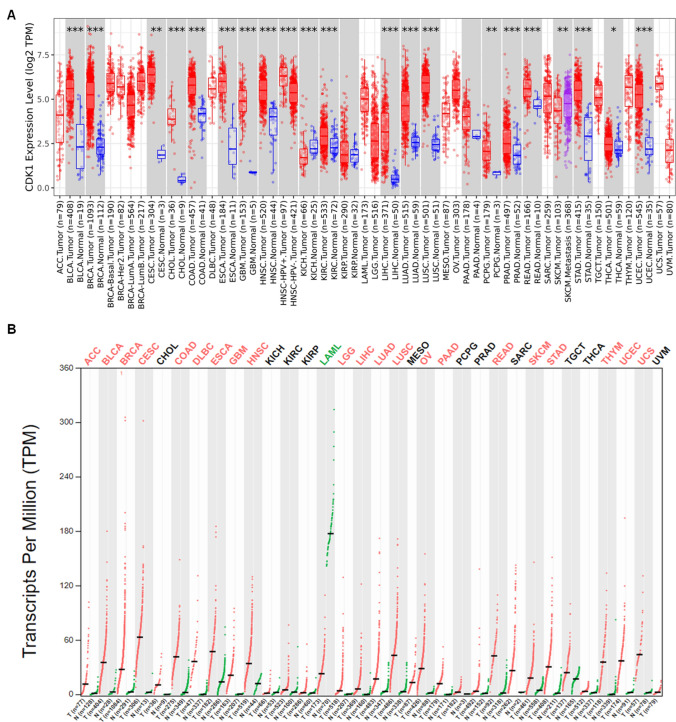
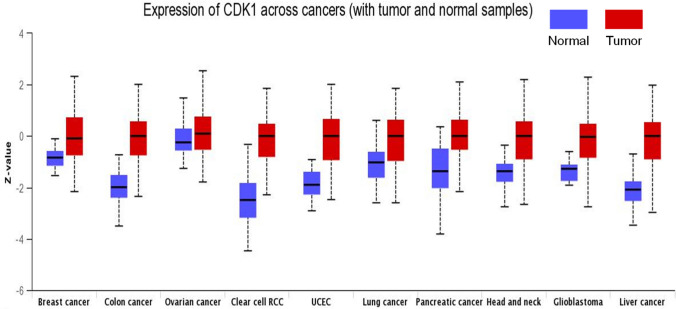
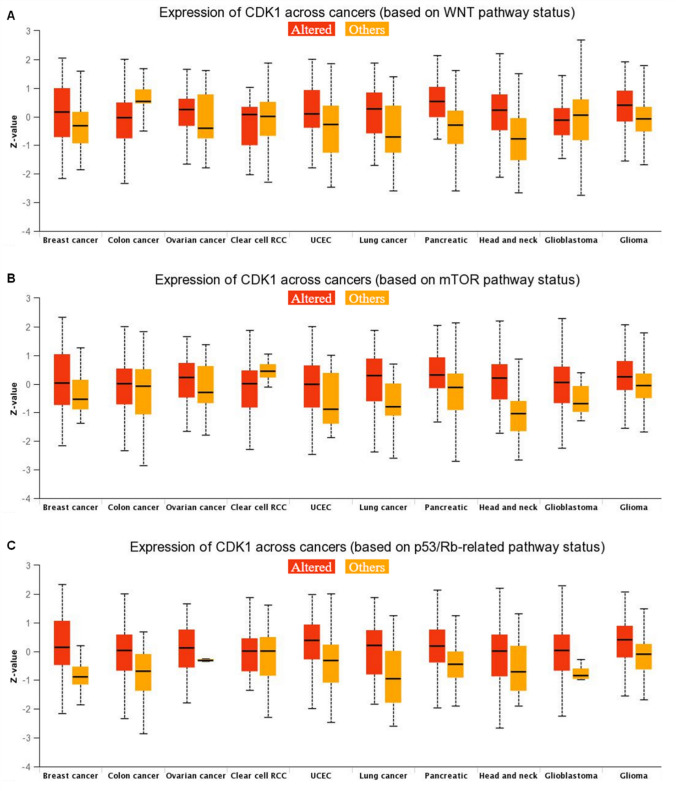
The TIMER 2.0 analysis of the CDK1 expression pattern showed upregulation of CDK1 across all TCGA datasets, as shown in the heatmap in Fig. 1A. We also evaluated the expression of CDK1 using the GEPIA 2 database across several malignancies. Our results illustrated that CDK1 is highly upregulated in several cancers, including BLCA, ESCA, BRCA, LIHC, HNSC, KIRC, LUAD, PAAD, OV, and STAD. Further, it was found that BLCA and STAD showed high mRNA levels of CDK1 in comparison to other cancers (Fig. 1B). The protein levels of CDK1 were evaluated using CPTAC samples at the UALCAN database. The study revealed that CDK1 protein levels are also highly upregulated, in consistent with CDK1 mRNA levels. The trend was observed across most of the malignancies with colon cancer, RCC, UCEC, glioblastoma, and lung cancer, showing enhanced CDK1 protein levels compared to normal samples (Fig. 2). We systematically assessed pathway-level somatic alterations in CDK1 (by small mutation or copy number alteration) across tumors with combined proteomic, whole-exome, and CNA data, involving key pathways across multiple cancer types, as shown in Fig. 3. It was found that cancer patients with overexpression of CDK1 show alterations in WNT signaling, mTOR signaling, and the p53-Rb pathway.
Fig. 1.
Expression analysis of CDK1 mRNA levels in cancer. A Differential expression of CDK1 between tumor and adjacent normal tissues across all TCGA tumors using TIMER 2.0 database. The box plots demonstrate that CDK1 is highly upregulated in several malignancies. The statistical significance was computed by the Wilcoxon test and is annotated by the number of stars (*p-value < 0.05; **p-value < 0.01; ***p-value < 0.001. B Expression pattern of CDK1 between tumor vs. paired normal samples in Gepia2 database by LIMMA analysis
Fig. 2.
Expression analysis of CDK1 levels in cancer. The protein levels of CDK1 across several malignancies were evaluated using the UALCAN database. It was found that CDK1 protein levels are augmented in tumors compared to paired normal samples, p-value < 0.01
Fig. 3.
Expression of CDK1 across pan-cancer based on pathway status. Utilizing the UALCAN database, the expression of CDK1 was evaluated for alteration in signaling pathways viz, A Wnt signaling, B mTOR pathway, and C p53/Rb-related pathway, p-value < 0.01
Prognostic significance of CDK1 in cancer
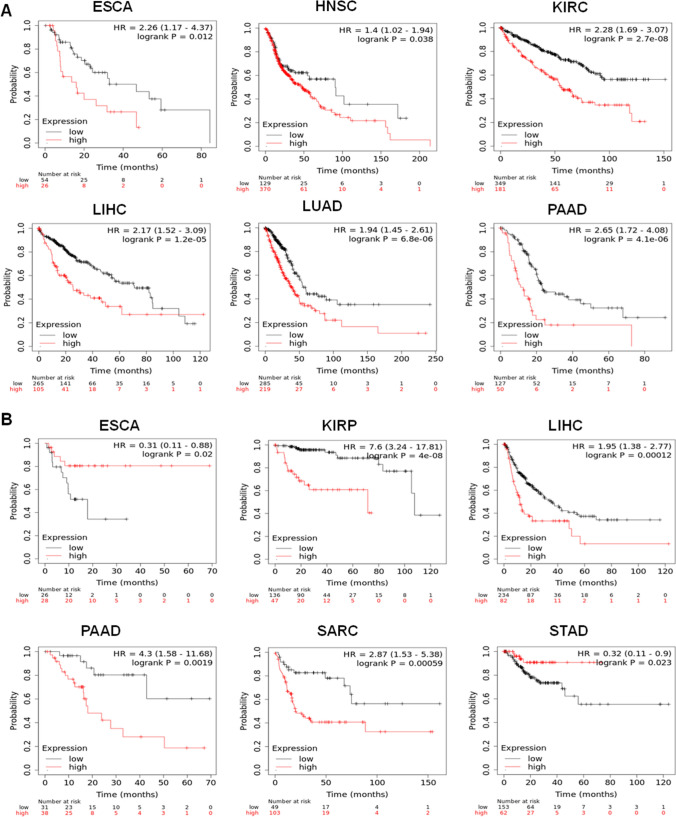
Next, we investigated the prognostic significance of CDK1 across pan-cancers using a KM-plotter. The cancer patients were grouped into two cohorts based on the median expression of CDK1, and its association with overall survival (OS) and relapse-free survival (RFS) was studied. The study highlighted that high expression profiles of CDK1 is associated with poor overall survival in ESCA, HNSC, KIRC, LUAD, LIHC, and PAAD (Fig. 4A). The hazard ratio across cancers ranged between 1.4 and 2.6, with the lowest in HNSC and the highest in PAAD. We also evaluated the association of CDK1 mRNA levels with relapse-free survival (RFS) across all tumors. The study revealed that high mRNA levels of the CDK1 gene is associated with worse RFS in KIRP, LIHC, PAAD, and SARC (Fig. 4B). The hazard ratio across the tumors ranged from 1.95 for LIHC to 7.6 for KIRP. The evaluation of expression pattern and prognostic significance indicated that CDK1 is a promising target for the effective management of cancer.
Fig. 4.
High expression of CDK1 is associated with poor overall survival (OS) and relapse-free survival (RFS). A OS plots of CDK1. High mRNA levels of CDK1 were associated with poor OS in ESCA, HNSC, KIRC, LIHC, LUAD, and PAAD. B RFS plots of CDK1. High mRNA expression of CDK1 in ESCA and STAD is associated with better RFS with HR = 0.31 and 0.32, respectively. However, high mRNA levels of CDK1 in KIRP, LIHC, PAAD, and SARC were found to be associated with worse RFS, with a hazard ratio reaching up to 7.6 in KIRP
Preparation of drug-like dataset
The 100 compounds retrieved from the PubChem were assessed for Lipinski's Rule of 5, trimming the list to 91 compounds with drug likeliness properties (Supplementary Table 1). Furthermore, ADMET evaluation was applied, and the number of compounds with drug likeliness properties was further reduced to 73 (Supplementary Table S2). These 73 compounds were selected for molecular docking to assess the binding affinity with CDK1 at the active site of the target.
Molecular docking analysis
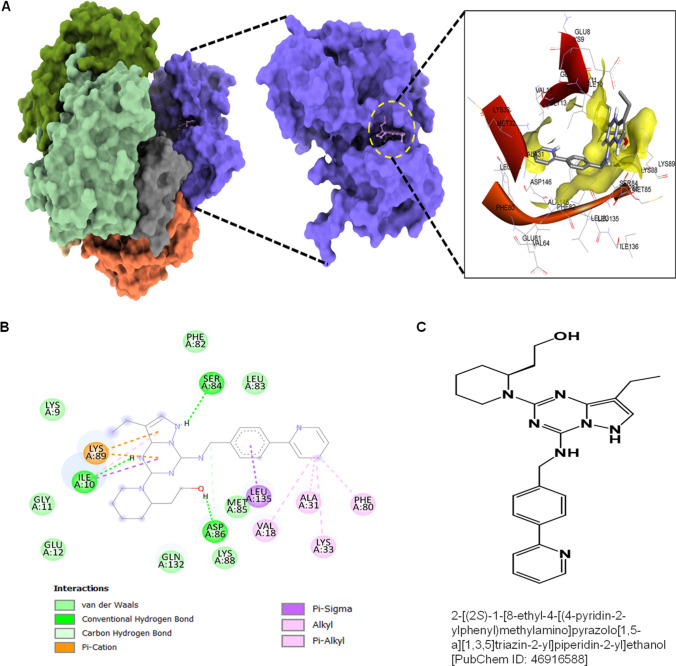
Molecular docking studies were performed to analyze the binding affinity of CDK1 with the 73 selected ligands screened after ADMET evaluation. Among the 73 ligands, several ligands showed moderate to modest binding affinity with CDK1 compared to dinaciclib, as shown in Table S2. However, two ligands with PubChem IDs: 46916588 and 67133456 exhibited increased binding affinity with CDK1. Molecular docking studies revealed that the 46916588-molecule bound significantly with CDK1 with the lowest binding energy − 9.3 kcal/mol and an inhibitory concentration (Ki) of 1.07 µM. The molecular surface view displayed the saddle-shaped binding cavity occupied by the ligand as depicted in Fig. 5A. Compound 46916588 formed pi-sigma interaction with Leu135, conventional hydrogen bond with Ile10, Ser84, and Asp86 residues (Fig. 5B). Other non-bonded interactions such as van der Waal’s interactions involved with LYS9, GLY11, GLU12, PHE82, LEU83, LYS88, and GLN132 were also observed as depicted in Fig. 5B. All the binding energy scores are calculated from the best cluster (95%) that fall within the lowest RMSD 0.25 Å.
Fig. 5.
Molecular docking evaluation of 46916588 with CDK1. A 3D schematic of 46916588 in the docked state with CDK1. B 2D plot of interactome between CDK1 and 46916588. C Structure of 46916588
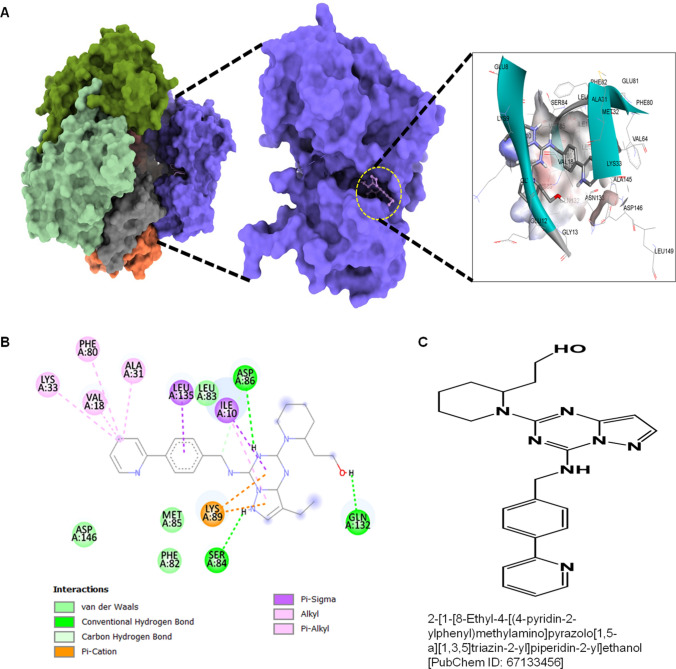
The second compound with PubChem ID: 67133456 exhibited significantly higher binding affinity with CDK1 protein with a binding energy of − 9.2 kcal/mol and an inhibitory concentration (Ki) of 2.1 µM. The molecular surface view also displayed the saddle-shaped binding cavity occupied by the ligand, as shown in Fig. 6A. The 67133456-molecule formed pi-sigma interaction with Ile10, Leu135, conventional hydrogen bond with Ser84, Asp86, Gln132 residues (Fig. 6B). Other non-bonded interactions, such as van der Waal’s interactions involved with PHE82, LEU83, and ASP146, are also depicted in Fig. 6B. All the binding energy scores were calculated from the best cluster (95%) that fall within the lowest RMSD 0.25 Å. The docking affinity studies found that 46916588 and 67133456 exhibited the highest binding affinity with CDK1 and thus considered for MD simulation studies among the screened molecules.
Fig. 6.
Molecular docking evaluation of 67133456 with CDK1. A 3D schematic of 67133456 in the docked state with CDK1. B 2D plot of interactome between CDK1 and 67133456. C Structure of 67133456
Molecular dynamics simulation (MD)
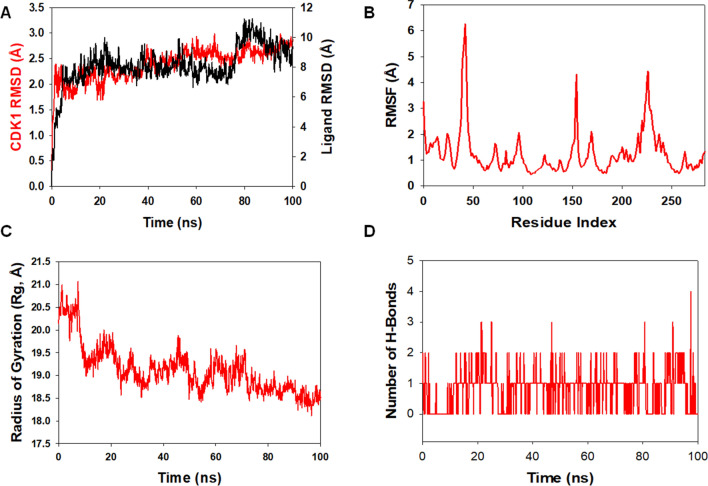
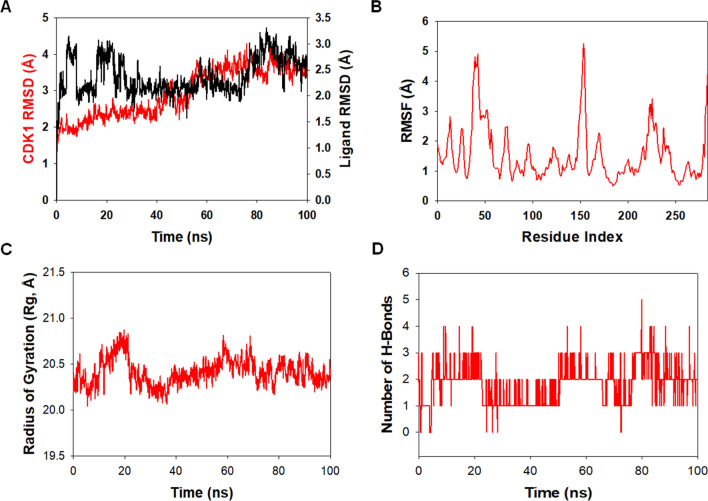
Molecular dynamics and simulation (MD) were carried out to assess the stability and coherence of CDK1 with 46916588 and 67133456 molecules. Simulation of 100 ns displayed stable conformation while comparing the root mean square deviation (RMSD) values. The RMSD of the Cα-backbone of CDK1 bound to 46916588-molecule exhibited a deviation of 0.2 Å (Fig. 7A). While Cα-backbone RMSD of CDK1 bound to 67133456 exhibited a deviation of 0.9 Å (Fig. 8A). Stable RMSD plots during simulation signify good convergence and stable conformations [38]. Therefore, it can be suggested that 46916588 or 67133456 molecules bound to CDK1 are quite stable in complex due to the higher affinity of the ligand. The plot for root means square fluctuations (RMSF) displayed small spikes of fluctuation in CDK1 protein bound to 46916588 except at residues 47, 155, and 220–240 might be due to higher flexibility of the residues conformed into the loop region. In contrast, the rest of the residues showed less fluctuation during the entire 100 ns simulation (Fig. 7B). RMSF of 67133456 bound complex displayed spikes of fluctuation at 25–60, 155, and 220–240 in CDK1 protein, indicating higher flexibility of the residues conformed into the loop region, while the rest of the residues less fluctuated during the entire 100 ns simulation (Fig. 8B) indicating the stable amino acid conformations during the simulation time. All these RMSF values were found to fall in the acceptable region. From the RMSF plots, it can be suggested that CDK1 has significant flexibility to accommodate the ligand at the biding pocket [38].
Fig. 7.
Analysis of MD simulation trajectories of 46916588 with CDK1 at 100 ns time scale. A RMSD plot displaying the molecular vibration of Cα backbone of CDK1 (red) and 46916588 (black). B RMSF plot showing the fluctuations of respective amino acids throughout the simulation time 100 ns for CDK1 + 46916588. C Radius of gyration plots to deduce compactness of protein CDK1 + 46916588. D Number of hydrogen bonds formed between CDK1 + 46916588 during 100 ns simulation time scale
Fig. 8.
Analysis of MD simulation trajectories of 67133456 with CDK1 at 100 ns time scale. A RMSD plot displaying the molecular vibration of Cα backbone of CDK1 (red) and 67133456 (black). B RMSF plot showing the fluctuations of respective amino acids throughout the simulation time 100 ns for CDK1 + 67133456. C Radius of gyration plots to deduce compactness of protein CDK1 + 67133456. D Number of hydrogen bonds formed between CDK1 + 67133456 during 100 ns simulation time scale
Next, we evaluated the radius of gyration of CDK1 bound to docked molecules. The radius of gyration (Rg) measures the compactness of the protein. CDK1 Cα-backbone bound 46916588 lowered gyration (Rg) radius from 20.5 to 18.6 Å (Fig. 7C). While CDK1 Cα-backbone bound to 67133456-molecule displayed a stable radius of gyration (Rg) from 20.25 to 20.24 Å (Fig. 8C). Significantly lowering and stable gyration (Rg) indicates a highly compact orientation of the protein in the ligand-bound state [39]. The comprehensive study of Rg signifies that the ligand-binding compels CDK1 to become more compact and less flexible.
The number of hydrogen bonds between protein and ligand suggests the significant interaction and stability of the complex. A significant number of hydrogen bonds developed between CDK1 with 46916588 throughout the simulation time of 100 ns (Average 1 number) (Fig. 7D). Consistent numbers of hydrogen bonds were also observed between CDK1 and 67133456 (Average 2 numbers) (Fig. 8D).
Followed by Rg analysis, higher stability of the complex was also analyzed in solvent accessible surface area (SASA) in both ligand unbound and bound states. In the unbound state of 46916588-molecule to CDK1, a high surface area was accessible to solvent. The SASA value of CDK1 lowered compared to the unbound state upon ligand binding (Fig. 9A). A similar pattern of accessible surface area was observed for 67133456 bounds to CDK1. Nevertheless, comparatively, less binding for 67133456 can be predicted from the SASA plot (Fig. 9B).
Fig. 9.
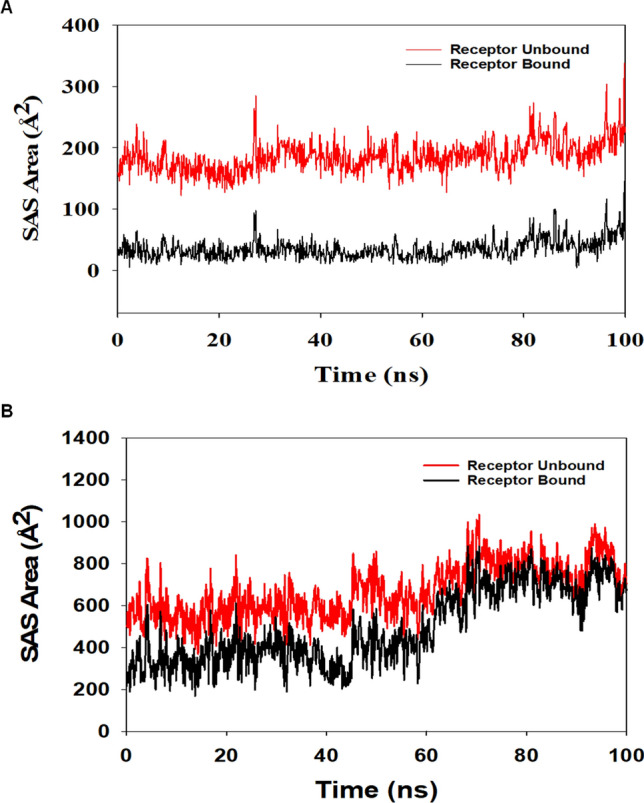
Solvent accessible surface area (SAS area). The SASA plot displays the area accessible to solvent in the unbound and bound form of the ligand, A 46916588 and B 67133456
Discussion
Cancer is a major public health problem globally and the second leading cause of death, with nearly one in six deaths [40–42]. Despite advances in diagnostics and therapy, prognosis remains poor due to emergence of drug resistance pointing toward identification of new therapeutic regimens for effective management of cancer [43–46]. During the last decade, combination therapy approach has minimized the drug resistance [47, 48], and also enhanced the prognosis of cancer patients [49, 50]. Moreover, therapeutic regimens used in combination may act in a synergistic manner reducing the drug dose required as well as reducing the toxicities associated with chemotherapy [51]. Targeting CDKs in combination with conventional therapeutic regimens is a promising approach, given the role of CDKs in cancer [12]. Thus, there is an urgent need to identify therapeutic agents specifically targeting CDKs in tumor cells. In the present study, we performed an in-silico validation of CDK1 as a therapeutic target and evaluation of novel dinaciclib analogs in inhibiting CDK1 as cancer therapeutics.
The activation of several CDKs is linked to cell cycle dysregulation, which is a hallmark of most malignancies [6]. CDKs are serine/threonine kinases that play a role in eukaryotic cell cycle progression and transcription [18]. Additionally, recent studies reveal high heterogeneity of CDKs in primary and metastatic tumors, indicating role in cancer metastasis [52–54]. CDKs are becoming appealing and potent targets for anti-cancer therapeutics because of their functional relevance [55]. As a result, creating highly selective and effective CDK inhibitors as novel therapeutic targets offers a compelling case for treating various malignancies.
This study showed that CDK1 is highly overexpressed across several malignancies. We utilized the TIMER 2.0 database and evaluated the expression pattern of CDK1 across all TCGA tumors. Differential gene expression analysis with normal samples demonstrated that tumors tend to overexpress CDK1. Given its role in regulating the cell cycle, a vital process for the proliferation of tumor cells, targeting CDK1 is a promising strategy. Gepia2 analysis also showed high expression of CDK1 in tumors except for Acute Myeloid Leukemia (LAML), wherein downregulation of CDK1 was observed. Among TCGA tumors, cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), Glioblastoma multiforme (GBM), Lymphoid Neoplasm Diffuse Large B-cell Lymphoma (DLBC), Bladder Urothelial Carcinoma (BLCA), Adrenocortical carcinoma (ACC), Breast invasive carcinoma (BRCA) showed significantly high mRNA levels of CDK1 compared to normal healthy controls.
In tandem with mRNA levels, protein levels of CDK1 were also found upregulated in cancer patients across several malignancies. Given the high levels of CDK1 protein across several malignancies, targeting CDK1 seems a promising therapeutic strategy for several malignancies. Next, we assessed the association between the expression pattern of CDK1 and alteration in key signaling cascades in cancer cells. The study demonstrated that high expression of CDK1 across several malignancies correlates with altered Wnt signaling, mTOR pathway, and p53/Rb pathway. These results indicate that CDK1 may play a key role in mediating tumorigenicity via the modulation of these pathways.
CDK1 expression pattern and survival outcome showed significant association in several malignancies with high hazard ratio. The study revealed that increased mRNA levels of CDK1 is associated with poor overall survival in ESCA. HNSC. KIRC, LIHC, LUAD, and PAAD. In other malignancies, CDK1 expression patterns did not significantly impact the overall survival of patients. In terms of relapse-free survival (RFS), high CDK1 mRNA levels were associated with worse RFS in KIRP, LIHC, PAAD, SARC, and STAD. These findings indicate that patients with low mRNA levels of CDK1 have better survival rates, and targeting CDK1 in patients with high CDK1 may prolong the survival rates.
Dinaciclib, a recently designed pan-can CDK inhibitor, including CDK1, has a substantially lower IC50 in the nano-molar range than existing CDK inhibitors [20, 56]. Dinaciclib exhibited outstanding anticancer activity in preclinical trials, outperforming earlier CDKIs, and suppressed the growth of a wide range of pre-clinical tumor models [22, 47]. Moreover, studies showed that dinaciclib was well tolerated in early trials and showed therapeutic benefit in patients with chronic lymphocytic leukemia and solid malignancies [21]. Given the orphan nature of dinaciclib, we screened dinaciclib analogs with potentially a better binding affinity toward CDK1. We utilized the PubChem database and in-silico methods to screen and evaluate dinaciclib analogs.
This mode of the in-silico approach has been utilized by various studies where the Molecular Docking and Molecular Dynamic Simulation analysis of the screened compounds have yielded the compounds having better properties than the already known compound [57–59]. For instance, the study done by Qazi et al. used the same approach to analyze potent inhibitors of SARS-CoV-2 RdRp and 3CLpro [60].
We retrieved 100 dinaciclib analogs from the PubChem database and subsequently analyzed for Lipinski’s Rule of 5 and further evaluated for ADMET Analysis. Among the screened compounds, 72 compounds showed characteristics acceptable for drug purposes.
After structural analysis and target preparation, the screened compounds were evaluated for binding affinity with CDK1. Among several compounds, two analogs showed a high binding affinity with CDK1 compared to dinaciclib and were selected for further evaluation using MD simulation studies. MD results revealed stable patterns during the entire simulation run, as is observed from the RMSD plots (Figs. 7A, 8A). These findings suggest that the compounds have not brought any abnormal behavior to the complex system. Higher flexibility of the resides in loop region of the protein may be responsible for the small spikes observed during RMSF. Most of the RMSF values were observed to fall in the acceptable region during the 100 ns. From the RMSF plots, it can be suggested that CDK1 has significant flexibility to accommodate the ligand at the biding pocket. The Rg plots also have shown that the protein remained compact during the simulation process. Delineating on the average recorded findings (Figs. 7C, 8C), it can be inferred that the protein backbone was compact. The residue fluctuations were analyzed during the simulations to recognize the behavior of the residues during the simulation run. Moreover, the accessible surface area of the target to solvent was lower upon ligand binding. In addition, the compound’s originality was determined by extracting their SMILES id and entering it into ChemSpider [61]. The results showed that these compounds had not been tested for CDK1 previously.
In conclusion, this study signifies the overexpression, copy number elevations, and prognostic association for CDK1 in human cancer specimens. In addition, our findings revealed the potential of dinaciclib analogs as a viable selective therapeutic approach for cancer treatment, either alone or in conjunction with other treatments (Table 1).
Table 1.
Binding affinity scores and pharmacokinetic properties of the selected compounds
| PubChem Id | Dock Score | Solubility | BBB | H-acceptors | H-donors | Molecular weight |
|---|---|---|---|---|---|---|
| 46926350 | − 8.5 | 3 | 3 | 6 | 2 | 396.5 |
| 46916588 | − 9.3 | 2 | 3 | 7 | 2 | 457.6 |
| 67133456 | − 9.2 | 2 | 3 | 7 | 2 | 457.6 |
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors are thankful to the school of Biological Sciences, University of Kashmir. Mr. Umar Mehraj is a senior research fellowship (SRF) recipient from UGC-CSIR, Govt. of India.
Abbreviations
- RFS
Relapse free survival
- OS
Overall survival
- BLCA
Bladder urothelial carcinoma
- ESCA
Esophageal carcinoma
- BRCA
Breast invasive carcinoma
- LIHC
Liver hepatocellular carcinoma
- HNSC
Head and neck squamous cell carcinoma
- KIRC
Kidney renal clear cell carcinoma
- STAD
Stomach adenocarcinoma
- LUAD
Lung adenocarcinoma
- PAAD
Pancreatic adenocarcinoma
- OV
Ovarian serous cystadenocarcinoma
- RCC
Renal cell carcinoma
- UCEC
Uterine corpus endometrial carcinoma
- KIRP
Kidney renal papillary cell carcinoma
- LIHC
Liver hepatocellular carcinoma
- SARC
Sarcoma
- MDS
Molecular dynamic simulation
- Rg
Radius of gyration
- SASA
Solvent accessible surface area
Author contributions
MAM designed and supervised the work. SS and UM performed the bioinformatic analysis, designed the figures and wrote the manuscript. MAM, MA, AA, HQ, and SA edited and revised the manuscript. All authors read and approved the manuscript.
Funding
This work was funded by the JK Science Technology & Innovation council DST India with Grant No. JKST&IC/SRE/885-87 to Dr. Manzoor Ahmad Mir.
Data availability
The breast cancer dataset utilized for the study is freely available at https://portal.gdc.cancer.gov/projects/TCGA-BRCA, and survival data can be accessed at https://kmplot.com/analysis/index.php?p=service&cancer=breast.
Declarations
Conflict of interest
The authors declare that they have no conflict(s) of interest in this research.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shazia Sofi and Umar Mehraj have contributed equally to this work.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Mir MA. Costimulation in lymphomas and cancers, developing costimulatory molecules for immunotherapy of diseases. Amsterdam: Elsevier; 2015. pp. 185–254. [Google Scholar]
- 3.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 4.Mehraj U, Dar AH, Wani NA, Mir MA. Tumor microenvironment promotes breast cancer chemoresistance. Cancer Chemother Pharmacol. 2021;87:1–12. doi: 10.1007/s00280-020-04222-w. [DOI] [PubMed] [Google Scholar]
- 5.Mehraj U, Ganai RA, Macha MA, Hamid A, Zargar MA, Bhat AA, Nasser MW, Haris M, Batra SK, Alshehri B. The tumor microenvironment as driver of stemness and therapeutic resistance in breast cancer: new challenges and therapeutic opportunities. Cell Oncol. 2021;44:1–21. doi: 10.1007/s13402-021-00634-9. [DOI] [PubMed] [Google Scholar]
- 6.Matthews HK, Bertoli C, de Bruin RAM. Cell cycle control in cancer. Nat Rev Mol Cell Biol. 2022;23(1):74–88. doi: 10.1038/s41580-021-00404-3. [DOI] [PubMed] [Google Scholar]
- 7.Sofi S, Mir M. Novel biomarkers in breast. Cancer. 2021 doi: 10.52305/DXSK7394. [DOI] [Google Scholar]
- 8.Panagopoulos A, Altmeyer M. The hammer and the dance of cell cycle control. Trends Biochem Sci. 2021;46(4):301–314. doi: 10.1016/j.tibs.2020.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Mehraj U, Sofi S, Alshehri B, Mir MA. Expression pattern and prognostic significance of CDKs in breast cancer: an integrated bioinformatic study. Cancer Biomark. 2022;34:1–15. doi: 10.3233/CBM-210186. [DOI] [PubMed] [Google Scholar]
- 10.Dang F, Nie L, Wei W. Ubiquitin signaling in cell cycle control and tumorigenesis. Cell Death Differ. 2021;28(2):427–438. doi: 10.1038/s41418-020-00648-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mehraj U, Aisha S, Sofi S, Mir MA. Expression pattern and prognostic significance of baculoviral inhibitor of apoptosis repeat-containing 5 (BIRC5) in breast cancer: a comprehensive analysis. Adv Cancer Biol. 2022 doi: 10.1016/j.adcanc.2022.100037. [DOI] [Google Scholar]
- 12.Sofi S, Mehraj U, Qayoom H, Aisha S, Asdaq SMB, Almilaibary A, Mir MA. Cyclin-dependent kinases in breast cancer: expression pattern and therapeutic implications. Med Oncol. 2022;39(6):1–16. doi: 10.1007/s12032-022-01731-x. [DOI] [PubMed] [Google Scholar]
- 13.Bury M, Le Calvé B, Ferbeyre G, Blank V, Lessard F. New insights into CDK regulators: novel opportunities for cancer therapy. Trends Cell Biol. 2021;31(5):331–344. doi: 10.1016/j.tcb.2021.01.010. [DOI] [PubMed] [Google Scholar]
- 14.Qayoom H, Wani NA, Alshehri B, Mir MA. An insight into the cancer stem cell survival pathways involved in chemoresistance in triple-negative breast cancer. Future Oncol. 2021;17(31):4185–4206. doi: 10.2217/fon-2021-0172. [DOI] [PubMed] [Google Scholar]
- 15.Mir M, Sofi S, Qayoom H. Targeting biologically specific molecules in triple negative breast cancer (TNBC) chapter-7. Amsterdam: Elsevier; 2022. pp. 245–277. [Google Scholar]
- 16.Izadi S, Nikkhoo A, Hojjat-Farsangi M, Namdar A, Azizi G, Mohammadi H, Yousefi M, Jadidi-Niaragh F. CDK1 in breast cancer: implications for theranostic potential. Anticancer Agents Med Chem. 2020;20(7):758–767. doi: 10.2174/1871520620666200203125712. [DOI] [PubMed] [Google Scholar]
- 17.Li M, He F, Zhang Z, Xiang Z, Hu D. CDK1 serves as a potential prognostic biomarker and target for lung cancer. J Int Med Res. 2020;48(2):0300060519897508. doi: 10.1177/0300060519897508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malumbres M, Barbacid M. Cell cycle CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9(3):153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 19.Kourea HP, Koutras AK, Scopa CD, Marangos MN, Tzoracoeleftherakis E, Koukouras D, Kalofonos HP. Expression of the cell cycle regulatory proteins p34cdc2, p21waf1, and p53 in node negative invasive ductal breast carcinoma. Mol Pathol. 2003;56(6):328. doi: 10.1136/mp.56.6.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saqub H, Proetsch-Gugerbauer H, Bezrookove V, Nosrati M, Vaquero EM, de Semir D, Ice RJ, McAllister S, Soroceanu L, Kashani-Sabet M. Dinaciclib, a cyclin-dependent kinase inhibitor, suppresses cholangiocarcinoma growth by targeting CDK2/5/9. Sci Rep. 2020;10(1):1–13. doi: 10.1038/s41598-020-75578-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar SK, LaPlant B, Chng WJ, Zonder J, Callander N, Fonseca R, Fruth B, Roy V, Erlichman C, Stewart AK. Dinaciclib, a novel CDK inhibitor, demonstrates encouraging single-agent activity in patients with relapsed multiple myeloma. Blood J Am Soc Hematol. 2015;125(3):443–448. doi: 10.1182/blood-2014-05-573741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parry D, Guzi T, Shanahan F, Davis N, Prabhavalkar D, Wiswell D, Seghezzi W, Paruch K, Dwyer MP, Doll R. Dinaciclib (SCH 727965), a novel and potent cyclin-dependent kinase inhibitor. Mol Cancer Ther. 2010;9(8):2344–2353. doi: 10.1158/1535-7163.MCT-10-0324. [DOI] [PubMed] [Google Scholar]
- 23.Martin MP, Olesen SH, Georg GI, Schönbrunn E. Cyclin-dependent kinase inhibitor dinaciclib interacts with the acetyl-lysine recognition site of bromodomains. ACS Chem Biol. 2013;8(11):2360–2365. doi: 10.1021/cb4003283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baig MH, Ahmad K, Rabbani G, Danishuddin M, Choi I. Computer aided drug design and its application to the development of potential drugs for neurodegenerative disorders. Curr Neuropharmacol. 2018;16(6):740–748. doi: 10.2174/1570159X15666171016163510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mehraj U, Qayoom H, Sofi S, Farhana P, Asdaq SMB, Mir MA. Cryptolepine targets TOP2A and inhibits tumor cell proliferation in breast cancer cells-an in vitro and in silico study. Anticancer Agents Med Chem. 2022 doi: 10.2174/1871520622666220419135547. [DOI] [PubMed] [Google Scholar]
- 26.Mehraj U, Alshehri B, Khan AA, Bhat AA, Bagga P, Wani NA, Mir MA. Expression pattern and prognostic significance of chemokines in breast cancer: an integrated bioinformatics analysis. Clin Breast Cancer. 2022 doi: 10.1016/j.clbc.2022.04.008. [DOI] [PubMed] [Google Scholar]
- 27.Agamah FE, Mazandu GK, Hassan R, Bope CD, Thomford NE, Ghansah A, Chimusa ER. Computational/in silico methods in drug target and lead prediction. Brief Bioinform. 2020;21(5):1663–1675. doi: 10.1093/bib/bbz103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, Li B, Liu XS. TIMER2 0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509–W514. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res Treat. 2010;123(3):725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 31.Almansouri S, Zwyea S. Early prognosis of human renal cancer with Kaplan–Meier plotter data analysis model. Bristol: IOP Publishing; 2020. p. 012051. [Google Scholar]
- 32.Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, Li Q, Shoemaker BA, Thiessen PA, Yu B. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019;47(D1):D1102–D1109. doi: 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 1997;23(1–3):3–25. doi: 10.1016/S0169-409X(96)00423-1. [DOI] [PubMed] [Google Scholar]
- 34.Veber DF, Johnson SR, Cheng H-Y, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem. 2002;45(12):2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 35.Brown NR, Korolchuk S, Martin MP, Stanley WA, Moukhametzianov R, Noble MEM, Endicott JA. CDK1 structures reveal conserved and unique features of the essential cell cycle CDK. Nat Commun. 2015;6(1):1–12. doi: 10.1038/ncomms7769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79(2):926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 37.Martyna GJ, Tobias DJ, Klein ML. Constant pressure molecular dynamics algorithms. J Chem Phys. 1994;101(5):4177–4189. doi: 10.1063/1.467468. [DOI] [Google Scholar]
- 38.Castrosanto MA, Abrera AT, Manalo MN, Ghosh A. In silico evaluation of binding of phytochemicals from bayati (Anamirta cocculus Linn.) to the glutathione-S-transferase of Asian Corn Borer (Ostrinia furnacalis Guenée) J Biomol Struct Dyn. 2022 doi: 10.1080/073911022036240. [DOI] [PubMed] [Google Scholar]
- 39.Ghosh A, Mukerjee N, Sharma B, Pant A, Mohanta YK, Jawarkar RD, Bakal RL, Terefe EM, Batiha GE-S, Mostafa-Hedeab G. Target specific inhibition of protein tyrosine kinase in conjunction with cancer and SARS-COV-2 by olive nutraceuticals. Front Pharmacol. 2021 doi: 10.3389/fphar.2021.812565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao W, Chen H-D, Yu Y-W, Li N, Chen W-Q. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin Med J. 2021;134(07):783–791. doi: 10.1097/CM9.0000000000001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qayoom BBUMUH, Mir MA. Rising trends of cancers in Kashmir valley: distribution pattern, incidence and causes. J Oncol Res Treat. 2020;5(150):2. [Google Scholar]
- 42.Mir MA, Agrewala JN. Signaling through CD80: an approach for treating lymphomas. Expert Opin Ther Targets. 2008;12(8):969–979. doi: 10.1517/14728222.12.8.969. [DOI] [PubMed] [Google Scholar]
- 43.Pucci C, Martinelli C, Ciofani G. Innovative approaches for cancer treatment: current perspectives and new challenges. Ecancer Med Sci. 2019 doi: 10.3332/ecancer.2019.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mehraj U, Qayoom H, Mir MA. Prognostic significance and targeting tumor-associated macrophages in cancer: new insights and future perspectives. Breast Cancer. 2021 doi: 10.1007/s12282-021-01231-2. [DOI] [PubMed] [Google Scholar]
- 45.Mir MA, Mehraj U, Sheikh BA, Hamdani SS. Nanobodies: the “magic bullets” in therapeutics, drug delivery and diagnostics. Hum Antibodies. 2020;28(1):29–51. doi: 10.3233/HAB-190390. [DOI] [PubMed] [Google Scholar]
- 46.Mir MA, Mehraj U, Sheikh BA. Recent advances in chemotherapeutic implications of deguelin: a plant-derived retinoid. Nat Prod J. 2021;11(2):169–181. [Google Scholar]
- 47.Mir MA, Qayoom H, Mehraj U, Nisar S, Bhat B, Wani NA. Targeting different pathways using novel combination therapy in triple negative breast cancer. Curr Cancer Drug Targets. 2020;20(8):586–602. doi: 10.2174/1570163817666200518081955. [DOI] [PubMed] [Google Scholar]
- 48.Mir M, Sofi S, Qayoom H. Different drug delivery approaches in combinational therapy in TNBC (Chapter-8) Amsterdam: Elsevier; 2022. pp. 278–311. [Google Scholar]
- 49.Mir M, Jan S, Mehraj U. Conventional adjuvant chemotherapy in combination with surgery, radiotherapy and other specific targets (chapter-4) Amsterdam: Elsevier; 2022. pp. 145–176. [Google Scholar]
- 50.Mir MA. Chapter 3—Costimulation immunotherapy in infectious diseases. In: Mir MA, editor. Developing costimulatory molecules for immunotherapy of diseases. London: Academic Press; 2015. pp. 83–129. [Google Scholar]
- 51.Mir M. Combinational therapy in triple negative breast cancer. 1. Amsterdam: Elsevier; 2021. [Google Scholar]
- 52.Knudsen ES, Kumarasamy V, Nambiar R, Pearson JD, Vail P, Rosenheck H, Wang J, Eng K, Bremner R, Schramek D. CDK/cyclin dependencies define extreme cancer cell-cycle heterogeneity and collateral vulnerabilities. Cell Rep. 2022;38(9):110448. doi: 10.1016/j.celrep.2022.110448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mehraj U, Mushtaq U, Mir MA, Saleem A, Macha MA, Lone MN, Hamid A, Zargar MA, Ahmad SM, Wani NA. Chemokines in triple-negative breast cancer heterogeneity: new challenges for clinical implications. Semin Cancer Biol. 2022 doi: 10.1016/j.semcancer.2022.03.008. [DOI] [PubMed] [Google Scholar]
- 54.Mir MA, Mehraj U. Double-crosser of the immune system: macrophages in tumor progression and metastasis. Curr Immunol Rev. 2019;15(2):172–184. doi: 10.2174/1573395515666190611122818. [DOI] [Google Scholar]
- 55.Choudhary ML, Sisodiya SS, Kumar N, Mishra SS. Cyclin dependent kinases: old target with new challenges for anti-cancer drugs. Int J Cancer Res. 2022;12(2):109–121. [Google Scholar]
- 56.Hafeez S, Urooj M, Saleem S, Gillani Z, Shaheen S, Qazi MH, Naseer MI, Iqbal Z, Ansari SA, Haque A. BAD, a proapoptotic protein, escapes ERK/RSK phosphorylation deguelin and siRNA-treated Hela cells. PLoS ONE. 2016;11(1):e0145780. doi: 10.1371/journal.pone.0145780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thillainayagam M, Malathi K, Ramaiah S. In-silico molecular docking and simulation studies on novel chalcone and flavone hybrid derivatives with 1,2,3-triazole linkage as vital inhibitors of Plasmodium falciparum dihydroorotate dehydrogenase. J Biomol Struct Dyn. 2018;36(15):3993–4009. doi: 10.1080/07391102.2017.1404935. [DOI] [PubMed] [Google Scholar]
- 58.Aamir M, Singh VK, Dubey MK, Meena M, Kashyap SP, Katari SK, Upadhyay RS, Umamaheswari A, Singh S. In silico prediction, characterization, molecular docking, and dynamic studies on fungal SDRs as novel targets for searching potential fungicides against Fusarium wilt in tomato. Front Pharmacol. 2018;9:1038. doi: 10.3389/fphar.2018.01038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bhat BA, Mir WR, Sheikh BA, Rather MA, Mir MA. In vitro and in silico evaluation of antimicrobial properties of Delphinium cashmerianum L., a medicinal herb growing in Kashmir, India. J Ethnopharmacol. 2022;291:115046. doi: 10.1016/j.jep.2022.115046. [DOI] [PubMed] [Google Scholar]
- 60.Qazi S, Das S, Khuntia BK, Sharma V, Sharma S, Sharma G, Raza K. In silico molecular docking and molecular dynamic simulation analysis of phytochemicals from indian foods as potential inhibitors of SARS-CoV-2 RdRp and 3CLpro. Nat Prod Commun. 2021;16(9):1934578X211031707. [Google Scholar]
- 61.Pence HE, Williams A. ChemSpider: an online chemical information resource. Washington: ACS Publications; 2010. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The breast cancer dataset utilized for the study is freely available at https://portal.gdc.cancer.gov/projects/TCGA-BRCA, and survival data can be accessed at https://kmplot.com/analysis/index.php?p=service&cancer=breast.