Abstract
In solfataric fields in southwestern Iceland, neutral and sulfide-rich hot springs are characterized by thick bacterial mats at 60 to 80°C that are white or yellow from precipitated sulfur (sulfur mats). In low-sulfide hot springs in the same area, grey or pink streamers are formed at 80 to 90°C, and a Chloroflexus mat is formed at 65 to 70°C. We have studied the microbial diversity of one sulfur mat (high-sulfide) hot spring and one Chloroflexus mat (low-sulfide) hot spring by cloning and sequencing of small-subunit rRNA genes obtained by PCR amplification from mat DNA. Using 98% sequence identity as a cutoff value, a total of 14 bacterial operational taxonomic units (OTUs) and 5 archaeal OTUs were detected in the sulfur mat; 18 bacterial OTUs were detected in the Chloroflexus mat. Although representatives of novel divisions were found, the majority of the sequences were >95% related to currently known sequences. The molecular diversity analysis showed that Chloroflexus was the dominant mat organism in the low-sulfide spring (1 mg liter−1) below 70°C, whereas Aquificales were dominant in the high-sulfide spring (12 mg liter−1) at the same temperature. Comparison of the present data to published data indicated that there is a relationship between mat type and composition of Aquificales on the one hand and temperature and sulfide concentration on the other hand.
Sulfide-rich hot springs with neutral or alkaline pH are relatively rare in most geothermal areas in the world. However, these types of hot springs are rather common in Iceland due to high ground water level and climatic conditions, i.e., from melting snow and rain. Bacteria that thrive in such springs often form long streamers or mats, but the appearance of the mats and the types of bacteria in them seem to vary depending on the sulfide concentration, pH, temperature, and other chemical and physical factors (6, 7, 10, 11, 12, 20, 33). Many Icelandic hot springs have sulfide concentrations as high as 30 mg liter−1 and, under such conditions, thick bacterial mats which can be spectacularly white or bright yellow from precipitated sulfur are formed.
The diversity of many microbial ecosystems has now been studied with different molecular methods, such as analysis of small-subunit (SSU) rRNA by sequencing, denaturing gradient gel electrophoresis, or restriction fragment length polymorphism analyses. These studies show that the diversity of microbial ecosystems is typically 100 to 1,000 times greater than that shown by cultivation alone (14, 15, 22, 23, 30, 31). The sequencing of rRNA genes from environmental samples is very informative, since it provides information on both the phylogenetic relationship and the population structure of the microbial community. With increased understanding of the role and importance of microbes in many ecosystems, the benefit of microbial diversity studies is being recognized. The practical value of these methods is already widespread, as they can be used to study the performance of wastewater treatment plants (27), monitor changes upon ecosystem perturbations (30), study grassland changes (19), and monitor the effects of genetically modified microorganisms in the environment (32).
The present study on the microbial diversity of one high-sulfide, neutral hot spring (sulfur mat) and one low-sulfide, Chloroflexus hot spring is part of an ongoing environmental assessment program aimed at evaluating the biological diversity of the Hengill geothermal areas in Iceland. In this study, we analyzed 316 SSU rRNA clones from a sulfur mat that develops at 67°C and 12 mg of sulfide liter−1 and 123 SSU rRNA clones from a Chloroflexus mat that forms at 65 to 70°C and 1 mg of sulfide liter−1. The molecular diversity analysis showed that the Bacteria diversity was lower in the sulfur mat than in the Chloroflexus mat. Aquificales were dominant in the sulfur mat, whereas Chloroflexus was the dominant mat organism in the low-sulfide spring. The majority of the sequences were >95% related to currently known sequences, but representatives of new divisions were found. The data were compared with results obtained from other types of hot springs.
MATERIALS AND METHODS
Study site and sample collection.
Mat or filamentous samples from two hot springs were collected and transferred to sterile flasks containing an equal volume of 5 M guanidine thiocyanate. The sulfur mat hot spring was located in a 2- to 3-m-high riverbank in Grensdalur, Iceland, and formed a V shape. The source temperature was 80°C, with a flow of about 1 liter min−1 and a pH of 6.7. The sulfide concentration was 20 mg liter−1. At 70 to 75°C (20 to 30 cm from the source), growth became visible as short filaments, and at 60 to 67°C, a thick mat had formed. The sample was taken at 67°C and at 12 mg of sulfide liter−1. At the outer surface of the mat, macroscopic white filaments were visible, but the inner part of the mat was gelatinous and dark grey. The bed of the hot spring at this site was made with sulfur-covered stones. The Chloroflexus mat hot spring was in the Badstofuhver area in Hveragerdi. The mat was a typical Chloroflexus mat, with long pink streamers on top and grey filaments at the edges. The temperature at the sample site was 65 to 70°C, the pH was 8.3, and the sulfide concentration was 1 mg liter−1. A small amount of sulfur precipitation was visible around the rims of the hot spring. Analysis of sulfide was done in the field by mercury-acetate titration (2).
DNA extraction.
The biomass was homogenized by using a mortar and a stomacher. After centrifugation at 180 × g for 10 min, isopropanol (1:1) was added to the supernatant. The sample was centrifuged again (17,000 × g), and DNA was isolated from the precipitate with an IsoQuick nucleic acid extraction kit according to the instructions of the manufacturer (ORCA Research).
Amplification and cloning of SSU rRNA.
Amplification of SSU rRNA genes from the sulfur mat was carried out by using both Bacteria- and Archaea-specific primer sets. PCRs were prepared for both sets by using different dilutions of DNA and three different annealing temperatures, 42, 47, and 52°C. PCR amplifications were performed with initial denaturation at 95°C for 5 min, 25 amplification cycles of 95°C for 50 s, 42, 47, or 52°C for 50 s, and 72°C for 2 min, and final extension for 7 min at 72°C to obtain A overhangs. PCR amplifications of SSU genes were performed by using DyNAzyme polymerase (Finnzymes) and Taq polymerase (QIAGEN) according to the instructions of the manufacturers. The Bacteria-specific primers used were F9 (5′-GAGTTTGATCCTGGCTCAG-3′; Escherichia coli positions 9 to 27) and R1544 (5′-AGAAAGGAGGTGATCCA-3′; E. coli positions 1544 to 1528). The Archaea-specific primer set used consisted of 23FPL and 1391R (5). One Archaea clone library and one Bacteria clone library were prepared by pooling PCR products obtained from the different PCRs. The pooled PCR products were run on 0.8% low-melt agarose gels in Tris-acetate-EDTA (TAE) buffer, the SSU rRNA bands were excised from the gels, and the slices were melted at 65°C before cloning. The PCR products were cloned directly by the TA cloning method by using a TOPO TA cloning kit according to the manufacturer's instructions (Invitrogen). Plasmid DNAs from single colonies were isolated and sequenced. A Bacteria SSU rRNA library was prepared in the same way for the Chloroflexus mat sample.
DNA sequencing.
The SSU rRNA genes from the sulfur mat were sequenced with an ABI 377 DNA sequencer by using a BigDye terminator cycle sequencing ready reaction kit according to the instructions of the manufacturer (PE Applied Biosystems). The following Bacteria sequencing primers were used (positions based on E. coli SSU rRNA numbering): F9, F338 (5′-ACICCTACGGGIGGCAGCAG-3′; 338 to 357), F515 (5′-GTGCCAGCAGCCGCGGTAATAC-3′; 515 to 536), F814 (814 to 830) (24), F1392 (1392 to 1406) (24), R357 (5′-CTGCTGCCICCCGTAGG-3′; 357 to 341), R805 (5′-GACTACCCGGGTATCTAATCC-3′; 805 to 785), R1195 (5′-GACGTCITCCCCICCTTCCTC-3′; 1195 to 1175), and R1544; in these sequences, “I” is inosine. The following Archaea sequencing primers were used: 23FPL, 765FA, R1391, 340RA, 744RA, and R805 (5). In addition, the universal reverse and forward M13 vector primers were used for sequencing of both Archaea and Bacteria SSU rRNA genes. Most of the SSU rRNA genes from the Chloroflexus mat sample were sequenced only with R805.
Phylogenetic analysis.
After BLAST searches, the sequences were manually aligned with closely related sequences obtained from the Ribosomal Database Project (RDP) (18). Phylogenetic trees were constructed for the sequences from the sulfur mat by using the ARB package from the Department of Microbiology, Technical University Munich, Germany (S. Strunk and W. Ludwig, http://www.mikro.biologie.tu-muenchen.de/pub/ARB/). Distance trees were constructed by using neighbor-joining algorithms, and maximum-likelihood trees were constructed by using the fastDNAml software included in the ARB package. Homologous nucleotide positions, based on the filter of the ARB database, were included in the alignment and used for the comparison analysis. The CHECK_CHIMERA program of the RDP server was used for searches of chimera artifacts (18).
The GenBank accession numbers of the SSU rRNA sequences of the organisms used in this analysis are as follows: GANI4 (AB005736), GANI3 (AB005735), NAK14 (AB005738), NAK9 (AB005737), Calderobacterium hydrogenophilum (Z30242), Hydrogenobacter thermophilus TK-6 (Z30214), EM17 (U05661), Thermocrinis ruber (AJ005640), Aquifex pyrophilus (M83548), Hydrogenobacter acidophilus (D16296), Nitrospira moscoviensis (X82558), Deinococcus radiodurans UWO 298 (M21413), Thermus sp. strain ZHGIB A.4 (L10071), Thermus filiformis (L09667), Thermus thermophilus HB-8 (X07998), Thermus aquaticus YT-1 (L09663), Thermus sp. strain YSPID A.1 (L10070), Thermus sp. strain ZFI A.2 (L09662), Thermus sp. strain NMX2 A.1 (L09661), EM19 (U05662), unidentified Thermotogales group OPB7 (AF027071), Thermotoga maritima (M21774), Fervidobacterium icelandicum (M59176), unidentified Aquificales OPB13 (AF027098), unidentified Thermodesulfobacterium OPT4 (AF027093), Thermodesulfovibrio sp. strain TGE-P1 (AB021302), unidentified Thermotogales OPB85 (AF027072), unidentified Thermotogales OPS66 (AF027074), candidate division OP5 clone OPS107 (AF027049), uncultured eubacterium clone sequence H1.43.f (AF005749), unidentified korarchaeote pJP78 (L25303), Thermofilum pendens (X14835), uncultured archaeon clone sequence WCHD3-02 (AF050616), Chloroflexus aurantiacus (M34116), Meiothermus cerbereus GY-1T (Y13594), Fervidobacterium gondwanalandicum (Z49117), Chlorogloeopsis sp. (X68780), Craurococcus roseus (D85828), Thiobacillus hydrothermalis (M90662), unidentified green nonsulfur bacterium OPB34 (AF027044), and Meiothermus ruber (Y13596).
Collector's curve for comparison of diversity in different environments.
To compare the bacterial diversity structure of the mat hot springs to that of a nonmat environment, data from the study of Hugenholtz and coworkers on the molecular diversity in a hot spring sediment in Yellowstone National Park were used (16). That study was done in a way very comparable to ours. A large number of clones were also analyzed, and the same cutoff value for operational taxonomic units (OTUs) (98%) was used. To determine if the number of clones analyzed in each of the studies was representative for the ecosystems and if there were differences in diversity between the different ecosystems, theoretical collector's curves were made from the data. A table was made in which each OTU was listed as many times as its observed frequency. The order of the OTUs in the table was randomized, and then the theoretical collector's curves were generated by plotting the cumulative number of OTUs against the cumulative number of sequences analyzed (21, 29). By repeating the randomization, several curves were generated for each data set. To check the reliability of the theoretical curves, real collector's curves were prepared for the results from the sulfur mat and the Chloroflexus mat hot springs and compared with the theoretical collector's curves.
Comparison of the ratios of different Aquificales branches in different hot springs.
To compare the frequency ratios of different branches of the Aquificales group in various hot springs, results were used from six different hot springs. In addition to the sulfur mat and the Chloroflexus mat hot springs studied here, results from four other hot springs were included (16, 26, 33; S. Hjörleifsdottir, S. Skirnisdottir, G. O. Hreggvidsson, O. Holst, and J. K. Kristjansson, submitted for publication). The hot springs were from different parts of the world and varied in temperature, pH, sulfide concentration, and mat type. The studies were done in such a way that we believe the data can be reliably compared. The initial step in the DNA extraction was mechanical disruption of cells with glass beads, a French pressure cell, a microwave, a mortar, or a stomacher. The commonly used phenol-chloroform method with slight modifications was used in the three published studies. In our laboratory, we have used the commercial kit IsoQuick, which in our experience is equally effective for extracting DNA from mat samples. The most conserved regions of the SSU rRNA genes were used as primer sets. Separate PCRs were always performed under several different conditions, i.e., variable annealing temperature, different primer sets, and different dilutions of DNA. After the PCRs, the amplified DNA was cloned, and a few different libraries were made. In two of the studies, only a few clones were analyzed; however, they still demonstrated reliably the main dominant types, especially since the dominance was confirmed by DNA hybridization using the sequences of the isolated clones as fluorescent probes (26, 33).
Nucleotide sequence accession numbers.
The SSU rRNA sequences from the sulfur mat (designated SRI) were deposited in the GenBank database under accession numbers AF255590 to AF255608. The sequences from the Chloroflexus mat were only approximately 400 bases long and thus were not deposited.
RESULTS
Cloning and sequence analysis of SSU rRNA.
Two SSU rRNA libraries were constructed for the sulfur mat using the 1.4- to 1.5-kb PCR fragments, one for the domain Archaea and one for the domain Bacteria. All clones were sequenced with M13 forward and reverse primers. The obtained partial sequences were aligned, and pairwise similarity values were calculated. A similarity of 98% was used as a cutoff value for grouping the sequences into different OTUs. Subsequently, at least one representative of each OTU was completely sequenced. Table 1 shows the frequencies and the phylogenetic positions of the sequences obtained from the sulfur mat. Figure 1 shows a phylogenetic tree (maximum likelihood) comprising the 14 OTUs obtained from the Bacteria library from the sulfur mat. Seven chimeric artifacts were detected in the libraries. The sequences obtained from the Chloroflexus mat were sequenced with R805 and grouped into OTUs by using 98% as a cutoff value. A total of 18 OTUs were found; the results are shown in Table 2. The majority of the sequences from the sulfur mat and the Chloroflexus mat showed more than 95% similarity to currently known sequences, but potentially novel divisions were also found in the libraries.
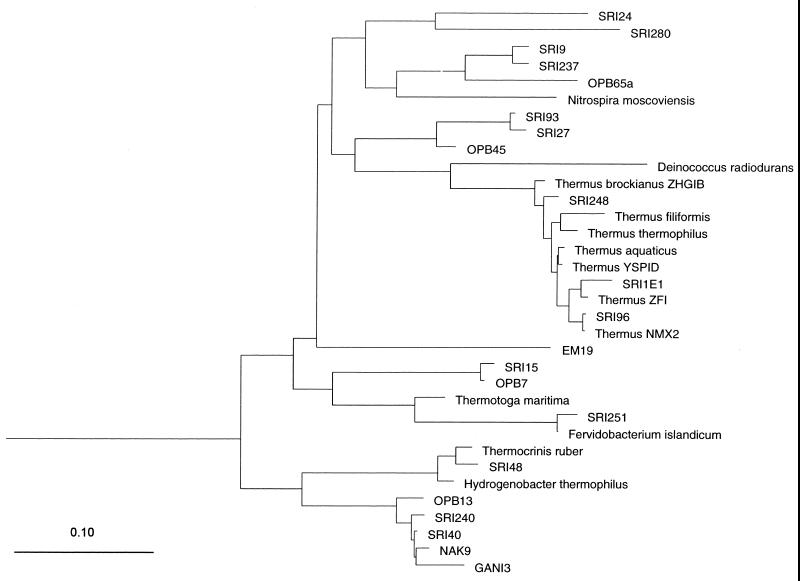
FIG. 1.
Evolutionary maximum-likelihood dendrogram of the bacterial type sequences (designated SRI) detected in the sulfur mat hot spring in Iceland in the context of currently recognized bacterial divisions in the RDP. Sulfolobus acidocaldarius was used as an outgroup. The scale bar is in nucleotide substitution per sequence position.
Phylogenetic analysis of the Bacteria library from the sulfur mat.
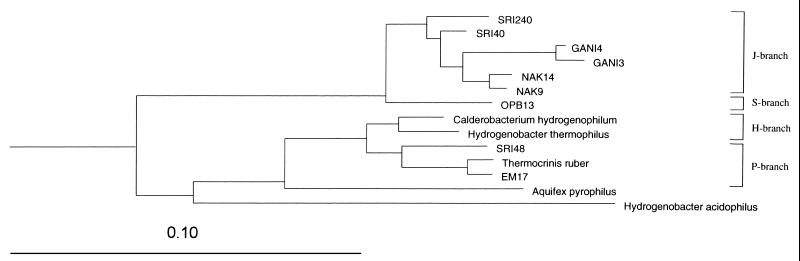
Representatives of the bacterial divisions Aquificales, Thermodesulfobacterium group, Thermus-Deinococcus group, Thermotogales, and Nitrospira group, as well as a potentially new division, were found. Representatives of the Aquificales group were dominant (68%) in the library, indicating that they are the main primary producers. They corresponded to three different OTUs (Table 1): SRI-40 was the most abundant (40%), but the other two, SRI-240 and SRI-48, were less frequent (19 and 9%, respectively). As shown in Fig. 1 and 2, SRI-40 and SRI-240 clustered close to the NAK sequences, from a sulfur mat in Japan (33). The third Aquificales OTU found in the sulfur mat, SRI-48, showed the highest sequence similarity (94%) to a recently cultured pink filament bacterium, Thermocrinis ruber (12).
FIG. 2.
Evolutionary-distance dendrogram of the Aquificales division, showing the OTUs detected in the sulfur mat hot spring in Iceland (designated SRI) in the context of currently recognized bacterial sequences in the RDP. Thermotoga maritima was used as an outgroup. The scale bar represents 10% sequence divergence.
Two OTUs (SRI-93 and SRI-27) comprising 18% of the Bacteria library were representatives of the sulfate reducer Thermodesulfobacterium. Representatives of the sulfate reducer Thermodesulfovibrio were also found at a low frequency (3%). Seven Thermus representatives were detected, belonging to three OTUs. Five of them (SRI-96) were within the T. scotoductus branch. The SRI-248 Thermus sequence was very different from the other sequences and branched deeply within the Thermus genus. This sequence is probably a representative of a new, uncultivated Thermus species. A few representatives of Thermotogales were also found.
Phylogenetic analysis of the Archaea library from the sulfur mat.
The sequences of the archaeal library were more homogenous than those of the bacterial library. Most (77%) of the sequences (SRI-306) were closely related to the Korarchaeota clone sequence pJP78, which was first found in Obsidian Pool in Yellowstone National Park (4, 5). The second most abundant archaeal OTU (19%; SRI-325) was a representative of Thermofilum pendens (99 to 100% similarity), which uses sulfur as an electron acceptor. Representatives of Desulfurococcus were found, but the known cultivated relatives are sulfur respiring.
Phylogenetic analysis of sequences representative of new divisions from the sulfur mat.
Bacterial sequence SRI-24 from the sulfur mat could not be placed within currently known bacterial divisions, as the closest database match was <82%, to the green nonsulfur bacterium clone sequence H1.43.f (8). Archaeal sequence SRI-298 showed <83% similarity to the clone sequence WCHD3-33 (9). SRI-24 and SRI-298 are therefore candidates for novel divisions. The bacterial OTU SRI-280 showed 98 to 99% sequence similarity to the division OP5 (16).
Phylogenetic analysis of the Bacteria library from the Chloroflexus mat.
Representatives of bacterial divisions different from those of the sulfur mat were retrieved from the Chloroflexus mat (Tables 1 and 2). As shown in Table 2, the majority (45%) of the sequences obtained from the Chloroflexus mat library were closely related to Chloroflexus aurantiacus, indicating that Chloroflexus is the primary producer in this ecosystem, instead of Aquificales, as in the sulfur mat. The frequencies of Thermus (and Meiothermus) and other heterotrophs (e.g., Thermotogales and Proteobacteria) were also higher. Four of the OTUs retrieved from the Chloroflexus mat (CHI-5 to CHI-8) could not be placed within currently known bacterial divisions.
Comparison of diversity in different environments.
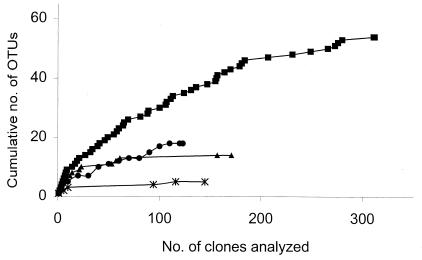
Collector's curves were generated to see if the collected sequences gave a good indication of the bacterial diversity in the different ecosystems (Fig. 3). The theoretical collector's curves from the sulfur mat and the Chloroflexus mat fitted perfectly with the real curves (data not shown), indicating that this method can be used for interpreting and comparing different data. The shape of the curves indicated that the number of clones analyzed from both hot springs in this study represented well the diversity of the ecosystems. As shown in the theoretical collector's curves represented in Fig. 3, bacterial diversity was much lower in the sulfur mat and the Chloroflexus mat hot springs analyzed in the present study than in the hot spring sediment (nonmat) in Obsidian Pool (16).
FIG. 3.
Theoretical collector's curves for results obtained from diversity studies by using culture-independent techniques from different environments. Symbols: ■, hot spring sediment (16); ▴, sulfur mat, bacterial library (this study); ∗, sulfur mat, archaeal library (this study); ●, Chloroflexus mat (this study).
Comparison of the ratios of different Aquificales branches in different hot springs.
Table 3 shows a comparison of the frequency ratios of different branches within the Aquificales group (Fig. 2) from six different hot springs. In addition to the sulfur mat spring and the Chloroflexus mat spring analyzed in this study, we included data from a third Icelandic spring (pink filaments) (Hjörleifsdottir et al., submitted), a sulfur mat spring in Japan (33), and two hot springs in Yellowstone National Park (sediment and pink filaments) (16, 26). The hot springs varied in pH, temperature, sulfide concentration, and mat type. Four different Aquificales branches were found in these six different hot springs from different parts of the world. The sediment sample from the nonmat hot spring in Yellowstone National Park had a unique type, OPB13, not found in the other springs (S branch) (16). The two high-sulfide springs, in Iceland and Japan, contained primarily related bacteria in the J branch (NAK and GANI). The two hot springs were quite similar in temperature and pH. Both hot springs were categorized as high-sulfide hot springs, although the sulfide content was three to four times higher in the Icelandic spring (Table 3). Both of the springs that contained pink filaments, in Iceland and Yellowstone National Park, had the P branch as the dominant type (EM17; Thermocrinis ruber). The springs were high in temperature and low in sulfide. The Chloroflexus mat hot spring was dominated by Chloroflexus (45%), with a low representation of Aquificales in the H (3%) and J (12%) branches. Bacteria in the H branch are closest to cultivated Hydrogenobacter and were found only in low ratios in some of the low-sulfide springs.
DISCUSSION
Microbial mats are communities of organisms that are selected by their habitat and by the population interaction within the community. They are typically characterized by few dominating organisms and are often formed in extreme environments, hypersaline lakes being a classical example (3, 28). A rich nutrient or energy source is a prerequisite for mat formation, and another major factor appears to be strong selective pressure. The different mat types investigated in this study and comparable studies from other geothermal areas demonstrate this principle very well (Table 3). All the different mats show a clear dominance of one OTU, but the compositions and relative ratios of the dominant organisms are very different. These findings may be explained by the patterns in physiochemical conditions and the availability, composition, and quantity of the relative energy sources. pH is unlikely to be the key discriminating factor, since out of the six hot springs compared, four are neutral (pH 6.7 to 7.6) and two are slightly alkaline (pH 8.3). The main differences between the four neutral hot springs compared are temperature and sulfide concentration. Pink filaments (P branch) are the dominant bacteria in both of the high-temperature and low-sulfide springs, although these springs differ only in pH (6.9 and 8.3). The upper temperature limit for the Chloroflexus mat and the sulfur mat is 72 to 74°C, while for the pink filament streamers it is much higher, 88 to 90°C. Chloroflexus is not detected in the high-sulfide spring (sulfur mat), although both the temperature and the pH should be suitable for it. Chloroflexus might be outcompeted in this mat community or inhibited by the high sulfide concentration. Both of the pink filament springs, however, have too high a temperature for Chloroflexus, although the low sulfide concentration should be suitable for it.
The difference in diversity between extreme and less extreme environments is clearly observed in these thermophilic mat environments. Thus, the most extreme mat habitats, in terms of sulfide or temperature, seem to have the overall lowest bacterial diversity (14 OTUs and 171 clones in the sulfur mat, 6 OTUs and 68 clones in pink filaments in Iceland, and 3 OTUs and 35 clones in pink filaments in Yellowstone National Park). The Chloroflexus mat is more diverse than the mats in the above springs, with 18 OTUs and 123 clones. However, the nonmat sediment community in Obsidian Pool is distinctively more diverse than all of the mat-type hot springs.
The collector's curve is a known method for describing and evaluating diversity by plotting the cumulative number of species against the cumulative number of individuals analyzed (21). The use of this method is twofold. First, from the shape of the curve, the differences in species ratio and richness can be compared between different ecosystems. Second, a plateau-shaped curve indicates that the sample size is large enough to give nearly complete coverage of a library. A valid comparison can be done only if the data used for the comparison are obtained by similar methods and effort (21, 29). Here we show that the method can also be used on published data by preparing theoretical collector's curves from frequency data.
The dominant bacteria in all but one of the hot springs studied here belong to the Aquificales. The only exception was the mat growing at 65 to 70°C and at a low sulfide concentration. It was dominated by the photosynthetic species Chloroflexus aurantiacus. Species belonging to the Aquificales are mainly obligately chemolithotrophic, aerobic bacteria using molecular hydrogen or reduced sulfur compounds as energy donors. They belong to one of the earliest branching orders of the domain Bacteria and can be subdivided into a few deep lineages. One of those, the genus Aquifex, appears to be confined to marine hydrothermal vents, but the other lineages have so far mostly been found in terrestrial hot springs (1, 12, 13, 17). The different terrestrial branches of Aquificales appear to be adapted to different environments (Table 3 and Fig. 2). Thus, bacteria in the P branch, Thermocrinis ruber and relatives, are highly dominant in the high-temperature, low-sulfide springs, whereas the bacteria in the J branch dominate in the high-sulfide springs (sulfur mats in Iceland and Japan). It is interesting that no bacteria in these two branches are found in the sediment of Obsidian Pool. The most dominant organisms there, represented by OPB13, belong to a special group, the S branch within the Aquificales, not found in any of the other spring types. Recently, the microbial diversity at 83°C in Calcite Springs, Yellowstone National Park, was published (25). There the investigators found a clone (pBB) which is closely related to OPB13 found in Obsidian Pool (S branch) (16). Both of these hot springs are rich in iron; therefore, iron may be an important factor for the growth and existence of the organisms that these clones represent (25). Interestingly, bacteria within the H branch are the closest relatives of the ubiquitous hydrogen-oxidizing genus, Hydrogenobacter. These bacteria are found only in low ratios in some of the low-sulfide springs. Hydrogenobacter is relatively easily isolated from hot springs by using hydrogen as an electron donor. This finding may indicate that species of the genus Hydrogenobacter use hydrogen preferentially and are outcompeted in relatively sulfide-rich habitats.
Most of the sequences of the Bacteria and Archaea libraries found in the sulfur mat belong to groups that are known to use reduced or oxidized sulfur compounds for growth. This finding is in a good agreement with the chemical composition of the hot spring, i.e., high sulfide. This information indicates that the sulfur cycle is very important in this ecosystem and is the main source of primary productivity. The high frequency of OTUs closely related to the Korarchaeota clone sequence pJP78 in the Archaea library from the sulfur mat may indicate that the korarchaeotes are also part of the sulfur cycle, although this notion cannot be confirmed without cultivation. The heterotrophic niches in this type of mat seem to be largely occupied by the sulfate-reducing bacteria Thermodesulfobacterium group, Nitrospira group, and Thermofilum pendens as well as the fermentative bacteria (Thermotogales). All of this information is consistent with the mat appearance, as by using both phase-contrast and transmission electron microscopy, we saw that the cells and filaments were often attached to sulfur particles and that sulfur deposits were also visible inside some of the cells. The conditions are favorable for aerobic sulfur- and hydrogen-oxidizing bacteria on top of the white sulfur mat and for anaerobic sulfur and sulfate reducers in the dark grey undermass.
Extreme ecosystems that are characterized by high dominance of particular organisms require a smaller sampling size to determine the main elements of their community structure than do less extreme environments. However, if the goal were to make an exhaustive mapping of the species composition in an extreme ecosystem, the sampling size needed would be much larger than is generally used. Such considerations may not be relevant for environmental assessments of extreme microbial environments like that in the present study but are very important to the new field of genetic bioprospecting.
TABLE 1.
Frequencies of the SSU rRNA sequences derived from the Archaea and Bacteria libraries obtained from a sulfur mat hot spring in Iceland
| Library and OTU | No. of clones | Bacterial division | Closest database match (%) |
|---|---|---|---|
| Bacteria | |||
| SRI-40 | 69 | Aquificales | NAK14 (96–97) |
| SRI-240 | 33 | Aquificales | NAK14 (96–97) |
| SRI-27 | 24 | Thermodesulfobacterium group | OPT4 (95) |
| SRI-48 | 15 | Aquificales | Thermocrinis ruber (94) |
| SRI-93 | 7 | Thermodesulfobacterium group | OPT4 (96) |
| SRI-251 | 6 | Thermotogales | Fervidobacterium islandicum (97) |
| SRI-96 | 5 | Thermus-Deinococcus | Thermus sp. strain NMX2 A.1 (99) |
| SRI-9 | 3 | Nitrospira group | Thermodesulfovibrio sp. strain TGE-P1 (95) |
| SRI-15 | 2 | Thermotogales | OPB85-OPS66 (98–99) |
| SRI-237 | 2 | Nitrospira group | Thermodesulfovibrio sp. strain TGE-P1 (95) |
| SRI-280 | 2 | OP5 | OPS107 (98) |
| SRI-248 | 1 | Thermus-Deinococcus | Thermus sp. strain YSPID A.1 (96) |
| SRI-1e | 1 | Thermus-Deinococcus | Thermus sp. strain ZFI A.2 (97) |
| SRI-24 | 1 | New division? | H1.43.f (82) |
| Total Bacteria clones | 171 | ||
| Archaea | |||
| SRI-306 | 112 | Korarchaeota | pJP78 (97–99) |
| SRI-325 | 27 | Thermofiliaceae | Thermofilum pendens (99–100) |
| SRI-465 | 4 | Desulfurococcaceae | Desulfurococcus mobilis (97) |
| SRI-370 | 1 | Thermofiliaceae | Thermofilum pendens (95) |
| SRI-298 | 1 | New division of Crenarchaeota? | WCHD3-02 (83) |
| Total Archaea clones | 145 |
TABLE 2.
Frequencies of OTUs within the Bacteria domain derived from the SSU rRNA sequences from a Chloroflexus mat hot spring in Iceland
| OTU | No. of clones | Bacterial division | Closest database match (%) |
|---|---|---|---|
| CHI-1 | 55 | Chloroflexaceae | Chloroflexus aurantiacus (99) |
| CHI-2 | 15 | Aquificales | NAK14 (98) |
| CHI-3 | 14 | Thermus-Deinococcus | Thermus sp. strain NMX2 A.1 (98–100) |
| CHI-4 | 11 | Meiothermus | Meiothermus cerbereus (96) |
| CHI-5–CHI-8 | 8 | New divisions? | Uncertain affiliation (<88) |
| CHI-9 | 5 | Nitrospira group | Thermodesulfovibrio sp. (97) |
| CHI-10 | 4 | Aquificales | Calderobacterium hydrogenophilum (96–97) |
| CHI-11 | 2 | Thermotogales | Fervidobacterium gondwanalandicum (97) |
| CHI-12 | 2 | Aquificales | Thermocrinis ruber (94) |
| CHI-13 | 2 | Stigonematales | Chlorogloeopsis sp. (99) |
| CHI-14 | 1 | Proteobacteria; alpha subdivision | Craurococcus roseus (90) |
| CHI-15 | 1 | Proteobacteria; gamma subdivision | Thiobacillus hydrothermalis (94) |
| CHI-16 | 1 | Green nonsulfur bacterium | OPB34 (97) |
| CHI-17 | 1 | Thermus-Deinococcus | Thermus sp. strain ZHGIB A.4 (97) |
| CHI-18 | 1 | Meiothermus | Meiothermus ruber (99) |
| Total clones | 123 |
TABLE 3.
Comparison of the ratios of major Aquificales branches in different types of hot springs
| Hot spring typea | Temp (°C) | pH | Sulfide (mg liter−1) | No. of clones analyzed | No. of Bacteria OTUs | % of total clones belonging to the following Aquificales branchb
|
Reference or source | |||
|---|---|---|---|---|---|---|---|---|---|---|
| H | P | J | S | |||||||
| Sulfur mat, Japan | 52–72 | 7.2–8.0 | 3–6 | 25 | No data | 0 | 0 | 48 | 0 | 33 |
| Sulfur mat, Iceland | 67 | 6.7 | 12 | 171 | 14 | 0 | 9 | 59 | 0 | This study |
| Pink filaments, OS YNP | 84–88 | 8.3c | 0.2c | 35 | 3 | 0 | 74 | 0 | 0 | 26 |
| Pink filaments, Iceland | 88 | 6.9 | 1.7 | 68 | 6 | 1.5 | 87 | 1.5 | 0 | Hjörleifsdottir et al., submitted |
| Sediment, OP YNP | 75–93 | 6.7/7.6d | No data | 312 | 54 | 0.6 | 0.6 | 0 | 26 | 16 |
| Chloroflexus mat, Iceland | 65–70 | 8.3 | 1 | 123 | 18 | 3 | 1.6 | 12 | 0 | This study |
ACKNOWLEDGMENTS
This work was supported by a grant from The National Power Company, a grant from Reykjavik Energy, grant 1377-98 from Nordtest, and grant 98.30.174-O from NorFA.
We thank N. R. Pace for critical reading of the manuscript.
REFERENCES
- 1.Aragno M. Aerobic, chemolithoautotrophic, thermophilic bacteria. In: Kristjansson J K, editor; Kristjansson J K, editor. Thermophilic bacteria. Boca Raton, Fla: CRC Press, Inc.; 1992. pp. 77–103. [Google Scholar]
- 2.Arnorsson, S. Isotopic and chemical techniques in geothermal exploration and use, in press. International Atomic Energy Agency.
- 3.Atlas R M, Bartha R. Microbial ecology: fundamentals and applications. 3rd ed. Redwood City, Calif: The Benjamin/Cummings Publishing Company; 1993. [Google Scholar]
- 4.Barns S M, Delwiche C F, Palmer J D, Pace N R. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc Natl Acad Sci USA. 1996;93:9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barns S M, Fundyga R E, Jeffries M W, Pace N R. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc Natl Acad Sci USA. 1994;91:1609–1613. doi: 10.1073/pnas.91.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brock T D. Thermophilic microorganisms and life at high temperatures. New York, N.Y: Springer-Verlag; 1978. [Google Scholar]
- 7.Caldwell D E, Caldwell S J, Laycock J P. Thermothrix thioparus gen. et sp. nov. a facultatively anaerobic facultative chemolithotroph living at neutral pH and high temperature. Can J Microbiol. 1976;22:1509–1517. doi: 10.1139/m76-223. [DOI] [PubMed] [Google Scholar]
- 8.Chandler D P, Brockman F J, Bailey T J, Fredrickson J K. Phylogenetic diversity of Archaea and Bacteria in a deep subsurface paleosol. Microb Ecol. 1998;36:37–50. doi: 10.1007/s002489900091. [DOI] [PubMed] [Google Scholar]
- 9.Dojka M A, Hugenholtz P, Haack S K, Pace N R. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl Environ Microbiol. 1998;64:3869–3877. doi: 10.1128/aem.64.10.3869-3877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorlenko V M, Bonch-Osmolovskaya E A, Kompantseva E I, Starynin D A. Differentiation of microbial communities in connection with a change in the physicochemical conditions in a thermophile spring. Mikrobiologiya. 1987;56:314–322. . (In Russian.) [Google Scholar]
- 11.Hiraishi A, Umezawa T, Yamamoto H, Kato K, Maki Y. Changes in quinone profiles of hot spring microbial mats with a thermal gradient. Appl Environ Microbiol. 1999;65:198–205. doi: 10.1128/aem.65.1.198-205.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huber R, Eder W, Heldwein S, Wanner G, Huber H, Rachel R, Stetter K O. Thermocrinis ruber gen. nov., sp. nov., a pink-filament-forming hyperthermophilic bacterium isolated from Yellowstone National Park. Appl Environ Microbiol. 1998;64:3576–3583. doi: 10.1128/aem.64.10.3576-3583.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huber R, Wilharm T, Huber D, Trincone A, Burggraf S, König H, Rachel R, Rockinger I, Fricke H, Stetter K O. Aquifex pyrophilus gen. nov. sp. nov., represents a novel group of marine hyperthermophilic hydrogen-oxidizing bacteria. Syst Appl Microbiol. 1992;15:340–351. [Google Scholar]
- 14.Hugenholtz P, Goebel B M, Pace N R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hugenholtz P, Pace N R. Identifying microbial diversity in the natural environment: a molecular phylogenetic approach. Trends Biotechnol. 1996;14:190–197. doi: 10.1016/0167-7799(96)10025-1. [DOI] [PubMed] [Google Scholar]
- 16.Hugenholtz P, Pitulle C, Hershberger K L, Pace N R. Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawasumi T, Igarashi Y, Kodama T, Minoda Y. Hydrogenobacter thermophilus gen. nov., sp. nov., an extremely thermophilic, aerobic, hydrogen-oxidizing bacterium. Int J Syst Bacteriol. 1984;34:5–10. [Google Scholar]
- 18.Maidak B L, Cole J R, Parker C T, Jr, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCaig A E, Glover L A, Prosser J I. Molecular analysis of bacterial community structure and diversity in unimproved and improved upland grass pastures. Appl Environ Microbiol. 1999;65:1721–1730. doi: 10.1128/aem.65.4.1721-1730.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Odintsova E V, Jannasch H W, Mamone J A, Langworthy T A. Thermothrix azorensis sp. nov., an obligately chemolithoautotrophic, sulfur-oxidizing, thermophilic bacterium. Int J Syst Bacteriol. 1996;46:422–428. doi: 10.1099/00207713-46-2-422. [DOI] [PubMed] [Google Scholar]
- 21.Odum E P. Principles and concepts pertaining to organization at the community level. In: Odum E P, editor; Odum E P, editor. Fundamentals of ecology. Philadelphia, Pa: Saunders College Publishing; 1971. pp. 140–161. [Google Scholar]
- 22.Pace N R. New perspective on the natural microbial world: molecular microbial ecology. ASM News. 1996;62:463–470. [Google Scholar]
- 23.Pace N R. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 24.Pitulle C, Yang Y, Marchiani M, Moore E R B, Siefert J L, Aragno M, Jurtshuk P, Jr, Fox G E. Phylogenetic position of the genus Hydrogenobacter. Int J Syst Bacteriol. 1994;44:620–626. doi: 10.1099/00207713-44-4-620. [DOI] [PubMed] [Google Scholar]
- 25.Reysenbach A L, Ehringer M, Hershberger K. Microbial diversity at 83°C in Calcite Springs, Yellowstone National Park: another environment where the Aquificales and “Korarchaeota” coexist. Extremophiles. 2000;4:61–67. doi: 10.1007/s007920050008. [DOI] [PubMed] [Google Scholar]
- 26.Reysenbach A L, Wickham G S, Pace N R. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl Environ Microbiol. 1994;60:2113–2119. doi: 10.1128/aem.60.6.2113-2119.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sekiguchi Y, Kamagata Y, Syutsubo K, Ohashi A, Harada H, Nakamura K. Phylogenetic diversity of mesophilic and thermophilic granular sludges determined by 16S rRNA gene analysis. Microbiology. 1998;144:2655–2665. doi: 10.1099/00221287-144-9-2655. [DOI] [PubMed] [Google Scholar]
- 28.Stal L J. Microbial mats in coastal environments. In: Stal L J, Caumette P, editors; Stal L J, Caumette P, editors. Microbial mats: structure, development and environmental significance. Berlin, Germany: Springer-Verlag KG; 1994. pp. 21–32. [Google Scholar]
- 29.Tipper J C. Rarefaction and rarefiction—the use and abuse of a method in paleoecology. Paleobiology. 1979;5:423–434. [Google Scholar]
- 30.Torsvik V, Sorheim R, Goksoyr J. Total bacterial diversity in soil and sediment communities—a review. J Ind Microbiol. 1996;17:170–178. [Google Scholar]
- 31.Ward D M, Ferris M J, Nold S C, Bateson M M. A natural view of microbial biodiversity within hot spring cyanobacterial mat communities. Microbiol Mol Biol Rev. 1998;62:1353–1370. doi: 10.1128/mmbr.62.4.1353-1370.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winding A, Kvaloy K, Hendriksen N B, Gustafsson K, Iversen T-G, Helgason E, Kolsto A-B. Procedures for risk identification and assessment of genetically modified microorganisms. NT technical report 381, NT project no. 1335-97. Espoo, Finland: Nordtest; 1998. [Google Scholar]
- 33.Yamamoto H, Hiraishi A, Kato K, Chiura H X, Maki Y, Shimizu A. Phylogenetic evidence for the existence of novel thermophilic bacteria in hot spring sulfur-turf microbial mats in Japan. Appl Environ Microbiol. 1998;64:1680–1687. doi: 10.1128/aem.64.5.1680-1687.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]