Abstract
A fluorogenic (TaqMan) PCR assay was developed to detect Ralstonia solanacearum strains. Two fluorogenic probes were utilized in a multiplex reaction; one broad-range probe (RS) detected all biovars of R. solanacearum, and a second more specific probe (B2) detected only biovar 2A. Amplification of the target was measured by the 5′ nuclease activity of Taq DNA polymerase on each probe, resulting in emission of fluorescence. TaqMan PCR was performed with DNA extracted from 42 R. solanacearum and genetically or serologically related strains to demonstrate the specificity of the assay. In pure cultures, detection of R. solanacearum to ≥102 cells ml−1 was achieved. Sensitivity decreased when TaqMan PCR was performed with inoculated potato tissue extracts, prepared by currently recommended extraction procedures. A third fluorogenic probe (COX), designed with the potato cytochrome oxidase gene sequence, was also developed for use as an internal PCR control and was shown to detect potato DNA in an RS-COX multiplex TaqMan PCR with infected potato tissue. The specificity and sensitivity of the assay, combined with high speed, robustness, reliability, and the possibility of automating the technique, offer potential advantages in routine indexing of potato tubers and other plant material for the presence of R. solanacearum.
Ralstonia solanacearum (Smith) (30) is the agent of bacterial wilt, infecting over 450 plant species, including many economically important crops (12). This species has been subclassified into biovars based on biochemical tests and host-dependent races. Biovar 2A (equivalent to race 3) is adapted to temperate climates, has a narrow host range, and is responsible for recent outbreaks of potato brown rot disease in several countries of Western Europe and elsewhere worldwide (13, 27). Although other biovars can also infect potatoes, biovar 2A is the most destructive phenotype in temperate areas.
R. solanacearum is listed as a quarantine organism in the European Union (EU) (2), where new legislation has been introduced to control and eradicate the organism (3). Latent infections in seed potato tubers (6) have lead to the spread of the organism, both locally and internationally, and effective control of brown rot is dependent on the reliability of detection of the pathogen at this latent stage. For practical purposes, a detection assay is required which is rapid, specific, and sensitive to levels lower than those occurring in naturally infected potatoes and should be applicable to a crude sample of the specimen of interest (25). Serological techniques such as immunofluorescence (IF) microscopy, the enzyme-linked immunosorbent assay (ELISA) (10, 15, 21), and molecular techniques involving the PCR (9, 24) have been described for detection of R. solanacearum. An EU control directive (3) allows for a variety of detection methods to be employed. Briefly, a primary screening test (i.e., IF and/or selective isolation) is conducted with extracts from vascular tissue sampled from 200 tubers per 25-tonne lot. To confirm the presence of the pathogen, presumptive cultures isolated on a semiselective medium (8) are identified (e.g., by fatty acid profiling) (16, 26), and pathogenicity is confirmed by a host test on tomato seedlings (15). This procedure is labor intensive and time-consuming. In addition, the primary screening techniques are not completely reliable, despite recent attempts to improve the specificity and sensitivity of R. solanacearum detection (8, 10, 29).
Fluorogenic PCR-based (TaqMan) assays have recently shown promise in the detection of a variety of organisms, including clinical bacteria (4, 5, 19) and plant pathogenic potato leaf roll virus (23). In addition, a TaqMan assay for detection of the potato ring rot bacterium, Clavibacter michiganensis subsp. sepedonicus, has also been reported (22). TaqMan PCR exploits the 5′ nuclease activity of Taq DNA polymerase (14) in conjunction with fluorogenic DNA probes (18). Each probe, designed to hybridize specifically to the target PCR product, is labelled with a fluorescent reporter dye and a quencher dye. During PCR amplification, the probe is digested by Taq DNA polymerase, separating the dyes, resulting in an increase in reporter fluorescence. Repeated PCR cycles result in exponential amplification of the PCR product and a corresponding increase in fluorescence intensity.
Because the TaqMan amplicon is generally between 60 and 70 bp, the reaction is also more efficient than a standard PCR in which target PCR products are required to be at least 200 bp in length to permit detection by electrophoretic separation techniques. This increase in efficiency and incorporation of a sequence-specific probe enhance sensitivity and specificity. Fluorescence can be measured throughout the PCR, providing real-time analysis of the reaction kinetics and allowing quantification of specific DNA targets. The measurement of fluorescence throughout the reaction by a fluorometer (ABI Prism 7700 Sequence Detection system; PE Biosystems, Foster City, Calif.) eliminates the need for post-PCR processing steps, such as gel electrophoresis and ethidium bromide staining of target DNA, easing automation of the technique and large-scale sample processing. Thus, there is reduced potential for contamination of the PCR mixture with target DNA because the reaction tubes remain closed throughout the assay. Interpretation of the fluorometric data can be presented as a simple qualitative conclusion as to the presence or absence of amplified DNA within minutes of the end of the PCR. Alternatively, real-time analysis can facilitate quantification of the amount of sample DNA present in the reaction by ascertaining when (i.e., during which PCR cycle) fluorescence in a given reaction tube exceeds that of a threshold (threshold cycle [CT]). Comparison between reaction tubes and/or known standards can quantify the amount of DNA template present in a given tube.
This paper reports the development of a fluorogenic PCR-based assay which uses a probe-primer set (RS) to detect all known strains of R. solanacearum and another set (B2) specific for the biovar 2A genotype. A multiplex (RS-B2) reaction is described in which each probe is labelled with a different reporter dye, thus providing a single tube assay for both tests. The sensitivity of the assay and the effect of potential TaqMan PCR inhibitors present in potato tissue are also quantified. Finally, we report the detection of R. solanacearum in infected potato tissue by using a multiplex reaction which incorporates a third probe-primer set (COX) designed to detect the potato cytochrome oxidase gene sequence as an internal control for the TaqMan reaction.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The R. solanacearum strains evaluated are listed in Table 1. Bacterial strains related to R. solanacearum or having previously cross-reacted with polyclonal antisera to R. solanacearum in IF or ELISA were also evaluated and are also listed in Table 1. Strains were grown (24 h at 27°C) on Casamino Acids-Peptone-glucose (CPG) agar (17). A single colony was then transferred to 100 μl of sterile nucleic acid-free water, vortexed, heated to >96°C for 4 min, and placed on ice. Samples were finally diluted in 900 μl of the sterile water and stored at −80°C until required. Previous work has indicated that this is an effective method for extracting DNA from Ralstonia spp. (8, 24).
TABLE 1.
R. solanacearum strains and strains related to R. solanacearum or having previously cross-reacted with polyclonal antisera to R. solanacearum in IF or ELISAa
| Species | Collectionb | Biovar | Origin | Yr isolated | Host or source | RS
|
B2
|
||
|---|---|---|---|---|---|---|---|---|---|
| Mean ΔRnc | Result | Mean ΔRnd | Result | ||||||
| Ralstonia solanacearum | NCPPB1824 | 2A | Egypt | 1966 | Potato | 1.12 | + | 1.43 | + |
| Ralstonia solanacearum | CSL465 | 2A | Egypt | 1990 | Potato | 0.68 | + | 1.36 | + |
| Ralstonia solanacearum | CSL1328 | 2A | Egypt | 1994 | Potato | 1.05 | + | 1.45 | + |
| Ralstonia solanacearum | CSL3467 | 2A | England | 1997 | River water | 0.96 | + | 1.51 | + |
| Ralstonia solanacearum | CSL3468 | 2A | England | 1997 | Tomato | 0.92 | + | 1.43 | + |
| Ralstonia solanacearum | NCPPB1584 | 2A | Cyprus | 1963 | Potato | 1.05 | + | 1.26 | + |
| Ralstonia solanacearum | NCPPB2505 | 2A | Sweden | 1972 | Potato | 1.08 | + | 1.48 | + |
| Ralstonia solanacearum | RVP502 | 2A | Belgium | 1989 | Potato | 1.07 | + | 1.39 | + |
| Ralstonia solanacearum | NCPPB 4069 | 2A | Slovenia | 1997 | Potato | 0.91 | + | 1.44 | + |
| Ralstonia solanacearum | CIP310 | 2A | Colombia | Unknown | Potato | 0.99 | + | 1.44 | + |
| Ralstonia solanacearum | NCPPB325 | 1 | United States | 1953 | Tomato | 0.97 | + | 0.01 | − |
| Ralstonia solanacearum | CIP226 | 2T | Brazil | Unknown | Potato | 0.85 | + | 0.01 | − |
| Ralstonia solanacearum | CIP49 | 3 | Australia | Unknown | Potato | 0.99 | + | 0.01 | − |
| Ralstonia solanacearum | R470 | 4 | Phillipines | Unknown | Ginger | 0.98 | + | 0.01 | − |
| Ralstonia solanacearum | R288 | 5 | China | Unknown | Mulberry | 0.94 | + | 0.19 | − |
| Ralstonia pickettii | NCPPB3839 | Unknown | Unknown | Unknown | 0.03 | − | 0.00 | − | |
| Ralstonia pickettii | NCPPB 4076 | Unknown | Unknown | Unknown | 0.05 | − | 0.00 | − | |
| Ralstonia pickettii | CSL3466 | Unknown | Unknown | Unknown | 0.05 | − | 0.00 | − | |
| Ralstonia pickettii | CFBP2459 | Unknown | Unknown | Unknown | 0.00 | − | 0.00 | − | |
| Pseudomonas sp. | NCPPB928 | Jamaica | 1959 | Sugarcane | 1.75 | + | 0.01 | − | |
| Pseudomonas sp. | NCPPB927 | United States | 1956 | Sugarcane | 0.00 | − | 0.00 | − | |
| Pseudomonas sp. | NCPPB1695 | Tanzania | 1965 | Sugarcane | 0.00 | − | 0.00 | − | |
| Ralstonia sp. | PD2778 | Unknown | Unknown | Unknown | 0.01 | − | 0.00 | − | |
| Banana blood disease bacterium | NCPPB3726 | South Sulawesi | 1988 | Banana | 1.96 | + | 0.00 | − | |
| Ralstonia syzygii | NCPPB3790 | North Sumatra | Unknown | Clove | 1.48 | + | 0.00 | − | |
| Ralstonia syzygii | NCPPB3791 | West Java | Unknown | Clove | 1.53 | + | 0.01 | − | |
| Ralstonia syzygii | NCPPB3792 | Central Java | Unknown | Clove | 1.56 | + | 0.01 | − | |
| Burkholderia andropogonis | NCPPB1127 | Zimbabwe | 1961 | Bougainvillea sp. | 0.01 | − | 0.00 | − | |
| Burkholderia caryophylli | NCPPB353 | United States | 1954 | Unknown | 0.02 | − | 0.00 | − | |
| Burkholderia cepacia | NCPPB945 | Unknown | 1961 | Unknown | 0.02 | − | 0.01 | − | |
| Burkholderia cepacia | CFBP2227 | Unknown | Unknown | Unknown | 0.00 | − | 0.00 | ||
| Burkholderia glumae | NCPPB3708 | Japan | 1967 | Rice | 0.05 | − | 0.00 | − | |
| Burkholderia plantarii | NCPPB3590 | Japan | 1982 | Rice | 0.06 | − | 0.00 | − | |
| Bacillus polymyxa | CFBP1954 | Unknown | Unknown | Unknown | 0.13 | − | 0.00 | − | |
| Pseudomonas marginalis subsp. marginalis | CFBP1538 | Unknown | Unknown | Unknown | 0.06 | − | 0.00 | − | |
| Pseudomonas chloraphis | NCPPB 4082 | Unknown | Unknown | Unknown | 0.03 | − | 0.00 | − | |
| Enterobacteriaceae | NCPPB 4078 | Unknown | Unknown | Unknown | 0.03 | − | 0.00 | − | |
| Ratinella aquatillis | NCPPB 4079 | Unknown | Unknown | Unknown | 0.14 | − | 0.21 | − | |
| Ochrobactrum anthropi | NCPPB 4080 | Unknown | Unknown | Unknown | 0.07 | − | 0.00 | − | |
| Unknown | CSL3461 | Unknown | Unknown | Unknown | 0.05 | − | 0.24 | − | |
| Unknown | CSL3462 | Unknown | Unknown | Unknown | 0.02 | − | 0.00 | − | |
| Unknown | CSL3463 | Unknown | Unknown | Unknown | 0.07 | − | 0.00 | − | |
| Unknown | CSL3464 | Unknown | Unknown | Unknown | 0.03 | − | 0.00 | − | |
| Unknown | PD2775 | Netherlands | Unknown | Unknown | 0.06 | − | 0.00 | − | |
Strains were evaluated and mean ΔRn values were generated during PCR with RS-I and RS-II primers plus the RS-P fluorogenic probe and B2-I and B2-II primers plus the B2-P fluorogenic probe.
NCPPB, National Collection of Plant Pathogenic Bacteria, Central Science Laboratory, Sand Hutton, York, United Kingdom; CSL, plant pathogenic bacteria culture collection, Central Science Laboratory; RVP, Rijkstation voor Plantenzickten, Merelbeke, Belgium; CFBP, Collection Française de Bacteries Phytopathogenes, INRA, Angers, France; R, culture collection IACR-Rothamsted, Harpenden, Hertsfordshire, United Kingdom; CIP, culture collection, International Potato Center, Lima, Peru; PD, Plantenziektenkundige Dienst, Wageningen, The Netherlands.
Concentrations: 300 nM F–300 nM R primer, 25 mM RS probe.
Concentrations: 300 nM F–300 nM R primer, 50 mM B2 probe.
TaqMan probe design.
The sequences of the primers and TaqMan probes used are given in Table 2. The probes and primers were designed with Primer Express version 1.0 software (PE Biosystems, Foster City, Calif.) by adapting previous R. solanacearum PCR protocols. The broad-host-range R. solanacearum probe (RS-P) is partially homologous to 16S rRNA gene primer OLI1 (24), with primers (RS-I and RS-II) flanking this region. The primers and TaqMan probe for the biovar 2A-specific assay were elucidated from the biovar 2A-specific DNA sequence described by Fegan et al. (9), retaining the original forward primer 631 (B2-II) and selecting the probe (B2-P) and reverse primer (B2-I) from the upstream region. The internal positive control primer-probe combination was designed according to the published sequence (20) of the abundant constitutive cytochrome oxidase (COX) gene. RS-P was covalently labelled at the 5′-terminal nucleotide with the FAM (6-carboxyfluorescein) reporter dye and at the 3′-terminal nucleotide with the TAMRA (tetra-methylcarboxyrhodamine) quencher dye. B2-P and COX-P were labelled with the VIC (PE Biosystems) reporter dye at the 5′-terminal nucleotide and again with the TAMRA quencher dye at the 3′-terminal nucleotide. TaqMan probes were synthesized by PE Biosystems.
TABLE 2.
Characteristics of primers and TaqMan probes used to detect R. solanacearum and potato cytochrome oxidase DNA
| Primer or probea | Sequence (5′→3′) | Length (nt) | Dye |
|---|---|---|---|
| RS-I-F | GCA TGC CTT ACA CAT GCA AGT C | 22 | |
| RS-II-R | GGC ACG TTC CGA TGT ATT ACT CA | 23 | |
| RS-Pb | AGC TTG CTA CCT GCC GGC GAG TG | 23 | FAM |
| B2-I-F | TGG CGC ACT GCA CTC AAC | 18 | |
| B2-IIc-R | AAT CAC ATG CAA TTC GCC TAC G | 22 | |
| B2-P | TTC AAG CCG AAC ACC TGC TGC AAG | 24 | VIC |
| COX-F | CGT CGC ATT CCA GAT TAT CCA | 21 | |
| COX-R | CAA CTA CGG ATA TAT AAG AGC CAA AAC TG | 29 | |
| COX-P | TGC TTA CGC TGG ATG GAA TGC CCT | 24 | VIC |
5′ Nuclease PCR assay.
The 5′ nuclease PCR with a fluorogenic probe is run under generic cycling conditions, and so requires the optimization of primer concentration to take into account real differences in primer melting temperature. Different forward and reverse primer concentrations for each probe were evaluated, to ascertain the effect on CT and endpoint fluorescence values. For each probe, a primer concentration of 300 nM was found to be most efficient, giving a high endpoint fluorescence and low CT (data not shown). This primer concentration was used, in an RS-B2 multiplex reaction, to test R. solanacearum strains and related species.
PCR was performed in 25-μl volumes using MicroAmp Optical 96-well reaction plates and MicroAmp Optical Caps (PE Biosystems) for each well. All reagents were obtained from the TaqMan Core PCR Reagent kit (PE Biosystems). The PCR mixture for the RS and B2 multiplex was as follows: 2.0 μl of cell lysate; 2.5 μl of 10× TaqMan buffer A; 200 nM (each) dATP, dCTP, and dGTP; 400 nM dUTP; 0.025 U of AmpliTaq Gold DNA polymerase per ml; 0.01 U of uracil-N-glycosylase per ml (AmpErase UNG); 300 nM (each) primers RS-I, RS-II, B2-I, and B2-II; 25 nM TaqMan probe RS-P; and 50 nM TaqMan probe B2-P. The RS-COX multiplex PCR mixture was as follows: 2.0 μl of infected tissue lysate; 2.5 μl of 10× TaqMan buffer A, 200 nM (each) dATP, dCTP, and dGTP; 400 nM dUTP; 0.025 U of AmpliTaq Gold DNA polymerase per μl; 0.01 U of uracil-N-glycosylase (AmpErase UNG) per ml); 150 nM (each) primers RS-I and RS-II; 100 nM (each) primers COX-F and COX-R; 25 nM TaqMan probe RS-P; and 50 nM TaqMan probe COX-P. An ABI Prism 7700 Sequence Detection system (PE Biosystems) was used for amplification and fluorescence measurement. All cycles began with 2 min at 50°C and then went to 10 min at 95°C, followed by 40 two-step cycles of 10 s at 95°C and then 1 min at 60°C.
Post-PCR analysis.
The fluorescent intensities of each dye were measured by the ABI Prism 7700 Sequence detection system (PE Biosystems) at every temperature step and cycle during the reaction. Data acquisition and analysis were handled by Sequence Detector version 1.6 software (PE Biosystems). Briefly a normalized reporter (Rn) value is defined for each reaction tube and ΔRn, an indication of the magnitude of signal generated by the PCR, is calculated. The CT value is the first cycle at which a statistically significant increase in ΔRn is detected and is based on average standard deviation of Rn during the early cycles, where no fluorescence is observed. Samples with a ΔRn value exceeding this threshold (and greater than 0.3) were considered positive.
Sensitivity of R. solanacearum TaqMan assays.
R. solanacearum biovar 2A isolate (CSL3468) was grown on CPG medium (24 h at 27°C) and suspended in 10 ml of sterile water (∼109 cells ml−1). A 10-fold dilution series was made from the suspension (109 to 100), and 100 μl of each dilution was heated above 96°C for 4 min. Total cell counts in each diluted suspension were estimated by indirect immunofluorescence microscopy with anti-R. solanacearum polyclonal antiserum IACR-278 and fluorescein isothiocyanate-conjugated antirabbit immunoglobulin G (Sigma F6005; Sigma-Aldrich Co., Ltd., Poole, Dorset, United Kingdom) according to the method of Janse (15). TaqMan PCR (RS and B2) was performed on the 107-to-100 dilutions. TaqMan PCR assays (RS and B2) were performed individually on the same dilution series. Three separate dilution series were tested.
Potato tissue extracts.
For the RS-COX multiplex reaction, a core of vascular tissue (∼0.1 g) was aseptically removed from the stolon end of a naturally infected potato tuber (cv. Cara) and homogenized in 1 ml of 50 mM phosphate buffer (pH 7.0). One hundred microliters of the resulting suspension was heated above 96°C for 4 min and cooled rapidly on ice. To this sample, 900 μl of sterile water was added to complete sample preparation. Ten-fold serial dilutions were made from this suspension.
The effect of inhibitors present in crude potato tissue extract was assessed by spiking a dilution series of R. solanacearum into extracts prepared in accordance with official testing methods (3). Briefly, a sample of 200 tuber stolon-end vascular tissue cores (cv. Cara) was macerated with 30 ml of 50 mM phosphate buffer (pH 7.0) in a sterile stomacher bag, allowed to stand for 30 min, and centrifuged at 180 × g for 10 min. The resultant supernatant was decanted and transferred to a fresh centrifuge tube before a second centrifugation step at 10,000 × g for 10 min. The pellet from this second centrifugation step was resuspended in 1 ml of 10 mM phosphate buffer (pH 7.2). The resuspended pellet was then used to prepare a decimal dilution series of R. solanacearum from 107 to 1 cell ml−1 in undiluted pellet or pellet diluted 1:10 and 1:100. A similar dilution series was also prepared in sterile nucleic acid-free water. Each of the samples was thoroughly mixed before being heated above 96°C for 4 min to lyse the cells.
RESULTS
TaqMan probe design and specificity.
For all R. solanacearum strains, ΔRn values between 0.68 and 1.75 were obtained by RS-P, indicating successful amplification (Table 1). The B2-P 5′ nuclease assay resulted in ΔRn values between 1.26 and 1.45 being obtained for all biovar 2A strains tested. With biovar 2T and biovar 1, 3, 4, and 5 strains, no fluorescence was detected during the B2-P assay, indicating that amplification had not occurred (Table 1). A range of closely related bacterial strains, including strains that had cross-reacted with R. solanacearum polyclonal antisera in IF or ELISA (J. Elphinstone, unpublished data), were also evaluated. Several of these strains produced fluorescence during the RS-P assay, namely the banana blood disease bacterium, Ralstonia syzygii strains, and one (from three tested) Pseudomonas sp. strain (Taxon B). No fluorescence was detected from the B2-P assay on any of these strains. No elevated ΔRn values were detected for any of the other reference bacteria from either assay.
Sensitivity of TaqMan PCR.
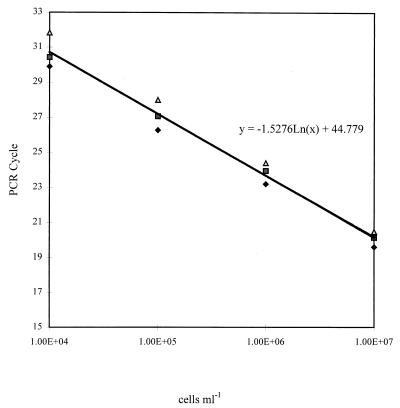
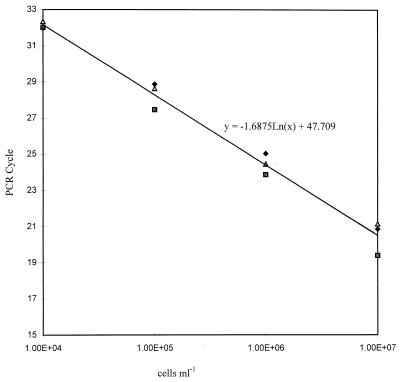
The sensitivities of the individual TaqMan PCR assays were measured with a 10-fold dilution series of R. solanacearum biovar 2A (isolate CSL3468). Target DNA was detectable in suspensions containing as few as 102 cells ml−1 (0.2 cells per reaction) when 300 nM F–300 nM R primer concentrations were used for both TaqMan probes. CT values increased at each dilution, demonstrating the validity of the assay as the target DNA concentration decreased and showing that quantification of target DNA is possible. For the RS-P fluorogenic probe, quantification was shown to be linear from 107 to 104 cells ml−1 for suspensions diluted in both water and 1:10-diluted potato extract (Fig. 1 and 2). Similar results were observed with the B2-P probe (data not shown).
FIG. 1.
Quantification of R. solanacearum biovar 2A cells (CSL3468) diluted in water from one experiment replicated three times with the RS-P probe. The trendline equation was calculated from the mean CT value by using Microsoft Excel. Primer concentration, 300 nM F–300 nM R; probe concentration, 50 nM.
FIG. 2.
Quantification of R. solanacearum biovar 2A cells (CSL3468) in diluted potato tissue, from one experiment replicated three times with RS-P probe. The trendline equation was calculated from the mean CT value by using Microsoft Excel. Primer concentration, 300 nM F–300 nM R; probe concentration, 50 nM.
TaqMan PCR from potato extracts.
Inhibition of TaqMan by high concentrations of potato extracts in the reaction mix was recorded (Table 3). Detection of R. solanacearum by using the generic TaqMan (RS) probe occurred with a lower limit of detection of 104 cells ml−1 in water and in potato pellet diluted 1:100. Detection with the biovar 2A-specific probe was approximately 10 times less sensitive, with a lower limit of detection of 105 cells ml−1. In those samples prepared with potato extract diluted 1:10, the minimum limits of detection were between 104 and 105 cells ml−1 with the R. solanacearum species-specific probe and 106 cells ml−1 with the biovar 2A-specific probe.
TABLE 3.
R. solanacearum biovar 2 dilution series, in water and potato extracts, and CT values generated during multiplex TaqMan PCR assay with the RS-I and RS-II primers plus RS-P flourogenic probe and B2-I and B2-II primers plus B2-P flourogenic probe
| No. of R. solanacearum cells ml−1 | CT value
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Water
|
Pellet
|
Pellet (diluted 1:10)
|
Pellet (diluted 1:100)
|
|||||
| RS (FAM) | B2 (VIC) | RS (FAM) | B2 (VIC) | RS (FAM) | B2 (VIC) | RS (FAM) | B2 (VIC) | |
| 107 | 19.31 | 20.76 | — | — | 22.59 | 25.74 | 19.46 | 21.64 |
| 106 | 22.08 | 24.08 | — | — | 26.89 | 31.18 | 24.36 | 26.13 |
| 105 | 26.17 | 28.36 | — | — | 28.67 | — | 27.56 | 29.45 |
| 104 | 29.37 | —a | — | — | — | — | 31.10 | — |
—, no fluorescence above threshold recorded.
Both probes were strongly inhibited by undiluted potato extract, with no positive reactions even at a cell concentration of 107 ml−1. TaqMan PCR was also completely inhibited in a set of samples prepared by lysing cells in 50 mM NaOH solution.
Conventional PCR was performed on the same spiked samples with the primers OLI-1 and Y-2 and the protocols of Seal et al. (24). With the samples prepared in water, a lower limit of detection of 103 cells ml−1 was recorded. Samples prepared with undiluted potato extract and potato extract diluted 1:10 and 1:100 all showed amplification when the concentration of cells was 104 cells ml−1 and greater.
Multiplex TaqMan PCR of infected potato extracts with the COX fluorogenic probe as an internal control simultaneously detected both R. solanacearum and potato cytochrome oxidase target DNA sequences (Table 4).
TABLE 4.
Brown rot-infected potato extract dilution series and CT values generated during multiplex TaqMan PCR assay with the RS-I and RS-II primers plus RS-P flourogenic probe and COX-F and COX-R primers plus COX-P flourogenic probe
| Potato tissue extract dilution | Mean CT value
|
|
|---|---|---|
| RS (FAM)a | COX (VIC)b | |
| Undiluted | 14.84 | 24.46 |
| 10−1 | 25.02 | 27.80 |
| 10−2 | 29.83 | 32.36 |
| 10−3 | —c | — |
| 10−4 | — | — |
Concentrations: 150 nM RS-F–150 nM RS-R primer, 25 mM RS-P probe. FAM, reporter dye.
Concentrations: 100 nM COX-F–100 nM COX-R primer, 50 mM COX-P probe. VIC, reporter dye.
—, no fluorescence above threshold recorded.
DISCUSSION
The results presented above demonstrate the development of a fluorogenic 5′ nuclease PCR-based (TaqMan) assay capable of detection and identification of R. solanacearum directly in potato tuber tissue with sensitivity and specificity equal to or greater than those achievable with existing PCR protocols (8, 9, 24). In routine laboratory studies, R. solanacearum can be detected in conventional PCR assays in aqueous suspensions ranging from 103 cells per ml (2 cells per PCR) by the method of Seal et al. (24) to 105 cells per ml (200 cells per PCR) by the method of Fegan et al. (9). However in potato extracts, detection limits for the former assay (24) have been shown to be increased to 106 cells per ml or higher (8).
The procedure is robust, rapid, automated, and quantitative, with high sample throughput potential, permitting analysis of up to 96 samples in 3 h. Avoidance of laborious post-PCR gel electrophoresis and greatly reduced opportunity for contamination of reaction mixtures with target DNA further increase the suitability of this assay for routine diagnostic testing.
With cells suspended in water, TaqMan PCR was able to detect R. solanacearum to 102 cells per ml−1 using both the RS-P and B2-P probes, comparable to the results of Oberst et al. (19) in detecting Escherichia coli by using a fluorogenic 5′ nuclease reaction. The lower CT values obtained from the RS-P assay, rather than those obtained from B2-P with the same extracts, indicate a higher concentration of RS target DNA than of B2 target DNA. This may be explained by RS target DNA being part of the multicopy 16S rRNA gene compared with the B2 target DNA, whose genomic location and function are not known (9).
For indexing potato tubers for freedom from R. solanacearum, two useful multiplex assays have been validated. The first of these, using RS and B2 probe-primer sets, permits identification of the biovar 2A (race 3) phenotype without the need for isolation, purification, or biochemical phenotyping (requiring up to 3 weeks for completion). The second assay, using RS or B2 with COX probe-primer sets, provides simultaneous internal PCR control, permitting elimination of false-negative results due to inhibition of the reaction, an essential requirement for routine diagnostic applications.
As expected, the RS TaqMan primer-probe set detected all bacteria tested with known homology to the specific region of the 16S rRNA gene used for probe and primer design (24, 28). These included isolates representing all biovars and other infrasubspecific strains of R. solanacearum as well as the closely related banana blood disease bacterium and R. syzygii (the distribution of which has not been recorded outside of Indonesia). These results are consistent with those of the PCR protocol described by Seal et al. (24) using the 16S rRNA sequence upon which the RS assay is based. In addition, one isolate (from three tested) of Pseudomonas sp. strain Taxon B, commonly associated with sugar cane (11), also produced a positive result during the RS assay. This bacterium is known to have cellular fatty acid profiles closely resembling those of R. solanacearum (26). Other known Ralstonia spp. (R. pickettii and R. eutropha) not sharing homology with this specific region of the 16S rRNA gene were not detected.
In contrast, the B2 TaqMan primer-probe set detected only isolates of the biovar 2A phenotype, including strains of both restriction fragment length polymorphism groups 26 and 27 as described by Cook and Sequeira (7). As predicted, the DNA sequence used for probe and primer design in this assay is apparently unique to the biovar 2A genotype (9). However, the sensitivity of detection of R. solanacearum biovar 2A was significantly higher when the TaqMan rather than the original PCR protocol was used (N. C. Smith, unpublished data). The B2 assay therefore ensures sensitivity and specificity of detection of the race 3 phenotype unattainable with other detection protocols (8–10, 15, 24, 29). Biovar 2T (which is believed to have originated in South America) has a significantly different genetic composition from that of other biovar 2A strains (7), accounting for the failure of the TaqMan probe B2-P to detect this strain.
TaqMan PCR using the COX-P probe as an internal PCR control and RS-P to detect the pathogen directly in infected potato extracts was demonstrated. Detection of R. solanacearum and potato cytochrome target DNA in 10-fold dilutions was observed to 10−2 of the original extract. During the experimental work, evidence was accumulated to suggest that high concentrations of potato extract inhibit the TaqMan PCR. As a result, it is unlikely that this assay could be used for indexing potato tubers in combination with currently recommended sampling and extraction procedures (1, 3). However, it should be possible to simplify extraction procedures and to exploit the high assay sensitivity by eliminating the need for concentration of extracts prior to testing. Should the problem of assay inhibition continue, further steps for purification of target DNA or the use of pathogen enrichment procedures (8) may be required.
For indexing of potato tubers for freedom from R. solanacearum, a rapid response is required in order to prevent costly disruption to trade and to limit loss of this perishable commodity during testing. TaqMan PCR offers reliable detection within 1 day of receipt of samples, whereas current EU testing protocols that demand isolation, purification, identification, and demonstration of pathogenicity can take several days or weeks. Our assay questions the need for such time-consuming protocols. Because post-PCR processing steps are not required, the assay can be easily automated, unlike other PCR protocols. Automation will lead to high sample throughput, which, together with the high specificity and sensitivity of the assay, offers significant advantages over other current R. solanacearum detection techniques. As more TaqMan assays are developed to test for other potato pathogens and further fluorescent reporter dyes are developed, the potential of testing for several potato diseases in a single tube reaction is created. The probes described in this report, used to detect pathogens and to act as an internal PCR control, could be included in such a reaction, improving seed indexing procedures—the primary method of control for many potato pathogens.
ACKNOWLEDGMENTS
This research was funded by the Plant Health Division, MAFF, project PH0154.
We thank David Howells of PE Biosystems for technical support during the project and Stephen Hill for critical reading of the manuscript.
REFERENCES
- 1.Anonymous. Quarantine procedure: Pseudomonas solanacearum, inspection and test methods. Bull OEPP/EPPO Bull. 1990;20:255–262. [Google Scholar]
- 2.Anonymous. Commission directive 95/4/EC. Off J Eur Communities. 1995;L-182:17–19. [Google Scholar]
- 3.Anonymous. Council directive 98/57/EC. Off J Eur Communities. 1998;L-235:1–39. [Google Scholar]
- 4.Bassler H A, Flood S J A, Livak K J, Marmaro J, Knorr R, Batt C A. Use of a fluorogenic probe in a PCR-based assay for the detection of Listeria monocytogenes. Appl Environ Microbiol. 1995;61:3724–3728. doi: 10.1128/aem.61.10.3724-3728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen S, Yee A, Griffiths M, Larkin C, Yamashiro C T, Behari R, Paszko-Kolva C, Rahn K, De Gradis S A. The evaluation of a fluorogenic polymerase chain reaction assay for the detection of Salmonella species in food commodities. Int J Food Microbiol. 1997;35:239–250. doi: 10.1016/s0168-1605(97)01241-5. [DOI] [PubMed] [Google Scholar]
- 6.Ciampi L, Sequeira L, French E R. Latent infection of potato tubers by Pseudomonas solanacearum. Am Potato J. 1980;57:377–386. [Google Scholar]
- 7.Cook D, Sequeira L. The use of subtractive hybridisation to obtain a DNA probe specific to Pseudomonas solanacearum. Mol Gen Genet. 1991;227:401–410. doi: 10.1007/BF00273930. [DOI] [PubMed] [Google Scholar]
- 8.Elphinstone J G, Hennessy J, Wilson J K, Stead D E. Sensitivity of different methods for the detection of Ralstonia solanacearum. Bull OEPP/EPPO Bull. 1996;26:674–678. [Google Scholar]
- 9.Fegan M, Holoway G, Hayward A C, Timmis J. Development of a diagnostic test based on the polymerase chain reaction (PCR) to identify strains of R. solanacearum exhibiting the biovar 2 genotype. In: Prior P H, Allen C, Elphinstone J G, editors. Bacterial wilt disease: molecular and ecological aspects. Berlin, Germany: Springer-Verlag; 1998. pp. 34–43. [Google Scholar]
- 10.Griep R A, van Twisk C, van Beckhoven J R C M, van der Wolf J M, Schots A. Development of specific recombinant monoclonal antibodies against the lipopolysaccharide of Ralstonia solanacearum race 3. Phytopathology. 1998;88:795–803. doi: 10.1094/PHYTO.1998.88.8.795. [DOI] [PubMed] [Google Scholar]
- 11.Hayward A C. Differentiation, taxonomy and nomenclature of the bacteria causing red stripe and mottled stripe diseases. Maurit Sugar Ind Res Inst Occas Paper. 1962;13:13–27. [Google Scholar]
- 12.Hayward A C. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu Rev Phytopathol. 1991;29:65–87. doi: 10.1146/annurev.py.29.090191.000433. [DOI] [PubMed] [Google Scholar]
- 13.Hayward A C, Elphinstone J G, Caffier D, Janse J D, Stefani E, French E R, Wright A J. Round table on bacterial wilt (Brown Rot) of potato. In: Prior P H, Allen C, Elphinstone J G, editors. Bacterial wilt disease: molecular and ecological aspects. Berlin, Germany: Springer-Verlag; 1998. pp. 420–430. [Google Scholar]
- 14.Holland P M, Abramson R D, Watson R, Gelfand D H. Detection of specific polymerase chain reaction product by utilising the 5′ to 3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janse J D. A detection method for Pseudomonas solanacearum in symptomless potato tubers and some data on its sensitivity and specificity. Bull OEPP/EPPO Bull. 1988;18:343–351. [Google Scholar]
- 16.Janse J D. Infra- and intraspecific classification of Pseudomonas solanacearum strains, using whole cell fatty acid analysis. Syst Appl Microbiol. 1991;12:335–345. [Google Scholar]
- 17.Kelman A. The relationship of pathogenicity in Pseudomonas solanacearum to colony appearance on the tetrazolium medium. Phytopathology. 1954;39:936–946. [Google Scholar]
- 18.Lee L G, Connell C R, Bloch W. Allelic discrimination by nick-translation PCR with fluorogenic probes. Nucleic Acids Res. 1993;21:3761–3766. doi: 10.1093/nar/21.16.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oberst R D, Hays M P, Bohra L K, Phebus R K, Yamashiro C T, Paszko-Kolva C, Flood S J A, Sargeant J M, Gillespie J R. PCR-based DNA amplification and presumptive detection of Escherichia coli O157:H7 with an internal fluorogenic probe and the 5′ nuclease (TaqMan) assay. Appl Environ Microbiol. 1998;64:3389–3396. doi: 10.1128/aem.64.9.3389-3396.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quinones V, Zanlungo S, Holuigue L, Litvak S, Jordana X. The cox1 initiation codon is created by RNA editing in potato mitochondria. Plant Physiol. 1995;108:1327–1328. doi: 10.1104/pp.108.3.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson-Smith A, Jones P, Elphinstone J G, Forde S M D. Production of antibodies to Ralstonia solanacearum, the causative agent of bacterial wilt. Food Agric Immunol. 1995;7:67–79. [Google Scholar]
- 22.Schaad N W, Berthier-Schaad Y, Sechler A, Knorr D. Detection of Clavibacter michiganensis subsp. sepedonicus in potato tubers by BIO-PCR and an automated real time fluorescence detection system. Plant Dis. 1999;83:1095–1100. doi: 10.1094/PDIS.1999.83.12.1095. [DOI] [PubMed] [Google Scholar]
- 23.Schoen C D, Knorr D, Leone G. Detection of potato leafroll virus in dormant potato tubers by immunocapture and a 5′ nuclease RT-PCR assay. Phytopathology. 1996;86:993–999. [Google Scholar]
- 24.Seal S E, Jackson L A, Young J P W, Daniels M J. Differentiation of Pseudomonas solanacearum, Pseudomonas syzygii, Pseudomonas pickettii and the blood disease bacterium by partial 16S rRNA sequencing: construction of oligonucleotide primers for sensitive detection by polymerase chain reaction. J Gen Microbiol. 1993;139:1587–1594. doi: 10.1099/00221287-139-7-1587. [DOI] [PubMed] [Google Scholar]
- 25.Seal S E, Elphinstone J G. Advances in identification and detection of Pseudomonas solanacearum. In: Hayward A C, Hartman G L, editors. Bacterial wilt: the disease and its causative agent, Pseudomonas solanacearum. Wallingford, United Kingdom: CAB International; 1994. pp. 35–58. [Google Scholar]
- 26.Stead D E. Grouping of plant pathogenic and some other Pseudomonas spp. by using cellular fatty acid profiles. Int J Syst Bacteriol. 1992;42:281–295. [Google Scholar]
- 27.Stead D E, Elphinstone J G, Pemberton A W. Conference Proceedings—Brighton Crop Protection Conference—Pests and Diseases 1996. Farnham, Surrey, United Kingdom: British Crop Protection Council; 1996. Potato brown rot in Western Europe; pp. 1145–1152. [Google Scholar]
- 28.Taghavi M, Hayward C, Sly L I, Fegan M. Analysis of the phylogenetic relationships of strains of Burkholderia solanacearum, Pseudomonas syzygii, and the blood disease bacterium of banana based on 16S rRNA gene sequences. Int J Syst Bacteriol. 1996;46:10–15. doi: 10.1099/00207713-46-1-10. [DOI] [PubMed] [Google Scholar]
- 29.Wullings B A, Van Beuningen A R, Janse J D, Akkermans A D L. Detection of Ralstonia solanacearum, which causes brown rot of potato, by fluorescent in situ hybridization with 23S rRNA-targeted probes. Appl Environ Microbiol. 1998;64:4546–4554. doi: 10.1128/aem.64.11.4546-4554.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yabuuchi E, Kosako Y, Yano I, Hotta H, Nishiuchi Y. Transfer of two Burkholderia and Alcaligenes species to Ralstonia gen. nov.—proposal of Ralstonia pickettii (Ralston, Palleroni & Doudororoff, 1973) comb. nov., Ralstonia solanacearum (Smith, 1896) comb. nov. and Ralstonia eutropha (Davis, 1969) comb. nov. Microbiol Immunol. 1995;39:897–904. doi: 10.1111/j.1348-0421.1995.tb03275.x. [DOI] [PubMed] [Google Scholar]