Abstract
As most erb‐b2 receptor tyrosine kinase 2 (HER2)‐positive breast cancer (BC) patients currently receive dual HER2‐targeting added to neoadjuvant chemotherapy, improved methods for identifying individual response, and assisting postsurgical salvage therapy, are needed. Herein, we evaluated the 41‐gene classifier trastuzumab advantage risk model (TRAR) as a predictive marker for patients enrolled in the NeoSphere trial. TRAR scores were computed from RNA of 350 pre‐ and 166 post‐treatment tumor specimens. Overall, TRAR score was significantly associated with pathological complete response (pCR) rate independently of other predictive clinico‐pathological variables. Separate analyses according to estrogen receptor (ER) status showed a significant association between TRAR score and pCR in ER‐positive specimens but not in ER‐negative counterparts. Among ER‐positive BC patients not achieving a pCR, those with TRAR‐low scores in surgical specimens showed a trend for lower distant event‐free survival. In conclusion, in HER2‐positive/ER‐positive BC, TRAR is an independent predictor of pCR and represents a promising tool to select patients responsive to anti‐HER2‐based neoadjuvant therapy and to assist treatment escalation and de‐escalation strategies in this setting.
Keywords: breast cancer, gene expression profile, HER2, pertuzumab, predictive biomarker, trastuzumab
TRAR‐low score is an independent predictor of pathological complete response (pCR) to anti‐HER2 agents in patients with HER2‐positive and ER‐positive breast cancer of the NeoSphere study. The higher risk of relapse observed in these patients likely depends on modification by treatment of tumor proliferation and ER activity. TRAR is a promising tool to assist escalation and de‐escalation of anti‐HER2‐based treatment strategies.
Abbreviations
- BC
breast cancer
- CI
confidence interval
- DEFS
distant event‐free survival
- ER
estrogen receptor
- GEP
gene expression profile
- H
trastuzumab
- HR
hazard ratio
- OR
odds ratio
- P
pertuzumab
- pCR
pathological complete response
- T
taxane
- TRAR
trastuzumab advantage risk model
1. Introduction
The combination of trastuzumab (H), pertuzumab (P), and chemotherapy has become the standard of care for patients with HER2 overexpressing and/or amplified, that is, HER2‐positive, breast cancer (BC) in the neoadjuvant setting. In the phase II randomized study NeoSphere, a 12‐week long neoadjuvant course with docetaxel (T), H and P significantly increased the rate of pathological complete response (pCR) in the breast as compared with TH, TP, or HP (45.8% versus 29.0%, 24.0%, 16.8%) [1], which led to FDA approval of dual antibody‐based HER2 blockade and chemotherapy in this patient population [2]. Since pCR correlates with long‐term outcome [3], increasing the proportion of patients who respond could have long‐term benefits. The early identification of patients who are unlikely to respond offers the potential to amend neoadjuvant treatment to obtain improved responses. On the other hand, undoubtedly highly effective antibody‐based HER2 blockade may be unnecessary in patients who already benefit from a single anti‐HER2 agent.
In addition to treatment, several biological features are implicated in response to HER2 targeting, including tumor intrinsic subtype, hormone receptor status, alterations in signaling pathways including PI3K, and host factors such as immune response [4, 5, 6]. Yet the recommendation regarding HER2‐targeted agents and chemotherapy in the (neo)adjuvant setting takes into account just HER2 status.
The development of biomarkers for tailoring HER2‐targeted therapy cannot ignore that HER2 targeting agents play their activity not only through the inhibition of HER pathway but also by their inherent antibody characteristics, which affect immune response [7, 8, 9, 10]. At Fondazione IRCCS Istituto Nazionale dei Tumori—Milano (INT), we developed the 41‐gene classifier TRAR, which is able to identify HER2‐positive BC patients with differential risk of relapse upon treatment with adjuvant H and provides reliable predictive information over established clinical factors in the neoadjuvant setting [11, 12]. The discriminatory capability of TRAR stands on its unique feature of including both genes related to HER2 and estrogen receptor (ER) signaling, (ERBB2, C17orf37, GRB7, ESR1), and to split tumors according to their immune infiltration and proliferation characteristics [11]. Here, we aimed to assess whether TRAR is associated with pCR and prognosis to single agent or dual antibody‐based HER2 blockade within the NeoSphere trial.
2. Materials and methods
2.1. Patients and samples
Details on the NeoSphere study (ClinicalTrials.gov number NCT00545688), and its results have been published elsewhere [1]. NeoSphere was a multicenter randomized phase II study in which patients with HER2‐positive BC were stratified by operable, locally advanced, and inflammatory disease and according to hormone receptor status and randomized to preoperative THP, TH, PH, or TP for 12 weeks. After surgery, all patients continued treatment with four cycles of AC followed by H to complete one year of treatment. The primary endpoint was pCR, that is, absence of invasive tumor cells in the breast [1]. The secondary endpoints were 5‐year progression‐free survival and disease‐free survival [3]. The study was conducted in accordance with the Declaration of Helsinki. Approval for the protocol was obtained from independent ethics committees. Written informed consent was obtained from all patients at study entry, which also covered future biomarker research.
2.2. Gene expression profile
RNA was extracted from baseline (pre‐treatment) formalin‐fixed paraffin‐embedded (FFPE) core biopsies and surgical FFPE specimens of patients with residual disease (post‐treatment), and gene expression profiling (GEP) was carried out with Affymetrix U133 Plus 2.0 gene chips as previously described [13]. Thirteen patients were not assessable for pCR. The 41‐gene classifier TRAR was computed as the sum + 5.856708 of the weighted logarithmic expression of 36 out of the 41 genes (Table S1) [11]. Proliferation metagene was computed as described [14].
2.3. Statistical analysis
The association of TRAR scores, measured on a continuous scale, with pCR as well as with other categorical clinico‐pathological variables, was evaluated by using the nonparametric Wilcoxon test. The strength of the association of TRAR scores with continuous variables was assessed by the Pearson correlation coefficient (r). Univariate logistic regression analysis, modeling pCR probability, was implemented for each variable of interest to estimate the odds ratio (OR) and its 95% confidence interval (CI). The predictive performance of TRAR with respect to pCR was further evaluated by resorting to a multivariate logistic regression model by taking into account treatment arm (TH arm as reference) and the available clinico‐pathological variables, that is, age (continuous), ER status (positive versus negative), and tumor type (operable as reference). TRAR was dichotomized according to the cutoff value identified by median value and association with other variables was evaluate by chi‐square test. Survival curves were estimated by the Kaplan–Meier method. The prognostic performance of TRAR with respect to distant event‐free survival (DEFS), which considers only distant relapse ignoring locoregional relapse from the surgery as event, was implemented estimating the hazard ratio (HR) and its 95% CI. All statistical analyses were carried out with r software (http://www.r‐project.org) by adopting a significance level of ≤ 0.05.
3. Results
3.1. Study cohorts
A total of 417 patients were enrolled in the NeoSphere trial, for 350 of them (84%) GEP data were obtained before treatment, as described [13]. Eighty‐seven patients (25%) were enrolled in the TH arm, 92 (26%) in the THP arm, 90 (26%) in the HP arm, and 81 (23%) in the TP arm. No significant differences in terms of baseline patient characteristics and treatment response were observed between the entire NeoSphere and the profiled patient population (pre‐treatment cohort) [13]. Of the 299 patients not attaining pCR in the NeoSphere trial, 193 (65%) had successful GEP of residual disease (post‐treatment cohort), including 166 with paired specimens (Fig. 1). TRAR was computed starting from GEP data using 36 out of the 41 genes in the signature because 5 genes (ARMET, C14orf173, C17orf37, C2orf48, WASH2P) were not available in the Affymetrix platform. The missing genes were not in the core of the signature [11] and TRAR scores calculated with and without these genes in already analyzed datasets [11, 12] were significantly highly correlated (Fig. S1).
Fig. 1.
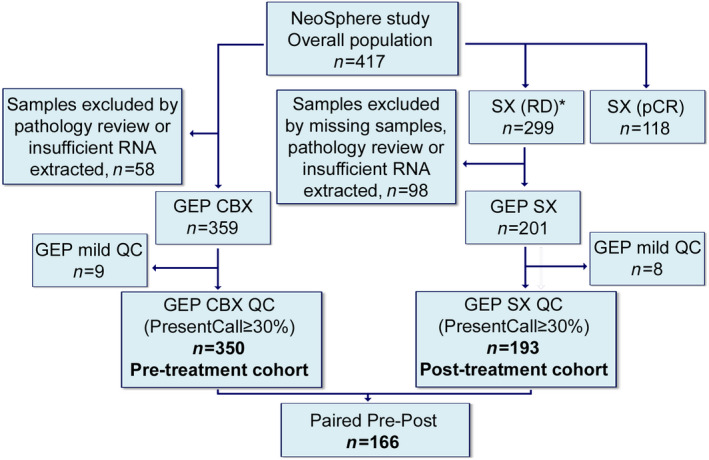
Consort diagram of patients and samples included in the analysis. GEP, gene expression profile; QC, quality check; CBX, core biopsies; SX, surgical samples; RD, residual disease; pCR, pathological complete response. *, includes unknown.
3.2. TRAR and treatment response in the overall cohort
In the pre‐treatment cohort, 104 (29.7%) patients achieved a pCR, specifically 27 (31%), 41 (45%), 14 (16%), and 22 (27%) following treatment with TH, THP, HP, and TP, respectively. As a continuous variable, TRAR was significantly lower in patients attaining a pCR overall (P < 0.0001) (Fig. 2A) and across the different treatment arms (Fig. S2). When TRAR was dichotomized, patients with TRAR‐low primary tumors (n = 175) resulted more likely to achieve a pCR as compared with those with TRAR‐high tumors (n = 175) (43% versus 17%, P < 0.0001, Fig. 2B). Univariate logistic regression analysis showed that the likelihood of attaining a pCR was 55% lower for each TRAR unit increment (OR: 0.45, 95% CI: 0.34‐0.60) (Table 1). In the multivariate analysis adjusted for relevant clinico–pathological characteristics including ER status, treatment arm, age and tumor type, TRAR and ER‐positive status remained each independently associated with lower pCR (OR 0.61 and 0.40, respectively), independently of other variables (Table 1).
Fig. 2.
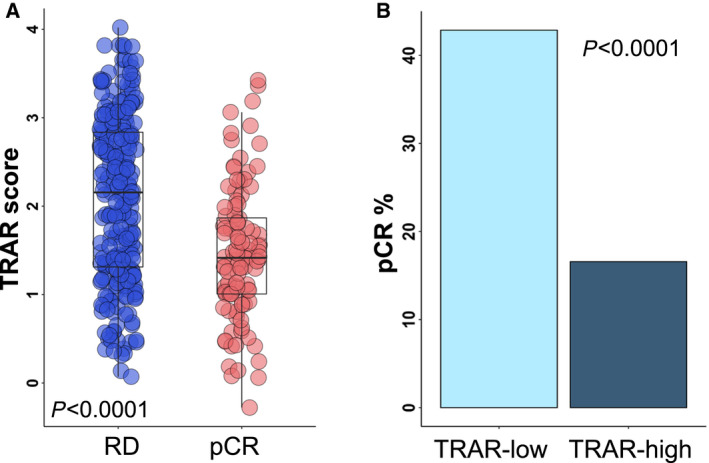
Predictive performance of TRAR. (A) Box‐plots of the distribution of TRAR score in patients with residual disease (RD) and pathological complete response (pCR) in the overall analyzed pre‐treatment cohort (n = 350). Shown are the 25th and the 75th percentiles of the distribution (box), the median (horizontal line), and the extreme values (whiskers). p‐value by Wilcoxon test. (B) Frequency of pCR in TRAR‐low and TRAR‐high subgroups. p‐value by chi‐square test.
Table 1.
Association of TRAR and clinico‐pathological variables with pathological complete response (pCR): Univariate and multivariate logistic regression model. OR, odds ratio; CI, confidence interval; ER, estrogen receptor; T, taxanes; H, trastuzumab; P, pertuzumab; LABC, locally advanced breast cancer; IBC, inflammatory breast cancer.
| All (n = 350) | Univariate | Multivariate* | ||
|---|---|---|---|---|
| Biomarker | OR (95% CI) | P‐value | OR (95% CI) | P‐value |
| TRAR | ||||
| TRAR (continuous) | 0.45 (0.34‐0.60) | 3.57E‐08 | 0.61 (0.43‐0.88) | 0.008 |
| ER IHC | ||||
| ER IHC (pos vs neg) | 0.24 (0.14‐0.40) | 7.04E‐08 | 0.40 (0.20‐0.77) | 0.006 |
| Arm | ||||
| THP (vs TH) | 1.79 (0.97‐3.30) | 0.063 | 1.80 (0.93‐3.48) | 0.084 |
| HP (vs TH) | 0.41 (0.20‐0.85) | 0.016 | 0.41 (0.19‐0.87) | 0.021 |
| TP (vs TH) | 0.83 (0.43‐1.62) | 0.581 | 0.79 (0.38‐1.61) | 0.512 |
| Age | ||||
| Age (continuous) | 0.98 (0.96‐1.01) | 0.161 | 0.98 (0.96‐1.01) | 0.188 |
| Type | ||||
| LABC (vs OPERABLE) | 1.28 (0.78‐2.09) | 0.336 | 0.10 (0.58‐1.72) | 0.993 |
| IBC (vs OPERABLE) | 0.94 (0.38‐2.35) | 0.894 | 0.95 (0.34‐2.58) | 0.909 |
| ER‐positive (n = 161) | Univariate | Multivariate** | ||
|---|---|---|---|---|
| Biomarker | OR (95% CI) | P‐value | OR (95% CI) | P‐value |
| TRAR | ||||
| TRAR (continuous) | 0.33 (0.17‐0.63) | 0.0008 | 0.40 (0.20‐0.79) | 0.009 |
| Arm | ||||
| THP (vs TH) | 1.83 (0.59‐5.66) | 0.292 | 1.56 (0.47‐5.17) | 0.472 |
| HP (vs TH) | 0.26 (0.05‐1.38) | 0.115 | 0.24 (0.04‐1.33) | 0.102 |
| TP (vs TH) | 1.03 (0.30‐3.53) | 0.960 | 0.75 (0.20‐2.83) | 0.668 |
| Age | ||||
| Age (continuous) | 0.96 (0.92‐1.03) | 0.070 | 0.97 (0.93‐1.02) | 0.279 |
| Type | ||||
| LABC (vs OPERABLE) | 1.14 (0.43‐2.98) | 0.797 | 0.99 (0.34‐2.87) | 0.990 |
| IBC (vs OPERABLE) | 0.00 (0.00‐Inf) | 0.988 | 0.00 (0.00‐Inf) | 0.992 |
| ER‐negative (n = 189) | Univariate | Multivariate** | ||
|---|---|---|---|---|
| Biomarker | OR (95% CI) | P‐value | OR (95% CI) | P‐value |
| TRAR | ||||
| TRAR (continuous) | 0.79 (0.52‐1.20) | 0.259 | 0.79 (0.51‐1.21) | 0.269 |
| Arm | ||||
| THP (vs TH) | 1.90 (0.86‐4.20) | 0.114 | 1.84 (0.82‐4.11) | 0.138 |
| HP (vs TH) | 0.45 (0.19‐1.08) | 0.075 | 0.44 (0.18‐1.69) | 0.070 |
| TP (vs TH) | 0.76 (0.33‐1.77) | 0.526 | 0.74 (0.31‐1.73) | 0.486 |
| Age | ||||
| Age (continuous) | 0.99 (0.96‐1.02) | 0.515 | 0.99 (0.96‐1.02) | 0.609 |
| Type | ||||
| LABC (vs Operable) | 1.06 (0.57‐1.93) | 0.864 | 0.98 (0.52‐1.86) | 0.959 |
| IBC (vs Operable) | 1.68 (0.53‐5.35) | 0.379 | 1.78 (0.53‐5.98) | 0.352 |
Multivariate analysis adjusted by (*) ER, treatment arm, age and type; (**) treatment arm, age and type.
3.3. TRAR and treatment response in ER‐positive and ER‐negative subgroups
TRAR was significantly associated with ER both as a continuous and as a dichotomized variable (Fig. 3). TRAR scores were significantly higher in ER‐positive (n = 161) as compared to ER‐negative (n = 189) cases (P < 0.0001) (Fig. 3A and D). TRAR scores were significantly associated with pCR in ER‐positive both as continuous (P = 0.0007) (Fig. 3B) and dichotomous variable (P = 0.01099) (Fig. 3E), but not in ER‐negative cases (Fig. 3C and F) (test for interaction, P = 0.0280 and P = 0.1547, respectively). In ER‐positive cases, TRAR was associated with lower pCR (OR: 0.33, 95% CI: 0.17–0.63) in univariate analysis, (Table 1). In multivariate analysis, a significant association with lower pCR was found only for TRAR (OR: 0.40, 95% CI: 0.20–0.79, Table 1). In ER‐negative tumors, no variables were significantly associated with pCR (Table 1). Similar results were obtained with TRAR dichotomous variable: TRAR‐high was associated with lower pCR only in ER‐positive cases both in univariate (OR: 0.26, 95% CI: 0.10–0.72) and in multivariate analyses (OR: 0.33, 95% CI: 0.11–0.96, Table S2).
Fig. 3.
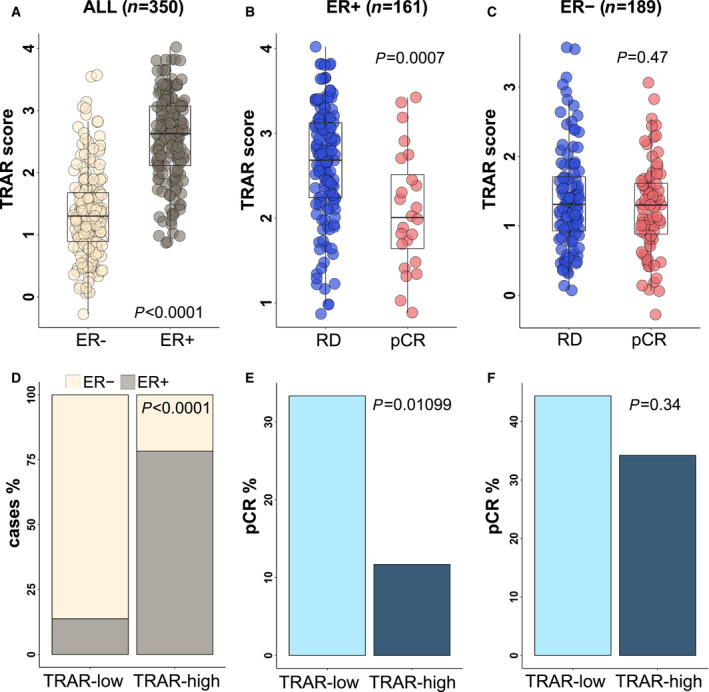
Predictive performance of TRAR according to tumor ER expression. (A) Box‐plots of the distribution of TRAR score in pre‐treatment patients (n = 350) with estrogen receptor‐positive (ER+) and estrogen receptor‐negative (ER−) tumors. P‐value by Wilcoxon test. (B‐C) Box‐plots of the distribution of TRAR score in patients with residual disease (RD) and pathological complete response (pCR) in the ER+ (n = 161, B) and ER− (n = 189, C) cohorts. P‐values by Wilcoxon test. (D) Frequency of ER status in TRAR‐low and TRAR‐high groups (n = 350). P‐value by chi‐square test. (E‐F) Frequency of pCR in patients with TRAR‐low and TRAR‐high and ER+ (n = 161, E) and ER− (n = 189, F) tumors. P‐values by chi‐square test.
3.4. Changes of TRAR score in patients not achieving pCR
The post‐treatment cohort (n = 193) differed from the pre‐treatment cohort (n = 350), with fewer TRAR‐low cases (24% vs 50%, P = 0.0005), as expected given high pCR rates among this subgroup, even among ER‐positive cases (8% vs 15%, P = 0.09). Comparison of matched pre‐ and post‐treatment tumors (n = 166) demonstrated a significant increase in TRAR scores in post‐treatment cohort overall, according to ER status (Fig. 4A) and treatment arms (Fig. S3). Specifically, a TRAR switch from low to high status occurred in 30/62 (48%) cases, while the opposite in 6/104 (6%) cases (P < 0.0001); among ER‐positive cases 8/13 (62%) turned to TRAR‐high, and 3/95 (3%) to TRAR‐low (P = 0.0005), while the corresponding rates were 22/49 (45%), and 3/9 (33%) in ER‐negative cases (n = 58) (Fig. 4B).
Fig. 4.
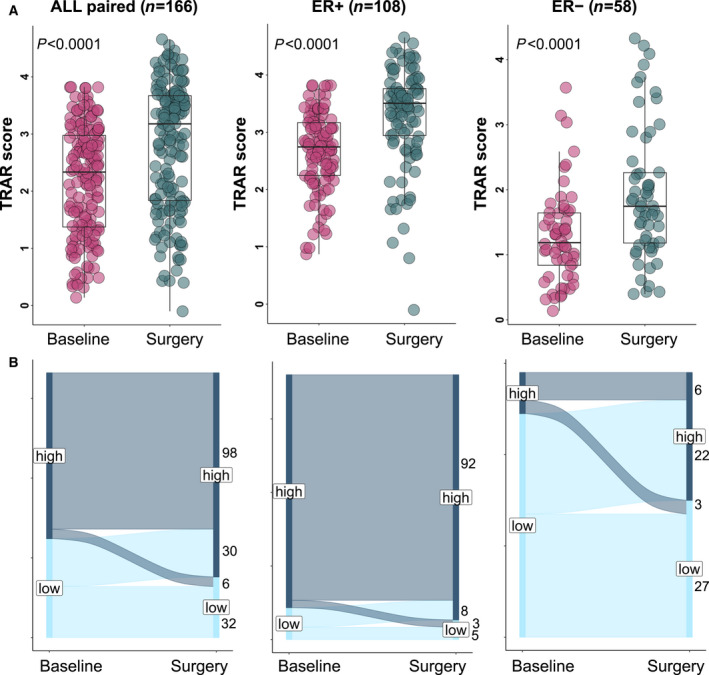
TRAR modulation by treatment in patients with residual disease at surgery. (A) Box‐plots of the distribution of TRAR score in pre‐treatment biopsies (baseline) and at surgery in the overall cohort (n = 166), in ER+ (n = 108) and in ER− (n = 58) subgroups. P‐values by Wilcoxon test. (B) Alluvial diagram of the change in TRAR classification between basal biopsies (Baseline) and samples at surgery in the overall cohort (n = 166), in ER+ (n = 108) and in ER− (n = 58) subgroups.
Of note, TRAR correlated with the expression levels of ESR1 and ERBB2 in both the pre‐treatment (r = 0.80, P < 0.0001 and r = −0.55, P < 0.0001, respectively) and post‐treatment (r = 0.74, P < 0.0001 and r = −0.68, P < 0.0001, respectively) cohorts. Nevertheless, while unresponsive ER‐positive cases classified as TRAR‐low at surgery tended to maintain the same low ESR1 and high ERBB2 expression levels of their pre‐treatment counterparts, those classified as TRAR‐high at surgery were more likely to show reduced levels of ERBB2 expression as compared to pre‐treatment counterparts (Fig. 5). TRAR–low tumors expressed significantly higher level of proliferation genes than TRAR‐high tumors (Fig. S4). Notably, while proliferation metagene levels were significantly decreased in TRAR‐high tumors at surgery compared to baseline (P < 0.0001), the opposite was observed in TRAR‐low cases with a significant increase in proliferation level at surgery (P = 0.0096) (Fig. S4).
Fig. 5.
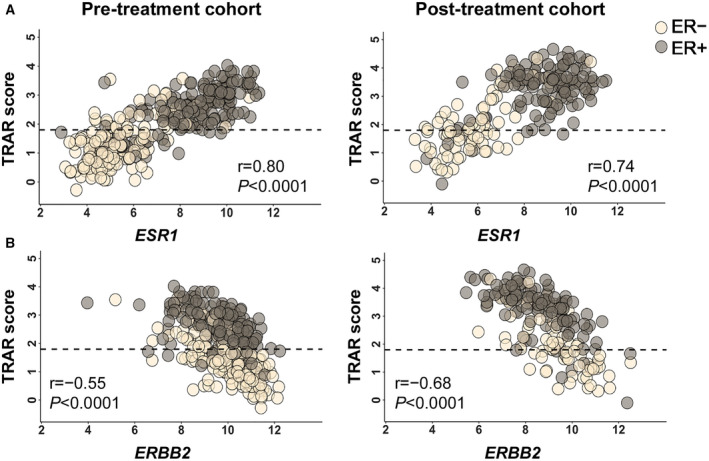
Correlation between TRAR score, ESR1 and ERBB2 genes. (A‐B) Pearson correlation analysis between TRAR score and ESR1 (A) and ERBB2 (B) in pre‐treatment (n = 350) and post‐treatment (n = 193) cohorts according to ER status. Horizontal dot lines separate TRAR‐low from TRAR‐high patients. r, Pearson correlation coefficients and related P‐values are shown.
3.5. TRAR and DEFS
At a median follow‐up of 61 months (range 60–63), no significant difference in terms of DEFS was found according to TRAR subgroups (Fig. S5A). Nevertheless, among ER‐positive cases, patients with TRAR‐high tumors at baseline in pre‐treatment cohort, despite achieving the lowest pCR rate (12%), tended to have a longer DEFS as compared to those with TRAR‐low tumors (HR: 0.41, 95% CI: 0.16–1.05, P = 0.06). The prognosis of patients with TRAR‐low tumors was especially dismal when baseline TRAR‐low status remained unaltered by neoadjuvant treatment (Fig. S5B), suggesting a potential intrinsic resistance to the HER2 targeted therapy. To further examine the relationship between TRAR and prognosis, we tested the outcome according to pCR (Fig. S5C). Remarkably, there was no difference in terms of DEFS among patients achieving a pCR, whereas in the absence of pCR, a trend toward worse outcome (P = 0.1259) was seen among TRAR‐low over TRAR‐high patients.
4. Discussion
Our findings confirm the predictive value of the TRAR classifier already identified in small retrospective studies [11] and in the large phase III NeoALTTO trial [12]. TRAR significantly associated with pCR independently of treatment arm, both as continuous and as dichotomized variable. Notably, the predictive value of TRAR showed a significant interaction with ER status. To our knowledge, this is the first report to suggest that a parsimonious gene assay might help to identify a subset of patients with nonmetastatic HER2‐ and ER‐positive disease that can benefit the most from HER2‐targeted based neoadjuvant chemotherapy.
TRAR score was significantly different in the ER‐positive and ER‐negative subgroups, supporting the described differences in gene expression pattern of HER2‐positive breast cancer according to ER expression [15, 16]. These differences may explain the lower pCR rates reported in ER‐positive as compared to ER‐negative cases in the NeoSphere and other trials, including NeoALTTO [17] and GEPARSIXTO [18]. Notably, TRAR is likely to identify a group of HER2‐positive and ER‐positive tumors that are not exquisitely driven by ER signaling and thus benefit the most from HER2 targeting therapies. This could be explained by the ability of TRAR to mirror not only ER expression/function through the expression of ESR1 and related genes but also HER2 and related genes also recapitulating their interplay, which is relevant in response determination [19, 20].
It is important to note that we could not exclude a benefit of applying TRAR to patients with ER‐negative tumors, as we recently reported in other dataset [12]. Indeed, contrary to other studies [21, 22, 23, 24], in the NeoSphere trial ERBB2, which is a core gene of TRAR score, was not significantly associated with pCR [25], and the PAM50 molecular classifier provided no predictive information overall [26]. Moreover, TRAR was previously tested in tumor biopsy of patients treated with chemotherapy in combination with trastuzumab [11] or other type of dual blockade, that is, trastuzumab plus lapatinib [12]. Therefore, the predictive value of TRAR only in ER‐positive tumors of NeoSphere study might be dependent on the anti‐HER2 drug used and limited to dual HER2‐blocking antibodies. Hence, prospective evaluation of the predictive findings of our study in a randomized trial is warranted, as no similar cohort are currently available for external validation.
Intriguingly, a trend toward lower DEFS was found for TRAR‐low cases, though the study was underpowered for survival. The paradox of higher sensitivity to neoadjuvant HER2‐targeted therapy and poor prognosis in the TRAR‐low subgroup could be explained by the high relapse among those with residual disease, as already observed for HER2‐enriched cases in CALGB 40601 and NeoSphere trials [21, 26]. Hence, it may be easier to achieve pCR in TRAR‐low tumors, especially in ER‐positive cases, but if pCR does not occur, then patients are more likely to relapse early. This finding is in line with the baseline aggressive features of TRAR‐low tumors, that is, high HER2 oncogene dependence, low ER activity, and high proliferation [11] and their poor prognosis if untreated [11].
From the clinical point of view, this finding leads to three major considerations. First, TRAR at surgery may reflect the importance of adjuvant endocrine therapy, which probably exerts its benefit in patients with TRAR‐high remnants, which increased luminal features after neoadjuvant therapy, rather than TRAR‐low remnants that being intrinsically resistant to anti‐HER2 therapies maintain low ESR1 expression and increase proliferation. Interestingly, higher pCR rate but also higher risk of relapse despite chemoendocrine treatments were predicted among ER‐positive/HER2‐negative patients by other genomic classifiers related to proliferation and ER activity [14, 27, 28], supporting a role for these features in explaining TRAR‐low poor prognosis. Next, genomic characterization of surgical specimens could help to identify actionable targets and to foster drug development for residual tumors after HER2‐targeted (neoadjuvant) therapy. In this sense, the HER2‐targeted therapy‐induced luminal phenotype has been already associated to increase sensitivity to CDK4/6 inhibition [29, 30]. Finally, if TRAR prognostic value will be validated, TRAR could be developed as a tool to aid salvage adjuvant treatment, which has already proven to ameliorate patient prognosis at least in a portion of cases not achieving pCR [31]. This adjuvant treatment escalation could be especially relevant for patients with TRAR‐low tumors at surgery.
Some caveats of our study require special consideration. The different platform did not allow us to test the validity of previous generated TRAR cutoff and the use of the median cutoff could have reduced its predictive value. Moreover, the small sample size of treatment arms, and the lack of a validation series did not allow to test whether the interaction between TRAR and ER is limited to the dual HER2‐blocking antibodies.
5. Conclusion
In conclusion, we independently confirm in NeoSphere trial, the clinical validity of TRAR in predicting pCR to anti‐HER2‐based chemotherapy beyond that provided by standard pathologic markers. TRAR represents a promising tool to stratify ER‐positive/HER2‐positive patients by likelihood of response to anti‐HER2‐based neoadjuvant therapy and to contribute to define treatment escalation and de‐escalation strategies in this setting. The clinical utility of this genomic test in predicting also long‐term benefit (i.e., DEFS and overall survival) warrants further investigation.
Conflict of interest
GB: Roche, MSD, Pfizer, AstraZeneca, Lilly, Novartis, Neopharm, Amgen, Chugai, Sanofi, Daiichi Sankyo, EISAI, Genomic Health (consultant/advisory board member); SDC: Novartis Pharma and Pierre Fabre (speaking fees); TP: Roche (honorarium fee, grants, travel grant); GVB: Novartis and Eli Lilly (consultant/advisory board member); MCL: Roche and Pfizer (advisory board member, travel grant); BB: Roche, MSD, Novartis (consultant/advisory board member), Roche, Novartis, MSD, Pfizer, Pierfabre (speakers’ bureau), Pfizer (travel grant); DYO: AstraZeneca, Novartis, Genentech/Roche, Merck Serono, Bayer, Taiho, ASLAN, Halozyme, Zymeworks, BMS/Celgene, BeiGene, Basilea, Turning Point (consultant/advisory board member), AstraZeneca, Novartis, Array, Eli Lilly, Servier, BeiGene, MSD, Handok (research grant); LG: ADC Therapeutics, Amgen, AstraZeneca, Biomedical Insights Inc, Celgene, Eli Lilly, G1 Therapeutics, Genentech, GENENTA, Genomic Health, Menarini Ricerche, METIS Precision Medicine, Merck Sharp & Dohme, Novartis, Oncolytics Biotech, Odonate Therapeutics, Onkaido Therapeutics, Revolution Medicines, Roche, Pfizer, Taiho Pharmaceutical, Hexal Sandoz, Seattle Genetics, selected programs of Forty Seven (CD47), Synthon, Synaffix, Zymeworks and Sanofi‐Aventis (consultant/advisory board member); TT, YHI, GB, MD, LDC, LMT,VS, GV, JdlHR, BP, PV and ET declare no competing interest.
Author contributions
TT, GB, SDC, LG, and ET conceived the work; TT, GB, BG, MD, LDC, and GV performed analyses; TT, GB, SDC, LG, and ET interpreted the results; TP, YHI, GVB, LMT, MCL, BB, VS, JdlHR, DYO, BP, and LG provide resources; GB and PV managed data; TT, SDC, and ET wrote the manuscript; GB, TP, YI, GVB, BG, MD, LDC, LMT, MCL, BB, VS, GV, JdlHR, DYO, BP, and PG critically revised the manuscript; all authors approved the final version of the manuscript.
Peer review
The peer review history for this article is available at https://publons.com/publon/10.1002/1878‐0261.13141.
Supporting information
Fig. S1. TRAR scores according to signature composition.
Fig. S2. Predictive performance of TRAR according to treatment arm.
Fig. S3. TRAR modulation according to treatment arm.
Fig. S4. Proliferation metagene according to TRAR.
Fig. S5. Prognostic performance of TRAR.
Table S1. Expression of genes used to calculate TRAR score and proliferation metagene.
Table S2. Association of TRAR and clinico‐pathological variables with pathological complete response (pCR): Univariate and multivariate logistic regression model.
Acknowledgements
The authors would like to thank F. Hoffmann—La Roche Ltd. for allowing these analyses in the NeoSphere study. This substudy was sponsored by Fondazione Associazione Italiana Ricerca sul Cancro (Fondazione AIRC) to ET (IG 15359) and SDC (IG 20774). In addition, the study was also partially supported by Fondazione AIRC (IG 21787) and Fondazione Michelangelo grant to GB and Breast Cancer Research Foundation (BCRF) grant to LG. Funding sources had no involvement in study design, data collection, analysis and interpretation, writing of the report, and decision to submit the article for publication.
Tiziana Triulzi, Giampaolo Bianchini, Serena Di Cosimo, Luca Gianni and Elda Tagliabue contributed equally to this study
Contributor Information
Luca Gianni, Email: luca.gianni@fondazionemichelangelo.org.
Elda Tagliabue, Email: elda.tagliabue@istitutotumori.mi.it.
Data accessibility
Gene expression data used to calculate TRAR score and proliferation metagene are available in the supplementary material of this article; clinical data are available upon reasonable request to Fondazione Michelangelo.
References
- 1. Gianni L, Pienkowski T, Im YH, Roman L, Tseng L‐M, Liu M‐C, Lluch A, Staroslawska E, de la Haba‐Rodriguez J , Im S‐A et al. (2012) Efficacy and safety of neoadjuvant pertuzumab and trastuzumab in women with locally advanced, inflammatory, or early HER2‐positive breast cancer (NeoSphere): a randomised multicentre, open‐label, phase 2 trial. Lancet Oncol 13, 25–32. [DOI] [PubMed] [Google Scholar]
- 2. Amiri‐Kordestani L, Wedam S, Zhang L, Tang S, Tilley A, Ibrahim A, Justice R, Pazdur R & Cortazar P (2014) First FDA approval of neoadjuvant therapy for breast cancer: pertuzumab for the treatment of patients with HER2‐positive breast cancer. Clin Cancer Res 20, 5359–5364. [DOI] [PubMed] [Google Scholar]
- 3. Gianni L, Pienkowski T, Im Y‐H, Tseng L‐M, Liu M‐C, Lluch A, Starosławska E, de la Haba‐Rodriguez J , Im S‐A, Pedrini JL et al. (2016) 5‐year analysis of neoadjuvant pertuzumab and trastuzumab in patients with locally advanced, inflammatory, or early‐stage HER2‐positive breast cancer (NeoSphere): a multicentre, open‐label, phase 2 randomised trial. Lancet Oncol 17, 791–800. [DOI] [PubMed] [Google Scholar]
- 4. Triulzi T, Bianchi GV & Tagliabue E (2016) Predictive biomarkers in the treatment of HER2‐positive breast cancer: an ongoing challenge. Future Oncol 12, 1413–1428. [DOI] [PubMed] [Google Scholar]
- 5. Di Modica M, Tagliabue E & Triulzi T (2017) Predicting the efficacy of HER2‐targeted therapies: a look at the host. Dis Markers 2017, 7849108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gingras I, Gebhart G, De Azambuja E & Piccart‐Gebhart M (2017) HER2‐positive breast cancer is lost in translation: time for patient‐centered research. Nat Rev Clin Oncol 14, 669–681. [DOI] [PubMed] [Google Scholar]
- 7. Spector NL & Blackwell KL (2009) Understanding the mechanisms behind trastuzumab therapy for human epidermal growth factor receptor 2‐positive breast cancer. J Clin Oncol 27, 5838–5847. [DOI] [PubMed] [Google Scholar]
- 8. Gennari R, Ménard S, Fagnoni F, Ponchio L, Scelsi M, Tagliabue E, Castiglioni F, Villani L, Magalotti C, Gibelli N et al. (2004) Pilot study of the mechanism of action of preoperative trastuzumab in patients with primary operable breast tumors overexpressing HER2. Clin Cancer Res 10, 5650–5655. [DOI] [PubMed] [Google Scholar]
- 9. Bianchini G & Gianni L (2014) The immune system and response to HER2‐targeted treatment in breast cancer. Lancet Oncol 15, e58–e68. [DOI] [PubMed] [Google Scholar]
- 10. Kroemer G, Senovilla L, Galluzzi L, André F & Zitvogel L (2015) Natural and therapy‐induced immunosurveillance in breast cancer. Nat Med 21, 1128–1138. [DOI] [PubMed] [Google Scholar]
- 11. Triulzi T, De Cecco L, Sandri M, Prat A, Giussani M, Paolini B, Carcangiu ML, Canevari S, Bottini A, Balsari A et al. (2015) Whole‐transcriptome analysis links trastuzumab sensitivity of breast tumors to both HER2 dependence and immune cell infiltration. Oncotarget 6, 28173–28182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Di Cosimo S, Triulzi T, Pizzamiglio S, De Cecco L , de Azambuja E , Fumagalli D, Putzai L, Harbeck N, Izquierdo M, Peña L et al. (2019) The 41‐gene classifier TRAR predicts response of HER2 positive breast cancer patients in the NeoALTTO study. Eur J Cancer 118, 1–9. [DOI] [PubMed] [Google Scholar]
- 13. Bianchini G, Pusztai L, Pienkowski T, Im Y‐H, Bianchi GV, Tseng L‐M, Liu M‐C, Lluch A, Galeota E, Magazzù D et al. (2015) Immune modulation of pathologic complete response after neoadjuvant HER2‐directed therapies in the NeoSphere trial. Ann Oncol 26, 2429–2436. [DOI] [PubMed] [Google Scholar]
- 14. Callari M, Cappelletti V, D'Aiuto F, Musella V, Lembo A, Petel F, Karn T, Iwamoto T, Provero P, Daidone MG et al. (2016) Subtype‐specific metagene‐based prediction of outcome after neoadjuvant and adjuvant treatment in breast cancer. Clin Cancer Res 22, 337–345. [DOI] [PubMed] [Google Scholar]
- 15. Konecny G, Pauletti G, Pegram M, Untch M, Dandekar S, Aguilar Z, Wilson C, Rong H‐M, Bauerfeind I, Felber M et al. (2003) Quantitative association between HER‐2/neu and steroid hormone receptors in hormone receptor‐positive primary breast cancer. J Natl Cancer I 95, 142–153. [DOI] [PubMed] [Google Scholar]
- 16. Brandão M, Caparica R, Malorni L, Prat A, Carey LA & Piccart M (2020) What is the real impact of estrogen receptor status on the prognosis and treatment of HER2‐positive early breast cancer? Clin Cancer Res 26, 2783–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Baselga J, Bradbury I, Eidtmann H, Di Cosimo S , de Azambuja E , Aura C, Gómez H, Dinh P, Fauria K, Van Dooren V et al. (2012) Lapatinib with trastuzumab for HER2‐positive early breast cancer (NeoALTTO): a randomised, open‐label, multicentre, phase 3 trial. Lancet 379, 633–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Von Minckwitz G, Schneeweiss A, Loibl S, Salat C, Denkert C, Rezai M, Blohmer JU, Jackisch C, Paepke S, Gerber B et al. (2014) Neoadjuvant carboplatin in patients with triple‐negative and HER2‐positive early breast cancer (GeparSixto; GBG 66): a randomised phase 2 trial. Lancet Oncol 15, 747–756. [DOI] [PubMed] [Google Scholar]
- 19. Giuliano M, Trivedi MV & Schiff R (2013) Bidirectional crosstalk between the Estrogen Receptor and Human Epidermal Growth Factor Receptor 2 signaling pathways in Breast Cancer: molecular basis and clinical implications. Breast Care 8, 256–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Montemurro F, Di Cosimo S & Arpino G (2013) Human epidermal growth factor receptor 2 (HER2)‐positive and hormone receptor‐positive breast cancer: new insights into molecular interactions and clinical implications. Ann Oncol 24, 2715–2724. [DOI] [PubMed] [Google Scholar]
- 21. Fernandez‐Martinez A, Krop IE, Hillman DW, Hillman DW, Polley M‐Y, Parker JS, Huebner L, Hoadley KA, Shepherd J, Tolaney S et al. (2020) Survival, pathologic response, and genomics in CALGB 40601 (Alliance), a Neoadjuvant Phase III trial of paclitaxel‐trastuzumab with or without lapatinib in HER2‐positive breast cancer. J Clin Oncol 38, 4184–4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fumagalli D, Venet D, Ignatiadis M, Azim HA, Maetens M, Rothé F, Salgado R, Bradbury I, Pusztai L, Harbeck N et al. (2017) RNA sequencing to predict response to neoadjuvant Anti‐HER2 therapy: a secondary analysis of the NeoALTTO randomized clinical trial. JAMA Oncol 3, 227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Denkert C, Huober J, Loibl S, Prinzler J, Kronenwett R, Darb‐Esfahani S, Brase JC, Solbach C, Mehta K, Fasching PA et al. (2013) HER2 and ESR1 mRNA expression levels and response to neoadjuvant trastuzumab plus chemotherapy in patients with primary breast cancer. Breast Cancer Res 15, R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schneeweiss A, Chia S, Hegg R, Tausch C, Deb R, Ratnayake J, McNally V, Ross G, Kiermaier A & Cortés J (2014) Evaluating the predictive value of biomarkers for efficacy outcomes in response to pertuzumab‐ and trastuzumab‐based therapy: an exploratory analysis of the TRYPHAENA study. Breast Cancer Res 16, R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bianchini G, Kiermaier A & Bianchi GV (2017) Biomarker analysis of the NeoSphere study: pertuzumab, trastuzumab, and docetaxel. Breast Cancer Res 19, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bianchini G, Parker J, Carey L, Sica L, Prat A, Pieńkowski T, Im Y‐H, Gv B, Ling‐Ming T, Liu M‐C et al. (2018) Research‐based PAM50 predicts risk of relapse in residual disease after anti‐HER2 therapies. Ann Oncol 29, viii58‐viii86. [Google Scholar]
- 27. Liedtke C, Hatzis C, Symmans WF, Desmedt C, Haibe‐Kains B, Valero V, Kuerer H, Hortobagyi GN, Piccart‐Gebhart M, Sotiriou C et al. (2009) Genomic grade index is associated with response to chemotherapy in patients with breast cancer. J Clin Oncol 27, 3185–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Goldstein LJ, Gray R, Badve S, Childs BH, Yoshizawa C, Rowley S, Shak S, Baehner FL, Ravdin PM, Davidson NE et al. (2008) Prognostic utility of the 21‐gene assay in hormone receptor‐positive operable breast cancer compared with classical clinicopathologic features. J Clin Oncol 26, 4063–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pernas S, Petit A & Climent F (2019) PAM50 Subtypes in Baseline and Residual Tumors Following Neoadjuvant Trastuzumab‐Based Chemotherapy in HER2‐Positive Breast Cancer: A Consecutive‐Series From a Single Institution. Front Oncol 9, 967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brasò‐Maristany F, Griguolo G, Pascual T, Paré L, Nuciforo P, Llombart‐Cussac A, Bermejo B, Oliveira M, Morales S, Martínez N et al. (2020) Phenotypic changes of HER2‐positive breast cancer during and after dual HER2 blockade. Nat Commun 11, 385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Von Minckwitz G, Huang CS, Mano MS, Loibl S, Mamounas EP, Untch M, Wolmark N, Rastogi P, Schneeweiss A, Redondo A et al. (2019) Trastuzumab emtansine for residual invasive HER2‐positive breast cancer. N Engl J Med 380, 617–628. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. TRAR scores according to signature composition.
Fig. S2. Predictive performance of TRAR according to treatment arm.
Fig. S3. TRAR modulation according to treatment arm.
Fig. S4. Proliferation metagene according to TRAR.
Fig. S5. Prognostic performance of TRAR.
Table S1. Expression of genes used to calculate TRAR score and proliferation metagene.
Table S2. Association of TRAR and clinico‐pathological variables with pathological complete response (pCR): Univariate and multivariate logistic regression model.
Data Availability Statement
Gene expression data used to calculate TRAR score and proliferation metagene are available in the supplementary material of this article; clinical data are available upon reasonable request to Fondazione Michelangelo.


