SUMMARY
RNA silencing (RNAi) has a well-established role in anti-viral immunity in plants. The destructive eukaryotic pathogen Phytophthora encodes suppressors of RNAi (PSRs), which enhance plant susceptibility. However, the role of small RNAs in defense against eukaryotic pathogens is unclear. Here, we show that Phytophthora infection of Arabidopsis leads to increased production of a diverse pool of secondary small interfering RNAs (siRNAs). Instead of regulating endogenous plant genes, these siRNAs are found in extracellular vesicles and likely silence target genes in Phytophthora during natural infection. Introduction of a plant siRNA in Phytophthora leads to developmental deficiency and abolishes virulence, while Arabidopsis mutants defective in secondary siRNA biogenesis are hypersusceptible. Notably, Phytophthora effector PSR2 specifically inhibits secondary siRNA biogenesis in Arabidopsis and promotes infection. These findings uncover the role of siRNAs as antimicrobial agents against eukaryotic pathogens and highlight a defense/counter-defense arms race centered on trans-kingdom gene silencing between hosts and pathogens.
Graphical Abstract

In Brief
The role of plant RNAi in defense against eukaryotic pathogens is unclear. Hou et al. report that Arabidopsis produces a reservoir of secondary siRNAs that confer resistance against the notorious pathogen Phytophthora, likely through trans-kingdom gene silencing. However, a Phytophthora effector defeats this defense by specifically inhibiting secondary siRNA biogenesis.
INTRODUCTION
Phytophthora are filamentous eukaryotic pathogens that exert major threats to food safety and human wellness (Kamoun et al., 2015). Hundreds of billions of dollars are lost each year due to destructive crop diseases caused by Phytophthora species. Battling Phytophthora diseases is a major challenge in agriculture.
Plants have evolved a complex immune system during an arms race with potential pathogens in the environment. However, successful pathogens are able to defeat host immunity, mainly through the function of secreted proteins, called effectors (Jones and Dangl, 2006). The study of pathogen effectors and their targets has yielded important insights into basic plant cell biology in general, and immune signaling in particular (Win et al., 2012). Similar to many other filamentous pathogens, Phytophthora establishes intimate symbiotic associations with host plants through infection structures called haustoria, which are invaginations of host plasma membrane induced by extensions of Phytophthora hyphae. Haustoria are believed to facilitate nutrient transportation from the host, and, more importantly, act as an essential interface for effector delivery from the pathogens (Petre and Kamoun, 2014). Each Phytophthora species is predicted to encode several hundreds to over 1,000 effectors that have diverse cellular functions in plant hosts (Pais et al., 2013). This remarkably large effector repertoire reflects a high level of complexity in the defense/counter-defense crosstalk between Phytophthora and their plant hosts.
Recent studies have revealed that some Phytophthora effectors can inhibit the RNA-silencing pathway in plants (Qiao et al., 2013). RNA silencing has a well-established role in anti-viral immunity, and viral RNA-silencing suppressors are indispensable for infection (Ding, 2010). Although Phytophthora suppressors of RNA silencing (PSR) enhance plant susceptibility (Qiao et al., 2013; Xiong et al., 2014), the role of RNA silencing in defense against eukaryotic pathogens is unclear. Importantly, how these pathogens can overcome RNAi-based defense mechanisms to establish successful infection remains unknown.
Gene silencing is mediated by small RNAs (sRNAs). Plants produce two major classes of sRNAs, microRNAs (miRNAs) and small interfering RNAs (siRNAs), which are distinctive in biosynthetic pathways and functions (Axtell, 2013). miRNAs are encoded from endogenous MIR loci, where the primary transcripts form foldback precursors that are subsequently processed (Papp et al., 2003). In contrast, the precursors of siRNAs are long double-stranded RNAs (dsRNAs) synthesized by RNA-dependent RNA polymerases (RDRPs) (Dalmay et al., 2000; Mourrain et al., 2000). A small number of miRNAs trigger the generation of secondary siRNAs, which are derived from a subset of miRNA-targeted transcripts (Peragine et al., 2004; Yoshikawa et al., 2005). Land plants retain a complex pathway to generate numerous secondary siRNAs with diverse sequences from both coding and non-coding transcripts, but their biological functions are largely unknown (Borges and Martienssen, 2015).
Among the RNA-silencing suppressors identified from Phytophthora, PSR2 was found to impair the accumulation of secondary siRNAs derived from the non-coding TAS1a/b/c and TAS2 transcripts in Arabidopsis (Qiao et al., 2013). This observation prompted us to investigate the role of the secondary siRNA pathway in plant immunity. Here, we report that Phytophthora infection induces the production of a pool of secondary siRNAs from specific transcripts in Arabidopsis. These siRNAs function as a collection of antimicrobial agents and silence target Phytophthora genes during infection. As a counter-defense mechanism, PSR2 blocks this host-induced gene silencing by suppressing the biogenesis of these antimicrobial siRNAs. Thus, hosts and pathogens are engaged in an arms race centered on cross-kingdom RNAi-based immunity.
RESULTS
Secondary siRNA Pathway Is Required for Arabidopsis Defense against Phytophthora
In Arabidopsis, RNA-dependent RNA polymerase 6 (RDR6) and Suppressor of Gene Silencing 3 (SGS3) are responsible for the synthesis of dsRNA precursors from miRNA-targeted transcripts and thereby are key components of secondary siRNA production (Adenot et al., 2006; Peragine et al., 2004). Infection assays using Phytophthora capsici strain LT263, which does not have a PSR2 homolog (Ye and Ma, 2016), showed that rdr6 and sgs3 mutants are hypersusceptible, with the rdr6 mutant exhibiting the most severe disease symptoms (Figures 1A and 1B). Similarly, a transgenic Arabidopsis line (PSR2-5) that constitutively expresses PSR2 (Xiong et al., 2014) also showed enhanced susceptibility (Figures 1A and 1B). These results support a role of the secondary siRNA pathway in plant defense during Phytophthora infection.
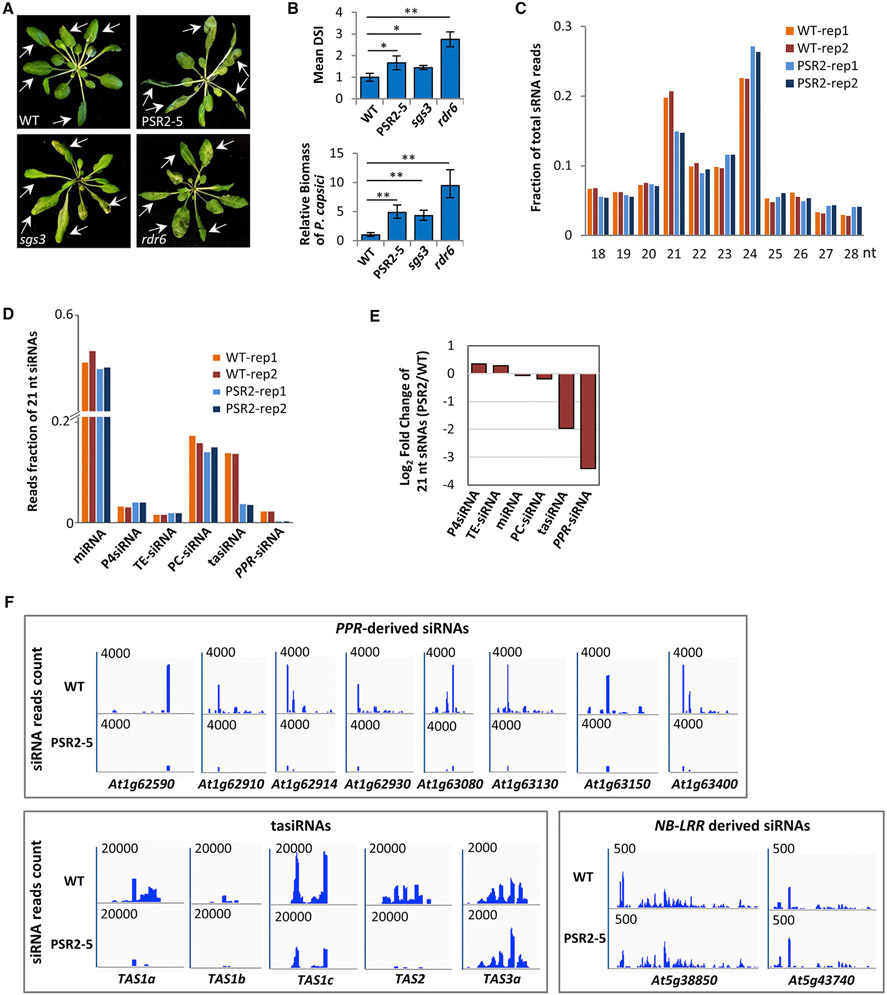
Figure 1. PSR2 Affects the Accumulation of Specific 21-nt siRNAs in Arabidopsis.
(A) Mutants defective in secondary siRNA production and a transgenic line expressing PSR2 (PSR2-5) are hypersusceptible to Phytophthora capsici. Arabidopsis plants were inoculated with zoospore suspensions of P. capsici isolate LT263. Photos were taken at 3 days post inoculation (dpi). Arrows indicate inoculated leaves. WT, wild-type Col-0.
(B) Disease severity index (DSI) and pathogen biomass in inoculated plants at 3 dpi. Values are mean ± SEM of three biological replicates (n ≥ 20 in each replicate). *p < 0.05, **p < 0.01 (Student’s t test).
(C) Size distribution of total sRNAs in WT and PSR2-5 plants. Data from two biological replicates are presented.
(D) Reads fraction of 21-nt sRNAs in WT and PSR2-5 plants. Percentage of reads count of miRNAs and siRNAs produced from Pol IV transcripts (P4siRNA), transposable elements (TE), protein-coding transcripts (PC), TAS and PPR transcripts are shown. Data from two biological replicates are presented.
(E) Changes in the abundance (in a log2 scale) of 21-nt sRNAs derived from different classes of transcripts in PSR2-5.
(F) Secondary siRNAs generated from PPR, TAS, and NB-LRR loci in WT and PSR2-5 plants. The number in each plot indicates the scale of sRNA reads count derived from each locus.
PSR2 Diminishes the Accumulation of Specific Secondary siRNAs
A genome-wide sRNA profiling of PSR2-5 revealed a significant reduction in the 21-nucleotide population (Figure 1C), which is mainly composed of miRNAs and secondary siRNAs. Further analysis of individual 21-nucleotide sRNA classes revealed a moderate (5%) reduction in the miRNA level of PSR2-5 but drastic decreases in the abundances of secondary siRNAs generated from transcripts of TAS1a/b/c, TAS2, and several pentatricopeptide-repeat protein (PPR)-encoding gene loci (Figures 1D-1F). In contrast, siRNAs produced from RNA polymerase IV-dependent transcripts, the non-coding TAS3 transcripts, or transcripts of other loci, including transposable elements and protein-coding genes other than PPR, remained mostly unchanged (Figures 1D-1F). These results demonstrate a specific inhibitory effect of PSR2 on siRNAs generated from PPR and TAS1/2 transcripts in Arabidopsis.
The largest reduction (>90%) in PSR2-5 was observed from PPR-derived secondary siRNAs (Figures 1D and 1E). PPR represents a gene family with approximately 450 members encoded in Arabidopsis (Schmitz-Linneweber and Small, 2008). The vast majority of PPR-siRNAs are produced from transcripts of 15 PPR loci and PSR2 inhibits siRNA production from all of them (Table S1). Thirteen siRNA-generating PPR transcripts contain target site(s) of microR161 (miR161), which is predicted to trigger sRNA production (Table S1). Another microRNA, miR173, can target TAS1/2 transcripts and trigger the production of trans-acting siRNAs (tasiRNAs) (Yoshikawa et al., 2005). Several TAS1/2-derived siRNAs have predicted targets sites in the siRNA-producing PPR transcripts and may also trigger siRNA production (Howell et al., 2007; Chen et al., 2007). Since tasiRNAs exhibit an approximately 75% reduction in PSR2-5 plants (Figures 1D and S1), the largely diminished PPR-derived siRNAs in PSR2-5 are likely attributed to inhibition of both miR161- and miR173-triggered siRNA production.
miR161 and PPR-siRNAs Are Induced during Phytophthora Infection
In order to investigate a potential contribution of miR161 and miR173 in plant defense, we examined the primary transcript levels of their corresponding MIR genes during P. capsici infection. Using qRT-PCR, we detected an induction of pri-miR161, whereas pri-miR173 remained unchanged (Figure S1A). Consistently, northern blotting of mature miRNAs showed that miR161 accumulation was increased during infection, especially at 6 and 24 hr post inoculation (hpi) (Figure 2A). A similar induction was also observed for miR393 (Figure S1B), which is known to be induced by bacterial flagella and contribute to plant basal defense (Navarro et al., 2006). In contrast to miR161 and miR393, the abundance of miR173 was unaltered, consistent with the unchanged pri-miR173 transcript levels (Figures S1A and S1B). We also quantified pri-miR390 transcripts and mature miR390, which is the trigger of siRNAs produced from the non-coding TAS3 transcripts (Adenot et al., 2006; Axtell et al., 2006). Similar to miR173, miR390 was not induced during P. capsici infection (Figures S1A and S1B).
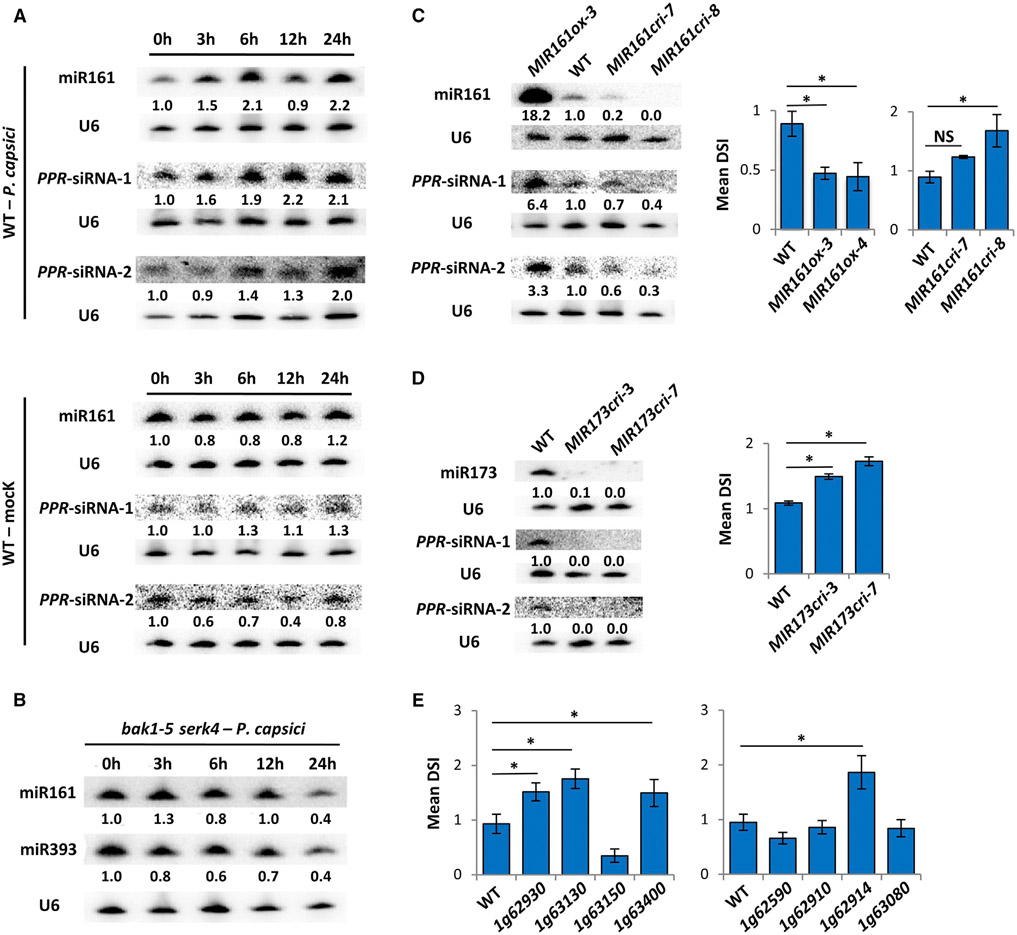
Figure 2. Secondary siRNAs Generated from a Cluster of PPR Genes Contribute to Arabidopsis Resistance to P. capsici.
(A) Northern blotting showing induced accumulation of miR161 and two representative PPR-siRNAs in WT Arabidopsis during P. capsici infection or mock treatment (water). Numbers represent relative signal intensities. U6 was used as a loading control. Similar results were obtained from two biological replicates.
(B) Northern blotting showing unchanged abundance of miR161 and miR393 in bak1 serk4 mutant after P. capsici inoculation.
(C) miR161 contributes to plant immunity. The abundance of miR161 and two PPR-siRNAs was determined in WT, MIR161ox, and MIR161cri plants by northern blotting. Disease severity (represented by DSI) was determined at 3 days after inoculation by P. capsici. Values are mean ± SEM of three biological replicates. *p < 0.05 (Student’s t test, n ≥ 20); NS, no statistical difference.
(D) MIR173cri mutants are hypersusceptible to P. capsici. The abundance of two PPR-derived siRNAs was evaluated in WT and MIR173cri plants. DSI was determined at 3 dpi. Values are mean ± SEM of three biological replicates. *p < 0.05 (Student’s t test, n ≥ 20).
(E) Secondary siRNA-producing PPR genes contribute to Arabidopsis resistance to P. capsici. DSI of eight PPR mutants was determined at 3 dpi. Values are mean ± SEM of three biological replicates. *p < 0.05 (Student’s t test, n ≥ 20).
See also Figures S1-S3.
The induction of miR161 at 6 hpi suggests that P. capsici elicits an immune response during an early infection stage in Arabidopsis. Plant immunity can be activated by “non-self” molecules called microbe-associated molecular patterns (MAMPs) (Jones and Dangl, 2006). Perception of MAMPs requires pattern recognition receptors and co-receptors located on the plant cell surface (Boutrot and Zipfel, 2017). In Arabidopsis, the co-receptors brassinosteroid insensitive 1-associated kinase 1 (BAK1) and somatic-embryogenesis receptor-like kinase 4 (SERK4) are required for the activation of plant immunity (Roux et al., 2011). The induction of miR161 or miR393 was abolished in bak1-5 serk4 plants inoculated with P. capsici (Figure 2B), indicating that the enhanced accumulation of miR161 is a defense response elicited upon perception of the pathogen.
As a major trigger of siRNA production from PPR transcripts, increased levels of miR161 could enhance accumulation of PPR-siRNAs. Indeed, northern blotting showed that, similar to miR161, the levels of two representative secondary siRNAs (named PPR-siRNA-1 and PPR-siRNA-2; Figure S1C) were also increased during P. capsici infection (Figure 2A). As a control, tasiRNAs dependent on miR173 did not exhibit differential accumulation, consistent with the unchanged levels of their trigger miRNA during infection (Figure S1D).
miR161 Contributes to Arabidopsis Defense to P. capsici by Triggering PPR-siRNA Production
We next determined whether miR161 contributes to Arabidopsis defense against P. capsici. For this purpose, we generated transgenic lines that either over-express MIR161 (MIR161ox) or have MIR161 knocked out (MIR161cri) using CRISPR/Cas9-based mutagenesis (Figures S2A and S2B). Northern blotting confirmed increased miR161 levels in MIR161ox lines or reduced levels in MIR161cri lines (Figure 2C). The MIR161ox plants showed enhanced resistance to P. capsici, whereas the MIR161cri mutants exhibited hypersusceptibility (Figures 2C and S2C). On the contrary, overexpression of miR173 or miR390 had no effect on Arabidopsis resistance to P. capsici (Figures S2D-S2G). These results suggest miR161 as a positive regulator of Arabidopsis defense.
Although the accumulation of PPR-siRNA-1 and PPR-siRNA-2 was decreased in MIR161cri lines (Figure 2C), it was not abolished, probably because PPR-siRNAs could also be triggered by miR173-dependent tasiRNAs1/2. Indeed, in MIR173 knockout lines (MIR173cri; Figure S2H), the abundance of PPR-derived siRNAs was diminished (Figure 2D), likely due to the largely reduced tasiRNA levels (Figure S2I). Similar to the MIR161cri mutants, MIR173cri plants also showed enhanced susceptibility to P. capsici (Figures 2D and S2J). Note that PPR-siRNA levels in MIR173ox lines were similar to those in wild-type plants, although the corresponding tasiRNA levels were significantly increased (Figure S2D). This agrees with the previous results that the susceptibility of MIR173ox lines to P. capsici remained unchanged.
These results prompted us to examine the function of the PPR-siRNAs in plant immunity by analyzing individual mutants of eight PPR genes in a cluster on Arabidopsis chromosome 1, from which secondary siRNAs are abundantly produced in an miR161- and tasiRNA-dependent manner (Addo-Quaye et al., 2008; Howell et al., 2007). Corresponding transcripts were largely reduced, if not diminished, in each of the transfer DNA insertion mutants (Figures S3A and S3B). Presumably, secondary siRNA production would also be abolished. Four of these eight mutants showed increased susceptibility to P. capsici (Figures 2E and S3C), providing further support that PPR-siRNAs contribute to plant immunity.
PPR-siRNAs Potentially Silence Phytophthora Transcripts and Confer Resistance
Secondary siRNAs are believed to amplify gene silencing, specifically by miRNAs that potentially regulate large gene families (Adenot et al., 2006). Arabidopsis encodes approximately 450 PPR genes (Barkan and Small, 2014). siRNAs derived from a small number of PPR transcripts may regulate additional family members (Fei et al., 2013). We conducted RNA sequencing analysis and found 249 genes to be upregulated in PSR2-5 and 366 downregulated (Figure S4A; Table S2). Interestingly, only 11 of the upregulated genes were predicted to have target site(s) of PPR-siRNAs (3,922 siRNAs in total), although 1,326 Arabidopsis genes, including 134 PPRs, have predicted target site(s) of this large pool of siRNAs (Table S3). Furthermore, only two of the 11 upregulated genes have PPR motifs (Figure S4B). These results indicate that the primary targets of the PPR-siRNA population may not be PPR genes.
Gene silencing in eukaryotic pathogens by plant hosts has been observed. The first example of this host-induced gene silencing (HIGS) during natural infection was reported in cotton, where two miRNAs regulate virulence-related genes in the fungal pathogen Verticillium dahliae (Zhang et al., 2016). Furthermore, transgenic plants expressing artificial RNAi-inducing dsRNAs can trigger specific gene silencing in fungal pathogens, Phytophthora, and other parasites (Hua et al., 2017; Baulcombe, 2015; Jahan et al., 2015). These observations indicate that host sRNAs can function, in part, to silence pathogen genes. We thus tested whether the PPR-siRNAs may be used by Arabidopsis to target genes in P. capsici. A prediction of potential targets in P. capsici using 3,922 distinct PPR-siRNA sequences revealed 437 siRNA-transcript pairs (Figure 3A), corresponding to 249 P. capsici genes as potential targets of PPR-siRNAs (Table S3).
Figure 3. A PPR-Derived siRNA Silences a Gene in Phytophthora to Confer Resistance.
(A) A flow chart describing the experimental procedure of the functional analysis of PPR-siRNAs.
(B) Base pairing of the PPR-derived siR1310 and siR0513 with their predicted target site in Phyca_554980 of P. capsici.
(C) qRT-PCR determining the transcript abundances of Phyca_554980 and Phyca_538731 (an off-target control) in P. capsici transformants harboring synthesized siR1310. P. capsici transformed with siRGFP was used as a negative control. Values are mean ± SEM of three biological replicates.
(D) Numbers of sporangia produced by WT or transformants of P. capsici harboring siR1310 or siRGFP. Sporangia (indicated by arrows) were numerated from four randomly selected fields of view under a microscope for each strain. Scale bars, 200 μm. Values are mean ± SEM of three biological replicates. One-way ANOVA and post hoc Tukey testing were used for statistical analysis. Different letters label significantly different values (p < 0.05).
(E) P. capsici transformants carrying siR1310 lost virulence activity. Mycelial plugs were used to inoculate detached leaves of N. benthamiana. Photos were taken at 3 dpi under UV to better visualize the lesions. Lesion sizes are presented as mean ± SEM of three replicates. One-way ANOVA and post hoc Tukey testing were used for statistical analysis. Different letters label significant different values (p < 0.05).
(F) qRT-PCR determining the abundances of siR1310 in leaves or EVs of WT and PSR2-5 Arabidopsis plants with or without P. capsici infection. Values are mean ± SEM of three biological replicates. *p < 0.05, **p < 0.01 (Student’s t test).
(G) Transcript abundances of Phyca_554980 determined by qRT-PCR in WT, MIR161ox, PSR2-5, and rdr6 plants inoculated with P. capsici. Values are mean ± SEM of three biological replicates. *p < 0.05, ***p < 0.001 (Student’s t test).
See also Figures S4 and S5.
To test whether some of the predicted targets could be silenced by the corresponding plant siRNAs, we directly introduced synthesized 21-bp sRNA duplexes into P. capsici, mimicking what may occur during natural infection (Figure 3A). For this purpose, we focused on a PPR-derived siRNA-1310 (hereafter siR1310), which is predicted to target the P. capsici gene Phyca_554980 (Figure 3B). Encoding a U2-associated splicing factor (Chen et al., 2013), Phyca_554980 is constitutively expressed. Homologs of Phyca_554980 are also present in other Phytophthora species (Figure S4C), consistent with a conserved function. In addition to siR1310, Phyca_554980 is predicted to be regulated by six other PPR-siRNAs (Table S3), including siR0513 (Figure 3B). Taken together, these results indicate that Phyca_554980 might be an important target of PPR-siRNAs in Phytophthora.
The synthesized siR1310 duplex was introduced in P. capsici together with a plasmid carrying a gene that confers resistance to the antibiotic G418. Transformants that gained G418 resistance potentially also took up the sRNAs. As a control, another sRNA duplex designed to target a GFP gene (siRGFP) was also synthesized and introduced into P. capsici. Seven of ten transformants potentially harboring siR1310 showed reduced abundance of Phyca_554980 transcripts (Figures 3C and S5A). This silencing effect is specific as Phyca_554980 transcript levels were not affected in strains transformed with siRGFP (Figures 3C and S5A). We further confirmed the silencing specificity of siR1310 by monitoring the transcript levels of Phyca_538731, which is predicted to be insufficient as a silencing target, although it contains a sequence that partially matches siR1310 (Figure S5B). The transcript abundance of Phyca_538731 was not reduced in transformants harboring either siR1310 or siRGFP (Figure 3C).
Since siR1310 has the potential to specifically silence target gene(s) in Phytophthora, we determined the consequence of this silencing event by analyzing the developmental phenotypes and virulence activities of the P. capsici transformants. Compared with wild-type and the transformants harboring siRGFP, transformants harboring siR1310 exhibited a moderate decrease in mycelial growth (Figures S5C and S5D) and significant defects in sporangia development (Figure 3D) and zoospore release (Figure S5E). Importantly, introduction of siR1310 nearly abolished the ability of P. capsici to cause disease in Nicotiana benthamiana (Figure 3E). Because the leaves were inoculated with mycelia plugs, the diminished virulence activity of these transformants cannot be fully attributed to sporulation defects. These data indicate that PPR-siRNAs have the potential to silence target gene(s) in P. capsici such as Phyca_554980, which is required for Phytophthora development and pathogenicity. As such, PPR-siRNAs may contribute to resistance to P. capsici.
PPR-siRNAs May Confer Cross-Kingdom Gene Silencing during Phytophthora Infection
To explore whether PPR-siRNAs could be transported from host plants to Phytophthora, we examined their presence in extracellular vesicles (EVs). A role of EVs in plant immunity has been proposed as they accumulate around haustoria of fungal pathogens (An et al., 2006a, 2006b; Micali et al., 2011) and have recently been shown to carry stress-response proteins (Rutter and Innes, 2017) and sRNAs (Cai et al., 2018). In addition, purified plant EVs were shown to be taken up by fungal hyphae in culture (Regente et al., 2017; Cai et al., 2018), indicating they could deliver sRNAs to pathogens.
Using qRT-PCR, we were able to detect siR1310 in EVs isolated from wild-type Arabidopsis leaves (Figure 3F). The abundance of siR1310 was significantly lower in EVs isolated from PSR2-5, consistent with an overall reduction of PPR-siRNAs in PSR2-5. Furthermore, P. capsici infection resulted in increased accumulation of siR1310 in EVs of wild-type plants, but this increase was abolished in PSR2-5 (Figure 3F). Similarly, siR0513, another PPR-siRNA that has a target site in Phyca_554980, could also be detected in EVs (Figure S5F). These results suggest that PPR-siRNAs are cargos of EVs and may contribute to host-induced gene silencing.
We next examined whether Phyca_554980 could be silenced by PPR-siRNAs during natural infection. Wild-type P. capsici was used to inoculate wild-type, PSR2-5, rdr6, or the MIR161ox-3 Arabidopsis plants and the transcript abundances of Phyca_554980 were determined. We observed increased levels of Phyca_554980 transcripts in infected tissues of PSR2-5 or rdr6 plants (Figure 3G), which have decreased accumulation of PPR-siRNAs compared with wild-type plants. In contrast, a decreased level of Phyca_554980 transcripts was observed in infected tissues of MIR161ox-3 plants (Figure 3G), which, at least in part, could be due to the enhanced silencing effect by the higher level of PPR-siRNAs. These results are consistent with the notion that Phyca_554980 expression in P. capsici may be manipulated by plant hosts during infection, potentially through the function of PPR-siRNAs in EVs such as siR1310.
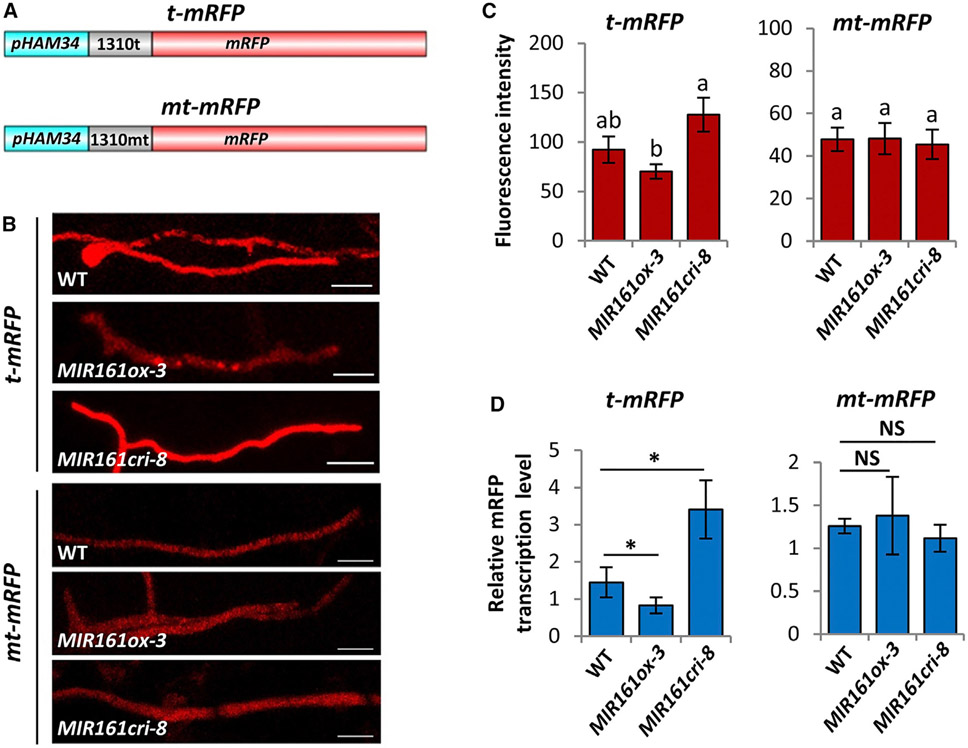
To further demonstrate that cross-kingdom gene silencing could occur during natural infection, we generated P. capsici strains carrying a reporter to monitor the silencing effect of siR1310. A siR1310 target site (t) and a mutant version (mt) were incorporated into the 5′ UTR sequence of an mRFP gene to generate t-mRFP and mt-mRFP, respectively (Figure 4A). These constructs were then introduced into P. capsici. Transformants with stable RFP expression were monitored for mRFP transcript levels during the infection of wild-type, MIR161ox, and MIR161cri plants, which accumulate different levels of siR1310 (Figure S5G). P. capsici(t-mRFP) showed higher fluorescence intensities in hyphae when infecting MIR161cri-8 compared with wild-type plants, whereas the lowest fluorescence signals were observed in hyphae infecting the MIR161ox plants (Figures 4B and 4C). In contrast, P. capsici(mt-mRFP) did not show observable differences in fluorescence intensities when infecting these lines (Figures 4B and 4C). A similar conclusion was made by quantifying mRFP transcripts using qRT-PCR (Figure 4D). Collectively, these results suggest that gene silencing by siR1310, and possibly other PPR-siRNAs, produced by Arabidopsis, may occur in P. capsici during natural infection.
Figure 4. PPR-Derived siR1310 Silences a Reporter Gene during Phytophthora Infection.
(A) Schematic illustration of the construction of mRFP reporters containing either a target site of siR1310 (t-mRFP) or a mutated target site (mt-mRFP).
(B and C) (B) Red fluorescence intensity was monitored during P. capsici infection of WT, MIR161ox, or MIR161cri Arabidopsis plants. Photos were taken at 2 dpi. Scale bars, 20 μm. Values shown in (C) are mean ± SEM and analyzed by one-way ANOVA and post hoc Tukey testing. Different letters label statistically different values (p < 0.05, n ≥ 8). Because t-mRFP and mt-mRFP constructs were independently transformed into P. capsici, their basal mRFP expression levels were different.
(D) mRFP transcript levels of P. capsici infecting WT, MIR161ox, and MIR161cri plants were determined by qRT-PCR. Values are mean ± SEM of four replicates. *p < 0.05 (Student’s t test,); NS, no statistical difference.
See also Figure S5.
PSR2 Interferes with Secondary siRNA Production by Associating with DRB4
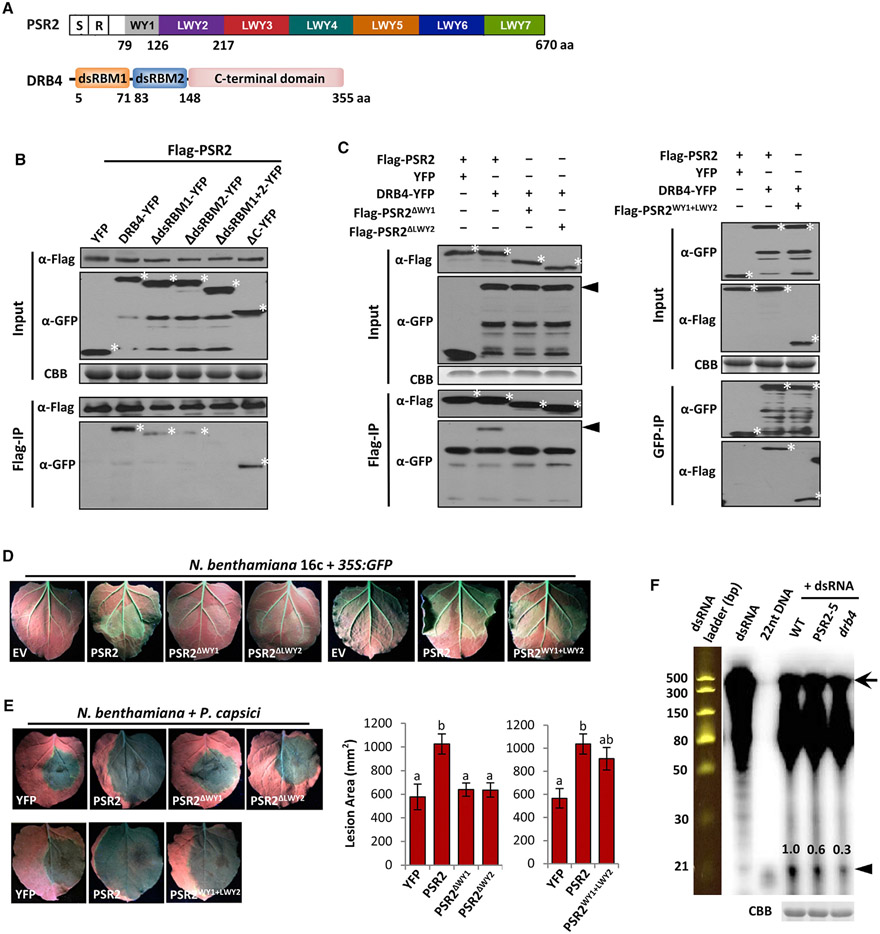
We next investigated the molecular basis underlying PSR2-mediated suppression of secondary siRNA accumulation in Arabidopsis by characterizing PSR2-interacting proteins. Candidates that may associate with PSR2 were identified using yeast two-hybrid screening of an Arabidopsis cDNA library (Table S4). Among them, we were particularly interested in dsRNA-binding protein 4 (DRB4), which has a known function in secondary siRNA biogenesis. DRB4 binds to dsRNA precursors through two dsRNA-binding domains (dsRBM1 and dsRBM2) and associates with Dicer-like 4 (DCL4), which processes the dsRNA substrates (Adenot et al., 2006; Fukudome et al., 2011). Co-immunoprecipitation of PSR2 with DRB4 was confirmed when both proteins were expressed in N. benthamiana (Figures 5A and 5B). The dsRBM domains of DRB4 are required for its interaction with PSR2.
Figure 5. PSR2 Associates with DRB4 in Arabidopsis.
(A) Schematic representation of the domain structure of PSR2 and DRB4. S, secretion signal; R, RxLR motif. Numbers indicate amino acid positions of the motifs.
(B) The two dsRNA-binding domains of DRB4 mediate interaction with PSR2. FLAG-PSR2, DRB4-YFP, and DRB4 truncates were transiently expressed in N. benthamiana. PSR2 was pulled down using anti-FLAG agarose. Enrichment of DRB4 or its truncated mutants in the agarose was detected by western blotting. Asterisk (*) labels corresponding protein band. Protein gel was stained with Coomassie brilliant blue (CBB) as a loading control.
(C) WY1 and LWY2 are necessary and sufficient for PSR2 interaction with DRB4. FLAG-tagged PSR2, PSR2ΔWY1, PSR2ΔLWY2, or PSR2WY1+LWY2 were expressed in N. benthamiana with DRB4-YFP. Enrichment of DRB4 in anti-FLAG agarose was detected by western blotting. Asterisk (*) labels corresponding protein band. The arrowhead labels DRB4.
(D) WY1 and LWY2 are necessary and sufficient for the transgene-silencing suppression activity of PSR2. Leaves of N. benthamiana 16c plants were co-infiltrated with Agrobacterium carrying 35S:GFP and 35S:PSR2 constructs. Pictures were taken 5 days after Agrobacterium infiltration. EV, empty vector.
(E) WY1 and LWY2 are necessary and sufficient for the virulence activities of PSR2. PSR2 or its derivatives were expressed in N. benthamiana leaves, which were subsequently inoculated with P. capsici strain LT263. YFP was used as a control. Lesions were examined at 3 dpi. Values are mean ± SEM of three biological replicates. One-way ANOVA and post hoc Tukey testing were used for statistical analysis. Different letters label significantly different values (p < 0.01).
(F) Reduced dsRNA cleavage in PSR2-5 and a drb4 plants. In vitro synthesized dsRNAs (510 bp in length) were labeled with 32P and incubated with protein extracts. Cleavage products were then analyzed by electrophoresis. A 32P-labeled 22-nt DNA and a dsRNA ladder were used as size markers. Arrow and arrowhead label dsRNA precursor and sRNA products respectively. Numbers represent the relative abundances of the sRNA products. Total proteins in the extracts were analyzed on SDS-PAGE and stained with CBB as a loading control.
See also Figure S6.
PSR2 protein has a modular architecture, containing seven imperfect tandem repeats (Figure 5A) (Ye and Ma, 2016). Repeats 2–7 each includes three motifs, which were named L, W, and Y after a conserved amino acid residue in their respective sequences (Jiang et al., 2008; Ye and Ma, 2016). Repeat 1 only contains the W and the Y motifs. Analysis of truncated mutants revealed that the first (WY1) and the second (LWY2) repeat units of PSR2 are required for interaction with DRB4 (Figure 5C). Consistent with this observation, the mutants PSR2ΔWY1 and PSR2ΔLWY2 lost the ability to suppress transgene silencing (Figures 5D and S6A) or promote Phytophthora infection in N. benthamiana (Figures 5E and S5B). In addition, a fragment of PSR2 (55–215 amino acids), which spans WY1 and LWY2, is sufficient for association with DRB4 (Figure 5C). This fragment is also sufficient, although with a slightly weaker activity than full-length PSR2, to suppress gene silencing (Figures 5D and S6A) and promote infection (Figures 5E and S6B). These results form a strong link between DRB4 interaction and the virulence function of PSR2.
Next, we explored how PSR2 may affect secondary siRNA biogenesis through its interaction with DRB4. Long dsRNAs (510 bp, corresponding to GFP sequence) were synthesized in vitro, labeled with 32P or biotin, and then incubated with crude protein extracts of leaf tissues collected from wild-type, PSR2-5, or drb4 plants. Reduced production of sRNAs, as cleavage products, was observed from drb4 and PSR2-5, suggesting that PSR2 interferes with the dicing of dsRNA substrates (Figures 5F and S6C). We further examined whether PSR2 can bind dsRNAs in plant cells. YFP-PSR2 and DRB4-YFP were expressed individually in N. benthamiana and pulled down using anti-GFP resins. The protein-bound resins were incubated with in vitro synthesized dsRNAs and those bound to the immunoprecipitated proteins were detected. Our results show that both PSR2 and DRB4 associates with dsRNAs (Figure S6D). This might be due to a direct binding of PSR2 with dsRNAs, which may lead to competition with DRB4 for binding to dsRNA substrates. Or, PSR2 may indirectly bind to dsRNAs by associating with the dicing complex. Either way, PSR2 interferes with dsRNA processing in plant hosts.
drb4 Phenocopies PSR2-5 Plants
To further demonstrate that DRB4 is a virulence target of PSR2, we examined the development and disease susceptibility phenotypes of an Arabidopsis drb4 mutant. Similar to PSR2-5, drb4 is hypersusceptible to P. capsici (Figures 6A and S6E). In addition, both PSR2-5 and drb4 exhibit a subtle developmental phenotype; i.e., narrow and curly leaves (Figure 6A). A similar, but more profound, phenotype was reported in rdr6 (Peragine et al., 2004), indicating that it is likely associated with secondary siRNA production. Genome-wide sRNA profiling further confirmed that, as all siRNA-producing PPR loci were affected by RDR6 and PSR2, most of them were also affected by DRB4 (Figure 6B). On the contrary, siRNAs produced from transcripts encoding nucleotide-binding site leucine-rich repeat proteins (NB-LRR) are unaffected in either PSR2-5 or drb4 plants, although their production is fully dependent on RDR6 (Table S5). Encoding canonical disease resistance proteins, NB-LRRs constitute another large gene family that can produce secondary siRNAs (Zhai et al., 2011). The observation that PSR2 does not have a major impact on NB-LRR-derived siRNAs is intriguing because reduced abundance of these siRNAs may lead to increased expression of disease resistance genes, which could be detrimental to the pathogen. Although it remains to be determined how PSR2 and DRB4 specifically affect PPR-derived but not NB-LRR-derived siRNAs, these results support DRB4 as a virulence target of PSR2 in Arabidopsis.
Figure 6. A drb4 Mutant Phenocopies PSR2-Expressing Plants.
(A) drb4 and PSR2-5 were hypersusceptible to P. capsici. Roots of 14-day-old seedlings were inoculated by zoospore suspensions and photos were taken at 3 dpi. This phenotype was complemented by introducing DRB4-YFP under its native promoter into the drb4 mutant.
(B) Five-week-old drb4 and PSR2-5 plants exhibited a similar curly/narrow leaf phenotype.
(C) Venn diagram showing PPR loci with reduced secondary siRNA production in rdr6, drb4, and PSR2-5 compared with WT Arabidopsis.
See also Figure S6.
DISCUSSION
Here, we show that siRNAs derived from endogenous plant transcripts are induced by Phytophthora and potentially silence transcripts of the pathogen during infection. Consistent with their importance as an antimicrobial strategy, the biogenesis of secondary siRNAs is specifically suppressed by the Phytophthora effector PSR2. RNAi-based immunity thus represents an important battleground in the host-pathogen arms race.
Host-derived sRNAs have been found to facilitate plant defense, especially to fungal pathogens (Zhang et al., 2016; Cai et al., 2018). In this study, we show secondary siRNAs are important executors of host-induced gene silencing in an oomycete pathogen that is evolutionarily distant from fungi (Kamoun et al., 2015). Perception of Phytophthora infection by Arabidopsis induces a transcriptional induction of miR161, which subsequently triggers an increased accumulation of secondary siRNAs derived from specific PPR transcripts. The diverse PPR-siRNA pool includes thousands of sequences, some of which may directly silence genes in P. capsici. Interestingly, induction of miR161 was also reported in Arabidopsis treated with bacterial flagellin (Li et al., 2010), indicating that PPR-siRNA production might be a general immune response. Indeed, 216 PPR-siRNA-target pairs can be predicted from the fungal pathogen Verticillium dahliae (Table S3), indicating that PPR-siRNAs may also target V. dahliae genes for silencing. Consistent with this notion, rdr6 mutants of Arabidopsis are hypersusceptible to V. dahlia as well as another fungal pathogen, Botrytis cinerea (Ellendorff et al., 2009; Cai et al., 2018).
Secondary siRNAs have been implicated in playing a role in host-parasite interactions. miRNAs produced by the parasitic plant Cuscuta campestris trigger siRNA production in host plants and manipulated host gene expression (Shahid et al., 2018). In our study, secondary siRNAs produced by a plant host function as antimicrobial agents. The overall abundance and sequence complexity of secondary siRNAs is much higher than their miRNA triggers, which may be of benefit to host defense. During host-pathogen co-evolution, it would be expected that pathogen genes targeted by host sRNAs are under strong selection to diversify, which could abolish sequence complementarity and thus evade silencing. Because MIR genes must maintain a foldback structure in their primary transcripts for processing, they are constrained in how rapidly they can evolve, which may compromise their utility as direct antimicrobial agents. The induction of a diverse pool of secondary siRNAs upon pathogen perception facilitates a co-evolutionary arms race with sequence changes in the targeted pathogen genes. This could be particularly robust when the siRNAs are generated from non-coding genes or genes within large families (such as PPR). Production of secondary siRNA from PPR transcripts is prevalent in eudicots, suggesting an ancient and potentially essential function (Xia et al., 2013). Most eudicot species encode over 400 PPR genes in their genomes (Barkan and Small, 2014), but only a small number produce siRNAs. These siRNA-producing PPRs constitute a rapidly evolving, monophyletic clade that has a distinct evolution dynamic from other family members (Dahan and Mireau, 2013), possibly driven by the arms race with pathogens. None of the PPR mutants in the secondary siRNA-generating cluster that we examined exhibit morphological defects in Arabidopsis, indicating that these genes may tolerate sequence changes.
The presence of PPR-siRNAs in EVs suggests a potential trafficking mechanism from plant hosts to pathogens. EVs mediate intercellular transport of sRNAs in mammals (Meldolesi, 2018), and also transport sRNAs from parasitic nematodes into mammalian host cells, where they suppress host immune responses (Buck et al., 2014). Notably, nematode EVs were recently demonstrated to be specifically enriched in secondary siRNAs (Chow et al., 2018). Global analysis of sRNA composition in plant EVs also shows that siRNAs are the major sRNA cargo in terms of abundance (Baldrich et al., 2018). These results, together, support secondary siRNAs as prominent executors for cross-kingdom silencing as a defense mechanism and set the foundation for manipulating this particular pathway as a strategy to enhance broad-spectrum resistance to plant disease.
Host defense mechanisms and pathogen virulence strategies are linked. Successful pathogens must defeat host immunity in order to establish infection. By investigating the function of PSR2, we demonstrate that Phytophthora pathogens have evolved effectors to suppress siRNA-based immunity. Since PSR2 is a conserved effector in Phytophthora, suppression of secondary siRNA production is likely a common virulence strategy of Phytophthora and may also be employed by other fungal and oomycete pathogens that are potentially targeted by host-induced gene silencing.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Wenbo Ma (wenbo.ma@ucr.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Arabidopsis thaliana
Arabidopsis thaliana ecotype Col-0 was used as wild-type and for generating transgenic lines. T-DNA insertion lines of AT1G62910 (SALK_152489), AT1G63130 (SAIL_119_G05), AT1G62930 (SAIL_18_E04), AT1G63080 (SALK_020638C), AT1G63400 (CS316928), AT1G63150 (SALK_152489), AT1G62590 (SALK_114012), and AT1G62914 (CS433098) were obtained from the Arabidopsis Biological Resource Center (ABRC). Plants were grown in a growth room at 22±2°C with a 12 h light/12 h dark photoperiod.
Nicotiana benthamiana
Plants were grown in a growth room at 22±2°C with a 12 h light/12 h dark photoperiod. WT and 16c (Ruiz et al., 1998) plants were used in this study.
Phytophthora capsici
P. capsici isolate LT263 were used in this study. P. capsici were grown on fresh 10% V8 plates at 25°C in the dark for mycelia growth. Zoospores release was based on a method descried in Method Details.
METHOD DETAILS
Cloning and Constructs
To construct miRNA overexpression lines in Arabidopsis, DNA sequences encoding the pri-miRNAs of miR161, miR173 and miR390 were amplified from cDNA of Arabidopsis Col-0 by PCR using gene-specific primers (listed in Table S6). The PCR products were inserted into pENTR/D-TOPO and subsequently pEG100 using Gateway cloning (Invitrogen). To generate the knockout lines of miR161 and miR173, the Yao promoter-driven CRISPR/Cas9 system was utilized (Yan et al., 2015). The sgRNA cassettes were cloned into pCAMBIA1300-pYAO:Cas9 into the SpeI site. To construct an mRFP reporter containing the target site of siR1310, sequences corresponding to the sense and antisense strands of the siR1310 target site or mutated target site were synthesized and annealed respectively. The DNA fragments were then ligated with pTOR-mRFP into the EcoRI site.
Phytophthora capsici Inoculation and Phenotypic Analysis
In most experiments, four-week-old Arabidopsis plants were inoculated with zoospore suspension of P. capsici isolate LT263. P. capsici was grown on a fresh 10% V8 plate at 25°C in the dark until mycelia covering the plate. Cut small pieces of agar with growing mycelia and grow them into 20 mL of 10% V8 broth at 25°C for 2 days in the dark. To release zoospores, wash the mycelium plugs with sterile water three times, then keep the plugs in sterile water at 25°C for 24 hours in the dark. Place the petri dish under illumination at room temperature for another 24 h. Zoospores were induced by incubating the mycelia at 4°C for 40 min, followed by illuminating for 20 min at room temperature. Zoospores were collected using one layer of miracloth and their concentration was determined under a microscope. The zoospore suspension (200–500 zoospores/μL) was used for inoculation. Eight Arabidopsis plants and 3-4 adult rosette leaves per plant were inoculated for each treatment. 20 μL zoospore suspension were applied to the abaxial side of each leaf. Sterile water was used as a mock control. The inoculated plants were placed in a growth chamber with high humidity (~85% RH) at 25°C. Disease symptoms were monitored three days after inoculation and disease severity was evaluated as Mean DSI (Wang et al., 2013). Biomass of P. capsici was also determined by qPCR using P. capsici specific primers (listed in Table S6). Each experiment was conducted with three independent biological replicates.
For root infection, roots of two-week old seedlings, grown on MS medium (Murashige and Skoog agar containing 1% (wt/vol) sucrose) were dipped in a zoospores suspension (100 zoospores/μL) for five seconds as described (Wang et al., 2013). The seedlings were immediately planted in soil and the disease symptoms were monitored at three days after inoculation. For inoculation of N. benthamiana, the abaxial sides of detached leaves were inoculated with fresh mycelial plugs (0.5 cm). Leaves were kept in sealed 0.8% water agar plates in the dark at 25°C. Lesions were observed under UV light three days after inoculation. Sizes of the lesion areas were analyzed using imageJ (https://imagej.net/).
Isolation of EVs
For each biological replicate, EVs were isolated from the pooled apoplastic fluid of 36 five-week old Arabidopsis plants grown at 22±2°C with a 9 h light/15 h dark photoperiod using a protocol including fractionation on an iodixanol density gradient (OptiPrep, Sigma Aldrich) (Rutter and Innes, 2017). Purified EVs were quantified using a ZetaView nanoparticle tracking analyzer from Particle-Metrix. Approximately 10 x 109 EVs from each replicate were used for RNA extraction.
RNA Extraction, Northern Blotting and Quantitative Real-Time PCR
Total RNAs were extracted from Arabidopsis leaves and EVs with or without infection at different time points using TRIzol reagent (Invitrogen). Small RNA northern blotting was performed as described using 5 μg of total RNA extract (Pall and Hamilton, 2008). U6 was used as a loading control. The results were visualized using a Typhoon phosphorimager and quantified with ImageQuant TL (GE). Sequences of the oligonucleotide probes are listed in Table S6. For quantitative RT-PCR, three biological repeats were performed, and relative expression levels were calculated using the 2−ΔΔCt equation. Actin2 was used as the internal control. Gene-specific primers used for qRT-PCR are listed in Table S6. For Figure S5G, siRNA levels were quantified using stem-loop qRT-PCR as described in Varkonyi-Gasic et al. (2007) starting with 1.0 μg total RNA from leaves. For Figures 3F and S5F, siRNA levels were quantified using the QuantiMir kit from System Biosciences (Mountain View, CA), starting with 2.0 μg total RNA from leaves, diluting the resulting cDNA 100 folds, and then using 2-4 μL of diluted cDNA for each 25 μL reaction. For both stem-loop and QuantiMir methods, the qRT-PCR step was performed using SYBR Green PCR Master Mix (Thermo Scientific) and using U6 as an internal control.
Transformation of sRNAs into Phytophthora Protoplasts
RNA oligonucleotides corresponding to siRGFP and siR1310 were synthesized and annealed to form siRNA duplex (listed in Table S6). A polyethylene glycol (PEG)-mediated protoplast transformation procedure was followed as described with modifications (Dou et al., 2008). Protoplasts of P. capsici isolate LT263 were prepared as described with a concentration of 2x104 protoplast per mL. 25 μg of pTOR plasmid DNA and 8 μg of siRNA duplex were added into 1 mL of protoplast suspension for transformation. Transformants were recovered in pea medium and then selected on V8 agar plates supplemented with 50 μg /mL of G418. Transformants with G418 resistance were sub-cultured and analyzed for development and virulence activity. Mycelia from each transformant were collected for RNA extraction and gene expression analysis.
Microscopy of Phytophthora capsici during Infection
Four-week-old Arabidopsis plants were inoculated with zoospore suspension (200-500 zoospores/μL) of P. capsici transformants expressing t-mRFP or mt-mRFP. Confocal images were captured at 2 dpi using a Leica SP5 confocal microscope under 40x lens. The average fluorescence strength of the entire hyphae in the images was estimated using Image J (http://imagej.nih.gov/ij/).
Co-immunoprecipitation Assays of PSR2 and DRB4
3xFlag-PSR2, DRB4-YFP and their derivatives were co-expressed in N. benthamiana using Agrobacterium-mediated infiltration. Total proteins were extracted using an IP buffer [10% (v/v) glycerol, 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 5 mM DTT, 1x protease inhibitor mixture (Roche), 1 mM PMSF, and 0.1% CA-630], and then incubated with either anti-Flag agarose (Sigma-Aldrich) or anti-GFP magnetic beads (Chromotek) at 4°C for one hour. The beads were washed for five times using the IP buffer, and PSR2 and DRB4 in the immune complexes were detected by western blotting using anti-Flag (Sigma Aldrich) or anti-GFP (Clontech) antibody respectively.
Transgene Silencing Suppression Assay Using N. benthamiana 16c Plants
PSR2 and its derivatives were cloned into the vector pEG100 and the recombinant plasmids were transformed into Agrobacterium tumefaciens strain GV3101. The bacteria were co-infiltrated into N. benthamiana 16c leaves together with Agrobacterium carrying 35S-GFP (Qiao et al., 2013). Green fluorescence was observed using a hand-held UV light at five days after Agro-infiltration. The protein levels of GFP were determined by western blotting using an anti-GFP antibody (Santa Cruz). The protein levels of PSR2 and its derivatives were examined by western blotting using an anti-PSR2 antisera generated in this study.
Double-Stranded RNA Binding and Cleavage Assays
Sense and antisense transcripts of GFP were synthesized from a plasmid template containing a partial GFP gene with 510 bp in length (oligos were listed in Table S6). In vitro transcription was conducted by incubating 0.1 μM plasmid DNA in a 100 μL reaction system with 0.5 μM of T7 RNA polymerase and 5 mM of NTP mix for 3 hours at 37°C. For internal labeling with [α-32P] UTP, 1 mM ATP/CTP/GTP, 0.05 mM cold UTP and 10 μL [α-32P] UTP were used. For internal labeling with Biotin-16, 5 mM ATP/CTP/GTP, 3 mM cold UTP and 2 mM Biotin-16-UTP were used. 2 U of Turbo DNase (Ambion) was added to the reaction mixture at 37°C for 15 minutes to remove the template DNA. Nucleotides and NTPs were also removed using Bio-Spin 6 columns (BioRad). Single-stranded RNAs were purified using 1:1 volume acidic phenol/chloroform (Ambion) and precipitated using 1:1 volume isopropanol and 1 μL glycogen (Thermo Scientific) in −20°C freezer overnight. RNA pellet was then dissolved in 30 μL RNase-free water. Equal amounts of the ssRNAs were annealed as previously described (Fukudome et al., 2011) and used for further analyses.
For protein-dsRNA binding assay, YFP-PSR2 and DRB4-YFP fusion proteins were transiently expressed in N. benthamiana and total proteins were extracted using an IP buffer containing 10% (vol/vol) glycerol, 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM DTT, 1x protease inhibitor mixture (Roche), 1 mM PMSF, and 0.1% CA-630. Double-stranded RNA was removed by adding 7 μL of RNaseIII (NEB), 10 μL of 10x RNaseIII buffer and 10 μL of 10x MnCl2 to 1 mL of protein extract. Samples were then centrifuged at 12,000 rpm, 4°C for 15 min, and the supernatant was incubated with anti-GFP magnetic beads (Chromotek) for two hours at 4°C. The beads were then incubated with the synthesized 510-bp dsRNAs (final concentration of ~33 nM) at 4°C in a dsRNA-binding buffer containing 30 mM Tris-HCl (pH 7.0), 10 mM NaCl, 20 mM MgCl2,0.1 mM EDTA, and 5 mM DTT for 30 minutes. The beads were then washed with binding buffer to remove the unbounded dsRNAs before the protein-bounded RNAs were extracted using Trizol/Chloroform (Ambion) and analyzed on 2% agarose gel with ethidium bromide staining.
For dsRNA cleavage assay, total proteins from 0.5 g leaves of four-week-old Arabidopsis plants were extracted using an extraction buffer containing 20 mM Tris-HCl (pH 7.5), 4 mM MgCl2, 5 mM DTT, 1x protease inhibitor mixture (Roche) and 1 mM PMSF. Double-stranded RNAs labeled with 32P (final concentration of ~1 nM) or Biotin-16 (final concentration of ~5 nM) were incubated with 15 μL of protein extracts in a 240 μL reaction mixture containing 4 μL of 5x cleavage buffer (150 mM Tris-HCl (pH 7.5), 20 mM MgCl2, 250 mM NaCl, 50 mM ATP, 10 mM GTP, 5 mM DTT, 0.5 μL of RNaseOUT (Invitrogen)). After incubation at 23°C for two hours, RNA cleavage products were purified by phenol/chloroform (Ambion) and precipitated using 1:1 volume of isopropanol supplemented with 1 μL of glycogen (Thermo Scientific) at −20°C overnight. The RNAs were then analyzed on 15% denaturing polyacrylamide gel containing 8 M urea. The 32P-labeled RNAs were detected by autoradiography and Biotin-16-labeled RNAs were detected using Chemiluminescent Nucleic Acid Detection Module Kit (Thermo Scientific).
RNA Sequencing and Data Analysis
sRNA libraries were single-end sequenced on the Illumina HiSeq4000 platform with read lengths of 50 bases. Adapter sequences were trimmed from fastq reads using Cutadapt v1.4.1 (Martin, 2011), the remaining sequences in the size range of 18- to 28-nt were mapped to the Arabidopsis thaliana reference genome annotation (TAIR10) using Bowtie v1.0.1 (Langmead et al., 2009), allowing all alignments (−a) and zero mismatch (−v 0) per read. After removing the reads associated with t/r/sn/snoRNAs, the rest of the perfectly matched 18-28 nt sRNAs were used for further analysis. The sequences of mature miRNAs were from miRbase (version 21), and eight TAS loci: TAS1a, TAS1b, TAS1c, TAS2, TAS3, TAS3b, TAS3c and TAS4 for trans-acting siRNAs identification. The list of Pol IV-dependent siRNA loci (P4siRNAs) has been previously described (Zhai et al., 2015). Protein-coding gene-derived siRNAs (PC-siRNAs) and transposon element derived siRNAs (TE-siRNAs) were calculated using Araport11 annotation (Cheng et al., 2017), with 27,655 protein-coding regions and 3,901 TEs. For normalization, the abundances of sRNAs in each library were normalized to transcripts per million (TPM), excluding t/r/sn/snoRNA-derived sequences. The abundance of sRNAs from each locus was summed by hits-normalized-abundance of all mapped reads from that region. Loci with normalized TPM >10 in WT were further analyzed for abundance changes in PSR2-5 (Table S1).
RNA-seq libraries were analyzed using paired-end sequencing on the Illumina HiSeq4000 platform with read lengths of 150 bases. Reads were mapped to the TAIR10 genome using HISAT2 v2-2.0.5 (Kim et al., 2015) allowing only one unique hit (−k 1) and length less than 5000 (−X 5000). PCR duplicates were further removed using SAMTools v1.4 (Li et al., 2009). FPKM (Fragments Per Kilobase of transcript per Million mapped reads) of each gene was calculated using StringTie v1.3.3b (Pertea et al., 2015), and edgeR (Robinson et al., 2010) was used to identify genes that were differentially expressed between PSR and WT replicates.
siRNA Target Prediction in Arabidopsis, Phytophthora capsici and Verticillium dahliae
3922 distinct PPR-derived siRNA sequences were used for target prediction using the psRNA Target web server (http://plantgrn.noble.org/psRNATarget) (Dai and Zhao, 2011). Default setting of Schema V1 was used. For target prediction in Arabidopsis, “Arabidopsis thaliana, transcript, JGI genomic project, Phytozome 12, 167_TAIR10” was chosen as the target file. For target prediction in P. capsici, “Phyca11_filtered_transcripts.fasta” was downloaded as the target file from JGI (https://genome.jgi.doe.gov/Phyca11/Phyca11.home.html). For target prediction in V. dahliae, “Verticillium dahliae v1.0” was downloaded as the target file from JGI (https://genome.jgi.doe.gov/Verda1/Verda1.home.html). Genes with expectation ≤ 2 were considered as potential targets of PPR-derived siRNAs.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data reported in this study were analyzed using JMP Pro v13.0 (SAS). The data are presented as mean ± SEM. For Phytophthora infection data, N represents inoculated leaves. For microscopy data, N represents the number of observed hyphae. When comparing a test group to a control group, a two-tailed Student’s t-test was used. The significance values are reported as follows: * = p < 0.05, ** = p < 0.01, and *** = p < 0.001. When comparing the means of multiple groups, a one-way ANOVA followed by Tukey’s HSD post hoc test was performed. Significant differences between groups (p < 0.05) are denoted with different letters.
DATA AND SOFTWARE AVAILABILITY
Data that support the findings of this study have been deposited in NCBI SRA with the accession codes SRA: SRP135923.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-Flag-HRP | Sigma-Aldrich | Cat# A8592; RRID: AB_439702 |
| Mouse anti-GFP | Clontech | Cat# 632375 |
| Rabbit anti-GFP | Santa Cruz | Cat# SC-8334; RRID: AB_641123 |
| Flag M2 Magnetic beads | Sigma-Aldrich | Cat# M8823; RRID: AB_2637089 |
| Rabbit anti-PSR2 | This paper | N/A |
| GFP Trap Magnetic beads | Chromotek | Cat# Gtma-20; RRID: AB_2631406 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| EDC N-(3-DIMETHYLAMINOPROPYL)-N’-ETHYLCAR | Sigma-Aldrich | Cat# E7750 |
| G418 Sulfate | Gold Biotechnology | Cat# G-418-5 |
| QuantiMir | System Biosciences | Cat# RA420A-1 |
| SYBR Green PCR Master Mix | Thermo Scientific | Cat# 4364346 |
| Easytide ATP-32P, [γ-32P] | PerkinElmer | Cat# BLU502A500UC |
| Easytide UTP-32P, [α-32P] | PerkinElmer | Cat# NEG007H250UC |
| Biotin-16-UTP | Roche | Cat# 11388908910 |
| Chemiluminescent Nucleic Acid Detection Module Kit | Thermo Scientific | Cat# 89880 |
| T4 PNK | Thermo Scientific | Cat# FEREK0032 |
| Cellulase from Trichodema reesei ATCC 26921 | Sigma-Aldrich | Cat# C5846 |
| Lysing Enzymes from Trichoderma harzianu | Sigma-Aldrich | Cat# L1412 |
| PEG4000 | Sigma-Aldrich | Cat# 81240 |
| Optiprep density gradient medium | Sigma-Aldrich | Cat# D1556 |
| TURBO DNAse | Ambion | Cat# AM2238 |
| Shortcut RNase III | New England Biolabs | Cat# N03635 |
| RNaseOUT | Invitrogen | Cat# 10777019 |
| Critical Commercial Assays | ||
| NEBNext Small RNA Library Prep Set for Illumina | NEB | Cat# E7300 |
| Chemiluminescent Nucleic Acid Detection Module Kit | Thermo Scientific | Cat# 89880 |
| Deposited Data | ||
| Raw data | This paper | SRA: SRP135923 |
| Experimental Models: Organisms/Strains | ||
| Nicotiana benthamiana: 16c | Ruiz et al., 1998 | N/A |
| Arabidopsis: PSR2-5 | Qiao et al., 2013 | N/A |
| Arabidopsis: sgs3-1: AT5G23570: G131A->W44STOP | Mourrain et al., 2000 | N/A |
| Arabidopsis: rdr6-11: AT3G49500 T-DNA insertion: CS24285 | ABRC | ID: CS24285 |
| Arabidopsis: drb4-1: AT3G62800 T-DNA insertion: SALK_000736 | ABRC | ID: SALK_000736 |
| Arabidopsis: drb4/pDRB4::DRB4 | This paper | NA |
| Arabidopsis: bak1-5/serk4 | Roux et al., 2011 | N/A |
| Arabidopsis: AT1G62910 T-DNA insertion: SALK_152489 | ABRC | ID: SALK_152489 |
| Arabidopsis: AT1G63130 T-DNA insertion: CS805864 | ABRC | ID: SAIL_119_G05 |
| Arabidopsis: AT1G62930 T-DNA insertion: CS800852 | ABRC | ID: SAIL_18_E04 |
| Arabidopsis: AT1G63080 T-DNA insertion: SALK_020638C | ABRC | ID: SALK_020638C |
| Arabidopsis: AT1G63400 T-DNA insertion: CS316928 | ABRC | ID: CS316928 |
| Arabidopsis: AT1G62590 T-DNA insertion: SALK_114012 | ABRC | ID: SALK_114012 |
| Arabidopsis: AT1G62914 T-DNA insertion: CS433098 | ABRC | ID: CS433098 |
| Arabidopsis: AT1G63150 T-DNA insertion: SALK_152489 | ABRC | ID: SALK_152489 |
| Arabidopsis: MIR161ox | This paper | N/A |
| Arabidopsis: MIR173ox | This paper | N/A |
| Arabidopsis: MIR390ox | This paper | N/A |
| Arabidopsis: MIR161cri | This paper | N/A |
| Arabidopsis: MIR173cri | This paper | N/A |
| Phytophthora capsici: LT263 | Wang et al., 2013 | N/A |
| Oligonucleotides | ||
| siRNA oligos for siR1310 see Table S6 | This paper | N/A |
| siRNA oligos for siRGFP see Table S6 | This paper | N/A |
| Probes for Northern blotting see Table S6 | This paper | N/A |
| Recombinant DNA | ||
| pEG100-35S::MIR161 | This paper | N/A |
| pEG100-35S::MIR173 | This paper | N/A |
| pEG100-35S::MIR390 | This paper | N/A |
| pCAMBIA-pYAO-Cas9-gRNA161 | This paper | N/A |
| pCAMBIA-pYAO-Cas9-gRNA173 | This paper | N/A |
| pTOR-1310t-mRFP | This paper | N/A |
| pTOR-1310mt-mRFP | This paper | N/A |
| pEG100-PSR2ΔWY1 | This paper | N/A |
| pEG100-PSR2ΔWY2 | This paper | N/A |
| pEG100-PSR2WY1+WY2 | This paper | N/A |
| pEG101-DRB4 | This paper | N/A |
| pEG101-DRB4ΔdsRBM1 | This paper | N/A |
| pEG101-DRB4ΔdsRBM2 | This paper | N/A |
| pEG101-DRB4ΔdsRBM1+2 | This paper | N/A |
| pEG101-DRB4ΔC | This paper | N/A |
| Software and Algorithms | ||
| ImageJ | NIH | Ver. 1.51j8 |
| ImageQuant TL | GE | Ver. 7.0 |
| JMP Pro | SAS | Ver. 13 |
Highlights.
Phytophthora infection increases production of a pool of secondary siRNAs in Arabidopsis
Secondary siRNAs from a PPR gene cluster contribute to defense against Phytophthora
PPR-siRNAs potentially silence Phytophthora transcripts to confer resistance
Phytophthora effector PSR2 suppresses the biogenesis of PPR-siRNAs to promote infection
ACKNOWLEDGMENTS
This research was supported by the US Department of Agriculture National Institute of Food and Agriculture grant #2014-67013-21554 to W.M., US National Science Foundation grants IOS-1340001 to W.M. and X.C. and IOS-1645745 to R.W.I., and the Thousand Talents Program for Young Scholars and the Program for Guangdong Introducing Innovative and Entrepreneurial Teams (2016ZT06S172) to J.Z. We are grateful to Dr. Qi Xie for providing the Yao promoter-driven CRISPR/Cas9 system, Drs. Ping He and Cyril Zipfel for providing bak1-5 serk4 Arabidopsis, and Dr. Howard S. Judelson for providing the pTOR vector.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures and six tables and can be found with this article online at https://doi.org/10.1016/j.chom.2018.11.007.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Addo-Quaye C, Eshoo TW, Bartel DP, and Axtell MJ (2008). Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol 18, 758–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouche N, Gasciolli V, and Vaucheret H (2006). DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol 16, 927–932. [DOI] [PubMed] [Google Scholar]
- An Q, Ehlers K, Kogel KH, van Bel AJ, and Huckelhoven R (2006a). Multivesicular compartments proliferate in susceptible and resistant MLA12-barley leaves in response to infection by the biotrophic powdery mildew fungus. New Phytol. 172, 563–576. [DOI] [PubMed] [Google Scholar]
- An Q, Huckelhoven R, Kogel KH, and van Bel AJ (2006b). Multivesicular bodies participate in a cell wall-associated defence response in barley leaves attacked by the pathogenic powdery mildew fungus. Cell Microbiol. 8, 1009–1019. [DOI] [PubMed] [Google Scholar]
- Axtell MJ (2013). Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol 64, 137–159. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Jan C, Rajagopalan R, and Bartel DP (2006). A two-hit trigger for siRNA biogenesis in plants. Cell 127, 565–577. [DOI] [PubMed] [Google Scholar]
- Baldrich P, Rutter BD, Karimi HZ, Podicheti R, Meyers B, and Innes RW (2018). Plant extracellular vesicles contain diverse small RNA species and are enriched in 10 to 17 nucleotide “tiny” RNAs. bioRxiv. 10.1101/472928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, and Small I (2014). Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol 65, 415–442. [DOI] [PubMed] [Google Scholar]
- Baulcombe DC (2015). VIGS, HIGS and FIGS: small RNA silencing in the interactions of viruses or filamentous organisms with their plant hosts. Curr. Opin. Plant Biol 26, 141–146. [DOI] [PubMed] [Google Scholar]
- Borges F, and Martienssen RA (2015). The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol 16, 727–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutrot F, and Zipfel C (2017). Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu. Rev. Phytopathol 55, 257–286. [DOI] [PubMed] [Google Scholar]
- Buck AH, Coakley G, Simbari F, McSorley HJ, Quintana JF, Le Bihan T, Kumar S, Abreu-Goodger C, Lear M, Harcus Y, et al. (2014). Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat. Commun 5, 5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Qiao L, Wang M, He B, Lin FM, Palmquist J, Huang SD, and Jin H (2018). Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 360, 1126–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HM, Li YH, and Wu SH (2007). Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proc. Natl. Acad. Sci. U S A 104, 3318–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XR, Xing YP, Li YP, Tong YH, and Xu JY (2013). RNA-seq reveals infection-related gene expression changes in Phytophthora capsici. PLoS One 8, e74588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CY, Krishnakumar V, Chan AP, Thibaud-Nissen F, Schobel S, and Town CD (2017). Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 89, 789–804. [DOI] [PubMed] [Google Scholar]
- Chow FW-N, Koutsovoulos G, Ovando-Vázquez C, Laetsch DR, Bermúdez-Barrientos J, Claycomb J, Blaxter M, Abreu-Goodger C, and Buck A (2018). An extracellular argonaute protein mediates export of repeat-associated small RNAs into vesicles in parasitic nematodes. BioRxiv. 10.1101/343772. [DOI] [Google Scholar]
- Dahan J, and Mireau H (2013). The Rf and Rf-like PPR in higher plants, a fast-evolving subclass of PPR genes. RNA Biol. 10, 1469–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, and Zhao PX (2011). psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 39, W155–W159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay T, Hamilton A, Rudd S, Angell S, and Baulcombe DC (2000). An RNA-Dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101, 543–553. [DOI] [PubMed] [Google Scholar]
- Ding SW (2010). RNA-based antiviral immunity. Nat. Rev. Immunol 10, 632–644. [DOI] [PubMed] [Google Scholar]
- Dou D, Kale SD, Wang X, Chen Y, Wang Q, Jiang RH, Arredondo FD, Anderson RG, Thakur PB, McDowell JM, et al. (2008). Conserved C-terminal motifs required for avirulence and suppression of cell death by Phytophthora sojae effector Avr1b. Plant Cell 20, 1118–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellendorff U, Fradin EF, de Jonge R, and Thomma BP (2009). RNA silencing is required for Arabidopsis defence against Verticillium wilt disease. J. Exp. Bot 60, 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Q, Xia R, and Meyers BC (2013). Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 25, 2400–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukudome A, Kanaya A, Egami M, Nakazawa Y, Hiraguri A, Moriyama H, and Fukuhara T (2011). Specific requirement of DRB4, a dsRNA-binding protein, for the in vitro dsRNA-cleaving activity of Arabidopsis Dicer-like 4. RNA 17, 750–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell MD, Fahlgren N, Chapman EJ, Cumbie JS, Sullivan CM, Givan SA, Kasschau KD, and Carrington JC (2007). Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 19, 926–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua C, Zhao JH, and Guo HS (2017). Trans-kingdom RNA silencing in plant-fungal pathogen interactions. Mol. Plant 11, 235–244. [DOI] [PubMed] [Google Scholar]
- Jahan SN, Asman AK, Corcoran P, Fogelqvist J, Vetukuri RR, and Dixelius C (2015). Plant-mediated gene silencing restricts growth of the potato late blight pathogen Phytophthora infestans. J. Exp. Bot 66, 2785–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang RH, Tripathy S, Govers F, and Tyler BM (2008). RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc. Natl. Acad. Sci. U S A 105, 4874–4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, and Dangl JL (2006). The plant immune system. Nature 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kamoun S, Furzer O, Jones JDG, Judelson HS, Ali GS, Dalio RJD, Roy SG, Schena L, Zambounis A, Panabieres F, et al. (2015). The top 10 oomycete pathogens in molecular plant pathology. Mol. Plant Pathol 16, 413–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R; 1000 Genome Project Data Processing Subgroup (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang Q, Zhang J, Wu L, Qi Y, and Zhou JM (2010). Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol. 152, 2222–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. [Google Scholar]
- Meldolesi J (2018). Exosomes and ectosomes in intercellular communication. Curr. Biol 28, R435–R444. [DOI] [PubMed] [Google Scholar]
- Micali CO, Neumann U, Grunewald D, Panstruga R, and O’Connell R (2011). Biogenesis of a specialized plant-fungal interface during host cell internalization of Golovinomyces orontii haustoria. Cell Microbiol. 13, 210–226. [DOI] [PubMed] [Google Scholar]
- Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, et al. (2000). Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101, 533–542. [DOI] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, and Jones JD (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, 436–439. [DOI] [PubMed] [Google Scholar]
- Pais M, Win J, Yoshida K, Etherington GJ, Cano LM, Raffaele S, Banfield MJ, Jones A, Kamoun S, and Saunders DGO (2013). From pathogen genomes to host plant processes: the power of plant parasitic oomycetes. Genome Biol. 14, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall GS, and Hamilton AJ (2008). Improved northern blot method for enhanced detection of small RNA. Nat. Protoc 3, 1077–1084. [DOI] [PubMed] [Google Scholar]
- Papp I, Mette MF, Aufsatz W, Daxinger L, Schauer SE, Ray A, van der Winden J, Matzke M, and Matzke AJ (2003). Evidence for nuclear processing of plant microRNA and short interfering RNA precursors. Plant Physiol. 132, 1382–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, and Poethig RS (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18, 2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, and Salzberg SL (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol 33, 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petre B, and Kamoun S (2014). How do filamentous pathogens deliver effector proteins into plant cells? PLoS Biol. 12, e1001801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y, Liu L, Xiong Q, Flores C, Wong J, Shi J, Wang X, Liu X, Xiang Q, Jiang S, et al. (2013). Oomycete pathogens encode RNA silencing suppressors. Nat. Genet 45, 330–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regente M, Pinedo M, San Clemente H, Balliau T, Jamet E, and de la Canal L (2017). Plant extracellular vesicles are incorporated by a fungal pathogen and inhibit its growth. J. Exp. Bot 68, 5485–5495. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux M, Schwessinger B, Albrecht C, Chinchilla D, Jones A, Holton N, Malinovsky FG, Tor M, de Vries S, and Zipfel C (2011). The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23, 2440–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz MT, Voinnet O, and Baulcombe DC (1998). Initiation and maintenance of virus-induced gene silencing. Plant Cell 10, 937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter BD, and Innes RW (2017). Extracellular vesicles isolated from the leaf apoplast carry stress-response proteins. Plant Physiol. 173, 728–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, and Small I (2008). Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 13, 663–670. [DOI] [PubMed] [Google Scholar]
- Shahid S, Kim G, Johnson NR, Wafula E, Wang F, Coruh C, Bernal-Galeano V, Phifer T, dePamphilis CW, Westwood JH, et al. (2018). MicroRNAs from the parasitic plant Cuscuta campestris target host messenger RNAs. Nature 553, 82–85. [DOI] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Wu R, Wood M, Walton EF, and Hellens RP (2007). Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Bouwmeester K, van de Mortel JE, Shan W, and Govers F (2013). A novel Arabidopsis-oomycete pathosystem: differential interactions with Phytophthora capsici reveal a role for camalexin, indole glucosinolates and salicylic acid in defence. Plant Cell Environ. 36, 1192–1203. [DOI] [PubMed] [Google Scholar]
- Win J, Chaparro-Garcia A, Belhaj K, Saunders DG, Yoshida K, Dong S, Schornack S, Zipfel C, Robatzek S, Hogenhout SA, et al. (2012). Effector biology of plant-associated organisms: concepts and perspectives. Cold Spring Harb. Symp. Quant. Biol 77, 235–247. [DOI] [PubMed] [Google Scholar]
- Xia R, Meyers BC, Liu Z, Beers EP, and Ye S (2013). MicroRNA superfamilies descended from miR390 and their roles in secondary small interfering RNA biogenesis in eudicots. Plant Cell 25, 1555–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Q, Ye W, Choi D, Wong J, Qiao Y, Tao K, Wang Y, and Ma W (2014). Phytophthora suppressor of RNA silencing 2 is a conserved RxLR effector that promotes infection in soybean and Arabidopsis thaliana. Mol. Plant Microbe Interact 27, 1379–1389. [DOI] [PubMed] [Google Scholar]
- Yan L, Wei S, Wu Y, Hu R, Li H, Yang W, and Xie Q (2015). High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol. Plant 8, 1820–1823. [DOI] [PubMed] [Google Scholar]
- Ye W, and Ma W (2016). Filamentous pathogen effectors interfering with small RNA silencing in plant hosts. Curr. Opin. Microbiol 32, 1–6. [DOI] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, and Poethig RS (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19, 2164–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Jeong DH, De Paoli E, Park S, Rosen BD, Li Y, Gonzalez AJ, Yan Z, Kitto SL, Grusak MA, et al. (2011). MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 25, 2540–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Bischof S, Wang H, Feng S, Lee TF, Teng C, Chen X, Park SY, Liu L, Gallego-Bartolome J, et al. (2015). A one precursor one siRNA model for pol IV-dependent siRNA biogenesis. Cell 163, 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Zhao YL, Zhao JH, Wang S, Jin Y, Chen ZQ, Fang YY, Hua CL, Ding SW, and Guo HS (2016). Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat. Plants 2, 16153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data that support the findings of this study have been deposited in NCBI SRA with the accession codes SRA: SRP135923.