Summary
Background:
Underlying biological mechanisms involved in sex differences in asthma status changes from pre- to post-adolescence are unclear. DNA methylation (DNAm) has been shown to be associated with the risk of asthma.
Objective:
We hypothesized that asthma acquisition from pre- to post-adolescence was associated with changes in DNAm during this period at asthma-associated cytosine-phosphate-guanine (CpG) sites and such an association was sex-specific.
Methods:
Subjects from the Isle of Wight birth cohort (IOWBC) with DNAm in blood at ages 10 and 18 years (n = 124 females, 151 males) were studied. Using a training-testing approach, epigenome-wide CpGs associated with asthma were identified. Logistic regression was used to examine sex-specific associations of DNAm changes with asthma acquisition between ages 10 and 18 at asthma-associated CpGs. The ALSPAC birth cohort was used for independent replication. To assess functional relevance of identified CpGs, association of DNAm with gene expression in blood was assessed.
Results:
We identified 535 CpGs potentially associated with asthma. Significant interaction effects of DNAm changes and sex on asthma acquisition in adolescence were found at 13 of the 535 CpGs in IOWBC (P-values <1.0 × 10−3). In the replication cohort, consistent interaction effects were observed at 10 of the 13 CpGs. At 7 of these 10 CpGs, opposite DNAm changes across adolescence were observed between sexes in both cohorts. In both cohorts, cg20891917, located on IFRD1 linked to asthma, shows strong sex-specific effects on asthma transition (P-values <.01 in both cohorts).
Conclusion and clinical relevance:
Gender reversal in asthma acquisition is associated with opposite changes in DNAm (males vs females) from pre- to post-adolescence at asthma-associated CpGs. These CpGs are potential biomarkers of sex-specific asthma acquisition in adolescence.
Keywords: ALSPAC, asthma acquisition, DNA methylation, IOWBC, sex-specificity
1 |. INTRODUCTION
Asthma is a common chronic condition that affects approximately 339 million people worldwide,1 causing substantial morbidity, reduced quality of life and substantial healthcare costs.2 Asthma predominantly originates in early childhood3 with an estimated 1.1 million children affected in the UK.4
There is a male predominance of asthma in early childhood. During adolescence, more boys remit asthma than girls, while more girls acquire asthma than boys, which results in gender reversal of asthma prevalence from pre- to post-adolescence5–14 with asthma becoming more prevalent and severer among females after puberty.9,15,16 However, the underlying biological mechanisms involved in these sex differences in the natural history of asthma across childhood and adolescence remain unclear.
Although the pathogenesis of asthma reflects a combination of inherited susceptibility and environmental exposures, the aetiology and biological mechanisms are poorly understood. The increase in prevalence of asthma in recent decades suggests an important role for environmental exposures in the development of asthma in genetically high-risk individuals, and a number of studies have highlighted the potential for a role of epigenetic programming in response to early life environmental exposures in asthma susceptibility.17–21 One of the most widely studied epigenetic mechanisms is DNA methylation (DNAm).22,23 DNAm at specific cytosine-phosphate-guanine (CpG) sites in DNA from both blood and lung tissue have been found to be associated with asthma24–38 and related phenotypes such as COPD.39–42
While these studies have established a clear association between DNAm patterns and asthma, they rely on asthma status determined at a single time point. Yet, as previously discussed, asthma phenotype within an individual can be dynamic, new incidence and clinical remission occurring across the life course with gender reversal in asthma prevalence observed in adolescence. In a candidate gene approach, we have previously investigated temporal changes of DNAm at CpG sites in genes encoding proteins in the Th2 pathway and the transition of asthma over adolescence.4 This study showed that the level of DNAm and the association between specific CpGs (and their interaction with DNA sequence variation) and asthma changes across adolescence. We therefore hypothesized that DNAm changes at specific sites across adolescence might explain the biological basis of sex differences in the natural history of asthma across adolescence and identify biomarkers of asthma acquisition in adolescence that would potentially be beneficial for prediction and prevention. To test this, we have used genome-wide DNAm data to assess sex-specific association of DNAm changes from pre- to post-adolescence with asthma acquisition at asthma-associated CpG sites.
2 |. METHODS
2.1 |. Study population
The Isle of Wight birth cohort (IOWBC) consists of children born between 1 January 1989 and 28 February 1990 on the Isle of Wight (IoW), United Kingdom 43 The IOWBC was established to investigate the natural history of allergic diseases among children residing on a semi-rural island near the UK mainland. Of the 1536 pregnancies in this period, 1456 parents consented for further follow-up with survey and clinical data collected at 1, 2, 4, 10 and 18 years.
2.2 |. Asthma acquisition
Detailed questionnaires that included the questions from the International Study of Asthma and Allergy in Childhood (ISAAC) were administered to parents/participants at 10 and 18 years. Asthma was defined as ‘ever had asthma’ and ‘wheezing or whistling in the chest in the last 12 months’ or ‘current treatment for asthma’. This study focuses on new incidence (ie acquisition) of asthma between 10 and 18 years defined as no asthma at 10 years but having asthma at 18 years. Subjects that had no asthma at both ages were used as a reference group.
Of the 1053 subjects who did not have asthma at 10 years, 275 subjects had DNAm measurements available from peripheral blood samples at both 10 and 18 years and were included in further analyses.
2.3 |. Covariates
Information regarding sex, birth weight, maternal and paternal disease status of asthma was assessed based on questionnaire data and hospital records collected at birth. Socio-economic status was defined based on household income, number of rooms and maternal education. Atopic status was assessed at 10 and 18 years using skin prick test (SPT) for 11 common allergens (house dust mite, cat dander, dog dander, grass pollen mix, tree pollen mix, Alternaria alternata, Cladosporium herbarium, cow’s milk, hen’s egg, peanut and cod), and change of atopic status from 10 to 18 years was recorded. Height and weight were measured at 10 and 18 years, and in cases that a participant did not visit the study centre, information was obtained by telephone interviews. Body mass index (BMI) was calculated based on height and weight, and relative changes in height and BMI were calculated for each subject, for instance, relative change in height of a subject is calculated as the difference in height from pre- to post-adolescence divided by their pre-adolescent height.
2.4 |. DNA methylation (DNAm)
DNAm was measured in peripheral blood with samples collected at 10 and 18 years using either the Infinium HumanMethylation450 BeadChips or MethylationEPIC BeadChips (illumina, Inc, San Diego, CA). Preprocessing of DNAm was carried out using the CPACOR pipeline.44 Details of DNAm data generation, quality control and preprocessing, as well as principal components (PC) analyses detecting latent variables for batch and technical variations, are in the Supplemental Material S1. After preprocessing, a total of 442 475 CpGs in common between the two platforms were included in the analyses.
Since blood is a mixture of functionally and developmentally distinct cell populations,45 adjusting cell-type compositions was needed in analyses to reduce confounding from cell heterogeneity in DNAm measured from blood samples.46 We estimated cell-type proportions using the method proposed by Jaffe and Irizarry,47 adapted from Houseman et al,48 using the Bioconductor minfi package.49 The estimated cell-type proportions of CD4+ T cells, natural killer cells, neutrophil, B cells, monocytes and eosinophil cells were included in the analyses as confounding factors.
2.5 |. Gene expression
Gene expression levels from peripheral blood samples collected at 26 years from IOWBC were determined using paired-end (2*75 bp) RNA sequencing. All samples were sequenced twice using the identical protocol and for each sample the output from both runs were combined. Normalized read count was calculated, and their log transformed values were used for data analysis. Details on RNA sequencing, transcript reading, mapping and assembly, and normalization are in the Supplemental Material S2.
2.6 |. Statistical analysis
To examine whether the subsample (n = 275) included in the study reasonably represented the complete cohort (n = 1053), one sample proportion tests and multinomial tests for categorical variables and one sample t tests for continuous variables were applied.
2.7 |. Screening analysis to identify asthma-associated CpGs
An R package, ttScreening, was implemented to screen for CpGs whose methylation (in M values) was associated with asthma cross-sectionally.50 The screening method implemented in this package has been shown to perform better than FDR-based and Bonferroni-based methods and has the potential of controlling both types I and II errors.50 Subjects with DNAm and asthma data at one or both ages were included in the screening. A CpG site showing statistical significance (at the .05 level) in at least 50% randomly selected training and testing data set pairs was treated as asthma-associated CpGs and included in subsequent analyses. This screening approach was cross-sectional and focused on asthma status rather than asthma transition, to avoid data double-dipping, that is avoid using the same or a very similar model in screening as well as in final data analyses with the same data.
2.8 |. Assessment of DNAm change across adolescence for asthma-associated CpGs
Cytosine-phosphate-guanines that passed screening were treated as potentially asthma-associated CpGs. At these sites, M values of DNAm at each CpG were regressed on 15 PCs obtained from control probes (Supplemental Material S1) and the 6 cell-type proportions48 to obtain batch and cell-type adjusted DNAm (residuals). This regression analysis was conducted at 10 and 18 years, respectively. At each of the asthma-associated CpGs, the difference in residuals between 10 and 18 years was then calculated for each subject to represent DNAm change from 10 to 18 years.
Logistic regressions via R function glm with a logit link were applied to evaluate the association of asthma acquisition (with asthma-free as the reference) with DNAm changes (independent variable) adjusted for covariates and confounders potentially associated with asthma: maternal and paternal history of asthma, sex, birth weight, socio-economic status, change of atopic status from 10 to 18 years, and relative changes in height and BMI from 10 to 18 years. Additionally, since multiple studies have demonstrated gender reversal on asthma prevalence from pre- to post-adolescence, interaction effects of CpGs and sex on asthma acquisition were assessed. Multiple testing was adjusted by controlling a false discovery rate (FDR) of 0.05.
2.9 |. Replication cohort—the Avon Longitudinal Study of Parents and Children (ALSPAC) cohort
CpGs shown to be associated with asthma acquisition in IOWBC were further assessed in an independent cohort, the ALSPAC.51 DNAm data at 7 and 15 or 17 years and asthma acquisition from 7 to 15 years were included in the replication analyses. Details of these data along with information on covariates are presented in the Supplemental Material S3. Please note that the study website contains details of all the data that is available through a fully searchable data dictionary and variable search tool (http://www.bristol.ac.uk/alspac/researchers/our-data/). A P-value <.05 was deemed as being statistically significant.
2.10 |. Association between DNAm and gene expression
For CpGs with DNAm changes showing consistent associations with asthma acquisition between the two cohorts, we evaluated their biological relevance. Genes annotated to the identified CpGs were extracted from the Illumina’s manifest file or SNIPPER (https://csg.sph.umich.edu/boehnke/snipper/) version 1.2. We tested the association between DNAm at these CpGs and gene expression in blood at 26 years using linear regressions. Gene expression (n = 136) was the dependent variable, and DNAm and sex were the independent variables. DNAm at 10 and 18 years were analysed separately. In addition, to assess sex-specificity of DNAm and expression association, an interaction term of DNAm × sex was also included in the model. Interaction effects were treated as being statistically significant with P-value <.05.
3 |. RESULTS
In IOWBC, the analytical subsample was representative of the complete cohort with respect to asthma transition status, demographic variables and other covariates (P-values >.05, Table 1). A sex difference in asthma acquisition was observed in the complete cohort; 10.8% of females acquired asthma from 10 to 18 years, as compared to only 7.1% of males (P-value = .03).
TABLE 1.
Asthma acquisition and non-asthma participants included in the present study compared to the participants in the complete cohort
| Females |
Males |
|||||
|---|---|---|---|---|---|---|
| Categorical Variables: N (%) | Subsample n = 124 | Complete cohort n = 544 | P-value | Subsample n = 151 | Complete cohort n = 509 | P-value |
| Asthma Transition | ||||||
| Acquisition | 12 (9.7%) | 59 (10.8%) | .78 | 14 (9.27%) | 36 (7.1%) | .37 |
| Non-asthma | 112 (90.3%) | 485 (89.1%) | 137 (90.7%) | 473 (92.9%) | ||
|
| ||||||
| Maternal asthma | ||||||
| Yes | 15 (12.1%) | 50 (9.3%) | .35 | 23 (15.4%) | 53 (10.4%) | .06 |
| No | 109 (87.9%) | 490 (90.7%) | 126 (84.5%) | 453 (89.5%) | ||
|
| ||||||
| Paternal asthma | ||||||
| Yes | 14 (11.5%) | 45 (8.4%) | .27 | 15 (10.1%) | 50 (9.9%) | 1 |
| No | 107 (88.4%) | 492 (91.6%) | 133 (89.8%) | 454 (90%) | ||
|
| ||||||
| Socio-economic status | ||||||
| Mid-High | 42 (33.8%) | 193 (36.4%) | .67 | 55 (36.4%) | 197 (40.2%) | .60 |
| Low-mid | 35 (28.2%) | 153 (28.8%) | 51 (33.7%) | 149 (30.4%) | ||
| Low-low | 47 (37.9%) | 184 (34.7%) | 45 (29.8%) | 143 (29.2%) | ||
|
| ||||||
| Change of atopic status from 10 to 18 y | ||||||
| Yes-No | 4 (3.6%) | 7 (2.37%) | .53 | 3 (2.5%) | 3 (1.4%) | .43 |
| No-Yes | 20 (18%) | 52 (17.6%) | 32 (26.8%) | 62 (28%) | ||
| No-No | 87 (78.3%) | 236 (80%) | 84 (70.5%) | 156 (70.5%) | ||
| Continuous variables: Mean (SD) | ||||||
| Birth weight | 3.32 (0.51) | 3.33 (0.50) | .88 | 3.45 (0.56) | 3.50 (0.52) | .51 |
| Relative change in height from 10 to 18 y | 0.18 (0.04) | 0.19 (0.04) | .27 | 0.28 (0.03) | 0.28 (0.04) | .39 |
| Relative change in BMI from 10 to 18 y | 0.29 (0.15) | 0.29 (0.16) | .64 | 0.29 (0.15) | 0.29 (0.15) | .88 |
To identify candidate CpGs potentially associated with asthma at 10 and 18 years for each sex, we applied ttScreening to 442 475 CpGs, stratified by sex. In total, 265 (220 for males, 45 for females) CpGs and 290 (40 for males, 250 for females) CpGs passed screening at 10 and 18 years, respectively. CpGs that passed screening at either age of males or females (535 CpGs in total; Table S1) were treated as asthma-associated CpGs and included in subsequent analyses.
For each CpG site that passed screening, whether asthma acquisition was associated with changes in DNAm from 10 to 18 years and whether such an association was sex-specific were tested using logistic regressions. Sex and DNAm changes, and their interaction, along with adjusting factors were included in the model. After controlling FDR at 0.05, statistically significant interaction effects were observed at 13 CpG sites (Table 2, left panel of Figure 1). All the coefficients for the interaction effects between sex and DNAm changes were positive. Combined with the estimates of main effect, a potential gender reversal with respect to the effects of DNAm changes on asthma acquisition was identified. For instance, at CpG site cg03269757, a larger increase in DNAm from 10 to 18 years was associated with an increased risk of acquisition in females (log-OR = 3.04), but a decreased risk of acquisition in males (log-OR = −1.34). Such opposite associations between males and females were observed at nine of the 13 CpGs. At the remaining four CpG sites, cg11814087, cg12587133, cg18278943 and cg22484084, the association of DNAm changes with asthma acquisition was much stronger in females with larger effect size (increased risk of acquiring asthma). For example, at cg11814087, the log-OR was 6.72 for females, much higher than the log-OR = 0.58 for males (interaction effect P-value 8.85 × 10−4 with 95% CI: 2.29, 10.01).
TABLE 2.
Association of DNA methylation (DNAm) change with asthma acquisition from pre- to post-adolescence at 13 cytosine-phosphate-guanine (CpG) sites that are sex-specific. Significant interactions between DNAm and sex on asthma acquisition from 10 to 18 y identified in the Isle of Wight birth cohort (IOWBC)a were further tested in the Avon Longitudinal Study of Parents and Children 11(ALSPAC) cohortb. Males are in the reference group
| CpG site | Est.c | IOWBC | Gene | CpG islands | Gene location | Chr.e | ALSPAC cohort | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Int.c | 95% CIc | P dRaw | P dFDR | Est.c | Int.c | P d Raw | ||||||
| cg03269757 | −1.34 | 4.38 | 1.93, 6.83 | 1.88 × 10−4 | .02 | ATL2 | N_Shore | Body | 2 | 0.03 | 0.59 | .19 |
| cg11295724 | −0.33 | 7.82 | 4.12, 11.52 | 5.35 × 10−6 | .003 | SIRPD | – | TSS200 | 20 | 0.06 | 0.76 | .22 |
| cg12587133 | 0.34 | 4.85 | 2.37, 7.32 | 1.03 × 10−5 | .003 | AGA | – | 4 | 0.01 | 0.24 | .5 | |
| cg15154628 | −0.67 | 3.23 | 1.14, 5.31 | 1.15 × 10−3 | .05 | CCDC146 | – | Body | 7 | 0.24 | 0.07 | .89 |
| cg16301989 | −0.48 | 4.42 | 1.89, 6.93 | 1.86 × 10−4 | .02 | FUNDC2P2f | – | 2 | 0.05 | 0.47 | .26 | |
| cg18278943 | 0.70 | 4.55 | 1.87, 7.24 | 4.36 × 10−4 | .03 | SLMAP | N_Shelf | 3 | −0.25 | 0.82 | .18 | |
| cg20885063 | −0.52 | 4.45 | 1.86, 7.05 | 3.78 × 10−4 | .03 | ATPAF2 | N_Shelf | Body | 17 | 0.007 | 0.4 | .36 |
| cg20891917 | −0.43 | 5.5 | 2.03, 8.96 | 9.86 × 10 −4 | .04s | IFRD1 | – | 5′UTR | 7 | −0.13 | 1.26 | .01 |
| cg21163444 | −2.22 | 11.13 | 4.31, 17.95 | 8.53 × 10−4 | .04 | ZNF385A | S_Shore | Body | 12 | 0.27 | 0.14 | .86 |
| cg22484084 | 0.29 | 7.18 | 2.69, 11.67 | 1.0 × 10−3 | .04 | PTPRV | – | TSS1500 | 1 | 0.13 | 0.84 | .39 |
| cg06492287 | −1.91 | 10.49 | 4.73, 16.25 | 7.78 × 10−5 | .01 | SNTG2 | – | Body | 2 | 0.002 | −0.04 | .96 |
| cg11770323 | −0.7 | 9.18 | 4.11, 14.25 | 7.92 × 10−5 | .01 | NDFIP2 | – | Body | 13 | −0.75 | −0.55 | .29 |
| cg11814087 | 0.58 | 6.14 | 2.29, 10.01 | 8.85 × 10−4 | .04 | ZFR | – | Body | 5 | 0.11 | −0.68 | .05 |
For the analyses in IoW, logistic regression models were adjusted for birth weight, sex, maternal and paternal disease status of asthma, socio-economic status (SES), change of atopic status, BMI, height from 10 to 18 y.
Analyses of ALSPAC used similar covariates: maternal disease status of asthma, sex, birth weight, SES, atopy status at 7 y, and changes of height, and BMI from 7 to 15/17 y.
Est., Estimated main effect; Int., interaction coefficient of the CpGs with the sex of a child. Interaction effects consistent between the two cohorts are with bold fonts; CI, confidence interval.
PRaw, raw P-value; PFDR, adjusted P-value for multiple testing by controlling a false discovery rate (FDR) of .05. All the P-values are for interaction effects.
Chr., Chromosome.
Genes closest to the CpG site annotated using the UCSC genome browser.
FIGURE 1.
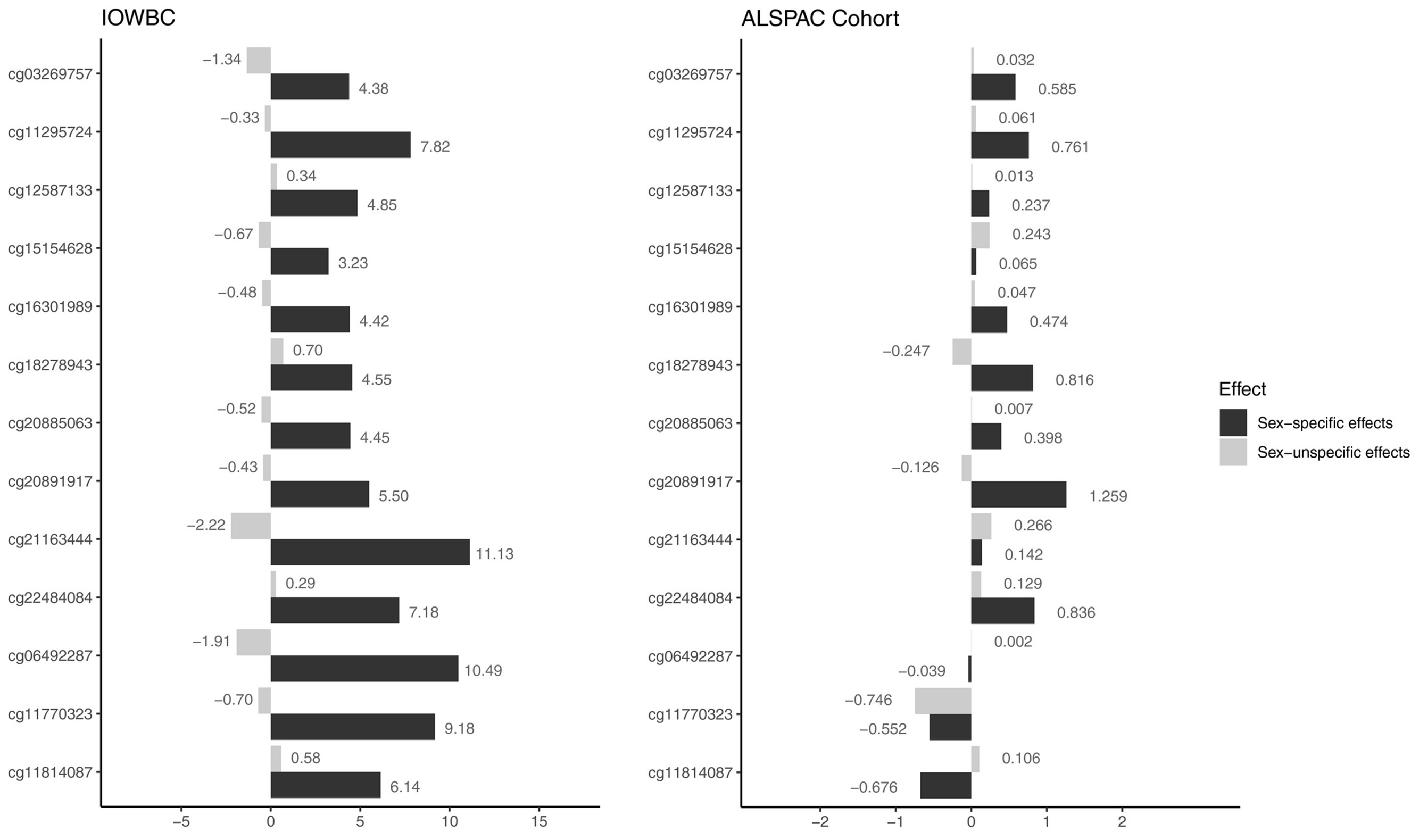
Effects of changes in methylation on asthma acquisition in adolescence for 13 cytosine-phosphate-guanines in Isle of Wight birth cohort (IOWBC) (left) and Avon Longitudinal Study of Parents and Children (right) cohorts, stratified by sex-specific (interaction effects with male as the reference group, black bars) and sex nonspecific effects (main effects, grey bars). X-axis is for the regression coefficients (main effects and interaction effects). For interaction effects (black bars) in the IOWBC, 95% confidence intervals are in Table 2
We further tested these 13 CpGs in the ALSPAC cohort. At 10 of the 13 CpG sites, consistent interaction effects with respect to the direction of effects were observed compared to those found in IOWBC, although only one of the 10 CpGs (cg20891917) showed a statistically significant effect (Table 2 and Figure 1). In addition, in the ALSPAC cohort, for eight of these 10 CpGs, the interaction effects were all much stronger than the main effects (Figure 1), the same pattern observed in IOWBC.
To explore underlying mechanisms of the observed interaction effects, for each of the 13 CpGs, we calculated average DNAm changes between 10 and 18 years in IOWBC, and between 7 and 15 or 17 years in the ALSPAC cohort, for males and females, separately (Figure 2A,B). In both cohorts, average DNAm changes in non-asthmatic subjects were all around zero at the 13 CpGs. However, for subjects in IOWBC who acquired asthma between 10 and 18 years, across all the 13 CpGs, the changes in DNAm were opposite between males and females with males showing decrease in DNAm from pre- to post-adolescence (negative differences in males but positive differences in females, Figure 2A). In the ALSPAC cohort, for the first 10 CpGs in Figure 2B, DNAm at these CpGs showed consistent directions of interaction effects with those in IOWBC (Table 2). The pattern of opposite changes in DNAm from age 7 to 15 or 17 years in ALSPAC between males and females was also observed at 7 of these 10 CpGs (Figure 2B).
FIGURE 2.
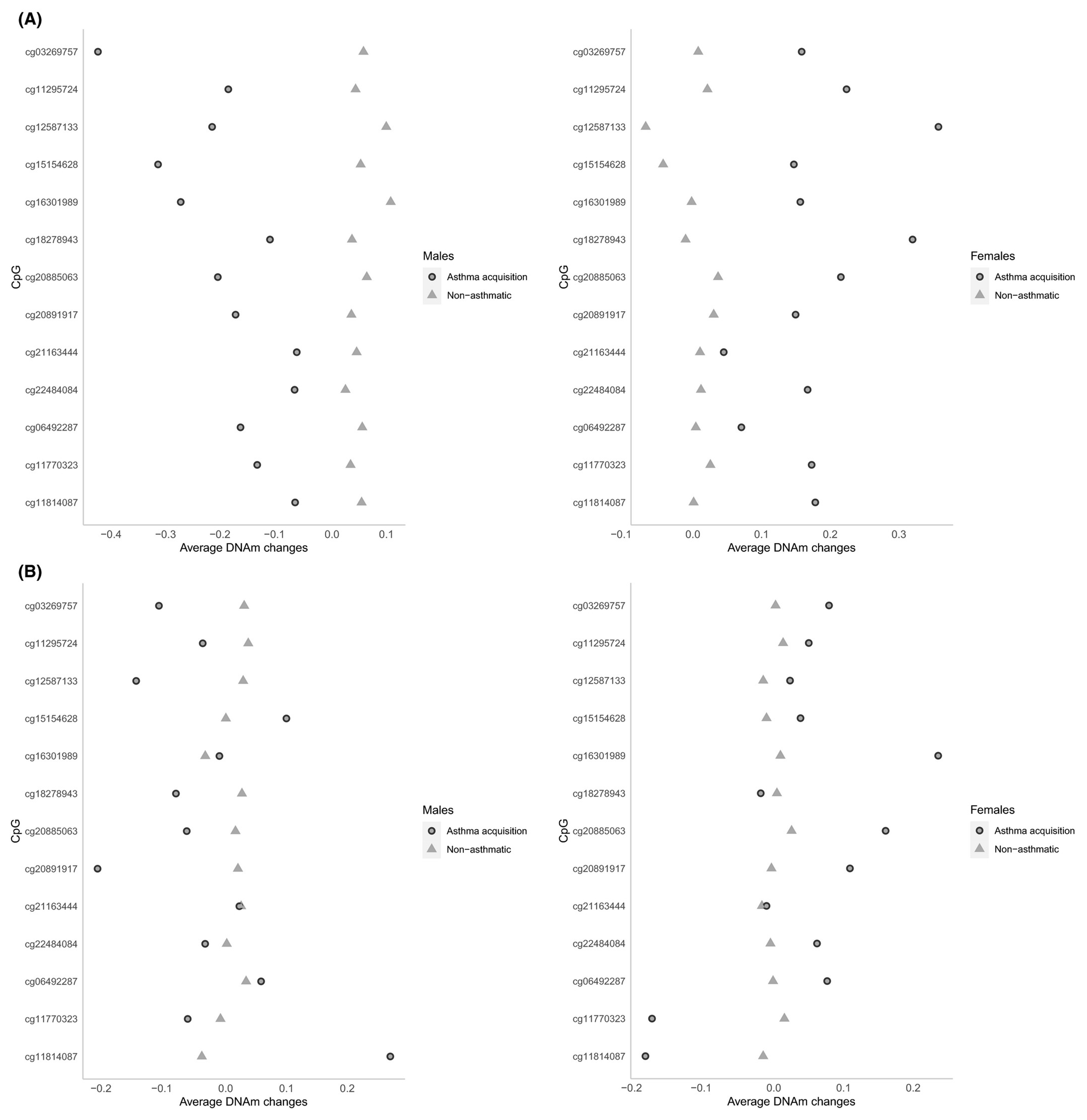
Scatter plots showing average DNA methylation (DNAm) changes from pre-adolescence to post-/late-adolescence, stratified by sex. A, Average DNAm changes between 10 and 18 y in Isle of Wight birth cohort. B, Average DNAm changes between 7 and 15 or 17 y in the Avon Longitudinal Study of Parents and Children cohort. In both panels, left figure is for males, and right for females
To assess the biological relevance of the 10 CpGs showing consistent sex-specificity between the two cohorts, we evaluated the association of DNAm at these CpGs with expression of their mapped genes and whether such associations were sex-specific. The 10 CpGs were mapped to 10 genes (Table 2). We did not have expression data for gene PTPRV. Statistically significant interaction effects were observed at five of the nine CpG sites (Table 3) with four genes identified based on age 10 DNAm and one based on age 18 DNAm. Combined with the estimates of main effect of DNAm, a potential gender reversal with respect to the association of DNAm with gene expression levels was found at all these five CpG sites (based on opposite signs of the estimated main and interaction regression coefficients). For instance, at CpG site cg11295724, an increase in DNAm at 10 years was associated with increased gene expression levels of SIRPD in males, but decreased expression in females. Similar opposite patterns were observed for the CpGs on CCDC146, SLMAP and ZNF385A. For cg03269757 on ATL2, although for both sexes, the regression coefficients were negative (−0.98 for males and −0.06 for females), the effect size for males was more than 16 times as that for females (which was close to zero), representing a potential gender reversal effect as well.
TABLE 3.
Association of DNAm at 5 CpGs with their mapped genes’ expression levels that are sex-specific
| DNAm effect |
Sex × DNAm interaction |
P-value (Sex × DNAm) |
DNAm effect |
Sex × DNAm interaction |
P-value (Sex × DNAm) |
||
|---|---|---|---|---|---|---|---|
| Gene | CpG | Age 10 y | Age 18 y | ||||
| SIRPD | cg11295724 | 0.89 | −1.39 | .022 | 0.24 | −0.42 | .45 |
|
| |||||||
| CCDC146 | cg15154628 | −0.40 | 0.72 | .041 | 0.44 | −0.43 | .39 |
|
| |||||||
| SLMAP | cg18278943 | −1.43 | 1.90 | .006 | −0.28 | 0.58 | .45 |
|
| |||||||
| ZNF385A | cg21163444 | −0.2 | 0.72 | .037 | 0.009 | −0.24 | .56 |
|
| |||||||
| ATL2 | cg03269757 | −0.66 | 0.80 | .09 | −0.98 | 0.92 | .040 |
Notes: Only results on CpGs showing statistically significant interaction effects of DNAm × Sex on gene expression were shown. Males are in the reference group. The P-values are for interaction effects, and P-values < .05 are in bold font.
4 |. DISCUSSION
We examined the sex-specificity on the association of changes in DNAm with asthma acquisition from pre-adolescence to late-(ALSPAC) or post-adolescence (IOWBC) in two birth cohorts. In IOWBC, 13 CpGs mapping to 13 genes were identified that showed statistically significant interaction effects with sex, of which 10 (77%) CpGs showed consistent directions of interaction effects in ALSPAC with one CpG (cg20891917) being statistically significant. In most of these CpGs, the effects of changes in DNAm on asthma acquisition during adolescence were opposite between males and females, showing gender reversal of DNAm effects. Accompanied by the opposite direction of changes in DNAm between males and females at most of the identified CpGs in both cohorts, this suggests that DNAm may represent a mechanism underlying the well-established gender reversal in asthma prevalence across adolescence.15,16
In addition, assessment of the biological relevance for 9 of the 10 CpGs indicated a potential of epigenetic regulatory functionality on gene activities. DNAm at 5 of the 9 CpGs showed sex-specific associations with gene expression, supporting the gender reversal phenomenon found in the association assessment between DNAm changes and asthma acquisition during adolescence. In addition, the association of gene expression with DNAm was overall stronger in males than in females, indicating a possibility of stronger influence of DNAm on gene activities in males. Previous studies have shown that ZNF385A is overexpressed and SLMAP is under expressed in asthma cases compared to controls,52 and gene CCDC146 is also identified as one of the differentially expressed genes in relation to asthma,53 although it is unclear whether such differentiation is different between males and females. The sex-specific association of DNAm with expression of these genes observed in our study may imply different underlying epigenetic regulations on gene activities between males and females. In addition, it will be interesting to examine how changes in DNAm are associated with changes in gene expressions, to further improve our understanding of epigenetic regulatory functionality on asthma acquisition.
The strength of this study is the availability of DNAm data and asthma status at two important time points, pre- and post-adolescence, enabling the possibility to examine changes in DNAm and its association with asthma acquisition during adolescence. To our knowledge, this is the first study to examine the epigenetics of asthma acquisition during adolescence regarding sex differences.
For each of the genes annotated to the identified CpGs, we performed a literature search for their possible roles that they played in the risk of asthma. Among the CpGs showing consistent direction of interaction effects, DNAm of gene ZNF385A and expression of IFRD1 have been reported as biomarkers of asthma.54 Our finding on the association of DNAm with expression of ZNF385A further strengthens its relationship with asthma. Lund et al demonstrated that NDFIP2, an IL-4 regulated gene, promoted IFN-γ production by the polarized human Th1 lymphocytes.55 One of our earlier studies in females showed that genes in the Th2 pathway were likely to contribute to an increased risk of asthma and be associated with the risk of asthma transition.4 Although in the present study we did not identify genes in the Th2 pathway, Th1 and Th2 cells work tightly and interact with other immune cells by regulating their functions with specific cytokine production, associated with the pathogenesis of asthma. As for gene ZFR, single nucleotide polymorphisms in ZFR have been shown to be associated with asthma or bronchial hyper-responsiveness.56 However, none of these studies have mentioned sex-specificity in these associations due to the focus of the study and the methods applied in the study. For instance, the study of Zhang et al4 only included females; while in the study of Kurz et al,56 the focus was to identify single nucleotide polymorphisms associated with asthma or bronchial hyper-responsiveness and the effects of sex were not considered. Furthermore, findings in the literature all focused on the risk of asthma instead of the risk of asthma acquisition, which might also explain the limited findings in the literature supporting the identified genes.
At most of the CpGs identified in IOWBC, consistent direction of interaction effects was found in ALSPAC. However, statistical significance was not observed at those CpG sites except for cg20891917. In the ALSPAC cohort, many subjects’ DNAm was assessed at 15 years and asthma status change was from 7 to 15 years. At the age of 15 years, it was likely that children were still in the period of pubertal transition, and thus sex-specificity might not be strong enough to be detected. In addition, we noticed that among the 10 CpG sites showing consistent sex-specificity between the two cohorts, associations in DNAm with expression of genes happened more often with DNAm at age 10 years. We do not have a specific biological explanation for this observation but postulate that this might have been due to larger variations in DNAm data at age 18 compared to DNAm at age 10, and thus we did not have enough power to detect the associations.
In this study, the candidate CpGs were identified based on their associations with asthma status at 10 and 18 years separately. With this approach, we were able to focus on asthma related CpG sites, which was the starting point of the study. On the other hand, we might have missed CpGs that were not related to asthma at ages 10 nor 18 years but were related to asthma acquisition from 10 to 18 years. However, screening candidate CpGs based on asthma acquisition had the risk of double-dipping the data. That is, the screening and final association analyses would share a similar analytical model applied to the same data, which in general is not encouraged. In addition, screening of CpGs and association analyses were applied to each individual CpG sites. CpG sites might be correlated and jointly impact asthma acquisition. Using our approach, correlated CpGs might have presented an issue that we were unable to address. Approaches analogous to linkage disequilibrium and haplotype identification in genetic studies deserve further investigations both methodologically and experimentally. We also would like to point out that the present study was based on a concurrent analysis (ie both DNAm changes and asthma acquisition were in the same period). The focus of the study was on associations rather than causality, and this analytical approach does not allow predictions or inferring causality.
Nevertheless, the consistency in the results between the two cohorts indicated that the identified CpGs are likely to play a role in the underlying mechanisms of sex-specific asthma acquisition. Furthermore, the sex-specific associations of DNAm at most of these CpGs with expressions of their mapped genes demonstrated their potential of biological relevance and supported our observed sex-specificity related to asthma acquisition. Although future studies are warranted to further examine the credibility of the identified CpGs, these CpGs have the potential to serve as candidate markers in subsequent mechanistic studies on gender reversal of asthma acquisition.
Supplementary Material
ACKNOWLEDGEMENTS
This study was supported by the National Institutes of Health research fund R01AI121226 (MPI: H Zhang and JW Holloway). Part of the methylation data generation was supported by R01AI091905 (PI: W Karmaus). The Isle of Wight Birth Cohort assessments have been supported by the National Institutes of Health USA (Grant no. R01HL082925, H. Arshad), Asthma UK (Grant no. 364. SH Arshad) and the David Hide Asthma and Allergy Research Trust.
The UK Medical Research Council and Wellcome (Grant ref: 217065/Z/19/Z) and the University of Bristol provide core support for ALSPAC. A comprehensive list of grants funding is available on the ALSPAC website (http://www.bristol.ac.uk/alspac/external/documents/grant-acknowledgements.pdf). Generation of methylation array data was specifically funded by NIH R01AI121226, R01AI091905, BBSRC BBI025751/1 and BB/1025263/1, MRC MC_UU_12013/1, MC_UU_12013/2, MC_UU_12013/8.
The authors are thankful to the nurses and staff at the David Hide Asthma & Allergy Research Centre, Isle of Wight, UK, for their help in recruitment and sample collections, and are thankful to all the cohort participants. Our special thanks also go to the High-Performance Computing facility provided by the University of Memphis.
For ALSPAC, DNA extraction and generation of illumina array data was carried out in the Bristol Bioresource Laboratories at the University of Bristol, UK. We are extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses.
Funding information
UK Medical Research Council and Wellcome, Grant/Award Number: 217065/Z/19/Z; National Institutes of Health, Grant/Award Number: R01AI091905, R01AI121226 and R01HL082925; Asthma UK, Grant/Award Number: 364; Biotechnology and Biological Sciences Research Council, Grant/Award Number: BBI025751/1 and BB/I025263/1; Medical Research Council, Grant/Award Number: MC_UU_12013/1, MC_UU_12013/2 and MC_UU_12013/8
Footnotes
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The IoW birth cohort study was approved by Isle of Wight, Portsmouth and Hampshire Local Research Ethics Committee (now known as the National Research Ethics Service, NRES Committee South Central—Hampshire A) (06/Q1701/34) and the IRB at the University of Memphis (FWA00006815). Written informed consent was obtained from parents during in-person visits. For participants assessed by phone interview, consent was documented on the consent form with the name of the person giving consent, and the name and signature of the person taking the form were recorded.
For ALSPAC, ethical approval for the study was obtained from the ALSPAC Ethics and Law Committee and the Local Research Ethics Committees and consent for collection of biological samples was provided in accordance with the Human Tissue Act (2004). For age seven years, United Bristol Healthcare Trust: E4168 (ALSPAC Hands on Assessments at Age Seven), Southmead Health Services: 67/98 (Avon Longitudinal Study of Pregnancy and Childhood (ALSPAC) - Hands on Assessments at Age Seven) and Frenchay Healthcare Trust: 98/52 (Avon Longitudinal Study of Pregnancy and Childhood (ALSPAC). Hands on Assessments at Age Seven). For age 15 years, Central & South Bristol Research Ethics Committee (UBHT): 06/Q2006/53 Avon Longitudinal Study of Parents and Children (ALSPAC), Hands on Assessments: Teen Focus 3 (Focus 15+), and for age 17 years, North Somerset & South Bristol Research Ethics Committee: 08/H0106/9 Avon Longitudinal Study of Parents and Children (ALSPAC), Hands on Assessments: Teen Focus 4 (Focus 17+). Full details of ethical approvals (local committees and approval numbers) are available at http://www.bristol.ac.uk/media-library/sites/alspac/documents/governance/Research%20Ethics%20Committee%20approval%20references.pdf
CONFLICT OF INTEREST
The authors declare no conflict of interest.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section.
DATA AVAILABILITY STATEMENT
The data sets analysed during the current study are not publicly available but are available from the corresponding author on reasonable request.
REFERENCES
- 1.The Global Asthma Report 2018: Global Asthma Network. 2018: Auckland, New Zealand. [Google Scholar]
- 2.Eder W, Ege MJ, von Mutius E. The asthma epidemic. N Engl J Med. 2006;355(21):2226–2235. [DOI] [PubMed] [Google Scholar]
- 3.Martinez FD, Wright AL, Taussig LM, Holberg CJ, Halonen M, Morgan WJ. Asthma and wheezing in the first six years of life. The Group Health Medical Associates. N Engl J Med. 1995;332(3):133–138. [DOI] [PubMed] [Google Scholar]
- 4.Zhang H, Tong X, Holloway JW, et al. The interplay of DNA methylation over time with Th2 pathway genetic variants on asthma risk and temporal asthma transition. Clin Epigenetics. 2014;6(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arathimos R, Granell R, Henderson J, Relton CL, Tilling K. Sex discordance in asthma and wheeze prevalence in two longitudinal cohorts. PLoS ONE. 2017;12(4):e0176293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuseini H, Newcomb DC. Mechanisms driving gender differences in asthma. Curr Allergy Asthma Rep. 2017;17(3):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Postma DS. Gender differences in asthma development and progression. Gend Med. 2007;4(Suppl B):S133–S146. [DOI] [PubMed] [Google Scholar]
- 8.Vink NM, Postma DS, Schouten JP, Rosmalen JGM, Boezen HM. Gender differences in asthma development and remission during transition through puberty: the TRacking Adolescents’ Individual Lives Survey (TRAILS) study. J Allergy Clin Immunol. 2010;126(3):498–504.e1-6. [DOI] [PubMed] [Google Scholar]
- 9.Koper I, Hufnagl K, Ehmann R. Gender aspects and influence of hormones on bronchial asthma - Secondary publication and update. World Allergy Organ J. 2017;10(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naeem A, Silveyra P. Sex differences in paediatric and adult asthma. Eur Med J (Chelmsf). 2019;4(2):27–35. [PMC free article] [PubMed] [Google Scholar]
- 11.Zein JG, Erzurum SC. Asthma is different in women. Curr Allergy Asthma Rep. 2015;15(6):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han Y-Y, Forno E, Celedón JC. Sex Steroid Hormones and Asthma in a Nationwide Study of U.S. Adults. Am J Respir Crit Care Med. 2020;201(2):158–166. 10.1164/rccm.201905-0996oc [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hohmann C, Keller T, Gehring U, et al. Sex-specific incidence of asthma, rhinitis and respiratory multimorbidity before and after puberty onset: individual participant meta-analysis of five birth cohorts collaborating in MeDALL. BMJ Open Respiratory Research. 2019;6(1):e000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soto-Ramírez N Ziyab AH, Karmaus W, et al. Epidemiologic methods of assessing asthma and wheezing episodes in longitudinal studies: measures of change and stability. J Epidemiol. 2013;23(6):399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pignataro FS, Bonini M, Forgione A, Melandri S, Usmani OS. Asthma and gender: the female lung. Pharmacol Res. 2017;119:384–390. [DOI] [PubMed] [Google Scholar]
- 16.Osman M, Hansell AL, Simpson CR, Hollowell J, Helms PJ. Gender-specific presentations for asthma, allergic rhinitis and eczema in primary care. Prim Care Respir J. 2007;16(1):28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Begin P, Nadeau KC. Epigenetic regulation of asthma and allergic disease. Allergy Asthma Clin Immunol. 2014;10(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durham AL, Wiegman C, Adcock IM. Epigenetics of asthma. Biochim Biophys Acta. 2011;1810(11):1103–1109. [DOI] [PubMed] [Google Scholar]
- 19.Ege MJ, Mayer M, Normand AC, et al. Exposure to environmental microorganisms and childhood asthma. N Engl J Med. 2011;364(8):701–709. [DOI] [PubMed] [Google Scholar]
- 20.Stein MM, Hrusch CL, Gozdz J, et al. Innate immunity and asthma risk in Amish and Hutterite farm children. N Engl J Med. 2016;375(5):411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang IV, Schwartz DA. Epigenetic mechanisms and the development of asthma. J Allergy Clin Immunol. 2012;130(6):1243–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joubert BR, Felix JF, Yousefi P, et al. DNA methylation in newborns and maternal smoking in pregnancy: genome-wide consortium meta-analysis. Am J Hum Genet. 2016;98(4):680–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joubert BR, Håberg SE, Nilsen RM, et al. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ Health Perspect. 2012;120(10):1425–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeVries A, Wlasiuk G, Miller SJ, et al. Epigenome-wide analysis links SMAD3 methylation at birth to asthma in children of asthmatic mothers. J Allergy Clin Immunol. 2017;140(2):534–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy TM, Wong CCY, Arseneault L, et al. Methylomic markers of persistent childhood asthma: a longitudinal study of asthma-discordant monozygotic twins. Clin Epigenetics. 2015;7:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arathimos R, Suderman M, Sharp GC, et al. Epigenome-wide association study of asthma and wheeze in childhood and adolescence. Clin Epigenetics. 2017;9:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gunawardhana LP, Gibson PG, Simpson JL, Benton ML, Lea RA, Baines KJ. Characteristic DNA methylation profiles in peripheral blood monocytes are associated with inflammatory phenotypes of asthma. Epigenetics. 2014;9(9):1302–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicodemus-Johnson J, Myers RA, Sakabe NJ, et al. DNA methylation in lung cells is associated with asthma endotypes and genetic risk. JCI Insight. 2016;1(20):e90151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicodemus-Johnson J, Naughton KA, Sudi J, et al. Genome-wide methylation study identifies an IL-13-induced epigenetic signature in asthmatic airways. Am J Respir Crit Care Med. 2016;193(4):376–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rastogi D, Suzuki M, Greally JM. Differential epigenome-wide DNA methylation patterns in childhood obesity-associated asthma. Sci Rep. 2013;3:2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang IV, Pedersen BS, Liu A, et al. DNA methylation and childhood asthma in the inner city. J Allergy Clin Immunol. 2015;136(1):69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang IV, Richards A, Davidson EJ, et al. The nasal methylome: a key to understanding allergic asthma. Am J Respir Crit Care Med. 2017;195(6):829–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rathod A, Duan J, Zhang H, et al. Interweaving between genetic and epigenetic studies on childhood asthma. Epigenet Insights. 2020;13:2516865720923395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forno E, Wang T, Qi C, et al. DNA methylation in nasal epithelium, atopy, and atopic asthma in children: a genome-wide study. Lancet Respir Med. 2019;7(4):336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neophytou AM, Oh SS, Hu D, et al. In utero tobacco smoke exposure, DNA methylation, and asthma in Latino children. Environ Epidemiol. 2019;3(3):e048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prunicki M, Stell L, Dinakarpandian D, et al. Exposure to NO(2), CO, and PM(2.5) is linked to regional DNA methylation differences in asthma. Clin Epigenetics. 2018;10:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reese SE, Xu CJ, den Dekker HT, et al. Epigenome-wide meta-analysis of DNA methylation and childhood asthma. J Allergy Clin Immunol. 2019;143(6):2062–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu CJ, Söderhäll C, Bustamante M, et al. DNA methylation in childhood asthma: an epigenome-wide meta-analysis. Lancet Respir Med. 2018;6(5):379–388. [DOI] [PubMed] [Google Scholar]
- 39.den Dekker HT, Burrows K, Felix JF, et al. Newborn DNA-methylation, childhood lung function, and the risks of asthma and COPD across the life course. Eur Respir J. 2019;53(4):1801795. 10.1183/13993003.01795-2018 [DOI] [PubMed] [Google Scholar]
- 40.Machin M, Amaral AFS, Wielscher M, et al. Systematic review of lung function and COPD with peripheral blood DNA methylation in population based studies. BMC Pulm Med. 2017;17(1):54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sundar IK, Yin Q, Baier BS, et al. DNA methylation profiling in peripheral lung tissues of smokers and patients with COPD. Clin Epigenetics. 2017;9:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wielscher M, Vierlinger K, Kegler U, Ziesche R, Gsur A, Weinhäusel A, et al. Diagnostic performance of plasma DNA methylation profiles in lung cancer, pulmonary fibrosis and COPD. EBioMedicine. 2015;2(8):929–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arshad SH, Holloway JW, Karmaus W, et al. Cohort profile: the Isle of Wight whole population birth cohort (IOWBC). Int J Epidemiol. 2018;47(4):1043–1044i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lehne B, Drong AW, Loh M, et al. A coherent approach for analysis of the Illumina HumanMethylation450 BeadChip improves data quality and performance in epigenome-wide association studies. Genome Biol. 2015;16:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reinius LE, Acevedo N, Joerink M, et al. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS ONE. 2012;7(7):e41361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koestler DC, Christensen B, Karagas MR, et al. Blood-based profiles of DNA methylation predict the underlying distribution of cell types: a validation analysis. Epigenetics. 2013;8(8):816–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jaffe AE, Irizarry RA. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 2014;15(2):R31. 10.1186/gb-2014-15-2-r31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13(1):86. 10.1186/1471-2105-13-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ray MA, Tong X, Lockett GA, Zhang H, Karmaus WJJ. An efficient approach to screening epigenome-wide data. Biomed Res Int. 2016;2016:2615348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boyd A, Golding J, Macleod J, et al. Cohort Profile: the ‘children of the 90s’-the index offspring of the Avon Longitudinal Study of Parents and Children. Int J Epidemiol. 2013;42(1):111–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bunyavanich SP, Gaurav; Schadt Eric. Nasal Biomarkers of Asthma, in International Bureau, W.I.P. Organization, Editor. 2017, Icahn School of Medicine at Mount Sinai: United States of America. [Google Scholar]
- 53.Maghsoudloo M, Azimzadeh Jamalkandi S, Najafi A, Masoudi-Nejad A. Identification of biomarkers in common chronic lung diseases by co-expression networks and drug-target interactions analysis. Mol Med. 2020;26(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Boever PL, Langie S. Epigenetic Markers for Respiratory Allergy, in International Bureau, W.I.P. Organization, Editor. 2016, Vito; NV. [Google Scholar]
- 55.Lund RJ, Löytömaki M, Naumanen T, et al. Genome-wide identification of novel genes involved in early Th1 and Th2 cell differentiation. J Immunol. 2007;178(6):3648. [DOI] [PubMed] [Google Scholar]
- 56.Kurz T, Hoffjan S, Hayes MG,et al. Fine mapping and positional candidate studies on chromosome 5p13 identify multiple asthma susceptibility loci .J Allergy Clin Immunol. 2006;118(2):396–402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets analysed during the current study are not publicly available but are available from the corresponding author on reasonable request.


