Abstract
With resistance to current agricultural fungicides rising, a great need has emerged for new antifungals with unexploited targets. In response, we report a novel series of diazaborines with potent activity against representative fungal plant pathogens. To identify their mode of action, we selected for resistant isolates using the model fungus Saccharomyces cerevisiae. Whole-genome sequencing of independent diazaborine-resistant lineages identified a recurring mutation in ERG25, which encodes a C-4 methyl sterol oxidase required for ergosterol biosynthesis in fungi. Haploinsufficiency and allele-swap experiments provided additional genetic evidence for Erg25 as the most biologically relevant target of our diazaborines. Confirming Erg25 as putative target, sterol profiling of compound-treated yeast revealed marked accumulation of the Erg25 substrate, 4,4-dimethylzymosterol and depletion of both its immediate product, zymosterol, as well as ergosterol. Encouraged by these mechanistic insights, the potential utility of targeting Erg25 with a diazaborine was demonstrated in soybean-rust and grape-rot models of fungal plant disease.
Introduction
Fungal plant pathogens pose a serious threat to global food security, as they can cause devastating yield loss in agriculturally important crops. Botrytis cinerea, the causative agent of gray mold, can infect more than 1,000 species of plants, including economically important fruits, such as grapes and strawberries.1 In harvested strawberries, up to 80% of the fruits can be lost to B. cinerea without fungicide treatment.2 Anthracnose, caused by Colletotrichum sublineolum, can have devastating effects on production of sorghum, one of the most widely produced cereal crops in the world as the yield loss can reach up to 86% in susceptible genotypes.3 In 2003, Phakopsora pachyrhizi, the causative agent of soybean rust, was estimated to have caused $2 billion in losses in Brazil.4 Clearly, these fungal pathogens pose a serious threat and require prompt action.
The primary approach to combating fungal plant pathogens has been deployment of agricultural fungicides.5 Among the limited number of essential processes currently targeted by such fungicides, the ergosterol biosynthetic pathway, a very highly conserved essential pathway in fungi, has been a major focus, with inhibitors in common use that target 14α-demethylase (encoded by ERG11, also known as CYP51), Δ14-reductase (encoded by ERG24), and Δ8 → Δ7-isomerase (encoded by ERG2).6,7 Unfortunately, resistance to these agents is becoming an increasingly common problem. Resistance to Erg11 inhibitors, owing to their wide deployment since the 1970s, has been well documented with a variety of resistance mechanisms reported, including ERG11 mutation or overexpression and increased drug efflux.6 In addition, alarming connections have been documented between the widespread use of Erg11 inhibitors in agriculture and selection for resistance in pathogens that can also cause devastating human disease, such as Aspergillus fumigatus. This issue is particularly problematic because Erg11 inhibitors are a mainstay of treatment for human fungal infections.8,9
Fenhexamide, a relatively recent addition to the agricultural fungicides used to control Botrytis species, inhibits 3-ketoreductase (Erg27) of the C-4 demethylation complex within the ergosterol biosynthetic pathway.10 This C-4 sterol demethylation complex is composed of three catalytic enzymes, Erg25, Erg26, and Erg27, and a transmembrane scaffold, Erg28. Together, they act to demethylate 4,4-dimethylzymosterol to yield the product, zymosterol.11,12 In French vineyards, where fenhexamide has been used as a Botrytis control agent since 2000, resistant B. cinerea isolates were detected in 2004. These resistant field isolates harbor mutations in ERG27 that confer resistance to fenhexamide.13 Given the emergence of resistance to even this newer class of agent, an urgent need exists to develop new fungicides with noncross-resistant modes of action.
Erg25 is a membrane bound C-4 methyl sterol oxidase in the C-4 demethylation complex that is conserved across eukaryotes.14 It initiates the demethylation reaction by converting a methyl group in 4,4-dimethylzymosterol to a carboxylic acid, which is then acted on by Erg26 and Erg27 to decarboxylate and reduce the sterol intermediate to 4α-methylzymosterol. This cycle is then repeated once more to produce zymosterol.7,14 Consistent with Erg25’s well-defined activity, an erg25 deletion mutant in Saccharomyces cerevisiae shows increased accumulation of the precursor metabolite, 4,4-dimethylzymosterol.14 Although Erg25 is essential under standard growth conditions, the activity of Erg 25 inhibitors has not been widely explored as an antifungal strategy. Of the few Erg25 inhibitors reported to date, PF1163A and 6-amino-2-n-pentylthiobenzothiazole (APB) have been most extensively characterized.15−17 Although APB was shown to inhibit dermatophyte growth and inhibit filamentation, a key virulence property in the human fungal pathogen Candida albicans, its utility as an antifungal for medical or agricultural applications has not been explored extensively, largely because of potency and specificity concerns.18,19
The diazaborine family of boron-containing heterocycles has been investigated for inhibition of diverse cellular targets.20 Boron’s ability to interact with proteins through either trigonal planar sp2 or tetrahedral sp3 states makes boron-containing compounds a unique source of novel inhibitors of many processes.20 Diazaborine-based compounds studied to date include inhibitors of bacterial enoyl-ACP reductase, human neutrophil elastase, and yeast AAA-ATPase.21−24 Here, we synthesized a small series of diazaborines with potent activity against several major plant pathogens in culture. We then used the model fungus S. cerevisiae to identify candidate cellular targets by whole-genome sequencing of resistant mutants. This approach identified a recurring mutation in ERG25 in independent resistant lineages, suggesting Erg25 as the most likely target of our diazaborines. This hypothesis was confirmed through haploinsufficiency and allele-swap experiments. Supporting the strong genetic evidence for Erg25 as putative target of our diazaborines, whole-cell sterol profiling of cells exposed to diazaborine showed marked accumulation of the Erg25 substrate, 4,4-dimethylzymosterol and depletion of its product, zymosterol, as well as depletion of ergosterol, the mature end-product of sterol biosynthesis in fungi. Encouraged by these mechanistic insights, we concluded by demonstrating the potential utility of targeting Erg25 for crop-protection in a soybean-rust model of plant disease.
Results and Discussion
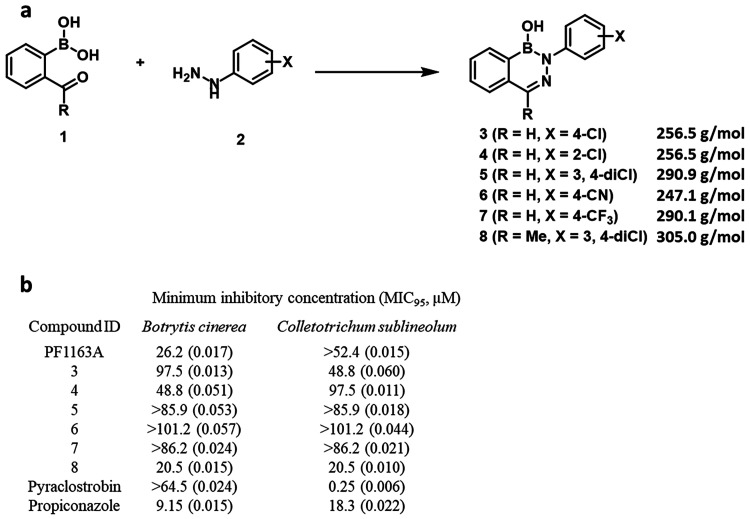
We identified six structurally related diazaborine analogues in a high-throughput screening campaign with a range of activity against plant fungal pathogens. These diazaborines were synthesized in acceptable yield under nonoptimized conditions by combining boronic acid 1 and the corresponding hydrazine 2 in aqueous media at RT (Figure 1a and Supplementary File 1)
Figure 1.
Synthesis and antifungal activity of diazaborines: (a) Reaction scheme for synthesis compounds. (b) Sensitivity of fungal pathogens to growth inhibition by diazaborines. Minimum concentration of compound required to inhibit growth by 95% (MIC95) after 3 days in culture was determined by averaging optical density (OD600) measurements from three technical replicates. Standard deviation of the mean is indicated in parentheses.
Compounds were stable for greater than one year as stock solutions in DMSO despite repeated freeze–thaw cycles, consistent with prior literature reporting stability of the scaffold to boiling in acid or base.25,26
All diazaborine analogues, along with PF1163A (Erg25 inhibitor), and the established agricultural fungicides pyraclostrobin (respiration inhibitor) and propiconazole (Erg 11 inhibitor) as comparators, were tested for in vitro antifungal activity against B. cinerea and C. sublineolum (Figure 1b). Compounds 3, 4, and 8 showed minimum inhibitory concentrations (MIC95) between 20.5 μM and 97.5 μM, whereas 5, 6, and 7 were inactive (MIC95 beyond highest concentrations tested).
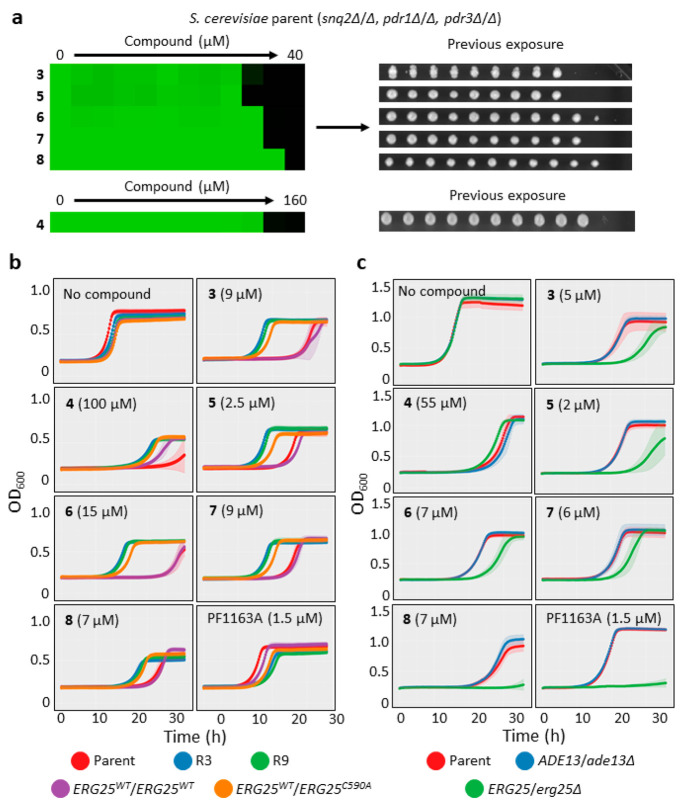
Encouraged by the promising antifungal activity of our diazaborines, we began to define their mode of action by selecting for clonal isolates resistant to 3, as resistance is often accompanied by mutations in the gene encoding a compound’s target or genes encoding proteins within the pathway targeted by a compound. We used the model fungus, S. cerevisiae because of its well-annotated genome and the wide variety of genomic tools available. To increase the likelihood of identifying gain-of-function mutations in the target gene and reduce the likelihood of nontarget related resistance mediated by increased efflux, we utilized a diploid strain with three genes related to ATP-binding cassette (ABC) transporter function deleted from its genome: the ABC transporter gene SNQ2, and the transcriptional regulators PDR1 and PDR3.27 This strain was 2-fold more sensitive to compound 3 when compared to its parent strain and sensitive to all diazaborines with MIC95 values between 80 μM to 10 μM in liquid culture (Supplementary File 4, Figure 2a, left). Furthermore, when yeast that had been exposed to compounds at concentrations exceeding the MIC95 were spotted onto compound-free growth medium, subsequent outgrowth was abolished. This result indicates a fungicidal mode of action for the compounds and is consistent with the known essentiality of ERG25 (Figure 2a, right).
Figure 2.
Genetic evidence for Erg25 as target of diazaborines: (a) Sensitivity of an efflux pump-compromised strain of the model fungus S. cerevisiae to diazaborines; the diploid parental strain harbors homozygous deletion of SNQ2, PDR1, and PDR3. (Left) Growth in liquid culture as measured by optical density (OD600) in the presence of a 2-fold dilution series of the indicated compounds. Relative growth is presented in heat-map format. Scale bar is provided below panel; dark green represents full growth (1) and black represents no growth (0) relative to no-compound control wells. Each colored box represents the mean of duplicate determinations. (Right) Following measurement of relative growth, cells were spotted on compound-free YPD agar to determine viability. The entire experiment was repeated once with quantitatively similar results. (b) Sensitivity testing of diazaborine-resistant S. cerevisiae mutants isolated by selection and an ERG25 mutant generated by recombinant engineering. Growth was monitored by measurement of OD600 over time in the presence or absence of the indicated concentrations of compounds using the parent strain, diazaborine-resistant mutants selected in culture (isolates R3 and R9), ERG25 point mutant constructed by recombinant technology, and its marker-matched control strain. Results are the average of three technical replicates and representative of two independent experiments. Error bars represent standard deviation of the technical replicates. (c) Reduced dosage of ERG25confers hypersensitivity to the diazaborines. Growth was monitored as described in (b) using the diploid parental strain and heterozygous deletion mutants constructed in the same background. Results are the average of three technical replicates and representative of two independent experiments. Error bars represent standard deviation of the technical replicates.
We recovered two independent lineages upon selection in the presence of 3 (R3 and R9). When we assessed relative resistance by performing growth curve assays, these lineages were resistant to all the diazaborines as compared with the parental strain, suggesting that the compounds likely share a common mode of action (Figure 2b). To identify mutations associated with the resistance phenotype, we sequenced the genomes of these two lineages. When compared to the genome sequence of the parental strain, we identified an identical heterozygous mutation, ERG25/ERG25C590A, in both resistant lineages leading to an Erg25S197Y substitution. We went on to demonstrate that the ERG25C590A mutation is sufficient to confer resistance by introducing the mutation into the parental strain. When compared to a hygromycin B-resistance marker-matched control strain, the mutation conferred a level of resistance similar to that seen in the resistant lineages isolated in selection experiments (Figure 2b). It did not confer resistance to the known inhibitor of Erg25, PF1163A, which suggests a divergent binding mode for this compound.17
To further assess the relevance of ERG25 to the activity of diazaborines, we tested for hypersensitivity of an ERG25/erg25Δ heterozygous deletion strain to our compounds and PF1163A. We predicted that if the diazaborines inhibit Erg25, reduced gene dosage would confer hypersensitivity. Compared with the parental strain or a control strain with a heterozygous deletion of a gene that is not involved in ergosterol biosynthesis (ADE13/ade13Δ), the ERG25/erg25Δ strain was hypersensitive to diazaborines and also PF1163A (Figure 2c). Taken together, genetic data strongly support Erg25 as the target responsible for the fungicidal activity of our diazaborines.
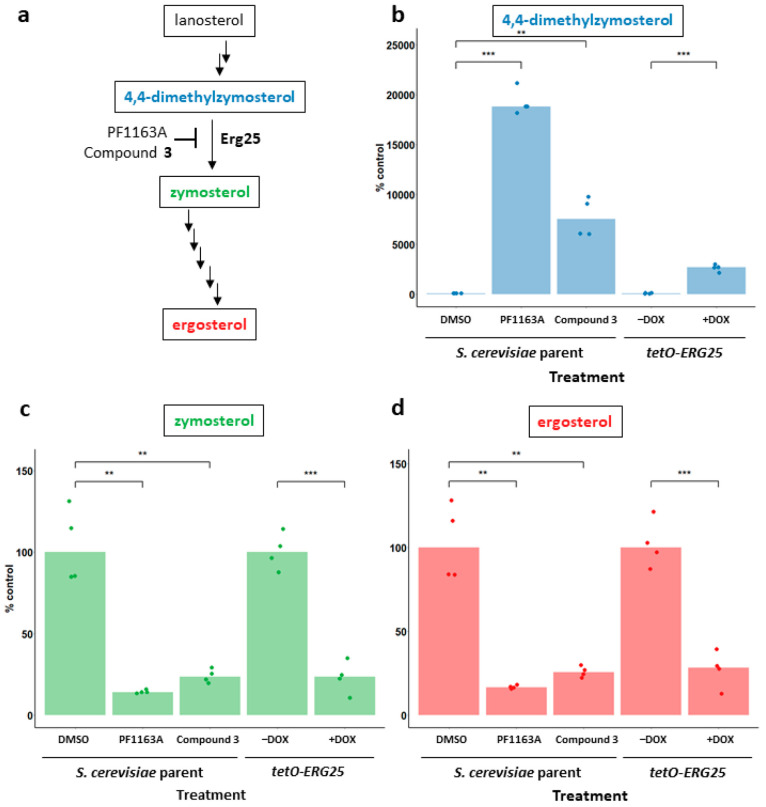
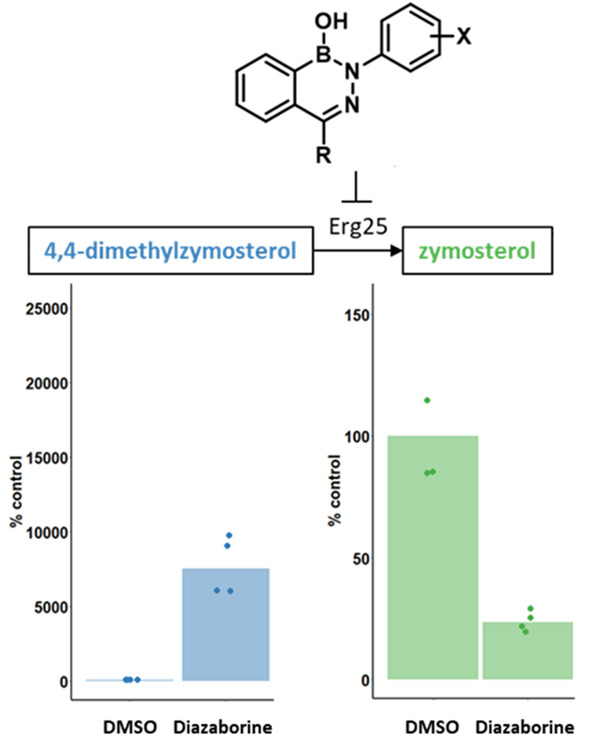
Erg25 initiates the C-4 demethylation of 4,4-dimethylzymosterol.14 If the diazaborines inhibit Erg25, the accumulation of 4,4-dimethylzymosterol would be expected, while the product of the C-4 demethylation complex, zymosterol, and the final product of the entire pathway, ergosterol, should be depleted (Figure 3a). To determine whether these effects do indeed occur upon diazaborine exposure, we treated S. cerevisiae with PF1163A or compound 3, one of the most bioactive compounds in S. cerevisiae, and we profiled the sterol content of extracts using gas chromatography–mass spectrometry (GC/MS). As a genetic control, we included a strain in which the sole copy of ERG25 is under the control of a tetracycline-repressible promoter (tetO) and examined its sterol profile following growth in the absence and presence of doxycycline (∓DOX).28 As expected based on our genetic findings, PF1163A and compound 3 treatment caused a clear accumulation of 4,4-dimethylzymosterol and depletion of both zymosterol and ergosterol compared with control vehicle-treated cells (Figure 3b–d). A similar pattern was seen with the tetO-ERG25 strain, where the repression of ERG25 expression by the addition of DOX also caused accumulation of 4,4-dimethylzymosterol and depletion of zymosterol and ergosterol.
Figure 3.
Diazaborine treatment causes accumulation of Erg25 substrate, 4,4-dimethylzymosterol, and depletion of downstream products, zymosterol and ergosterol: (a) Simplified schematic of the ergosterol biosynthesis pathway in fungi to highlight the reaction catalyzed by Erg25. Relative cellular levels of (b) 4,4-dimethylzymosterol, (c) zymosterol, and (d) ergosterol were calculated by normalizing peak area of the indicated sterols to a cholesterol internal standard and the wet cell weight of the sample prior to processing. The effect on sterol levels of growth in the presence of each compound is presented relative to sterol levels in cells grown in the presence of vehicle alone (% control). The median value of two technical replicates from two biological replicates is depicted by the height of each bar and values from each experimental determination are indicated by dots. * p < 0.05, ** p < 0.01, *** p < 0.001, t test.
To begin investigating the potential utility of diazaborines as agricultural fungicides, we examined the ability of compound 3 to limit disease progression in models of infection caused by P. pachyrhizi, an obligate parasite of soybean leaves, and B. cinerea which causes grape-rot.29 To do so, we sprayed soybean plants and grapes with 125 μg/mL of compound 3 or 675 μg/mL propiconazole, as per the manufacturer’s recommendations, and then inoculated the subject plant materials with P. pachyrhizi or B. cinerea respectively. The soybean leaves and grapes were monitored for disease post inoculation, and relative disease inhibition was determined as previously described.30 Compound 3 was able to inhibit damage caused by P. pachyrhizi at a concentration lower than that required for the conventional agent propiconazole to achieve a similar level of protection in soybean leaves. It also provided protection against grape-rot caused by B. cinerea (Figure 4).
Figure 4.
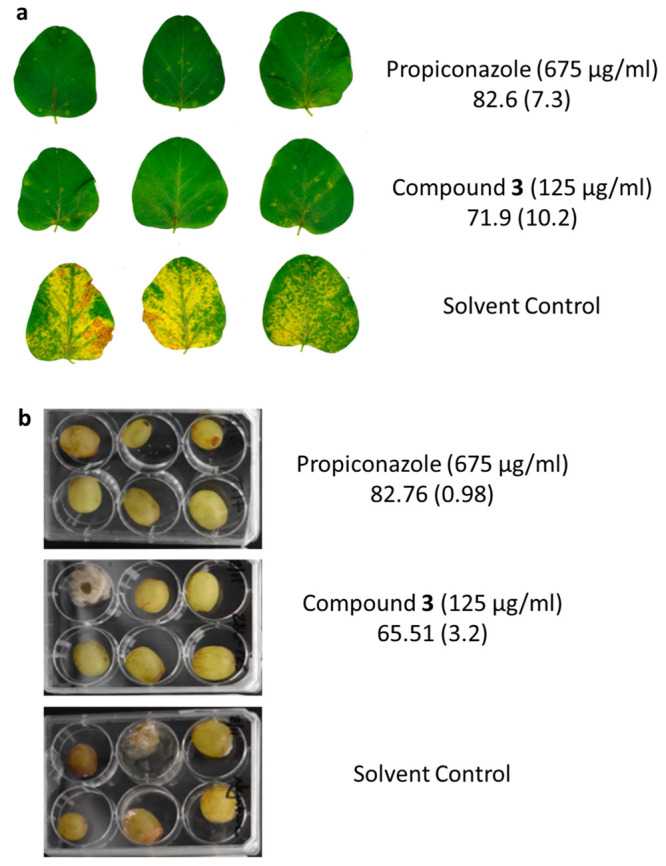
Compound 3 inhibits disease progression caused by fungal plant pathogens. Numbers below treatment indicate percent disease inhibition relative to solvent control with standard deviation indicated in parentheses. (a) P. pachyrhizi on soybean leaves: Representative images of soybean leaves postinfection at the time of scoring for disease severity. (b) Images of B. cinerea infected grapes in a fruit-rot model: Representative images of grapes at the time of scoring.
While we did not see evidence for acute toxicity in our plant models, C-4 methyl sterol oxidases are reported as essential for developmental processes in plants.31 The lack of observed toxicity could reflect the fact that compound 3 was only applied to mature tissues or that compound 3 selectively inhibits fungal Erg25 compared with its counterpart in plants. Characterization of another Erg25 inhibitor, APB, has shown that the compound is 500-fold more effective at inhibiting the fungal C-4 methyl sterol oxidase over its plant homologue, suggesting that achieving fungal selectivity at this target is possible and could provide a promising avenue for developing Erg25 inhibitors as agricultural fungicides.15
Although compounds targeting distinct steps in the ergosterol biosynthetic pathway of fungi are known, only a few chemotypes, as represented by PF1163A and APB have been characterized as Erg25 inhibitors. Both these compounds suffer from significant liabilities and neither is being developed for use as an agricultural fungicide or clinical antifungal.15−17 Given the urgent need for discovery of antifungal compounds with novel targets, the diazaborines reported here could provide a new lead for further development of an effective Erg25 inhibitor. Beyond their fungicidal activity as single agents, diazaborine Erg25 inhibitors also offer the potential for combination with established agents to target the biosynthetic pathway for ergosterol at distinct points. Precedent for this type of approach to increase efficacy and reduce the emergence of resistance to antimicrobials is well established, most notably in the practice of combining folate pathway inhibitors to treat malaria and bacterial infections.32,33 Multiple candidates for combination with a diazaborine are already used in agriculture including azoles (Erg11 inhibitors), morpholines (Erg24 inhibitors) and fenhexamid, as mentioned above, an inhibitor of Erg27.
Here, we harnessed the power of yeast genetics to establish that the diazaborines inhibit Erg25. We observed differences in the MIC values of our six diazaborines, despite their sharing of a common benzodiazaborine core structure (Figure 1a). For example, the ortho-substituted chlorobenzene group in 4 diminished its whole cell activity against S. cerevisiae by 8-fold compared with the para-substituted chlorobenzene group in 3 (Figure 1b). Interestingly, 4 did not show a similar pattern of diminished activity against B. cinerea or C. sublineolum. The differences observed in relative activity between the analogues could be due to diverse factors such as variant target affinity, compound metabolism, permeability, and/or efflux in fungal plant pathogens compared to our pump-compromised S. cerevisiae strain. These factors could be deconvoluted in future development work through in vitro enzymatic assays and measurement of intracellular compound accumulation. Taken together, our results highlight the promise of novel diazaborines as inhibitors of the relatively underexplored target, Erg25 and with further development, their potential as agricultural fungicides.
Experimental Section
General Chemical Methods
Commercial reagents and solvents were used as supplied. Low-resolution mass spectroscopy was carried out using liquid chromatography/mass spectrometry (LC/MS) on an Agilent instrument using electrospray ionization (ESI). (1H NMR) magnetic resonance spectra were obtained on a Bruker instrument in CDCl3 or DMSO-d6 at 400 MHz at 298 K unless otherwise noted. Compounds used in biological studies were >95% pure, as determined by HPLC based on ultraviolet detection at 210 or 254 nm. Details regarding synthesis and characterization of compounds are provided in Supporting Information File 1.
Fungal Plant Pathogen MIC Testing
The minimal inhibitory concentration (MIC) was determined using a modified broth microdilution protocol. MIC was defined as the lowest concentration that inhibited fungal growth by greater than 95% (determined as relative absorbance using the Bio-Tek Synergy H1 microplate reader at 600 nm) relative to the corresponding antifungal-free control.
Sensitivity Testing of S. cerevisiae Strains
Cells (1000/well) were inoculated into 2-fold compound gradients in 384-well plates (final assay volume: 40 μL). Plates were incubated at 30 °C for 48 h before measurement of OD600 using SpectraMax M2e (Molecular Devices). Relative growth was calculated by normalizing measurements against no-compound control wells. Results were plotted in heat-map format using R. To assess cell viability, after reading OD600, cultures were spotted on YPD agar and incubated at 30 °C for 24 h before imaging.
Selection and Whole-Genome Sequencing of Resistant Yeast Isolates
Approximately 1 × 108 yeast were plated on YPD agar containing 80 μM of Compound 3 and incubated at 30 °C for 48 h prior to isolation and subculture of resistant colonies. Whole-genome sequencing of resistant clones was performed as described in Supporting Information File 5.
Soybean Rust P. pachyrhizi Greenhouse Assay
Twelve soybean plants were grown for each compound tested. After emergence (Day 14, V2 stage), compound 3 was dissolved in a 50:50 acetone to water mixture and applied evenly to each plant at 125 μg/mL and propiconazole was applied at 675 μg/mL per label instructions by the manufacturer via track spray booth (Devries Manufacturing Inc., Hollandale, MN). After drying for 24 h, plants were inoculated by soaking to runoff with a 1 × 105 spores/ml spore suspension of P. pachyrhizi. To prepare inoculum, spores were harvested from live plants using a 0.1% Tween 20 solution. The second true leaf of each plant was given a disease rating based on % coverage by rust pustules at 14 days postinoculation. Scores were transformed into % disease inhibition relative to the solvent-control plants.
B. cinerea Gray Mold Ex Vivo Assay
Six green grapes (cultivars vary based on grocery store availability) were sterilized in 10% sodium hypochlorite, then triple-rinsed in ddH2O and patted dry with sterilized paper towel. Compounds were dissolved in a 50:50 acetone to water mixture, then applied at 125 ppm, after which each grape was placed in an individual well of a 6-well polystyrene plate and left to dry. Five hours after application of compound, each grape was inoculated with 0.5 mL (1 × 106 spores/ml) of a B. cinerea spore suspension, prepared from cultures grown in potato dextrose broth. Plates were incubated at 20 °C for 7 days and then assessed for percent infection of each grape. The severity of gray mold disease was assessed on a scale of 0 (healthy) to 10 (grape covered in mycelium, wrinkled, and shrunken).34 These scores were then transformed into percent disease inhibition relative to grapes treated with the solvent control.
Yeast Growth Curve Analyses
Approximately 1000 cells/well of appropriate heterozygous deletion strains in YPD medium were incubated with a single compound concentration in 384-well format (final volume 40 μL/well) and sealed with Breathe-Easy Sealing Film (Diversified Biotech). The plates were incubated at 30 °C in an Infinite 200 PRO plate reader (TECAN) and OD600 was measured every 15 min for 36 h. A growth curve based on OD600 was plotted using R.
Quantification of Sterols Using GC-MS
After sterol extraction and derivatization for GC-MS (Supplementary File 5), measurements were obtained using a GC/MS system (HP7890/HP5975, Agilent Technologies) coupled with Masshunter Quantitative Analysis software including the NIST Library. Sterol identification was based on the comparison of mass spectra and retention time data with available standards in the NIST Library. Additional details are provided in Supplementary File 5.
Acknowledgments
We thank Cowen lab members for helpful discussions, and the Donnelly Sequencing Centre for whole genome sequencing. We thank the Boone lab for providing the drug-sensitized S. cerevisiae strain and the heterozygous deletion library in the same background. We also thank the Hughes lab for providing the S. cerevisiae tetracycline-repressible promoter library.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acschembio.2c00257.
Supporting Information File 1: This file contains descriptions of the methods used to synthesize the six compounds that are presented in this study, their chemical structures and the results of their characterization by NMR and LC/MS. It also contains a table summarizing the LC/MS method that was used to characterize the compounds. Supporting Information File 2: This file contains NMR spectra of the six compounds that were synthesized for this study. Supporting Information File 3: This file contains LC-UV spectra obtained at 214 and 254 nm for the six compounds that were synthesized for this study. It also contains MS spectra for the same compounds. Supporting Information File 4: This file contains the results of testing BN100286 (Compound 3) in MIC assays and spotting assays for fungicidal activity against wild-type and drug-sensitized (snq2Δ pdr1Δ pdr3Δ) strains of S. cerevisiae. Supporting Information File 5: This file describes in detail the experimental methods used for culture of fungi, MIC testing of fungi, yeast strain construction, whole genome sequencing of compound-resistant isolates. It also provides culture conditions and methods for sterol extraction, derivatization and quantification by GC-MS. Supporting Information File 6: This file contains a tabular summary of the fungal strains used in this study including their genotypes and references to their source if not newly constructed in support of this work. It also provides a description of how new strains were constructed, and how their genotypes were confirmed by PCR and Sanger sequencing. Descriptions of the plasmids and oligonucleotides used in support of strain construction are also provided. Supporting Information File 7: This file provides complete references to all literature cited within these supplementary files (PDF)
S.H.K. is supported by a Mitacs Accelerate Fellowship (IT13564) in association with Boragen Inc.; L.E.C. is supported by the Canadian Institutes of Health Research Foundation Grant (FDN- 154288); L.E.C. is a Canada Research Chair (Tier 1) in Microbial Genomics & Infectious Disease and codirector of the CIFAR Fungal Kingdom: Threats & Opportunities program.
The authors declare the following competing financial interest(s): L.E.C. and L.W. are co-founders and shareholders in Bright Angel Therapeutics, a platform company for development of novel antifungal therapeutics.
Supplementary Material
References
- Williamson B.; Tudzynski B.; Tudzynski P.; Van Kan J. A. L. Botrytis cinerea: The Cause of Grey Mould Disease. Mol. Plant Pathol. 2007, 8 (5), 561–580. 10.1111/j.1364-3703.2007.00417.x. [DOI] [PubMed] [Google Scholar]
- Petrasch S.; Knapp S. J.; van Kan J. A. L.; Blanco-Ulate B. Grey Mould of Strawberry, a Devastating Disease Caused by the Ubiquitous Necrotrophic Fungal Pathogen Botrytis cinerea. Mol. Plant Pathol. 2019, 20 (6), 877–892. 10.1111/mpp.12794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutts L. R.; Vermerris W. Elucidating Anthracnose Resistance Mechanisms in Sorghum—A Review. Phytopathology® 2020, 110 (12), 1863–1876. 10.1094/PHYTO-04-20-0132-RVW. [DOI] [PubMed] [Google Scholar]
- Yorinori J. T.; Paiva W. M.; Frederick R. D.; Costamilan L. M.; Bertagnolli P. F.; Hartman G. E.; Godoy C. V.; Nunes J. Epidemics of Soybean Rust (Phakopsora pachyrhizi) in Brazil and Paraguay from 2001 to 2003. Plant Dis. 2005, 89 (6), 675–677. 10.1094/PD-89-0675. [DOI] [PubMed] [Google Scholar]
- Lucas J. A.; Hawkins N. J.; Fraaije B. A. The Evolution of Fungicide Resistance. Advances in applied microbiology 2015, 90, 29–92. 10.1016/bs.aambs.2014.09.001. [DOI] [PubMed] [Google Scholar]
- Ziogas B. N.; Malandrakis A. A.. Sterol Biosynthesis Inhibitors: C14 Demethylation (DMIs). In Fungicide Resistance in Plant Pathogens; Ishii H., Hollomon D., Eds.; Springer Japan: Tokyo, 2015; pp 199–216. 10.1007/978-4-431-55642-8_13. [DOI] [Google Scholar]
- Debieu D.; Leroux P.. Sterol Biosynthesis Inhibitors: C-4 Demethylation. In Fungicide Resistance in Plant Pathogens; Ishii H., Hollomon D., Eds.; Springer Japan: Tokyo, 2015; pp 217–231. 10.1007/978-4-431-55642-8_14. [DOI] [Google Scholar]
- Berger S.; El Chazli Y.; Babu A. F.; Coste A. T. Azole Resistance in Aspergillus fumigatus: A Consequence of Antifungal Use in Agriculture?. Front. Microbiol. 2017, 8, 1024. 10.3389/fmicb.2017.01024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meis J. F.; Chowdhary A.; Rhodes J. L.; Fisher M. C.; Verweij P. E. Clinical Implications of Globally Emerging Azole Resistance in Aspergillus fumigatus. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371 (1709), 20150460. 10.1098/rstb.2015.0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debieu D.; Bach J.; Hugon M.; Malosse C.; Leroux P. The Hydroxyanilide Fenhexamid, a New Sterol Biosynthesis Inhibitor Fungicide Efficient against the Plant Pathogenic Fungus Botryotinia fuckeliana (Botrytis cinerea). Pest Manag. Sci. 2001, 57 (11), 1060–1067. 10.1002/ps.394. [DOI] [PubMed] [Google Scholar]
- Baudry K.; Swain E.; Rahier A.; Germann M.; Batta A.; Rondet S.; Mandala S.; Henry K.; Tint G. S.; Edlind T.; et al. The Effect of the Erg26–1 Mutation on the Regulation of Lipid Metabolism in Saccharomyces cerevisiae. J. Biol. Chem. 2001, 276 (16), 12702–12711. 10.1074/jbc.M100274200. [DOI] [PubMed] [Google Scholar]
- Mo C.; Valachovic M.; Randall S. K.; Nickels J. T.; Bard M. Protein-Protein Interactions among C-4 Demethylation Enzymes Involved in Yeast Sterol Biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 2002, 99 (15), 9739–9744. 10.1073/pnas.112202799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillinger S.; Leroux P.; Auclair C.; Barreau C.; Al Hajj C.; Debieu D. Genetic Analysis of Fenhexamid-Resistant Field Isolates of the Phytopathogenic Fungus Botrytis cinerea. Antimicrob. Agents Chemother. 2008, 52 (11), 3933–3940. 10.1128/AAC.00615-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard M.; Bruner D. A.; Pierson C. A.; Lees N. D.; Biermann B.; Frye L.; Koegel C.; Barbuch R. Cloning and Characterization of ERG25, the Saccharomyces cerevisiae Gene Encoding C-4 Sterol Methyl Oxidase. Proc. Natl. Acad. Sci. U. S. A. 1996, 93 (1), 186–190. 10.1073/pnas.93.1.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnet S.; Rahier A. Enzymological Properties of Sterol-C4-Methyl-Oxidase of Yeast Sterol Biosynthesis. Biochim. Biophys. Acta - Mol. Cell Biol. Lipids 2003, 1633 (2), 106. 10.1016/S1388-1981(03)00093-3. [DOI] [PubMed] [Google Scholar]
- Kuchta T.; Barková K.; Kubinec R. Ergosterol Depletion and 4-Methyl Sterols Accumulation in the Yeast Saccharomyces cerevisiae Treated with an Antifungal, 6-Amino-2-n-Pentylthiobenzothiazole. Biochem. Biophys. Res. Commun. 1992, 189 (1), 85–91. 10.1016/0006-291X(92)91529-Y. [DOI] [PubMed] [Google Scholar]
- Nose H.; Fushimi H.; Seki A.; Sasaki T.; Watabe H.; Hoshiko S. PF1163A, a Novel Antifungal Agent, Inhibit Ergosterol Biosynthesis at C-4 Sterol Methyl Oxidase. J. Antibiot. (Tokyo). 2002, 55 (11), 969. 10.7164/antibiotics.55.969. [DOI] [PubMed] [Google Scholar]
- Bujdáková H.; Múčková M.; Klobušický M.; Sidóová E. Efficacy of 6-Amino-2-n-Pentylthiobenzothiazole on Trichophyton in vitro and in vivo. Mycopathologia 1995, 130 (3), 141–145. 10.1007/BF01103096. [DOI] [PubMed] [Google Scholar]
- Fábry S.; Gáborová S.; Bujdáková H.; Klobušický M.; Volleková A.; Kuchta T. Inhibition of Germ Tube Formation, Filamentation and Ergosterol Biosynthesis in Candida albicans Treated with 6-Amino-2-n-Pentylthiobenzothiazole. Folia Microbiol. (Praha). 1999, 44 (5), 523–526. 10.1007/BF02816254. [DOI] [PubMed] [Google Scholar]
- Fernandes G. F. S.; Denny W. A.; Dos Santos J. L. Boron in Drug Design: Recent Advances in the Development of New Therapeutic Agents. Eur. J. Med. Chem. 2019, 179, 791. 10.1016/j.ejmech.2019.06.092. [DOI] [PubMed] [Google Scholar]
- Högenauer G.; Woisetschläger M. A Diazaborine Derivative Inhibits Lipopolysaccharide Biosynthesis. Nature 1981, 293 (5834), 662. 10.1038/293662a0. [DOI] [PubMed] [Google Scholar]
- Baldock C.; Rafferty J. B.; Sedelnikova S. E.; Baker P. J.; Stuitje A. R.; Slabas A. R.; Hawkes T. R.; Rice D. W. A Mechanism of Drug Action Revealed by Structural Studies of Enoyl Reductase. Science (80-.). 1996, 274 (5295), 2107. 10.1126/science.274.5295.2107. [DOI] [PubMed] [Google Scholar]
- António J. P. M.; Gonçalves L. M.; Guedes R. C.; Moreira R.; Gois P. M. P. Diazaborines as New Inhibitors of Human Neutrophil Elastase. ACS Omega 2018, 3 (7), 7418. 10.1021/acsomega.8b00702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loibl M.; Klein I.; Prattes M.; Schmidt C.; Kappel L.; Zisser G.; Gungl A.; Krieger E.; Pertschy B.; Bergler H. The Drug Diazaborine Blocks Ribosome Biogenesis by Inhibiting the AAA-ATPase Drg1. J. Biol. Chem. 2014, 289 (7), 3913. 10.1074/jbc.M113.536110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stress C. J.; Schmidt P. J.; Gillingham D. G. Comparison of Boron-Assisted Oxime and Hydrazone Formations Leads to the Discovery of a Fluorogenic Variant. Org. Biomol. Chem. 2016, 14 (24), 5529–5533. 10.1039/C6OB00168H. [DOI] [PubMed] [Google Scholar]
- Dilek O.; Lei Z.; Mukherjee K.; Bane S. Rapid Formation of a Stable Boron–Nitrogen Heterocycle in Dilute, Neutral Aqueous Solution for Bioorthogonal Coupling Reactions. Chem. Commun. 2015, 51 (95), 16992–16995. 10.1039/C5CC07453C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski J. S.; Li S. C.; Deshpande R.; Simpkins S. W.; Nelson J.; Yashiroda Y.; Barber J. M.; Safizadeh H.; Wilson E.; Okada H.; et al. Functional Annotation of Chemical Libraries across Diverse Biological Processes. Nat. Chem. Biol. 2017, 13 (9), 982–993. 10.1038/nchembio.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mnaimneh S.; Davierwala A. P.; Haynes J.; Moffat J.; Peng W. T.; Zhang W.; Yang X.; Pootoolal J.; Chua G.; Lopez A. Exploration of Essential Gene Functions via Titratable Promoter Alleles. Cell 2004, 118 (1), 31. 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Twizeyimana M.; Hartman G. L. Culturing Phakopsora pachyrhizi on Detached Leaves and Urediniospore Survival at Different Temperatures and Relative Humidities. Plant Dis. 2010, 94 (12), 1453–1460. 10.1094/PDIS-02-10-0131. [DOI] [PubMed] [Google Scholar]
- Franceschi V. T.; Alves K. S.; Mazaro S. M.; Godoy C. V.; Duarte H. S. S.; Del Ponte E. M. A New Standard Area Diagram Set for Assessment of Severity of Soybean Rust Improves Accuracy of Estimates and Optimizes Resource Use. Plant Pathol. 2020, 69 (3), 495–505. 10.1111/ppa.13148. [DOI] [Google Scholar]
- Rahier A. Dissecting the Sterol C-4 Demethylation Process in Higher Plants. from Structures and Genes to Catalytic Mechanism. Steroids. 2011, 76, 340. 10.1016/j.steroids.2010.11.011. [DOI] [PubMed] [Google Scholar]
- Estrada A.; Wright D. L.; Anderson A. C. Antibacterial Antifolates: From Development through Resistance to the Next Generation. Cold Spring Harb Perspect Med. 2016, 6 (8), a028324. 10.1101/cshperspect.a028324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell A. N.; Winzeler E. A. The genomic architecture of antimalarial drug resistance. Brief Funct Genomics. 2019, 18 (5), 314–328. 10.1093/bfgp/elz008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Glawe D. A.; Kramer E.; Weller D.; Okubara P. A. Biological Control of Botrytis cinerea: Interactions with Native Vineyard Yeasts from Washington State. Phytopathology. 2018, 108 (6), 691–701. 10.1094/PHYTO-09-17-0306-R. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.