Abstract
Background:
Breast Cancer (BC), the second leading cause of cancer mortality after lung cancer and varied across the world due to genetic and environmental factors. In this study, we evaluated the interaction between the polymorphisms in genes encoding enzymes of folate metabolism: methylenetetrahydrofolate reductase (MTHFR), methionine synthesis reductase (MTR) with the BC prognostic factors.
Methods:
This study was conducted on 160 Egyptian subjects, 60 controls and 100 cases. Sequencing, RFLP analysis in addition to statistical analysis including Chi‐squared test, haplotype analysis was used to evaluate associations with BC risk and its clinicopathological parameters. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated using unconditional logistic regression.
Results:
Strong significant association with breast cancer risk was observed for the haplotype (T-C-G) of MTHFR C677T/ MTHFR A1289C and MTRA2576G and hormonal receptor expression (ER-/PR-/HER2+), bigger and advanced tumor and metastatic lymph nodes. However, no significant difference was observed for age.
Conclusion:
The combination of SNPs from MTHFR and MTR genes has a more synergistically genetic effect on BC disease progression. These SNPs could be used as tumor aggressiveness markers among Egyptian females with BC and could help in saving money and time.
Key Words: Breast cancer, Methionine synthesis reductase, MTHFR, PCR-RFLP, SNPs
Introduction
Globally, Breast cancer (BC) is one of the main causes of mortality among females (1). In Egypt, BC ranks the second most common cancer after liver cancer, accounting for 15.4% of all cancers (2). In spite of its high prevalence, the pathogenesis and the etiology of BC are still poorly understood. Although obesity, radiation and hormone replacement therapy were already identified as prospective risk factors of developing BC, other factors such as familiar history, genetic factors, age of menopause, changes in estrogen level, and smoking could be involved (3).
Epidemiological evidence suggests that folate metabolism imbalance may be involved in predisposition to cancer. Folate metabolism is a series of communicated pathways which is essential for nucleic acid, amino acid synthesis and DNA repair through folate and methionine cycles. Genome-wide association studies (GWASs), indicate several genetic variations in different genes involved in critical pathways that responsible for diseases onset, mortality, and survival rate (4). Genetic polymorphisms of folate metabolism enzymes such as Methionine synthase reductase (MTRR), and 5,10‐methylenetetrahydrofolate reductase (MTHFR) could result in altered folate level (5,6). Furthermore, Methionine synthase (MTR), an enzyme-dependent on vitamin B12, plays a significant role in the folate–methylation cycle. These DNA methylation reactions are critical for gene expression and genome integrity. Failure in folate metabolizing genes and the impairment of methylation leads to the activation of proto-oncogenes, down regulation of tumor suppressor genes, and chromosomal segregation abnormalities during cellular division. All these processes are involved in carcinogenesis and may be associated with the risk of breast cancer (5, 7).
In this study, our focus is to examine the interaction of the Polymorphisms in genes encoding enzymes of folate metabolism as MTHFR C677T and A1298C with MTR A2756G. These enzymes interact through their substrates with an imbalance due to the combination of their polymorphisms. This can be more effective than that of single polymorphisms and might help in determining the routes of DNA synthesis, repair, or methylation. This would help in giving better explanation of the relevance of these polymorphisms and their association with BC and its aggressive clinical effects in a prior study among the Egyptian females. This could reveal their utility as genetic markers of BC.
Materials and Methods
Subjects
This study was executed on 160 Egyptian females, 60 controls and 100 cases, who were clinically and histologically diagnosed with breast cancer at National Cancer Institute, Cairo University]. Demographic data, breast clinical risk factors and laboratory indexes were collected from their medical files including age, family history of cancer tumor size and stage, metastasis and involvement of cancer in lymph nodes. In addition to the expression of classical immunohistochemistry (IHC) markers estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (Her2 neu). These markers were classified into: Luminal A (ER+/PR+/HER2-); Luminal B (ER+/PR+/HER2-/+); HER2-overexpression (ER-/PR-/HER2+). Clinical staging of breast cancer patients was carried out according to the American Joint Committee on Cancer (AJCC). Patients with other malignancy or cancer directed treatment (hormonal therapy, radiotherapy, or chemotherapy) were excluded from the study. Most cases (66%) were sporadic and with post-menopausal status (60%) with mean age (50.44±11.45).
Controls included sixty (60) healthy volunteers who were blood bank donors with mean age (45.00 ± 8.65). All participants were genetically unrelated and from the same geographical region (Egypt), and they were matched to cases with reference to gender, no history of malignancy, liver, or renal diseases at the time of ascertainment. All patients and healthy individuals agreed to participate in the study and signed an informed written consent form in accordance with the principles of the Helsinki II Declaration. This study has received the ethical approval no. 16472 by the ethical committee of the National Research Center, Cairo, Egypt.
Ethical Information
All patients and healthy individuals agreed to participate in the study and signed an informed written consent form in accordance with the principles of the Helsinki II Declaration. This study has received the ethical approval no. 16472 by the ethical committee of the National Research Center, Cairo, Egypt.
DNA extraction
Genomic DNA was extracted soon after blood sampling, using salting-out extraction procedure (8). DNA purity and concentration was detected using the NanoDrop1000-Detector (Nano Drop-Technologies, Wilmington, DE). DNA quality was determined by 1% agarose gel electrophoresis.
Genotyping
The MTHFR C677T (rs1801133), MTHFR A1298C (rs1801131) polymorphisms were determined by PCR-RFLP (5). Also, Genomic variants of MTR A2756G (rs1805087) genotypes were detected by RFLP technique through amplification by polymerase chain reaction (PCR) using sequence specific primers shown in Table 1. The Thermal cycling protocol comprised: initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 1 minute, annealing at 60 °C for 1 minute and 30 s, and extension at 72 °C for 1 minute followed by final extension at 72 °C for 10min. Success¬fully amplified products of 211 bp were digested with 10 units of HaeII (New England Biolabs) at 37 °C overnight. Restricted fragments were resolved on 3% agarose gel electrophore¬sis parallel with a DNA size marker (Amersham Pharmacia-Biotech) were shown in (Fig. 1).
Table 1.
Primer sequences of studied genes.
| Gene | SNP | Primers | Product (bp) |
|---|---|---|---|
| MTR | A2756G (rs1805087) | (F): 5'-TGTTCCAGACAGTTAGATGAAAATC-3' | 211 |
| (R): 5'-GATCCAAAGCCTTTTACACTCCTC-3'. | |||
| MTHFR | C677T (rs1801133) | (F): 5′- TGAAGGAGAAGGTGTCTGCGGGA-3′ | 198 |
| (R): 5′-AGGACGGTGCGGTGAGAGTG -3′ | |||
| A1289C (rs1801131) | (F): 5′-AAGGAGGAGCTGCTGAAGATG-3′ | 237 | |
| (R): 5′-CTTTGCCATGTCCACAGCATG-3′. |
MTR: Methonine synthase, MTHFR: 5,10 Methylenetetrahydrofolate reductase, SNP: single nucleotide polymorphism.
Fig. 1.
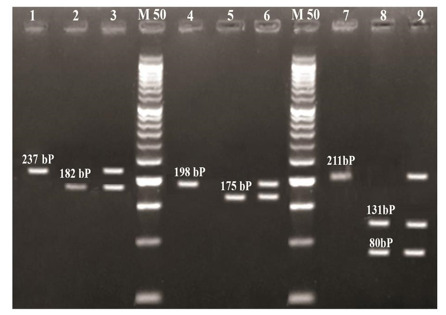
Agarose Gel Electrophoresis of Digested MTHFR C677T, MTHFR A1289C & MTR A2756G Amplicons.
Lanes M: represent 50bp molecular weight marker.
Lanes 1, 2, 3 represent digested MTHFR A1289C Ampli¬cons:
Lane1shows no cut 237 bp (AA), lane 2shows homozygous cut 182bp (CC), lane 3 shows heterozygous cut (AC).
Lanes 4, 5, 6 represent digested MTHFR C677T Amplicons:
Lane 4shows no cut 198bp (CC), lane 5 shows homozygous cut 175bp (TT), lane 6 shows heterozygous cut (CT).
Lanes 7, 8, 9 represent digested MTR A2756G Amplicons:
Lane 7 shows no cut 211 (AA), lane 8 shows homozygous cut (GG), lane 9 shows heterozygous cut (AG).
Sequencing Analysis
DNA sequence analysis was done by using the Sanger dideoxy nucleotide chain termination method and the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems Inc, Carlsbad, CA, United States). Forward primers were used for the PCR products of MTR (A2576G) while the Reverse primer for the MTHFR polymorphisms of (C677T & A1298C) (Fig. 2). The sequenced samples were analyzed in the Au¬tomated Sequencer “ABI Prism 3500 Genetic Analyzer”. The sequences were aligned with the consensus sequences that were retrieved from Gene Bank using the program ClustalX implemented in the Bioedit package (9).
Fig. 2.
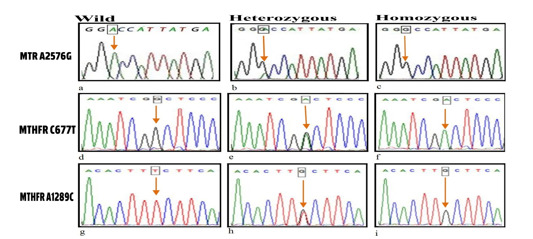
Sequencing analysis of MTHFR C677T, A1298C & MTR A2756G genotypes.
Polymorphisms of MTR A2756G using forward primer: (a) wild type AA genotype, (b) heterozygous AG genotype, (c) homozygous mutant GG genotype.
Polymorphisms of MTHFR C677T using reverse primer: (d) wild type CC genotype, (e) heterozygous CT genotype, (f) homozygous mutant TT genotype.
Polymorphisms of MTHFR A1289C using reverse primer: (g) wild type AA genotype, (h) heterozygous AC genotype, (i) homozygous mutant CC genotype.
Statistical analysis
The Statistical software program SPSS (Statistical Package for Social Science) was used for data analysis. Data was expressed as mean±SD of percent (%) of each genotype among subject population. Comparison between difference groups was done using Independent samples T-Test and Chi-Square Test with Odds ratio (OR) and corresponding 95% CI calculations. The allelic frequencies of each SNP that compared between cases and controls and the wide-type genotype was regarded as the reference group. Haplotype frequency distributions were deduced from genotype data and the most common haplotype was selected as the reference. Odds ratios and 95% CI were calculated to estimate the degree of the association between haplotypes and the risk of breast cancer. The difference between groups was considered to be significant if p< 0.05 (5, 10).
Results
Clinical features and Demographic Data of BC Patients
Table 2 shows the clinicopathological characteristics of Egyptian females with BC. Most of the patients, tumors (80%) were invasive ductal carcinoma (IDC) that grows into the surrounding tissues, and (81%) with big tumor size (T2-T4). Histopathologically, most of cases, tumors (83%) were of grade 2 and (57%) had advanced tumor stage (III-IV). Also, shows data concerning the clinical subtypes of the 100 BC that were detected according to the expression of ER, PR and HER2neu hormones and it was as follows: (31%) of Luminal A (ER+/PR+/HER2-), (20%) of Luminal B (ER+/PR+/HER2-/+), (49%) of Her-2 overexpressing (ER-/PR-/HER2+). Furthermore, the metastatic lymph nodes were involved in (66%) of cases.
Table 2.
Tumor criteria of breast cancer patients.
| Parameters | Breast cancer patients N= 100 (%) |
|---|---|
| Pathology | |
| IDC | 80 (80%) |
| ILC | 20 (20%) |
| Tumor size | |
| T1 | 19 (19%) |
| T2-T4 | 81 (81%) |
| Lymph nodes | |
| N0 | 34 (34%) |
| N1-N3 | 66 (66%) |
| Stage | |
| I-II | 43 (43%) |
| III-IV | 57 (57%) |
| Grade | |
| 2 | 83 (83%) |
| 3 | 17 (17%) |
| Immunohistochemical subtypes | |
| Luminal A (ER+/PR+/HER2-) | 31 (31%) |
| Luminal B (ER+/PR+/HER2-/+) | 20 (20%) |
| HER2-enriched (ER-/PR-/HER2+) | 49 (49%) |
IDC: Invasive ductal carcinoma, ILC: Invasive lobular carcinoma, HER2Neu: Human epidermal growth factor2, ER: Estrogen Receptor, PR: Progesterone Receptor.
Allelic and Genotypic Distribution of MTR and MTHFR Gene Polymorphisms
Table 3 shows a strong significant association of the SNPs of A/G genotypes of MTR A2756G, C/T genotypes of MTHFRC677T and the A/C genotypes of MTHFRA1289C with BC with highly significant p value (p= 0.001). Furthermore, it is clarified that the patients with breast cancer had a significant association with the (G) allele of MTR A2756G, (T) allele of MTHFRC677T and with the C allele of MTHFRA1289C, showing highly significant values (p= 0.001).
Table 3.
Allelic and Genotypic Distribution of MTR and MTHFR Gene Polymorphisms.
| Genotypes/Alleles frequencies | Cases N (100) (%) | Controls N (60) (%) | OR (95% CI) | p value |
|---|---|---|---|---|
| MTR A2576G | ||||
| AA | 18 (18%) | 48 (80%) | 1.00 (Reference) | |
| AG | 58 (58%) | 12 (20%) | 12.889 (5.651 - 29.396) | 0.001** |
| GG | 24 (24%) | 0 (0%) | 3.667 (2.473 - 5.437) | 0.001** |
| AG+GG | 82 (82%) | 12 (20%) | 18.222 (8.085 - 41.071) | 0.001** |
| A | 94 (47%) | 108 (90%) | 5.712 (3.460 | 0.001** |
| G | 106 (53%) | 12 (10%) | ||
| MTHFR C677T | ||||
| CC | 18 (18%) | 40 (66.7%) | 1.00 (Reference) | |
| CT | 55 (55%) | 20 (33.3%) | 6.111 (2.870 - 13.014) | 0.001** |
| TT | 27 (27%) | 0 (0%) | 3.222 (2.196 - 4.729) | 0.001** |
| CT+TT | 82 (82%) | 20 (33.3%) | 9.111 (4.345 - 19.106) | 0.001** |
| C | 91 (45.5%) | 100 (83.3%) | 5.989 (3.438 - 10.432) | 0.001** |
| T | 109 (54.5%) | 20 (16.7%) | ||
| MTHFR A1289C | ||||
| AA | 11 (11%) | 32 (53.3%) | 1.00 (Reference) | |
| AC | 43 (43%) | 24 (40%) | 5.212 (2.233 - 12.167) | 0.001** |
| CC | 46 (46%) | 4 (6.7%) | 33.455 (9.778 - 114.467) | 0.001** |
| AC+CC | 89 (89%) | 28 (46.7%) | 9.247 (4.130 - 20.703) | 0.001** |
| A | 65 (32.5%) | 88 (73.3%) | 5.712 (3.460 - 9.429) | 0.001** |
| C | 135 (67.5%) | 32 (26.7%) | ||
OR: odds ratio, CI: confidence interval, NS (Not significant),
highly significant.
Haplotypes distribution of MTHFR 677 C, MTHFR 1298 A>C > and MTR 2576 A>G in BC patients and controls.
Table 4 reveals the most haplotype frequencies of MTHFR 677 C>T, MTHFR 1298 A>C and MTR 2576 A>G that are associated with elevated risk of BC cases as follows: those in the heterozygous state (CT/AC/AG), and the homozygous state (TT/CC/GG), in addition to the other haplotypes(CT/AC/GG), (CT/CC/GG), (TT/AC/AG), (TT/AC/ GG), and (TT /CC/AG) giving highly significant value p= 0.001.
Table 4.
Haplotypes distribution of MTHFR C677T, MTHFR A1298C and MTR A2756G in breast cancer patients and controls.
| MTHFR C677T | MTHFR A1298C | MTR A2756G | Cases N (100) | Controls n (60) | OR/RS (95% CI) | p value |
|---|---|---|---|---|---|---|
| CC | AA | AA | 1 (1%) | 16 (26.7%) | 1.00 (Reference) | |
| AG | 5 (5%) | 6 (10%) | 13.333 (1.280 - 138.845) | 0.013 * | ||
| GG | 1 (1%) | 0 (0%) | 17.000 (2.539 - 113.824) | 0.004 ** | ||
| AC | AA | 6 (6%) | 12 (20%) | 8.000 (0.847 - 75.555) | 0.042 NS | |
| AG | 1 (1%) | 2 (3.3%) | 8.000 (0.347 - 184.364) | 0.144 NS | ||
| GG | 0 (0%) | 0 (0%) | ---------- | -------- | ||
| CC | AA | 4 (4%) | 4 (6.7%) | 16.000 (1.381 - 185.405) | 0.010 ** | |
| AG | 0 (0%) | 0 (0%) | ---------- | -------- | ||
| GG | 0 (0%) | 0 (0%) | ---------- | -------- | ||
| CT | AA | AA | 2 (2%) | 7 (11.7%) | 4.571 (0.354 - 59.106) | 0.215NS |
| AG | 2 (2%) | 3 (5%) | 10.667 (0.718 - 158.505) | 0.050 * | ||
| GG | 0 (0%) | 0 (0%) | ---------- | -------- | ||
| AC | AA | 3 (3%) | 9 (15%) | 5.333 (0.481 - 59.144) | 0.141 NS | |
| AG | 20 (20%) | 1 (1.7%) | 320.000 (18.534 - 5524.894) | 0.001 ** | ||
| GG | 2 (2%) | 0 (0%) | 17.000 (2.539 - 113.824) | 0.001 ** | ||
| CC | AA | 1 (1%) | 0 (0%) | 17.000 (2.539 - 113.824) | 0.004 ** | |
| AG | 19 (19%) | 0 (0%) | 17.000 (2.539 - 113.824) | 0.001 ** | ||
| GG | 6 (6%) | 0 (0%) | 17.000 (2.539 - 113.824) | 0.001 ** | ||
| TT | AA | AA | 0 (0%) | 0 (0%) | ---------- | -------- |
| AG | 0 (0%) | 0 (0%) | ---------- | -------- | ||
| GG | 0 (0%) | 0 (0%) | ---------- | -------- | ||
| AC | AA | 0 (0%) | 0 (0%) | ---------- | -------- | |
| AG | 5 (5%) | 0 (0%) | 17.000 (2.539 - 113.824) | 0.001 ** | ||
| GG | 6 (6%) | 0 (0%) | 17.000 (2.539 - 113.824) | 0.001 ** | ||
| CC | AA | 1 (1%) | 0 (0%) | 17.000 (2.539 - 113.824) | 0.004 ** | |
| AG | 6 (6%) | 0 (0%) | 17.000 (2.539 - 113.824) | 0.001 ** | ||
| GG | 9 (9%) | 0 (0%) | 17.000 (2.539 - 113.824) | 0.001 ** |
OR: odds ratio, CI: confidence interval,
(Not significant)
highly significant.
Correlation of MTHFR C677T, MTHFR A1298C and MTR A2576G haplotypes with clinical pathological features of BC patients
Table 5 demonstrates a strong significant association between the haplotype TCG of MTHFR (C677T), (A1289C) and MTR (A 2576G) polymorphisms with the aggressive clinical features of bigger tumor size (T2-T4), advanced stage (III and IV), and involvement of cancer in lymph nodes (N1-N3) with P value= 0.001. However, there was no significant correlation found with other tumor criteria including age, and metastasis.
Table 5.
Association of MTHFR C677T, A1298C and MTR A2576G haplotypes with clinical characteristics of breast cancer patients.
| Haplotype (MTHFRC677T -MTHFRA1298C-MTR A2576G) | Age (n= 100) | OR (95% CI) | p value | |
|---|---|---|---|---|
| > 45 | <= 45 | |||
| C-A-A | 54 (31.0%) | 59 (40.4%) | 1.00 (Reference) | |
| C-A-G | 2 (1.1%) | 13 (8.9%) | 0.168 (0.036 - 0.779) | 0.011 |
| C-C-A | 32 (18.4%) | 22 (15.1%) | 1.589 (0.824 - 3.064) | 0.165 |
| C-C-G | 7 (4.0%) | 2 (1.4%) | 3.824 (0.761 - 19.212) | 0.083 |
| T-A-A | 9 (5.2%) | 5 (3.4%) | 1.967 (0.620 - 6.235) | 0.244 |
| T-A-G | 8 (4.6%) | 3 (2.1%) | 2.914 (0.735 - 11.550) | 0.114 |
| T-C-A | 11 (6.3%) | 10 (6.8%) | 1.202 (0.473 - 3.054) | 0.699 |
| T-C-G | 51 (29.3%) | 32 (21.9%) | 1.741 (0.979 - 3.097) | 0.058 |
| Tumor Size(n= 100) | ||||
| T2 - T4 | T1 | |||
| C-A-A | 30 (18.5%) | 11 (28.9%) | 1.00 (Reference) | |
| C-A-G | 2 (1.2%) | 7 (18.4%) | 0.105 (0.019 - 0.583) | 0.004 |
| C-C-A | 22 (13.6%) | 12 (31.6%) | 0.672 (0.251 - 1.802) | 0.429 |
| C-C-G | 7 (4.3%) | 0 (0.0%) | 1.367 (1.135 - 1.645) | 0.119 |
| T-A-A | 5 (3.1%) | 2 (5.3%) | 0.917 (0.155 - 5.432) | 0.924 |
| T-A-G | 7 (4.3%) | 1 (2.6%) | 2.567 (0.283 - 23.309) | 0.389 |
| T-C-A | 11 (6.8%) | 1 (2.6%) | 4.033 (0.465 - 34.993) | 0.178 |
| T-C-G | 78 (48.1%) | 4 (10.5%) | 7.150 (2.112 - 24.203) | 0.001 ** |
| Lymph nodes (n= 100) | ||||
| N1 - N3 | N0 | |||
| C-A-A | 23 (17.4%) | 11 (28.9%)18 (26.5%) | 1.00 (Reference) | |
| C-A-G | 2 (1.5%) | 7 (10.3%) | 0.224 (0.041 - 1.210) | 0.066 |
| C-C-A | 15 (11.4%) | 19 (27.9%) | 0.618 (0.247 - 1.544) | 0.302 |
| C-C-G | 5 (3.8%) | 2 (2.9%) | 1.957 (0.339 - 11.281) | 0.447 |
| T-A-A | 3 (2.3%) | 4 (5.9%) | 0.587 (0.116 - 2.963) | 0.516 |
| T-A-G | 7 (5.3%) | 1 (1.5%) | 5.478 (0.617 - 48.666) | 0.095 |
| T-C-A | 10 (7.6%) | 2 (2.9%) | 3.913 (0.760 - 20.146) | 0.087 |
| T-C-G | 67 (50.8%) | 15 (22.1%) | 3.496 (1.520 - 8.041) | 0.003** |
| Stage (n= 100) | ||||
| III - IV | ||||
| C-A-A | 15 (13.2%) | 26 (30.2%) | 1.00 (Reference) | |
| C-A-G | 2 (1.8%) | 7 (8.1%) | 0.495 (0.091 - 2.698) | 0.410 |
| C-C-A | 13 (11.4%) | 21 (24.4%) | 1.073 (0.419 - 2.745) | 0.883 |
| C-C-G | 6 (5.3%) | 1 (1.2%) | 10.400 (1.141 - 94.835) | 0.015* |
| T-A-A | 3 (2.6%) | 4 (4.7%) | 1.300 (0.256 - 6.610) | 0.751 |
| T-A-G | 7 (6.1%) | 1 (1.2%) | 12.133 (1.359 - 108.364) | 0.008** |
| T-C-A | 9 (7.9%) | 3 (3.5%) | 5.200 (1.216 - 22.234) | 0.019 |
| T-C-G | 59 (51.8%) | 23 (26.7%) | 4.446 (2.003 - 9.871) | 0.001** |
| Breast Subtypes (n=100) | ||||
| Her2 Enriched | ||||
| C-A-A | 12(12.2%) | 21(33.9%) | 1.00 (Reference) | |
| C-A-G | 2(2.0%) | 6(9.7%) | 0.583(0.101-3.358) | 0.546 |
| C-C-A | 12(12.2%) | 16(25.8%) | 1.312(0.468-3.681) | 0.605 |
| C-C-G | 5(5.1%) | 0(0.0%) | 18.920 (0.963-371.679) | 0.053 |
| T-A-A | 3(3.1%) | 3(4.8%) | 1.750(0.304-10.075) | 0.531 |
| T-A-G | 5(5.1%) | 2(3.2%) | 4.375(0.733-26.116) | 0.105 |
| T-C-A | 8(8.2%) | 2(3.2%) | 7.000(1.274-38.475) | 0.025 |
| T-C-G | 51(52.0%) | 12(19.4%) | 7.437(2.882-19.191) | <0.001** |
OR: odds ratio, CI: confidence interval,
= highly significant.
Luminal A (ER+/PR+/HER2-); HER2Neu: Human epidermal growth factor2, ER: Estrogen Receptor, PR: Progesterone Receptor.
Discussion
Recently, increased attention had been drawn to the role of single nucleotide polymorphisms (SNPs) in identifying genes that contribute to disease pathogenesis. Such SNP alleles could cause alterations in gene function or regulation that directly contribute to the disease progression (11, 12). Several studies have been reported on the association between the SNPs of the folate metabolizing genes Methylenetetrahydrofolate reductase (MTHFR) and Methionine synthase (MTR) genes with the changes that alter the primary structure of each protein, decrease enzymatic activity. This causes modification in DNA methylation profiles, abnormal gene expression and inactivation of tumor suppressor genes (4, 13).
In the present study we focused on clarifying the role of genetic predisposition and gene –gene interaction of the functional SNPs of MTR A2576G, MTHFR C677T, MTHFR A1298C in the incidence of breast cancer among Egyptian females and their association within the clinical features and its prognosis. These SNPs could be used as genetic markers in disease progression.
Our descriptive analysis on the two compared groups (cases versus healthy controls) highlighted significant differences within higher percentages of some variables among cases such as: (81%) of bigger tumor size (T2-T4) with (57%) advanced stage (III-IV), (80%) of Invasive ductal carcinoma (IDC), (49%) of HER2-overexpression with dual negativity of both ER and PR hormones (ER-/PR-/HER2+), and (66%) metastatic lymph nodes (N1-N3). These data are in accordance with previous studies (5, 14). In contrast to what should have been expected no significant differences was observed for age. However, it has been stated before the role of aging in cancer development that it is attributed to the acquisition of mutations that favors carcinogenesis (15).
Our analysis revealed a significant difference between the two studied groups as most of the breast cancer patients had the combination of genotypes of 82% (AG+GG) of MTR A2576G, 82% (CT+TT) of MTHFR C677T, 89% (AC+CC) of MTHFR A1298C that could obviously influence the risk of breast cancer. We also noted a higher frequency of the major alleles that predominates in the control group: 90% of (A allele) of MTR A2576G, 83.3% of (C allele) of MTHFR C677T, 88% of (A allele) of MTHFR A1298C. On the other side, the frequency of the minor (mutant) alleles was extremely high in the breast cancer group as follows: 53% of (G allele) of MTR, 54.5% of (T allele) of MTHFR C677T, 67.5% of (C allele) of MTHFR A1298C.
Moreover, this would stratify those women having the latest mutant alleles significantly increased the risk of BC. In agreement to our data, it was shown that the frequency of MTHFR C677T (TT) genotype was significantly higher in patients with breast cancer among Kurdish, Moroccan females. Additionally, it increases the cervical dysplasia and the development of immune-mediated inflammatory disorders and higher incidence of cardiovascular comorbidities (16, 17, and 18).
Furthermore, our results were in accordance with previous studies which demonstrated that carriers of MTHFR A1298C (CC) genotype were significantly associated with a higher incidence of major depressive disorders, lacunar stroke, ischemic stroke, and sporadic breast cancer in Iranian, Brazilian, and Italian Population (19- 22).
Also, our data is supported by the previously studies showing significant association (G allele) of MTR with the breast cancer output among North Brazilian, Iranian, and Chinese populations (20, 22, and 23).
On the other side, other studies reported no significant association with susceptibility to breast cancer (24), while others demonstrated that it may be a reduced risk factor for lymphoblastic leukemia (ALL), colorectal cancer and gastric cancer (25, 26). This conflict in data between the different studies may be attributed to geographic, ethnic variations and several factors such as a measurement sample, hormones, environmental factors, differences in dietary and intake of nutrients (26, 27).
In consequence, it is shown that studying genetic association by approach of gene-gene interaction generates more powerful analysis. In the present work we analyzed gene-gene interaction through determination frequencies of haplotypes generated by the combination of MTHFR C677T, MTHFR A1289C, and MTR A2576G genotypes and our findings revealed that there is a high significance association of haplotype generated by the presence of heterozygous genotypes of the studied genes (CT-AC-AG) to the breast cancer development that increases by about 320.00 fold in BC women harboring it (p= 0.001, OR= 320.000, CI= 18.534 - 5524.894). This demonstrates that the presence of polymorphic alleles elevated the breast cancer risk through the generation of less active thermo liable enzymes leading to global hypomethylation and chromosomal breaks. These findings are in agreement with (28).
Also, the interaction of these polymorphisms results in reduction in methylation pattern, allowing the activation of oncogenes which initiated the tumor genesis process (29). These results are in accordance with a study conducted on the population in Northeast Brazil which reported that the risk increased with the presence of CT/AG of MTHFR C677T and MTR A2576G, while CC/AG of MTHFR A1298C and MTR A2576G had a less effect with AC/AG of MTHFR A1298C and MTR A2576G. On the other side, other studies did not find any association (17, 24).
Meanwhile, our results were related to other reported studies on other diseases clarifying those patients harboring the MTR*G and the MTHFR *C variants were associated with 11-fold increased risk for bladder cancer development. Similarly in patients with lung cancer, a significant interaction was also observed for individuals having the gene interaction of MTHFR 677 CT/TT and MTHFR 1298 AC/CC genotypes which led to an increase in the risk of developing lung cancer (30, 31).
Additionally, other studies have investigated gene-gene interactions in cancer susceptibility, as in acute lymphoblastic leukemia demonstrating that subjects with the MTHFR 677CT/TT and MS 2756AG/GG genotypes revealed a 3.6-fold (25).
The correlation of the SNPs, the mutant alleles and molecular pathogenesis of BC disease is largely unidentified and there are many related factors, as hormone factors and tumor pathology which plays changeable roles in the pathogenesis of BC, progression, and survival. The understanding of these factors could be helpful to the management of patients with breast cancer (32-34).
Thus, the present study represented a prior study among Egyptian females of investigating the gene-gene interactions of the haplotypes of MTHFR C677T/ MTHFR A1289C and MTRA2576G genotypes with the prognostic clinicopathological parameters of breast cancer. Our results provided a strong significant association between the T-C-G haplotype with bigger tumor size tumor size (T2-T3), advanced stage (III-IV) (p= 0.001). In accordance with our data, concerning C677T and A1289C polymorphism gave a very highly significance with bigger tumor size (19, 32, 33). On the other hand, no significant association was reported in other studies (24).
Moreover, it is observed that the haplotype T-C-G plays a high role in metastatic lymph nodal status with strong significant association (p= 0.003). In a support from (16, 33) to our data, it is reported that there were tendencies for CRC patients with mutant genotypes to have more positive lymph nodes. However, other studies showed no significant difference in genotype distribution among different phases of metastatic lymph nodes (20).
Furthermore, this work demonstrated a strong association between the haplotype (T-C-G) of MTHFR C677T/ MTHFR A1289C and MTRA2576G and hormonal receptor expression and it can be seen that there is a tendency towards the association between aggressiveness tumor type (ER-/PR-) and the mutant alleles. In confirmation of (20, 29) to our data, the risk of BC with an aggressive biophenotype is two or three-fold higher in patients with a mutant allele of MTHFR C677T and A1298C. On the other hand, another study did not detect any relation (17, 30).
Overall, our results are supported by the evidence that breast carcinomas with aneuploidy, positive axillary nodes, and high proliferative activity led to higher levels of DNA hypomethylation which were not only observed during early stages of tumorgenesis but were also strongly associated with tumor progression (4, 33).
To sum up, we conclude strong connection of MTHFR C677T, MTHFR A1289C, and MTR A2576G haplotype with the clinical markers of the patients revealing that T-C-G haplotype is significantly contributed to the developing of more severity tumors. This supports the concept that the combination of more SNPs from different genes has a more synergistically powerful genetic effect and contributes to increased disease risk and progression. These SNPs could be used as tumor aggressiveness markers among BC patients. Extensive studies should be warranted to demonstrate the biological mechanisms of these associations.
Acknowledgements
The authors would like to thank the medical staff at the Department of clinical and chemical pathology, National Cancer Institute, and the technical staff at Microbial Biotechnology Department, national Research Center.
This study was supported financially by Academy of Scientific Research through National Research Centre.
References
- 1.Sharif MR, Karimian M. MTR-A2756G and breast cancer risk: a study of Iranian women with a meta-analysis. Bioscience Biotechnololgy Research Communication. 2016;9(4):795–803. [Google Scholar]
- 2.Awwad N, Yousef AM, Abuhaliema A, Abdalla I, Yousef M. Relationship between genetic polymorphisms in MTHFR (C677T, A1298C and their Haplotypes) and the incidence of breast cancer among Jordanian females–case-control study. Asian Pac J Cancer Prev. 2015;16(12):5007–11. doi: 10.7314/apjcp.2015.16.12.5007. [DOI] [PubMed] [Google Scholar]
- 3.Bonaventure A, Goujon-Bellec S, Rudant J, Orsi L, Leverger G, Baruchel A, et al. Maternal smoking during pregnancy, genetic polymorphisms of metabolic enzymes, and childhood acute leukemia: the ESCALE study (SFCE). Cancer Causes Control. 2012;23(2):329–45. doi: 10.1007/s10552-011-9882-9. [DOI] [PubMed] [Google Scholar]
- 4.Castiglia P, Sanna V, Azara A, et al. Methylenetetrahydrofolate reductase (MTHFR) C677T and A1298C polymorphisms in breast cancer a Sardinian preliminary case-control study. Int J Med Sci. 2019;16(8):1089–1095. doi: 10.7150/ijms.32162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Omran MH, Fotouh BE, Shousha WG, Ismail A, Ibrahim NE, Ramadan SS. Strong Correlation of MTHFR Gene Polymorphisms with Breast Cancer and its Prognostic Clinical Factors among Egyptian Females. Asian Pac J Cancer Prev. 2021;22(2):617–626. doi: 10.31557/APJCP.2021.22.2.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carvalho DC, Wanderley AV, Mello Junior FAR, Santos AMRD, Leitão LPC, Souza TP, et al. Association of genes ARID5B, CEBPE and folate pathway with acute lymphoblastic leukemia in a population from the Brazilian Amazon region. Leukemia Research Reports. 2019;27(13):100188. doi: 10.1016/j.lrr.2019.100188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi SW, Mason JB. Folate and carcinogenesis: an integrated scheme. J Nutr. 2000;130(2):129–32. doi: 10.1093/jn/130.2.129. [DOI] [PubMed] [Google Scholar]
- 8.Omran MH, Ibrahim NE, Youssef SS, et al. Relation of interleukin-1β gene to treatment response in chronic patients infected with HCV genotype 4. The Journal of Infection in Developing Countries. 2013;7(11):851–8. doi: 10.3855/jidc.3823. [DOI] [PubMed] [Google Scholar]
- 9.Omran MH, Nabil W, Youssef SS, El-Sayed M, El Awady MK. Heterogeneity and new epitopes of hepatitis C virus genotype 4. Hepatitis Monthly. 2013;2013(8):e10521. doi: 10.5812/hepatmon.10521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Solé X, Guinó E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinform. 2006;22(15):1928–9. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 11.Omran MH, Khamis M, Nasr N, Massoud AA, Youssef SS, Bader El Din NG, et al. A Study of CC-Chemokine Receptor 5 (CCR5) Polymorphism on the Outcome of HCV Therapy in Egyptian Patients. Hepat Mon. 2013;13(12):e13721. doi: 10.5812/hepatmon.13721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan H, Fu M, Yang X, Huang K, Ren X. Single nucleotide polymorphism of MTHFR rs1801133 associated with elevated Hcy levels affects susceptibility to cerebral small vessel disease. PeerJ. 2020;8:e8627. doi: 10.7717/peerj.8627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ward M, Hughes CF, Strain JJ, Reilly R, Cunningham C, Molloy A, et al. Impact of the common MTHFR 677C→T polymorphism on blood pressure in adulthood and role of riboflavin in modifying the genetic risk of hypertension: evidence from the JINGO project. BMC Med. 2020;18(1):318. doi: 10.1186/s12916-020-01780-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donepudi M, Kondapalli K, Amos S, Venkanteshan P. Breast cancer statistics and markers. J Cancer Res Ther. 2014;10(3):506–11. doi: 10.4103/0973-1482.137927. [DOI] [PubMed] [Google Scholar]
- 15.Vijg J, Suh Y. Genome instability and aging. Annu Rev Physiol. 2013;75:645–68. doi: 10.1146/annurev-physiol-030212-183715. [DOI] [PubMed] [Google Scholar]
- 16.Rahimi Z, Bozorgi M, Rahimi Z, Shakiba E, Yari K, et al. MTHFR C677T Polymorphism Is Associated with the Risk of Breast Cancer Among Kurdish Population from Western Iran. Int J Cancer Manag. 2019;12(3):e67895. [Google Scholar]
- 17.Hardi H, Melki R, Boughaleb Z, et al. Significant association between ERCC2 and MTHR polymorphisms and breast cancer susceptibility in Moroccan population: genotype and haplotype analysis in a case-control study. BMC Cancer. 2018;18:292. doi: 10.1186/s12885-018-4214-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chita DS, Tudor A, Christodorescu R, Buleu FN, Sosdean R, Deme SM, et al. MTHFR gene polymorphisms prevalence and cardiovascular risk factors involved in cardioembolic stroke type and severity. Brain Sci. 2020;10(8):476. doi: 10.3390/brainsci10080476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar A, Sharma R, Misra S, Nath M, Kumar P. Relationship between methylenetetrahydrofolate reductase (MTHFR) gene (A1298C) polymorphism with the risk of stroke: A systematic review and meta-analysis. Neurol Res. 2020;42(11):913–922. doi: 10.1080/01616412.2020.1798107. [DOI] [PubMed] [Google Scholar]
- 20.Karimian M, Rezazadeh N, Khamehchian T. Association analysis of methylenetetrahydrofolate reductase common gene polymorphisms with breast cancer risk in an Iranian population: A Case-Control Study and a Stratified Analysis. Asian Pac J Cancer Prev. 2020;21(9):2709–2714. doi: 10.31557/APJCP.2020.21.9.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pepe C, Guidugli L, Sensi E, Aretini P, D'Andrea E, Montagna M. Methyl group metabolism gene polymorphisms as modifier of breast cancer risk in Italian BRCA1/2 carriers. Breast Cancer Res Treat. 2007;103(1):29–36. doi: 10.1007/s10549-006-9349-y. [DOI] [PubMed] [Google Scholar]
- 22.Carvalho Barbosa R, da Costa DM, Cordeiro DE, Vieira AP, Rabenhorst SH. Interaction of MTHFR C677T and A1298C, and MTR A2756G gene polymorphisms in breast cancer risk in a population in Northeast Brazil. Anticancer Res. 2012;32(11):4805–11. [PubMed] [Google Scholar]
- 23.Niu Z, Zhao H, Hou X. Association of MTHFR, MTRR and MTR polymorphisms with breast cancer risk: A study in Chinese females. International Journal of Clinical and Experimental Pathology. 2017;10(6):7059–7066. [Google Scholar]
- 24.Zhao Y, Chen Z, Ma Y, Xia Q, Zhang F, Fu D, et al. Lack of association between methionine synthase A2756G polymorphism and digestive system cancer risk: Evidence from 39327 subjects. PLoS ONE. 2013;8(4):e61511. doi: 10.1371/journal.pone.0061511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mo LM, Yang HP, Yang XW, Ruan LH. Methionine synthase A2756G polymorphism influences pediatric acute lymphoblastic leukemia risk: a meta-analysis. Biosci Rep. 2019;39(1):BSR20181770. doi: 10.1042/BSR20181770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Marchand L, Donlon T, Hankin JH, Kolonel LN, Wilkens LR, Seifried A. B-vitamin intake, metabolic genes, and colorectal cancer risk (United States). Cancer Causes Control. 2002;13(3):239–48. doi: 10.1023/a:1015057614870. [DOI] [PubMed] [Google Scholar]
- 27.Gong Z, Yao S, Zirpoli G, David Cheng TY, et al. Genetic variants in one-carbon metabolism genes and breast cancer risk in European American and African American women. Int J Cancer. 2015;137(3):666–677. doi: 10.1002/ijc.29434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lajin B, Alhaj Sakur A, Ghabreau L, Alachkar A. Association of polymorphisms in one-carbon metabolizing genes with breast cancer risk in Syrian women. Tumour Biol. 2012;33(4):1133–9. doi: 10.1007/s13277-012-0354-y. [DOI] [PubMed] [Google Scholar]
- 29.Floris M, Sanna D, Castiglia P, Putzu C, Sanna V, Pazzola A, et al. MTHFR, XRCC1 and OGG1 genetic polymorphisms in breast cancer. a case-control study in a population from North Sardinia. BMC Cancer. 2020;20(1):234. doi: 10.1186/s12885-020-06749-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piskac-Collier AL, Monroy C, Lopez MS, Cortes A, Etzel CJ, Greisinger AJ, et al. Variants in folate pathway genes as modulators of genetic instability and lung cancer risk. Genes Chromosomes Cancer. 2011;50(1):1–12. doi: 10.1002/gcc.20826. [DOI] [PubMed] [Google Scholar]
- 31.Poodineh M, Saravani R, Mirhosseini M, Sargazi S. Association of two methylenetetrahydrofolate reductase polymorphisms (rs1801133, rs1801131) with the risk of type 2 diabetes in South-East of Iran. Rep Biochem Mol Biol. 2019;8(2):178–183. [PMC free article] [PubMed] [Google Scholar]
- 32.Arpino G, Milano M, De Placido S. Features of aggressive breast cancer. Breast. 2015;24(5):594–600. doi: 10.1016/j.breast.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 33.Waseem M, Hussain SR, Kumar S, et al. Association of MTHFR (C677T) gene polymorphism with breast cancer in North India. Biomark Cancer. 2016;8:111–117. doi: 10.4137/BIC.S40446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nourolahzadeh Z, Houshmand S M, Mostafa Mohammad F, Ghorbian S. Correlation between Lsp1 (Rs3817198) and Casc (Rs4784227) Polymorphisms and the Susceptibility to Breast Cancer. Reports of biochemistry & molecular biology. 2020;9(3):291–296. doi: 10.29252/rbmb.9.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]


