Abstract
Introduction:
The current prevalence of the metabolically healthy obesity is about 3%. Genetic and nutrition are influencers of such phenotypes. The main goal of this study was to assess the interaction between Dietary Total Antioxidant Capacity (DTAC) and the genotypes of MC4R and Insulin resistance in metabolically healthy/unhealthy overweight and obese women in Iran.
Material And Methods:
This cross-sectional study was conducted on 237 overweight-obese women with a mean age of 36. The value of Dietary total antioxidant capacity (DTAC) was calculated using the following indices: Total reactive antioxidant potential (TRAP), Trolox equivalent antioxidant capacity (TEAC), and ferric reducing ability of plasma (FRAP). The Metabolic health status was evaluated using the Karelis criteria. Melanocortin 4 receptor single nucleotide polymorphisms were determined by the restriction fragment length polymorphism (PCR-RFLP) method. Also, insulin resistance was evaluated through homeostasis model assessment (HOMA).
Result:
Our data noted that 72.96% of participants presented Unhealthy Metabolically and 26.94% Healthy Metabolically including 33.5% of the total had T/T genotype, 23.8% had the C/T genotype, and 42.5% had the C/C genotype (P = .05). A linear regression model test showed that the probability of metabolically healthy obesity was significantly higher in patients with the T/C genotype. The test value was statistically significant (95% CI: 0.000-0.001; P = .056, β = 0). No statistically significant relation was observed between study parameters and DTAC values. HOMA-Index was higher in all unhealthy subjects significantly.
Conclusions:
The findings indicated that there are significant associations between genotypes of rs1333048 SNP and DTAC. The C/C genotype subjects with higher DTAC had a better lipid profile and were metabolically healthier.
Keywords: Diet, melanocortin type 4 receptor (MC4R), MHO, MUO, dietary total antioxidant capacity, Karelis criteria, HOMA index
Introduction
The current global prevalence of obesity is 30% to 40% which lead to the onset and development of metabolic complications and chronic diseases such as insulin resistance, dyslipidemia, hypertension, inflammation, cardiovascular diseases (CVD), type 2 diabetes mellitus (D, M2) and eventually death.1-4
Interestingly 3% of the obese population do not have the burden of any metabolic disorder.5-7 This metabolically healthy obese group has a higher risk of coronary heart disease and cerebrovascular disease compared to normal-weight individuals without metabolic alterations.8-10 Although the mechanisms underlying obesity-induced metabolic alterations are still not clearly understood, evidence suggests that genetic predisposition and lifestyle factors can influence such phenotypes. 11
Identification of genetic factors contributing to multifactorial obesity has been a necessity. Rare mutations in the melanocortin-4 receptor (MC4R), the most common single nucleotide polymorphisms) SNPs (for obesity, were associated with lower energy expenditure binge eating, hyperphagia, and food-seeking behavior.12,13 The common SNP rs17782313 near the MC4R gene was significantly associated with higher intakes of total energy and dietary fat. Moreover. it was significantly related to the increased risk of obesity among subgroups of Europeans and East Asians, adults, and children.10,13,14
According to estimates, polymorphisms in this gene are responsible for 2.4% to 4% of significant obesity.15-17 There are also some variants in the MC4R gene, which may have preventive effects on obesity.18,19 Several variants in the MC4R gene have been identified, and rs17782313 polymorphism (C) is one of those frequently described.20-25
Findings suggest that the difference between the metabolically healthy obese (MHO) and metabolically unhealthy obese (MUO) phenotypes may be partly attributed to specific genetic traits modulating body fat distribution in different regional fat depots which distributed in different regional fat depots, held diverse biological properties and functions. 17
On the other hand, oxidative stress plays an important role in the prevalence of metabolic complications. Dietary total antioxidant capacity (DTAC) is an indicator of diet quality and is typically used to estimate the cumulative effect of antioxidants in the diet overall. It can also provide insight into synergisms that may exist between diverse antioxidants in a mixed diet.18,19 Some previous studies have shown that higher dietary antioxidant intakes have favorable effects on metabolic disorders.26,27
The overproduction of reactive oxygen species may impair insulin signaling pathways and lead to endothelial damage. This causes insulin resistance and promotes acceleration of the atherogenic process.20,21 Meanwhile, IR is known to be a major risk factor for metabolic syndrome, obesity, type 2 diabetes, and cardiovascular diseases. Therefore, the assessment of insulin resistance (IR) is important due to its key role in the pathophysiology of listed diseases.22,23
The relationship between DTAC and MC4R polymorphism has been investigated in limited studies mostly with healthy people. To our knowledge, there have been few human-based studies in Middle East countries with different genotype structures and diet patterns, evaluating this relationship regarding MHOW-OB. With the high incidence of metabolic disorders in our country, this cross-sectional study aimed to assess the interaction between DTAC and MC4R and Insulin resistance in metabolically healthy/unhealthy overweight and obese women in Iran.
Materials and Methods
Study population
In this cross-sectional study 237 healthy overweight and obese women who were referred to community health centers of Tehran University of medical science, volunteers were randomly selected for the study based on the following inclusion criteria: (1) body mass index (BMI) 25 to 40 kg/m2—(2) aged 18 to 50 years old. The primary exclusion criteria included the following: (1) menopause (2) pregnancy (3) cardiovascular diseases (4) diabetes (5) Cancer (6) kidney disease (7) thyroid disease (8) acute or chronic diseases (9) use of dietary supplements for weight loss (10) follow up diet during the past year (11) use of lipid-lowering drugs (12) use of blood glucose-lowering drugs. This study was carried out following the recommendations of the institution’s ethics committee (Ethical number IR.TUMS.VCR.REC.1398.619) with written informed consent from all participants. Based on the study protocol, we assessed demographic characteristics and anthropometric measurements. Demographic questions collect data about the characteristics such as age, medical history, taking medication supplements, and medical history.
Anthropometric assessment
For each participant, height, weight, as well as waist, and hip circumferences were measured. These measurements were made in compliance with WHO recommendations and the assessments were performed by an experienced nutritionist. The equipment used was a solar digital scale and a free-standing portable height meter. Participants were weighed without shoes and heavy outdoor clothing and weight was recorded to the last 0.2 kg. Standing height was measured with a precision of 0.5 cm without shoes. The body mass index, (BMI) is considered to be the best available population marker for monitoring trends in overweight and obesity. BMI was calculated by weight (kg)/height (m) squared. 11
Dietary assessment and DTAC evaluation
Detailed dietary information was obtained by trained interviewers at the health centers in Tehran through the use of a semi-quantitative food frequency questionnaire (FFQ) that was validated. 28 The FFQ includes 147 items with a standard serving size commonly consumed by Iranians. Items are defined by a series of foods or beverages which are categorized into 9 major food groups. Completing a questionnaire aimed at capturing the total dietary intake usually requires 30 to 60 minutes. Food frequency categories “daily/weekly/monthly” were used for assessing the total dietary intake in which participants would be asked to report their consumption frequency of each food item. The reported frequency for each food item was then converted to a daily intake. Nutrient content of foods was computed by the Nutritionist 4 software based on the United States Department of Agriculture (USDA) food composition table modified for Iranian foods.
The value of DTAC was calculated using the following indices: total reactive antioxidant potential (TRAP), Trolox equivalent antioxidant capacity (TEAC), and the ferric reducing ability of plasma (FRAP). Since no databases were available to calculate the quantity of antioxidants in Iranian foods, given the available resources, we chose databases that contained most of the foods that are consumed by the Iranian population. TRAP and TEAC values were obtained from published databases for Italian foods. For FRAP, we used a database developed by Halvorsen et al Total antioxidant capacity values for each food item in the FFQ were matched to an equivalent food in each of the databases. If any food was not directly matched with a corresponding food in a database, a proxy estimation was used based on the mean value of similar food or the value of raw food as a substitute for the cooked food.23,24 DTAC for every participant was obtained by multiplying the daily intake of each selected food item by its corresponding antioxidant value per food portion and summing the final values. Antioxidants from supplements were not included in the calculation of DTAC. The DTAC values of the individuals were reported as micromole of Trolox Equivalents per day (μmol TE/day).
Biochemical analysis
Venous blood samples were drawn from the antecubital veins at 8:00 AM to avoid variations due to circadian rhythm and after a fast equal to or>12 hours. The samples were collected in 2 tubes, 1 containing sodium citrate and the other without anticoagulant. Samples were centrifuged at 3000 rpm for 15 minutes at 4°C, and aliquots of plasma and serum were prepared for testing. Glucose, total cholesterol, HDL-C, and triglycerides levels were determined in serum through the semiautomatic chemical analyzer Ekem KontroLab. LDL-C serum concentration was calculated with Friedewald’s formula. Insulin resistance was evaluated through homeostasis model assessment (HOMA): HOMA-IR = insulin (Mu/mL) × fasting glucose (mmol/L)/22.5. 6 C-reactive protein (CRP) was measured using an immunoturbidimetric assay in a BN ProSpec nephelometer.
Definition of metabolically healthy and unhealthy phenotypes
Metabolic health status was evaluated using the Karelis criteria which were adapted for the present study. In the case of the Karelis criteria: Total cholesterol ⩽200 mg/dL, Triglycerides ⩽150 mg/dL (⩽1.7 mmol/L), High-density lipoprotein cholesterol (HDL_C) ⩾50 mg/dL and no treatment, Low-density lipoprotein cholesterol (LDL_C) ⩽100 mg/dL (⩾1.3 mmol/L) and no treatment, and HOMA-IR ⩽2.8 and CRP ⩽3.0 mg/L; with ⩾4 criteria, comprised a diagnosis of metabolic health. Thus, participants were classified into 4 groups according to metabolic health: Metabolically Healthy Over-Weight (MHOW), Metabolically Unhealthy Over-Weight(MUOW), Metabolically Healthy Obese (MHO), and Metabolically Unhealthy Obese (MUO).10,11
The Genotype determination
The MC4R rs17782313 and rs1333048 SNPs (genotypes C&T) was genotyped by the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP)technique. The MC4R Genomic DNA was extracted from 200 mL of whole blood using the GeneAll Mini Columns Type kit (GeneAll, South Korea). The extracted DNA was used to assess 2 reported SNPs near the theMC4R gene, rs17782313, and rs17700633 SNPs. Polymerase chain reaction (PCR) was performed on the rs17782313 and rs17700633 SNPs using the following primers: 89 forward primer 5AAGTTCTACCTACCATGTTCTTGG-3 and reverse primer 5-90TTCCCCCTGAAGCTTTTCTTGTCATTTTGAT-3. The PCR was carried out on a total volume of 20 μL, which included 10 pmol of each primer, 50 ng/mL of DNA, and 1M of dimethyl sulfide. The amplification protocol consisted of a primary denaturation step at 94°C for 5 minutes, followed by 35 cycles of denaturation at 60°C for 1 minute, annealing at 94°C for 45 seconds, and extension at 72°C for 1 minute, and final extension at 72°C for 10 minutes. To auscultate the SNPs rs4588 (Asp, 416, Glu) and rs7041 (Thr, 420, Lys), StyI and HaeIII enzymes were used as follows: for HaeIII, PCR product 5 μL, HaeIII 1 μL, Buffer Y. Tango 10 × 1 μL, and distilled water (D.W.) 8 μL, and for StyI, PCR product 5 μL, StyI 1 μL, Buffer Y. Tango 10 × 1 μL, and D.W. 8 μL. The digestion products were stained with ethidium bromide on a 2% agarose gel and imaged. Ten percent of the samples were directly sequenced for confirmation of the PCR-RFLP results. The sequencing process was performed using the ABI PRISM 3730 automated sequencer (Applied Biosystems, Foster City, CA, USA). 29 The fragments containing 3 genotypes were distinguished: CC, CT, and TT.
Statistical analyses
Data and statistical analyses were performed using the SPSS 20 statistical package (SPSS Inc., Chicago, IL, USA). The sample size was estimated using a binary logistic equation. Normal data distribution was determined using the Kolmogorov-Smirnov test. A general linear model and ANOVA were used to assess biochemical measurements and characteristics differences between MHO and MUO groups, DTAC and rs7041, and haplotype groups. A general linear model adjusted for MC4R, DTACT as confounder effects was used. An independent-samples t-test was used to assess differences between the 3 rs4588 groups in biochemical measurements and characteristics. Coding of the SNPs was performed using an additive model. The study population characteristics are reported as mean ± standard deviation (SD). Statistical significance was defined as P ⩽ .05 for all analyses.
Results
The mean age of the participants was 36.23 ± 8.2 years, the youngest woman was 18 years old and the oldest was 50 years. The characteristics of participants are summarized in Table 1. The mean BMI was 30.69 ± 3.69 kg/m2.
Table 1.
Characteristics of the study participants.
| Mean ± SD | Min | Max | |
|---|---|---|---|
| Age (y) | 36.23 ± 8.22 | 18 | 50 |
| BMI (kg/m2) | 30.69 ± 3.69 | 25.00 | 40.70 |
| WHR | 0.93 ± 0.05 | 0.81 | 1.08 |
| TG (mg/dL) | 120.94 ± 68.69 | 37.00 | 512.00 |
| HDL (mg/dL) | 46.36 ± 10.58 | 18.00 | 82.00 |
| LDL (mg/dL) | 94.42 ± 23.81 | 34.00 | 156.00 |
| Insulin (μIU/mL) | 15.56 ± 6.02 | 6.67 | 65.89 |
| HOMA index | 3.40 ± 1.52 | 1.29 | 16.59 |
| DTAC (mmol TE/100 g) | 1167.28 ± 764.66 | 9.11 | 4179.96 |
Abbreviations: SD, standard deviation; Min, minimum; Max, maximum; BMI, body mass index; HDL, high-density lipoprotein cholesterol; LDL, low-density lipoprotein cholesterol; HOMA, homeostatic model assessment; TG, triglycerides; DTAC, Dietary total antioxidant capacity.
Our data noted that 43.4% of participants presented MUOB, 6.3%MHOB, 29.5% MUOW, and MHOW 20.6%, according to Karelis criteria. As expected, subjects in the Metabolically Unhealthy groups had higher BMI, TG, LDL_c, HOMA-Index, Insulin, and lower HDL_c levels than those in the Metabolically Healthy groups (Table 2). Remarkably, the HDL_c values were lower and HOMA-Index was higher in all unhealthy subjects in comparison with healthy subjects(P < .05).
Table 2.
The metabolic characteristics of healthy and unhealthy groups.
| MH-OW | MU-OW | MHO | MUO | Groups | |
|---|---|---|---|---|---|
| BMI (kg/m2) | 27.55 ± 1.60 | 27.72 ± 1.36 | 32.42 ± 2.80 | 33.33 ± 2.69 | Mean ± SD |
| .000 | P-value | ||||
| HDL (mg/dL) | 80.20 ± 26.50 | 117.05 ± 49.38 | 80.00 ± 20.84 | 149.13 ± 84.10 | Mean ± SD |
| .000 | P-value | ||||
| TG (mg/dL) | 51.69 ± 9.97 | 43.98 ± 8.05 | 53.00 ± 7.33 | 44.47 ± 11.60 | Mean ± SD |
| .000 | P-value | ||||
| LDL (mg/dL) | 86.10 ± 19.84 | 95.97 ± 22.93 | 81.75 ± 18.77 | 99.73 ± 25.06 | Mean ± SD |
| .001 | P-value | ||||
| Insulin (μIU/mL) | 11.73 ± 3.62 | 15.92 ± 4.68 | 12.33 ± 3.92 | 17.67 ± 7.13 | Mean ± SD |
| .000 | P-value | ||||
| HOMA index | 2.45 ± 0.85 | 3.38 ± 1.03 | 2.52 ± 0.89 | 3.96 ± 1.84 | Mean ± SD |
| .000 | P-value | ||||
Abbreviations: SD, standard deviation; MH-OWBMI, body mass index; HDL, high-density lipoprotein cholesterol; LDL, low-density lipoprotein cholesterol; HOMA, homeostatic model assessment; TG, triglycerides; MH-OW, metabolically healthy overweight; MU-OW, metabolically unhealthy overweight; MHO, metabolically healthy obesity; MUO, metabolically healthy overweight, (P ⩽ .05).
The mean DTAC level calculated in our study was 1167.28 ± 764.66 mEq. The levels of selected metabolic parameters were assessed depending on the tested DTAC. Although TG, LDL, HOMA-INDEX values were inversely related to DTAC values, no statistically significant relation was observed between them (with P = .807, P = .758, P = .909 respectively) (Table 3).
Table 3.
The mean ± SD of metabolic values in DTAC tertile groups.
| Variables | DTAC tertile | Mean ± SD | P-value |
|---|---|---|---|
| BMI (kg/m2) | Low | 30.32 ± 3.59 | .61 |
| Median | 30.87 ± 3.42 | ||
| High | 30.68 ± 3.92 | ||
| Total | 30.62 ± 3.64 | ||
| TG (mg/dL) | Low | 122.86 ± 72.58 | .80 |
| Median | 117.78 ± 73.38 | ||
| High | 115.12 ± 67.06 | ||
| Total | 118.51 ± 70.67 | ||
| HDL (mg/dL) | Low | 47.17 ± 10.52 | .75 |
| Median | 45.92 ± 10.77 | ||
| High | 47.12 ± 11.73 | ||
| Total | 46.75 ± 11.00 | ||
| LDL (mg/dL) | Low | 93.45 ± 24.74 | .97 |
| Median | 92.75 ± 22.51 | ||
| High | 92.64 ± 23.04 | ||
| Total | 92.94 ± 23.33 | ||
| Insulin (μIU/mL) | Low | 15.76 ± 5.07 | .88 |
| Median | 15.21 ± 4.69 | ||
| High | 15.45 ± 8.38 | ||
| Total | 15.48 ± 6.32 | ||
| HOMA index | Low | 3.43 ± 1.20 | .90 |
| Median | 3.31 ± 1.31 | ||
| High | 3.37 ± 2.14 | ||
| Total | 3.37 ± 1.618 |
Abbreviations: SD, standard deviation; min, minimum; max, maximum; BMI, body mass index; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; HOMA, homeostatic model assessment; IR, insulin resistance; TG, triglycerides; DTAC, Dietary total antioxidant capacity, (P ⩽ .05).
The T/T genotype was found in 33.5% of the total, 23.8% had the C/T genotype, and 42.5% had the C/C genotype. There was a statically significant association between MC4R genotype and Insulin(P < .041) and HOMA-Index(P < .022). Likewise, the T/C genotype had higher TG, LDL_c and HOMA-index compared to 2 other genotypes although it was not statically significant(Table 4).
Table 4.
The mean ± SD of variables in MC4R rs17782313 polymorphism groups.
| Variables | MC4R polymorphism | Mean ± SD | P-value |
|---|---|---|---|
| BMI (kg/m2) | TT | 30.74 ± 3.65 | .91 |
| TC | 30.51 ± 3.61 | ||
| CC | 30.74 ± 3.70 | ||
| TG (mg/dL) | TT | 125.09 ± 81.36 | .33 |
| TC | 126.60 ± 68.60 | ||
| CC | 110.71 ± 55.15 | ||
| HDL (mg/dL) | TT | 45.34 ± 10.34 | .33 |
| TC | 47.30 ± 11.32 | ||
| CC | 47.45 ± 10.96 | ||
| LDL (mg/dL) | TT | 91.36 ± 23.04 | .34 |
| TC | 97.00 ± 26.07 | ||
| CC | 95.31 ± 24.93 | ||
| Insulin (μIU/mL) | TT | 14.63 ± 4.58 | .04 |
| TC | 17.25 ± 9.21 | ||
| CC | 15.65 ± 4.96 | ||
| HOMA index | TT | 3.14 ± 1.08 | .02 |
| TC | 3.88 ± 2.42 | ||
| CC | 3.37 ± 1.25 |
Abbreviations: SD, standard deviation; min, minimum; max, maximum; BMI, body mass index; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; HOMA, homeostatic model assessment; IR, insulin resistance; TG, triglycerides; DTAC, Dietary total antioxidant capacity; MC4R.
Gene, melanocortin-4 receptor polymorphism, (P ⩽ .05).
To assess the relationship between particular polymorphisms and the incidence of MUO and MHO more accurately, a linear model was applied(Table 5). Three models were tested: model (1)—crude data, model (2)—data adjusted for MC4R, and model (3)—data adjusted for DTAC. The probability of MHO was higher in patients with the T/C genotype. The test value was statistically significant (95% CI: 0.000-0.001; P = .056, β = 0). Significant interactions were observed between DTAC value and rs17782313 SNP in terms of metabolic health (Figure 1).
Table 5.
The interaction between MC4R gene polymorphism and DTAC.
| Independent variables | B | Std. error | CI | P-value |
|---|---|---|---|---|
| MC4R TT* DTAC | .000 | 0.0003 | −0.001-0.000 | .28 |
| MC4R TC * DTAC | −.001 | 0.0003 | −0.001-4.796 | .05 |
Liner model test, reference = mc4r cc.
Dependent variable: Karlis.
Model: (intercept) mc4r*dtac, model1: mc4r tt*dtac, model2: mc4r tc * dtac, *for interactions, P < .1 was considered significant.
Figure 1.
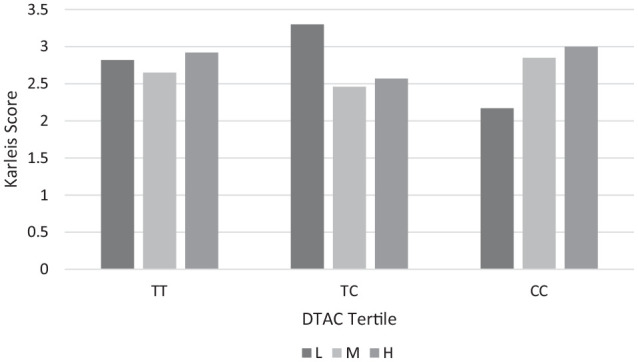
The association between DTAC and the chromosome MC4R SNPs Polymorphism.
L, low level of DTAC; M, the median level of DTAC; H, high level of DTAC, P-value = .05.
Discussion
Results of this cross-sectional study showed that 26.9% of participants are metabolic health phenotype. This figure was higher than other studies which had been reported to be 2% to 3% according to Karelis criteria.6,10 Furthermore, we observed that the prevalence of MC4R SNPs in this group was similar to the results of the Mollahosseini et al study with 52.85% of the C allele in Iranian. 3 The prevalence the of T/T genotype was 39.0 in the European study. 19
Moreover, results showed that the probability of MHOW/OB was higher in patients with the T/C genotype. In another study, MHO diagnosed based on the Karelis was observed in 39.3% of women with the T/T genotype which agrees with our results. 24
The most remarkable result of this study was providing evidence for a significant effect between DTAC level and MC4R polymorphism. The C/C genotype subjects with lower DTAC were metabolically unhealthy. Conversely, those carrying T/C genotype with lower DTAC values were metabolically healthy. It had observed that women with the C/X genotype had higher levels of total cholesterol, TG, LDL-C, and apolipoprotein B. 24 Studies showed that higher DTAC values affected obesity-related disorders prevention.10,25
Furthermore, we observed that TG, LDL, HOMA-Index values were inversely related to DTAC values, but no statistically significant relation was found. Similarly in another study, high DTAC was significantly associated with a reduced odds ratio for the prevalence of MetS components. 30 Whereas, the findings of the longitudinal Tehran Lipid and Glucose study showed that higher DTAC was positively associated with a lower occurrence of Metabolic complications hence being healthy. 26 The European Prospective Investigation into Cancer and Nutrition study showed that the DTAC may play an important role in reducing the risk of diabetes in women. 31
There was also a close connection between this gene’s SNP and an increased BMI (P = .002). Interestingly, some studies showed that the BMI of the C allele carriers was higher by 0.2 kg.m2 on average.7,13 Comparable results were reported by German authors, who claimed that the presence of the C allele entailed a considerably higher BMI (P = .0013). They showed that SNPs had an impact on the total mass of adipose tissue (P = .022).19,24
In line with previous studies, we observed that there was a statically significant association between the MC4R genotypes and Insulin and HOMA-Index.6,10 Tschritter et al’s 32 study demonstrated that in subjects carrying the rare allele of rs17782313 in the homozygous (TC) or the heterozygous form (CC), the insulin response of the brain was significantly reduced as compared to carriers of the wild-type allele (TT). Thus, insulin resistance may be one mechanism through which the MC4R polymorphisms affect the metabolism of people and be associated with being metabolically healthy and obese which decrease with dietetic interventions. 33 MC4R SNPs phenotypic expression is influenced by lifestyle factors, such as diet and physical activity. 24 The C/C genotypes with higher DTAC whilst had lower TG, LDL_c and HOMA-index were metabolically healthier. Unexpectedly, results showed that the T/C genotype subjects with lower DTAC values were metabolically healthier. The HOMA index reflecting insulin resistance could be a useful marker to early identify individuals at a high risk of developing metabolic syndrome and cardiovascular events. 34
The main strengths of this study include the study assessment of MC4R SNPs regarding Metabolic Health OW-OB in Iranian women (A Middle East country). Furthermore, metabolic health was assessed separately in overweight and obese women groups.
The limitation of the study was the small sample that it was not possible to examine the exact odds ratios. Hence, it is suggested that other scientists perform similar studies with larger sample sizes and case-control method. Another limitation was the homogender and cross-sectional method of study.
Conclusion
The evidence from this study indicated that the C/C genotype people which had lower HOMA-Index and higher DTAC were metabolically healthier. Our research highlighted the importance of lifestyle modification, as the consumption of an antioxidant-rich diet to prevent metabolic complications and so well-being. Additionally, it is suggested that to consider the genetics which will allow therapeutic focused interventions.
Acknowledgments
The authors are grateful to participants in the study for their contribution.
Footnotes
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by a Tehran University of Medical Sciences Grant (Grant Ids: 95-04-161-33833, 97-03-161-41144, 97-02-161-38999).
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Availability of Data and Material: The data that support the findings of this study are available from the Corresponding author, upon reasonable request.
Ethical Approval: This study was carried out following the recommendations of the institution’s ethics committee (Ethical number IR.TUMS.VCR.REC.1398.619) with written informed consent from all participants.
Code availability (Software Application or Custom Code): There was not any new software application or custom code in our manuscript.
ORCID iD: Maryam ElhamKia  https://orcid.org/0000-0002-6230-8698
https://orcid.org/0000-0002-6230-8698
References
- 1. Gonçalves CG, Glade MJ, Meguid MM. Metabolically healthy obese individuals: key protective factors. Nutrition. 2016;32:14-20. [DOI] [PubMed] [Google Scholar]
- 2. Hamer M, Stamatakis E. Metabolically healthy obesity and risk of all-cause and cardiovascular disease mortality. J Clin Endocrinol Metab. 2012;97:2482-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mollahosseini M, Rahimi MH, Yekaninejad MS, Maghbooli Z, Mirzaei K. Dietary patterns interact with chromosome 9p21 rs1333048 polymorphism on the risk of obesity and cardiovascular risk factors in apparently healthy Tehrani adults. Eur J Nutr. 2020;59:35-43. [DOI] [PubMed] [Google Scholar]
- 4. Powell-Wiley TM, Poirier P, Burke LE, et al. Obesity and cardiovascular disease: a scientific statement from the American Heart Association. Circulation. 2021;143:e984-e1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van Vliet-Ostaptchouk JV, Nuotio ML, Slagter SN, et al. The prevalence of metabolic syndrome and metabolically healthy obesity in Europe: a collaborative analysis of ten large cohort studies. BMC Endocr Disord. 2014;14:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Basurto L, Sánchez L, Díaz A, Valle M, Robledo A, Martínez-Murillo C. Differences between metabolically healthy and unhealthy obesity in PAI-1 level: fibrinolysis, body size phenotypes and metabolism. Thromb Res. 2019;180:110-114. [DOI] [PubMed] [Google Scholar]
- 7. Gao L, Wang L, Yang H, Pan H, Gong F, Zhu H. MC4R single nucleotide polymorphisms were associated with metabolically healthy and unhealthy obesity in Chinese Northern Han populations. Int J Endocrinol. 2019;2019:4328909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Iacobini C, Pugliese G, Blasetti Fantauzzi C, Federici M, Menini S. Metabolically healthy versus metabolically unhealthy obesity. Metabolism. 2019;92:51-60. [DOI] [PubMed] [Google Scholar]
- 9. Alberti KG, et al. Harmonizing the metabolic syndrome: a joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of obesity. Circulation. 2009;120:1640-1645. [DOI] [PubMed] [Google Scholar]
- 10. Hinnouho GM, Czernichow S, Singh-Manoux A. Response to comment on Hinnouho et al. Metabolically healthy obesity and risk of mortality: does the definition of metabolic health matter? Diabetes care 2013;36:2294-2300. Diabetes Care. 2014;37:e105-e300. [DOI] [PubMed] [Google Scholar]
- 11. Ramos-Lopez O, Riezu-Boj JI, Milagro FI, Cuervo M, Goni L, Martinez JA. Genetic and nongenetic factors explaining metabolically healthy and unhealthy phenotypes in participants with excessive adiposity: relevance for personalized nutrition. Ther Adv Endocrinol Metab. 2019;10:2042018819877303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Young EH, Wareham NJ, Farooqi S, et al. The V103I polymorphism of the MC4R gene and obesity: population based studies and meta-analysis of 29 563 individuals. Int J Obes. 2007;31:1437-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Qi L, Kraft P, Hunter DJ, Hu FB. The common obesity variant near MC4R gene is associated with higher intakes of total energy and dietary fat, weight change and diabetes risk in women. Hum Mol Genet. 2008;17:3502-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chambers JC, Elliott P, Zabaneh D, et al. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat Genet. 2008;40:716-718. [DOI] [PubMed] [Google Scholar]
- 15. Xi B, Chandak GR, Shen Y, Wang Q, Zhou D. Association between common polymorphism near the MC4R gene and obesity risk: a systematic review and meta-analysis. PLoS One. 2012;7:e45731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Manriquez V, Aviles J, Salazar L, et al. Polymorphisms in genes involved in the leptin-melanocortin pathway are associated with obesity-related cardiometabolic alterations in a southern Chilean population. Mol Diagn Ther. 2018;22:101-113. [DOI] [PubMed] [Google Scholar]
- 17. Gilardini L, Zambon A, Soranna D, Croci M, Invitti C. Predictors of the transition from metabolically healthy obesity to unhealthy obesity. Eat Weight Disord. 2018;23:739-744. [DOI] [PubMed] [Google Scholar]
- 18. Adan RA, Tiesjema B, Hillebrand JJ, la Fleur SE, Kas MJ, de Krom M. The MC4 receptor and control of appetite. Br J Pharmacol. 2006;149:815-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zujko ME, Waśkiewicz A, Witkowska AM, et al. Dietary total antioxidant capacity and dietary polyphenol intake and prevalence of metabolic syndrome in Polish adults: a nationwide study. Oxid Med Cell Longev. 2018;2018:7487816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sheikhansari G, Soltani-Zangbar MS, Pourmoghadam Z, et al. Oxidative stress, inflammatory settings, and microRNA regulation in the recurrent implantation failure patients with metabolic syndrome. Am J Reprod Immunol. 2019;82:e13170. [DOI] [PubMed] [Google Scholar]
- 21. Mozaffari H, Daneshzad E, Larijani B, Surkan PJ, Azadbakht L. Association of dietary total antioxidant capacity to anthropometry in healthy women: a cross-sectional study. Nutrition. 2020;69:110577. [DOI] [PubMed] [Google Scholar]
- 22. Mohammadabadi FZV, Seyed Mehran H, Mohammad A, Eshghinia S. Assessment of insulin resistance with two methods: HOMA-IR and TyG Index in Iranian obese women. Iran J Diab Obes. 2014;6:23-27. [Google Scholar]
- 23. Lim J, Kim J, Koo SH, Kwon GC. Comparison of triglyceride glucose index, and related parameters to predict insulin resistance in Korean adults: an analysis of the 2007-2010 Korean National Health and Nutrition Examination Survey. PLoS One. 2019;14:e0212963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brodowski J, Szkup M, Jurczak A, et al. Searching for the relationship between the parameters of metabolic syndrome and the rs17782313 (T>C) polymorphism of the MC4R gene in postmenopausal women. Clin Interv Aging. 2017;12:549-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aghamohammadi V, Sajjadi SF, Jarrahi F, Abdollahi A, Mirzaei K. The association between total antioxidant capacity and resting metabolic rate (RMR) / respiratory quotient (RQ) in overweight and obese woman. Diabetes Metab Syndr. 2019;13:2763-2767. [DOI] [PubMed] [Google Scholar]
- 26. Bahadoran Z, Golzarand M, Mirmiran P, Shiva N, Azizi F. Dietary total antioxidant capacity and the occurrence of metabolic syndrome and its components after a 3-year follow-up in adults: Tehran lipid and glucose study. Nutr Metab. 2012;9:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gantenbein KV, Kanaka-Gantenbein C. Mediterranean diet as an antioxidant: the impact on metabolic health and overall wellbeing. Nutrients. 2021;13:1951-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mirmiran P, Esfahani FH, Mehrabi Y, Hedayati M, Azizi F. Reliability and relative validity of an FFQ for nutrients in the Tehran lipid and glucose study. Public Health Nutr. 2010;13:654-662. [DOI] [PubMed] [Google Scholar]
- 29. Pooyan S, Rahimi MH, Mollahosseini M, et al. A high-protein/low-fat diet may interact with vitamin D-Binding protein gene variants to moderate the risk of depression in apparently healthy adults. Lifestyle Genom. 2018;11:64-72. [DOI] [PubMed] [Google Scholar]
- 30. Sotoudeh G, Abshirini M, Bagheri F, Siassi F, Koohdani F, Aslany Z. Higher dietary total antioxidant capacity is inversely related to prediabetes: a case-control study. Nutrition. 2018;46:20-25. [DOI] [PubMed] [Google Scholar]
- 31. Mancini FR, Affret A, Dow C, et al. Dietary antioxidant capacity and risk of type 2 diabetes in the large prospective E3N-EPIC cohort. Diabetologia. 2018;61:308-316. [DOI] [PubMed] [Google Scholar]
- 32. Tschritter O, Haupt A, Preissl H, et al. An obesity risk SNP (rs17782313) near the MC4R gene is associated with cerebrocortical insulin resistance in humans. J Obes. 2011;2011:283153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mazidi M, Katsiki N, Kengne AP, Mikhailidis DP, Banach M. Adiposity mediates the association between whole grain consumption, glucose homeostasis and insulin resistance: findings from the US NHANES. Lipids Health Dis. 2018;17:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jin JL, Cao YX, Wu LG, et al. Triglyceride glucose index for predicting cardiovascular outcomes in patients with coronary artery disease. J Thorac Dis. 2018;10:6137-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]


