Abstract
Photoreceptors are the key functional cell types responsible for the initiation of vision in the retina. Phototransduction involves isomerization and conversion of vitamin A compounds, known as retinoids, and their recycling through the visual cycle. We demonstrate a functional readout of the visual cycle in photoreceptors within stem cell-derived retinal organoids and mouse retinal explants based on spectral and lifetime changes in autofluorescence of the visual cycle retinoids after exposure to light or chemical stimuli. We also apply a simultaneous two- and three-photon excitation method that provides specific signals and increases contrast between these retinoids, allowing for reliable detection of their presence and conversion within photoreceptors. This multiphoton imaging technique resolves the slow dynamics of visual cycle reactions and can enable high-throughput functional screening of retinal tissues and organoid cultures with single-cell resolution.
1. Introduction
Photoreceptors are the primary light responsive cells in the retina, and phototransduction in photoreceptor cells is an active area of research in developmental biology, physiology, retinal disease, and regenerative therapy [1–4]. The standard test for phototransduction in photoreceptor cells is electrophysiology of single cells through perforated patch clamping or suction electrode recording during light stimulus [5–7]. This direct approach is sensitive to photoreceptor hyperpolarization in response to light stimuli and provides excellent (sub-millisecond) time resolution, but is also labor-intensive and extremely low-throughput. New approaches with higher throughput include indirect measurement of photoreceptor activity by mapping the downstream neural signaling of retinal ganglion cells in response to light stimuli using a microelectrode array [8], and direct measurement of photoreceptor outer segment length change due to photostimulation using adaptive optics optical coherence tomography (AO-OCT) in the living human retina [9–12].
To sustain their phototransduction function, photoreceptors rely on supporting cells like retinal pigment epithelium (RPE) and Müller glia to regenerate their photopigments. The recycling of the vitamin A compounds that form the light-sensitive chromophores in the eye is an integral part of vision that is implicated in many degenerative retinal diseases (e.g., RDH12- or RPE65- associated inherited retinal degeneration in Leber congenital amaurosis, retinitis pigmentosa) and has been studied using imaging techniques including two-photon fluorescence microscopy [13–17].
Here, we describe a multiphoton autofluorescence lifetime imaging assay of light stimulus response in photoreceptor cells that is indirectly related to phototransduction through downstream reactions involved in visual pigment regeneration. Retinal cells such as photoreceptors and retinal pigment epithelium exhibit autofluorescence from retinoids and their condensation products (e.g., bisretinoids, A2E, esters, lipofuscin, etc.) [18]. Retinoids play a critical role in phototransduction, as absorption of visible light by a photosensitive isomer of retinaldehyde (11-cis-RAL) that is covalently bound to photoreceptor opsin and its subsequent fast [19] isomerization to all-trans retinaldehyde (AT-RAL) leads to activation of the opsin and initiation of the signaling cascade that registers light (Fig. 1(a)) [20]. AT-RAL is then reduced to all-trans retinol (AT-ROL) by the photoreceptor in a reaction that is mediated by retinal dehydrogenase (RDH) enzymes (Fig. 1(a)) [21,22]. This is the first step in a relatively slow chain of reactions, known as the visual cycle, involving photoreceptors and RPE cells that regenerates the visual pigments (Fig. 1(a)). 11-cis-RAL and AT-RAL have virtually identical fluorescence spectra and lifetimes, making it difficult to distinguish them via spectral fluorescence imaging or fluorescence lifetime imaging microscopy (FLIM). However, the slow conversion of AT-RAL to AT-ROL by photoreceptors can be detected from an increase in autofluorescence intensity [23–25]. The fluorescence lifetimes of AT-RAL (∼1 ps [26], 56 ps for RAL Schiff base in ethanol [27]) and AT-ROL (3 ns in ethanol [28], 3.4 ns [29]) differ, and the emission spectrum of AT-ROL is slightly blue-shifted from AT-RAL [29,30]. Therefore, we hypothesized that differences in fluorescence lifetimes between AT-ROL and AT-RAL can be imaged to assess visual cycle function in photoreceptor cells.
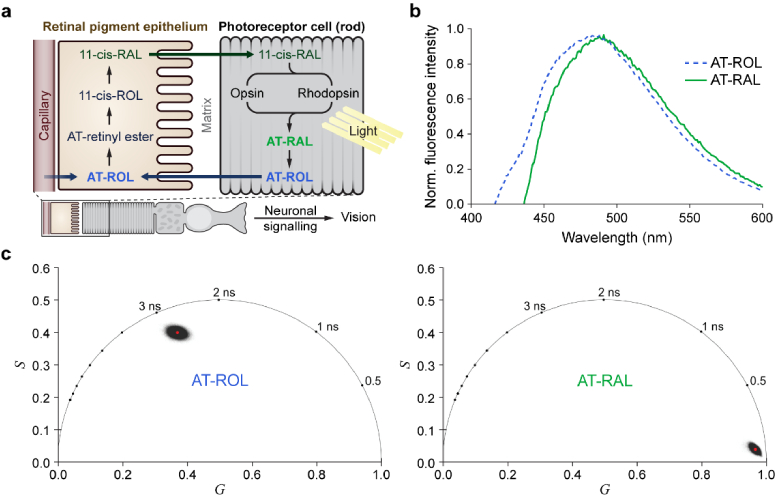
Fig. 1.
The visual cycle includes AT-ROL and AT-RAL with overlapping fluorescence emission spectra and distinct fluorescence lifetimes. (a) For phototransduction, photoreceptors rely on the fast (sub-picosecond) isomerization of 11-cis-RAL to all-trans-RAL after light absorption. The visual cycle is responsible for clearing the resulting AT-RAL and converting it back to the light-sensitive 11-cis-RAL through several enzymatic steps. This is a slow process (several minutes) that starts with conversion of AT-RAL to AT-ROL in the photoreceptors, followed by transfer of AT-ROL to the retinal pigment epithelium where it is converted to 11-cis-RAL and transported back to photoreceptor outer segments. Autofluorescence of these retinoids provides an endogenous signal for probing light response in retinal photoreceptor cells. (b) Normalized fluorescence emission spectra (excited at 350 nm) of AT-RAL and AT-ROL dissolved in ethanol. The emission spectra are broad and overlapping, with AT-ROL slightly blue-shifted relative to AT-RAL. (c) Phasor representation of fluorescence lifetime (two-photon Ex: 760 nm, Em: 550/100 nm) of pure AT-ROL or AT-RAL in ethanol shows significant separation of AT-ROL and AT-RAL. AT-RAL has a short lifetime of <0.1 ns while AT-ROL has a long lifetime of nearly 3 ns.
In this study, we characterized the fluorescence spectrum and lifetime of AT-ROL and AT-RAL in solution to provide reference measurements for photoreceptor imaging. Next, we identified a three-photon excitation scheme to increase the contrast between AT-ROL and AT-RAL. This approach was used to image the dynamics of fluorescence change after light exposure in a known light-sensitive sample, i.e., freshly-excised mouse retina explants. Finally, we imaged the response of human induced pluripotent stem cell (hiPSC)-derived retinal organoids to chemical stimuli. This non-invasive autofluorescence imaging assay can streamline studies of the visual cycle in model systems and screen culture methods to generate functionally mature photoreceptors from hiPSCs.
2. Materials and methods
2.1. Retinoid measurements in solution
Solutions of AT-RAL and AT-ROL in ethanol were prepared from crystalline solid (Cayman Chemical). A stock 35 mM solution of each retinoid was made by dissolving 10 mg of solid retinoid in 1 mL anhydrous ethanol under nitrogen gas. To protect AT-ROL from oxidizing to AT-RAL, 1 mg of sodium borohydride (NaBH4, a reducing agent, Sigma) was also added to the AT-ROL stock solution. Fluorescence spectra of AT-RAL and AT-ROL were measured on a spectrofluorometer (Quantamaster C-60/2000, Photon Technology Instruments) after diluting either solution to 10 µM in anhydrous ethanol. To characterize multiphoton excitation and fluorescence lifetime of the visual cycle retinoids in solution, 1 mL aliquots of 1 mM solution were prepared by diluting the stock solutions using 70% EtOH. 1 mM solutions of AT-RAL and AT-ROL and their mixtures were imaged on the FLIM microscope using the multiphoton excitation and detection scheme described below.
2.2. Multiphoton excitation and FLIM imaging of retinoids
FLIM imaging was performed on a custom multiphoton microscope (Bruker Fluorescence Microscopy) built around an inverted Nikon Ti-E microscope, using a Ti:Sapphire tunable laser (Insight DS, Spectra Physics), GaAsP photomultiplier tubes (H7422PA-40, Hamamatsu), bandpass emission filters (Semrock), and time-correlated single photon counting electronics (SPC-150, Becker & Hickl) for lifetime measurement. Baseline two-photon excited FLIM imaging was performed using the tunable output of the Ti:Sapphire source at a wavelength of 760 nm using 2-5 mW average laser power (80 MHz repetition rate, pulse width <120 fs). To achieve FLIM contrast between AT-RAL and AT-ROL while exciting and collecting fluorescence emission from both species simultaneously, near-infrared multiphoton excitation from the 1040 nm fixed-wavelength output of the laser source was used. To avoid photothermal damage to the sample, 1040 nm laser power (80 MHz repetition rate, pulse width <200 fs) was limited to a maximum of 15 mW at the sample. A 550/50 nm filter was used for the green emission channel and a 440/80 nm filter for the blue emission channel. The blue emission channel exclusively transmits three-photon-excited fluorescence. The green emission channel transmits two- and three-photon-excited fluorescence, although two-photon excitation is an intrinsically more probable event. These filter choices also reject second and third harmonic signals from the 1040 nm excitation. Excitation at 1040 nm with emission at 550/50 nm and 440/80 nm is advantageous because the different multiphoton excitation spectra of AT-RAL and AT-ROL [31,32] achieves contrast between these retinoids when mixed in solution and in photoreceptor cells. Specifically, three-photon excitation preferentially excites AT-ROL while two-photon excitation excites both AT-RAL and AT-ROL.
2.3. Mouse retina explant preparation
To characterize the response of functional retina and photoreceptor cells to light exposure or chemical stimulus, dark-adapted mouse retina explants were imaged. Wild-type C57Bl6 mice between 1 and 2 months old were dark-adapted overnight, sacrificed under isoflurane anesthesia according to IACUC-approved guidelines, and the eyes were harvested. Dissection was performed under infra-red illumination and night-vision optics. The retina was cut away from the anterior segment of the eye and the vitreous was cleared away [33]. The eye cup was flattened with four radial cuts. The sclera and retinal pigment epithelium (RPE) were carefully removed from the photoreceptor layer by mechanical dissociation with fine-tip forceps and the tip of a hypodermic needle. This process damages the outer segments of the photoreceptors that are attached to the apical processes of the RPE. However, sufficient outer segments remain to ensure light responsiveness, as verified by patch clamp measurements on similarly prepared explants [6]. The retina explant was placed in a 35 mm glass-bottom imaging dish (Mattek) with the photoreceptors facing down on the glass and suspended in Ames’ medium [34] to prolong the retina responsiveness. A horseshoe-shaped titanium weight with fine nylon wires bridging its 1-cm opening was placed over the retina explant to immobilize and flatten it against the glass. The dish was placed in a dark incubator (at 37°C and 5% CO2) for 30 minutes prior to imaging.
2.4. Mouse retinal explant stimulation and FLIM imaging
The imaging dish with the retinal explant was placed in a stage-top incubator (Tokai Hit) at 37°C and 5% CO2, and the inverted objective lens was focused on the photoreceptor layer using dim red light for bright-field view finding. A baseline FLIM image was acquired while the sample was dark-adapted, using the multiphoton excitation and detection scheme described above for retinoids. A long-working-distance 20× /1.0 NA water-immersion objective lens (Zeiss) was used, and the 500 µm × 500 µm field of view was imaged with a resolution of 512 × 512 pixels with a pixel dwell time of 4.7 µs and a total integration time of 90 seconds per image. Next, the sample was exposed to intense white light from the microscope condenser 100W tungsten halogen lamp at maximum brightness for one minute. The lamp produces a luminous flux of 3600 lm of broad-spectrum light, resulting in an estimated irradiance of 100 mW/cm2 for the entire visible spectrum (350-700 nm) at the sample plane. This light exposure bleaches most of the rhodopsin photopigments in the photoreceptor cells and results in isomerization of 11-cis-RAL to all-trans-RAL and its release from the outer segment membranes into the cytosol [20,23]. Following light exposure, FLIM images were acquired immediately and every few minutes thereafter for ∼1 hour. Next, additional exogenous AT-RAL from a 1 mM aliquot in Ames’ medium + 1% bovine serum albumin (Sigma) was added to the medium in the imaging dish for a final concentration of 50 µM. FLIM imaging was continued thereafter for an additional hour.
2.5. iPSC-derived retinal organoid stimulation and FLIM imaging
Human iPSC-derived retinal organoids were differentiated as described previously [35] and grown for over 200 days in culture until they presented photoreceptors with mature stage 3 morphology, including an outer nuclear layer and inner and outer segments. Individual retinal organoids were suspended in standard retinal differentiation medium (RDM) in 35 mm glass-bottom imaging dishes and dark-adapted in the incubator for 48 hours. Individual dishes were placed in the stage-top incubator (at 37°C and 5% CO2) and a baseline FLIM image was acquired using a long-working-distance 20× /1.0 NA water-immersion objective lens (Zeiss). The 500 µm × 500 µm field of view was imaged with a resolution of 512 × 512 pixels with a pixel dwell time of 4.7 us and a total integration time of 90 seconds per image. Next, each organoid was exposed to one minute of white light from the 100W tungsten halogen lamp at maximum brightness, followed by FLIM imaging at 8-minute intervals for one hour. Next, exogenous AT-RAL from a 1 mM aliquot in RDM medium was added to the imaging dish for a final concentration of 50 µM. FLIM imaging was continued thereafter for an additional hour with 8-minute intervals.
2.6. Phasor analysis of fluorescence lifetimes
Fit-free phasor analysis [36] of FLIM images was performed in MATLAB (MathWorks). First, neighboring pixels were binned to improve signal to noise ratio. The two-photon-excited green channel was binned by a factor of 3 or 4 (i.e., 7×7 or 9×9 pixel windows), and the lower-intensity three-photon-excited blue channel was binned by a factor of 10 (i.e., 21 × 21 pixel window). Cosine and sine transform of the resulting decays were calculated at the laser pulse repetition frequency (i.e., PRF = 80 MHz, period T = 12.5 ns), after normalizing each decay by its intensity, to yield raw phasor coordinates, and . The microscope instrument response function (IRF) was measured from the instantaneous second harmonic generation (SHG) of urea crystals in either emission channel. A correction transformation (scale and rotation) based on the phasor representation of the IRF in either channel was applied to the raw phasor coordinates of the image pixels in that channel, and the resulting corrected pixel phasor coordinates were used to generate phasor plots. Contour maps were overlaid on the phasor representation of each image to better visualize the concentration of pixels in different regions of the phasor plot. For supplementary time-lapse videos, a false colormap was applied to intensity images based on the phasor representation of the lifetimes in the images. Color assignment was performed by projecting the phasor representation of each image pixel onto a false color axis that spans the line connecting the phasor locations of pure retinoid species, as measured in solution.
2.7. Immunocytochemistry
Organoids were fixed in 4% paraformaldehyde (Electron Microscopy Sciences) at room temperature (RT) with gentle agitation for 40 min and washed with PBS before cryopreservation in 30% sucrose. Organoids were cryosectioned (15- or 30-µ;m sections), blocked in 10% normal donkey serum (NDS), 5% bovine serum albumin, and 0.5% Triton X-100 for 1 hour at RT, and incubated with primary antibody at 4°C overnight. [1:100 goat anti-ARR3, Novus (NBP1-37003); 1:100 mouse anti-RHO, Millipore (MABN15); 1:100 rabbit anti-RDH8, Aviva (ARP65373_P050) or 1:50 rabbit anti-RDH12, Aviva (ARP52972_P050)]. Sections were incubated with species-specific, fluorophore-conjugated secondary antibodies at 1:500 for 30 min in the dark at RT (Alexa Fluor 488, AF546, AF594 and AF647; Thermo Fisher). Samples were imaged on a Nikon A1R-Si laser scanning confocal microscope.
3. Results
3.1. Fluorescence lifetime separates spectrally overlapping AT-RAL and AT-ROL
Figure 1(a) diagrams the visual cycle reactions in the retinal pigment epithelium and photoreceptor cell. After isomerization of 11-cis-RAL to AT-RAL upon absorption of visible light, AT-RAL is converted to AT-ROL in the photoreceptor cell. AT-ROL is then transferred to the retinal pigment epithelium where it is converted to 11-cis-RAL. Both AT-RAL and AT-ROL have broad fluorescence emission spectra with peaks just under 500 nm that are largely overlapping, as measured in anhydrous ethanol solutions on a spectrofluorometer using 350 nm excitation (Fig. 1(b)). AT-ROL has only a small blue-shift in fluorescence emission spectrum compared to AT-RAL, so spectral discrimination of AT-ROL and AT-RAL is difficult. However, FLIM of pure retinoids in 70% EtOH solution (two-photon excitation at 760 nm, emission 500-600 nm) reveals that AT-RAL fluorescence has a short lifetime of under 100 ps, while AT-ROL has a long lifetime of nearly 3 ns (Fig. 1(c)). This significant lifetime difference could be used to discriminate AT-ROL and AT-RAL to detect the presence and conversion of retinoids in cells.
3.2. Simultaneous three- and two-photon excitation improves spectral separation of AT-ROL and AT-RAL
Multiphoton excitation at 1040 nm allows for simultaneous two- and three-photon excitation of AT-ROL and AT-RAL in two emission channels. Thus, each emission channel transmits fluorescence excited at a different energy level, as shown in Fig. 2(a), in contrast to excitation at 760 nm, where emission in both channels is excited at the same two-photon energy level. Excitation at 1040 nm is compared to two-photon excitation alone (760 nm) in Fig. 2(a) for a 100 µM solution of AT-RAL in 70% EtOH. Fluorescence intensity in two emission channels (blue: 400-480 nm, green: 525-575 nm) are plotted versus average laser power on a log-log scale to determine the slope of a linear fit. A slope of 2 and 3 indicates two- and three-photon excitation, respectively. Excitation at 1040 nm indicates three-photon excited fluorescence in the blue channel and two-photon excited fluorescence in the green channel. Excitation at 760 nm indicates two-photon excited fluorescence in both the blue and green channels. Intensity measurements of 100 µM solutions of pure AT-RAL and AT-ROL in 70% EtOH at 1040 nm excitation show that the blue emission channel receives disproportionately more (i.e., about 12 times) signal from AT-ROL compared to AT-RAL (Fig. 2(b)). In the green emission channel, two-photon excitation at either 760 nm or 1040 nm result in roughly equal contribution from AT-RAL and AT-ROL (Fig. 2(c)). One caveat is that higher average laser powers (15 mW for 1040 nm 3P vs. 2-5 mW for 760 nm 2P) are needed for 1040 nm three-photon excitation to match the fluorescence intensity of 760 nm two-photon excitation, which requires trade-offs between signal to noise ratio and photothermal damage to the sample [17] be taken into consideration for long imaging sessions.
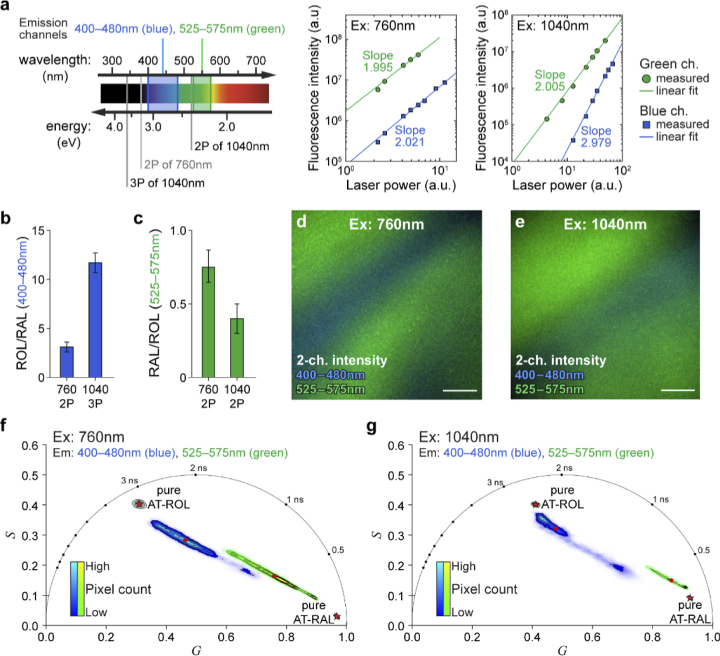
Fig. 2.
Simultaneous three- and two-photon excitation at 1040 nm increases spectral separation of AT-ROL and AT-RAL compared to two-photon excitation at 760 nm. (a) Comparison of multiphoton excitation at 760 nm and 1040 nm. Using 760 nm excitation, both emission channels (blue: 400-480 nm, green: 525-575 nm) collect fluorescence from two-photon excitation as indicated by the slope of the linear regression fit (slope≈2) on the log-log plot of fluorescence intensity vs. laser power. However, at 1040 nm excitation, the blue channel exclusively collects three-photon excited fluorescence (slope≈3), while the green channel primarily collects two-photon excited fluorescence (slope≈2). (b) Relative intensity of AT-ROL compared to AT-RAL in the blue emission channel is significantly increased under three-photon 1040 nm excitation (ROL/RAL≈12) compared to two-photon 760 nm excitation (ROL/RAL≈3). (c) Relative intensity of AT-RAL compared to AT-ROL in the green emission channel (RAL/ROL≈0.5) is not drastically different under either 1040 nm or 760 nm excitation. Ratios are calculated from intensity images of pure 100 µM solutions in 70% EtOH. (d) Two-channel fluorescence intensity image of a non-homogeneous mixture of 1 mM AT-RAL and 1 mM AT-ROL in ethanol, excited at 760 nm. (e) Two-channel intensity image of the same non-homogeneous mixture of AT-RAL and AT-ROL, excited at 1040 nm. Signal gradients within the images imply that different pixels emit different ratios of AT-RAL and AT-ROL fluorescence. (f) Phasor representation of the 760nm-excited image shows a mixture of AT-ROL (long lifetime) and AT-RAL (short lifetime) contributing to the emission in either channel. Phasor points fall on the axis connecting the pure retinoid locations (shown as red pentagrams). (g) Phasor representation of the 1040nm-excited image shows improved separation between AT-ROL and AT-RAL due to preferential three-photon excitation of AT-ROL compared to AT-RAL in the blue channel. The phasor points fall closer to the location of pure species. Red dots represent intensity-weighted phasor centroid in either channel. Red pentagrams represent phasor locations of the pure species. Scale bar: 100 µm.
To simulate the cell environment where mixtures of different retinoids are present, and to explore the utility of the new excitation scheme, a non-homogeneous mixture of 1 mM solutions of AT-RAL and AT-ROL in 70% EtOH was prepared by slowly pipetting each solution into the imaging dish from opposite directions and allowing them to slowly diffuse (without any mechanical mixing) over several minutes. FLIM was performed with simultaneous detection of emission in the blue and green channels by alternating the excitation wavelength between 1040 nm and 760 nm in one-minute intervals. Two-channel intensity images of the mixture (Fig. 2(d-e)) qualitatively show gradients of AT-RAL and AT-ROL within the field of view. Therefore, different pixels emit different ratios of AT-RAL and AT-ROL fluorescence. The phasor representation of the associated lifetimes reveals that every pixel (both 1040 nm and 760 nm excitation, in either emission channel) falls onto the linear axis connecting the phasor locations of the pure species, indicating that the fluorescence decay is comprised of a linear combination of AT-RAL and AT-ROL (Fig. 2(f-g)). For both excitation wavelengths, the blue channel phasor clusters near the AT-ROL location and the green channel phasor clusters near the AT-RAL location, reflecting the spectral separation of AT-ROL and AT-RAL emissions. However, the 760 nm phasor spans a wide portion of the common axis, with pixels distributed closer to the middle, indicating substantial bleed-through (Fig. 2(f)). Conversely, the 1040 nm phasor of the two emission channels separate further. Particularly, the three-photon excited blue channel preferentially excites the AT-ROL fraction of the mixture at each pixel location. Therefore, the phasor representation of nearly all image pixels in the 3P-excited blue channel fall close to the phasor location of pure AT-ROL, regardless of the mixture ratio (Fig. 2(g)).
3.3. Simultaneous three- and two-photon excitation provides image and lifetime contrast in mouse retina explants
To further validate the new multiphoton excitation scheme and to verify in-situ presence of the lifetime differences indicated by solution experiments, mouse retinal explants separated from the underlying RPE were imaged. Figure 3 shows the two-channel multiphoton-excited autofluorescence intensity image and the associated phasor plot of the photoreceptor layer of a mouse retina explant, one hour after exposure to bright white light, using either 760 nm (Fig. 3(a-b)) or 1040 nm (Fig. 3(c-d)) excitation.
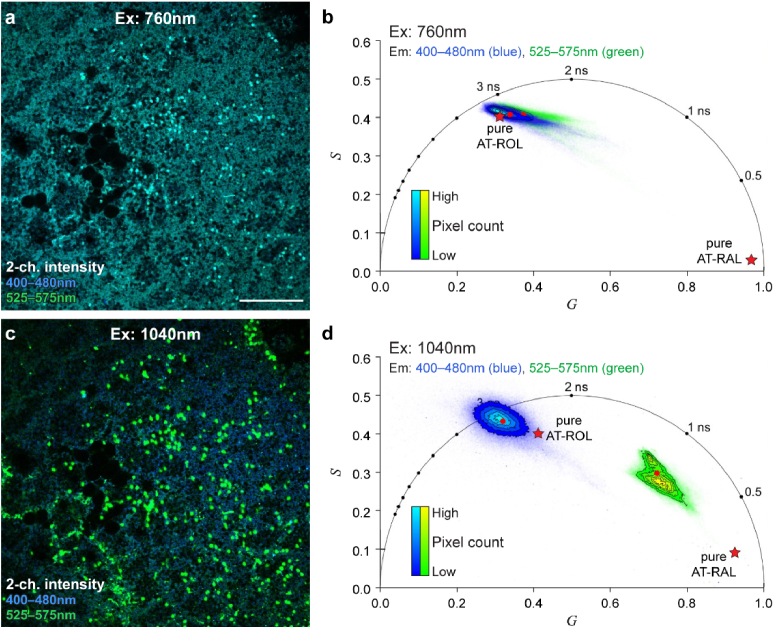
Fig. 3.
Three- and two-photon excitation at 1040 nm in mouse retina improves image and lifetime contrast compared to two-photon excitation at 760 nm. (a) Two-channel autofluorescence intensity image of the photoreceptor layer of a light-exposed mouse retina explant, excited at 760 nm. (b) Phasor representation of the 760 nm excitation image in (a) reveals that the long lifetime species, AT-ROL, is the dominant signal in both emission channels (blue, 400-480 nm; green, 550-600 nm). (c) Two-channel autofluorescence intensity image of the same field of view as (a), excited at 1040 nm, provides increased image contrast. Cone photoreceptors are prominent in the green emission channel and bleached rod photoreceptors are prominent in the blue emission channel. (d) Phasor representation of the 1040 nm excitation image in (c) reveals the long lifetime of AT-ROL in the blue emission channel, and a mixture of the short lifetime of AT-RAL with longer lifetimes of AT-ROL and metabolic fluorophores (FAD and NAD(P)H) in the green emission channel. Red dots represent intensity-weighted phasor centroid in either channel. Red pentagrams represent phasor locations of the pure species measured in solution. Scale bar: 100 µm.
The white-light photobleaching ensures that all the endogenous 11-cis-RAL from the photopigments is isomerized to AT-RAL, and the one-hour time delay ensures that AT-RAL is converted to AT-ROL by the photoreceptors. The phasor at 760 nm excitation (Fig. 3(b)) reveals the long lifetime of AT-ROL as the dominant signal in both emission channels, and the intensity image (Fig. 3(a)) does not show contrast between the green and blue emission channels. The phasor at 1040 nm (Fig. 3(d)) shows the long lifetime of AT-ROL in the three-photon excited blue channel, but the two-photon excited green channel includes shorter fluorescence lifetimes. The intensity image at 1040 nm excitation (Fig. 3(c)) shows the blue AT-ROL signal originating from the predominant rod photoreceptors and the green signal with shorter lifetime originating from the dispersed cone photoreceptors. Therefore, 1040 nm excitation provides improved contrast between blue and green emission channels compared to 760 nm excitation.
3.4. FLIM captures the dynamics of visual cycle reactions in mouse retina following light exposure
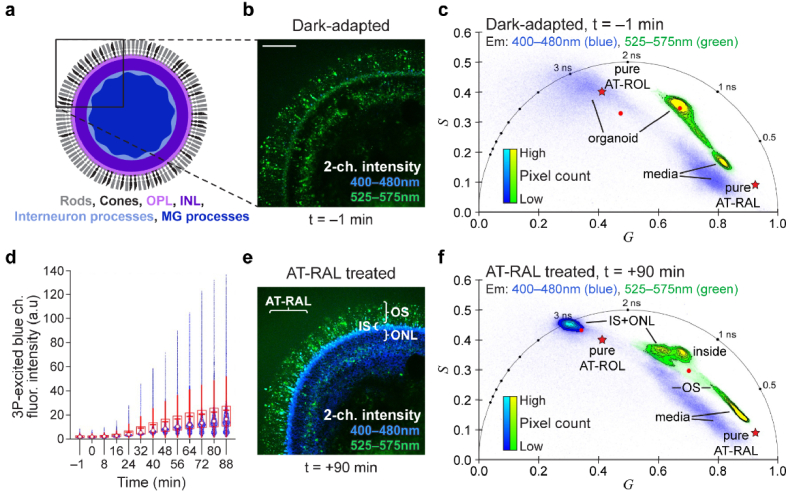
Given that three-photon excitation at 1040 nm can acquire fluorescence specifically from AT-ROL, we next characterized light response in a dark-adapted mouse retinal explant. FLIM images at 1040 nm excitation were acquired from a mouse retinal explant while dark-adapted and after white light exposure at 8-minute intervals up to one hour (Fig. 4).
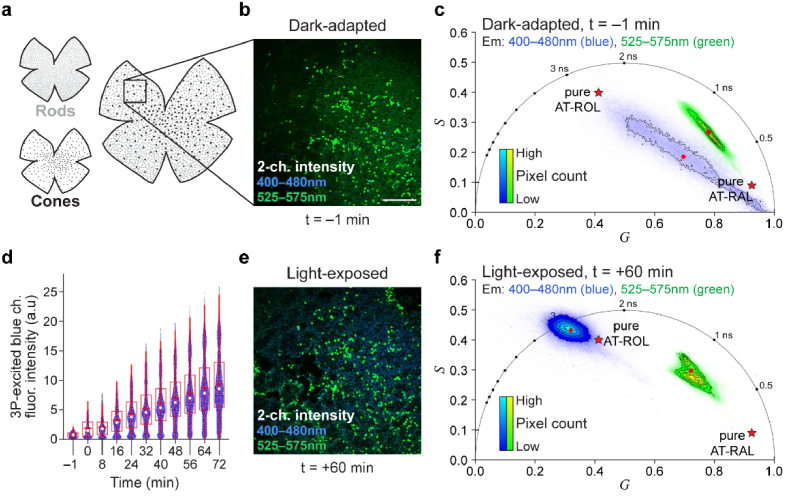
Fig. 4.
FLIM at 1040 nm excitation captures visual cycle dynamics in mouse retina explant following white light exposure. (a) Schematic of the mouse retina explant flat mount with RPE and sclera dissociated, and the photoreceptor layer preserved. Box shows microscopy field of view. (b) Two-channel fluorescence intensity image of the retina sample while dark-adapted. (c) Phasor representation of dark-adapted image reveals short lifetimes of RAL species and the absence of long lifetime of AT-ROL in the blue emission channel. The green channel phasor shows a mixture of RAL and metabolic fluorophores (FAD and NAD(P)H). (d) Following exposure to bright white light for 1 minute, the intensity of the three-photon (3P) excited fluorescence in the blue emission channel increases over 1 hour. White dot shows the median; red horizontal line shows the mean; box encompasses 25th to 75th percentile range; whiskers extend from the box to 1.5 times the interquartile range. Change in mean intensity with time is significant according to the linear trend test (t-statistic = 33, p = 8.7e-11). (e) 60 min after white light exposure, intensity image shows an increase in blue signal due to isomerization of 11-cis-RAL to AT-RAL and subsequent conversion of AT-RAL to AT-ROL. (f) Phasor representation of the image at one hour after light exposure reveals the long lifetime of AT-ROL. Red dots represent intensity-weighted phasor centroid in either channel. Red pentagrams represent phasor locations of the pure species measured in solution. Scale bar: 100 µm.
Figure 4(a) illustrates the mouse retinal explant and the microscopy field of view. Figure 4(b) shows the two-channel autofluorescence intensity image of the dark-adapted mouse retina. Figure 4(c) shows the phasor of the dark-adapted FLIM image in Fig. 4(b). The long lifetime of AT-ROL is absent in the blue channel (Fig. 4(c)) and the fluorescence intensity in the blue channel is low (Fig. 4(b)). However, after white light exposure, the intensity of autofluorescence in the three-photon excited blue channel increases over the course of one hour (Fig. 4(d)). Figure 4(e) shows the intensity image at the final timepoint (1 hour) and Fig. 4(f) shows the corresponding phasor. The more ubiquitous rod photoreceptors have increased intensity and long lifetime of AT-ROL in the blue channel, while the dispersed cone photoreceptors appear prominent in the green emission channel and maintain a shorter fluorescence lifetime. This could be indicative of the privileged access of cone cells to photopigment regeneration pathways via Müller glia [37–39] to maintain bright light vision.
Supplemental videos (Visualization 1 (15MB, avi) and Visualization 2 (70.3MB, avi) ) show two-photon excited two-channel autofluorescence intensity time series of a mouse retina explant, along with phasor plots and lifetime-based false colored images that visualize the formation of AT-ROL in photoreceptor cells after light exposure. Visualization 3 (16.5MB, avi) shows that the mouse retina explant photoreceptors can convert exogenously supplied AT-RAL to AT-ROL, resulting in an increase in fluorescence intensity over time.
3.5. Three-photon FLIM of visual cycle function in human iPSC-derived retinal organoids with AT-RAL treatment
3D-cultured hiPSC-derived retinal organoids older than 200 days in culture have developed outer retina-like photoreceptors with laminar organization including inner segments (IS), outer segments (OS), and an outer nuclear layer (ONL) [35]. Retinal organoids were dark adapted for 48 hours in the incubator and imaged at 1040 nm excitation with FLIM before and after one minute of white light exposure and again after treatment with 50 µM exogenous AT-RAL, similar to the mouse retina experiments. Figure 5(a) illustrates the retinal organoid structure and the microscopy field of view. Figure 5(b) shows the 2-channel autofluorescence intensity image of a dark-adapted retinal organoid at day 240 in culture. The corresponding phasor is shown in Fig. 5(c). The outer segments are prominent in the green channel with short fluorescence lifetime. Other endogenous fluorophores (e.g., metabolic coenzymes FAD and NAD(P)H) also contribute to the green channel which increases its lifetime to about 1 ns. The long lifetime of AT-ROL is sparsely present in the blue channel (primarily in the inner segments) with low image intensity, indicating low abundance.
Fig. 5.
FLIM at 1040 nm excitation captures visual cycle dynamics in a stem cell-derived retinal organoid. (a) Schematic of the 3D-cultured stem cell-derived retinal organoid. Box shows microscopy field of view. OPL: outer plexiform layer, INL: inner nuclear layer, MG: Müller Glia. (b) Two-channel autofluorescence intensity image of a dark-adapted retinal organoid at day 240 in culture (blue: 400-480 nm, green: 525-575 nm). (c) Phasor representation of dark-adapted image in (b) shows short fluorescence lifetimes of the media surrounding the organoid as well as intermediate lifetimes of metabolic fluorophores (FAD, NAD(P)H) from the organoid in the green channel and sparse long lifetime of AT-ROL in the blue channel. (d) The organoid was treated with exogenous 50 µM AT-RAL, which simulates photobleaching of visual pigments, and the intensity of the three-photon excited fluorescence in the blue emission channel increases over 1 hour at a rate similar to the mouse retina. White dot shows the median; red horizontal line shows the mean; box encompasses 25th to 75th percentile range; whiskers extend from the box to 1.5 times the interquartile range. Change in mean intensity with time is significant according to the linear trend test (t-statistic = 27, p = 1.9e-11). (e) Two-channel fluorescence intensity image of the retinal organoid 90 min after treatment with AT-RAL shows an increase in the blue channel signal, due to conversion of AT-RAL to AT-ROL, in the photoreceptor cells, particularly in the inner segments (IS), outer nuclear layer (ONL) and the outer segments (OS). (f) Phasor representation of the treated organoid image in (e) reveals abundant long lifetime of AT-ROL in the blue channel, while the exogenous AT-RAL in the media (i.e., background seen at the upper left corner of the intensity image) appears with its characteristic short lifetime in the green channel of the phasor plot. Red dots represent intensity-weighted phasor centroid of the organoid in either channel. Red pentagrams represent phasor locations of the pure species measured in solution. Scale bar: 100 µm.
Unlike the mouse retina, white light exposure did not induce a consistent signal intensity and lifetime increase in retinal organoids. This may reflect a lack of photoreceptor outer segment maturity and/or the absence of any prior association with an organized RPE layer that would allow sufficient intrinsic production of 11-cis-RAL for signal detection. However, when the retinal organoid was treated with exogenous AT-RAL, which simulates the photobleaching of photopigments and isomerization of 11-cis-RAL to AT-RAL, the fluorescence intensity of the three-photon excited blue emission channel increases over the course of an hour (Fig. 5(d)). The time series of intensity images can be seen in supplementary Visualization 4 (28.5MB, avi) . The fluorescence intensity image at the end of the time series is shown in Fig. 5(e) and the corresponding phasor is given in Fig. 5(f). The long lifetime of AT-ROL is prominent in the blue channel, with the highest fluorescence intensity in the photoreceptor IS and ONL layers, respectively. The better formed OS includes some AT-ROL signal, but the concentration is low, such that the abundant exogenous AT-RAL fluorescence dominates the AT-ROL signal, resulting in only a small lifetime increase for the OS in the blue channel. Supplementary Fig. S1 shows that organoid layers can be masked based on the pixel locations in the phasor plot. A time series of phasor-based false colored intensity images (supplementary Visualization 5 (66.8MB, avi) ) visualizes the absorption of AT-RAL with short lifetime, followed by its conversion to AT-ROL with long lifetime in the organoid photoreceptors. In a control experiment (Fig. S2), the conversion of AT-RAL to AT-ROL was inhibited [21,23] by pre-treatment of a dark-adapted organoid with 100 µM all-trans retinoic acid (ATRA) for 30 minutes. Subsequent treatment with 50 µM AT-RAL resulted in a minimal increase in AT-ROL as seen in the intensity and lifetime changes in the three-photon excited blue emission channel (Fig. S2 d-f).
These results suggest that photoreceptors within hiPSC-derived retinal organoids demonstrate visual cycle function, and that this function can be noninvasively monitored by FLIM at 1040 nm excitation. Note that while the lifetime data is aggregated at the image level in the presented results, the analysis can be done at the single cell or subcellular level (OS, IS, ONL) as presented in supplementary Fig. S1.
3.6. Immunocytochemistry (ICC) of retinal organoid photoreceptor cell expression of RDH enzymes involved in the visual cycle
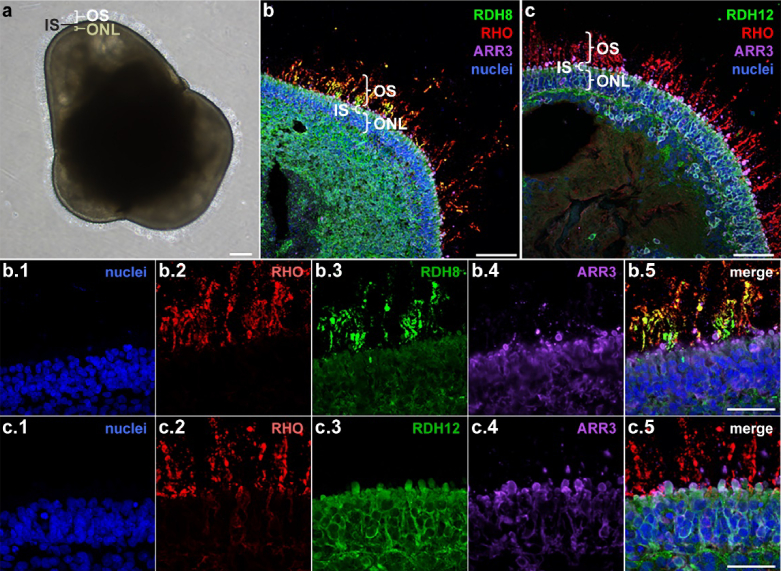
The data presented in Fig. 5 suggests that human retinal organoid derived photoreceptors have a functional visual cycle. To further investigate this finding, we looked for the presence of RDH enzymes in retinal organoid inner and outer segments by ICC. Figure 6 presents ICC images of mature hiPSC-derived retinal organoids (260 days in culture) showing the photoreceptor organization, including expression of rhodopsin (RHO) in the rod outer segments, expression of ARR3 in cone photoreceptors, and the presence of a stratified outer nuclear layer. Importantly, the retinal dehydrogenase enzyme RDH8 is expressed in the photoreceptor outer segments and the RDH12 enzyme is expressed in the inner segments and outer nuclear layer of the photoreceptor cells and some Müller glia. These enzymes catalyze the reduction reaction that converts AT-RAL to AT-ROL. The location of RDH expression in ICC images of fixed organoids coincides with the location of the three-photon excited AT-ROL signal in the FLIM images of live organoids.
Fig. 6.
Immunocytochemistry (ICC) of fixed retinal organoids reveals expression of retinal dehydrogenase (RDH) enzyme in photoreceptor cells. (a) Bright field image of a retinal organoid after 260 days in culture. (b) ICC indicates presence of outer segment (OS)-localized RDH8 enzyme [see b3] in photoreceptor outer segments as well as its expression in ectopic photoreceptor cells deeper inside the organoid. (c) ICC indicates presence of inner segment (IS)-localized RDH12 [see c3] in photoreceptor inner segments (IS) and the outer nuclear layer (ONL). The location of the RDH expression in fixed organoids coincides with the location of the three-photon excited AT-ROL fluorescence signal in the blue channel of live retinal organoids. RHO: the rod photopigment rhodopsin; RDH8: OS-localized retinal dehydrogenase; RDH12: IS- and ONL-localized retinal dehydrogenase; ARR3: cone arrestin; DAPI: nuclei. OS: outer segments; IS: inner segments; ONL: outer nuclear layer. Scale bar: 50 µm.
4. Discussion and conclusions
High-throughput and noninvasive functional measurements of phototransduction and visual cycle are needed to screen treatments in vitro for retinal disease (e.g., biologics, gene therapy, genome editors [40–42]) and to optimize iPSC-derived retinal cells in vitro for regenerative cell transfer therapy. Here, we developed autofluorescence methods to monitor a critical step in retinoid recycling in photoreceptor cells based on retinoid fluorescence intensity, spectral, and lifetime changes in the slow reactions of the visual cycle. We used a new multiphoton excitation scheme that leverages spectral differences in AT-RAL and AT-ROL at two- and three-photon excitation with 1040 nm, combined with fluorescence lifetime differences between AT-RAL and AT-ROL to resolve the dynamics of the visual cycle with cellular resolution.
Simultaneous two- and three-photon excitation of retinoids increased spectral contrast between the emission channels (Fig. 2–3) due to higher specificity of 3P excitation for AT-ROL. However, the 2P-excited channel is not specific for AT-RAL and collects a combination of fluorescence signals from AT-ROL, AT-RAL, and other endogenous fluorophores such as metabolic coenzymes FAD and NAD(P)H. In mouse retina samples, this contrast appears to highlight a difference between rod and cone photoreceptors (Fig. 3–4). This observation is supported by the different function, morphology, and organization of cones relative to rods in mouse retina [43]. Cones are responsible for photopic vision and can regenerate photopigments at a faster rate than rods by using a second visual cycle involving Müller glia [37]. This could explain the absence of a detectable concentration of AT-ROL in cones following light exposure. Cones comprise 3% of total photoreceptors and have larger inner segments with more mitochondria, which could explain the higher intensity for cones in the green channel of the 1040 nm excited images (Fig. 4). Additionally, cone somas are confined to the outer edge of the ONL adjacent to the IS layer, while rod somas exclusively form the deeper ONL [43]. This description is consistent with our depth-resolved imaging of the mouse retina explants (see supplementary Fig. S3). However, further investigation of this contrast with direct comparison to immunofluorescence labeled histology is needed.
While 3P excitation results in higher specificity for AT-ROL, it also provides lower fluorescence intensity and, therefore, lower signal to noise ratio (SNR) compared to 2P excitation. This is due to the lower 3P action cross section ( ) of AT-ROL by a ratio factor of compared to its 2P action cross section ( ), assuming similar quantum efficiency at the two wavelengths, as estimated in [44]. Given that the ratio of 3P- and 2P-excited fluorescence is proportional to , and provided a typical excitation laser intensity ( ) on the order of at the sample plane for a few mW of femtosecond-pulsed laser power, it is expected that the 1040 nm 3P-excited fluorescence intensity may be up to 3 orders of magnitude smaller than the 760 nm 2P-excited fluorescence for the same average laser power. This is consistent with the results in Fig. 2(a). The SNR trade-off was justified in our experiments for the increased specificity and contrast that 3P excitation provided for identifying the visual cycle fluorophores. Although 3P-excited fluorescence SNR increases with the third power of the excitation intensity, we limited the 1040 nm laser power to 15 mW to avoid photothermal damage to the sample.
The lifetime measurements in solution established the short and long fluorescence lifetime of AT-RAL and AT-ROL, respectively. However, in live cell images, AT-ROL presents with slightly longer fluorescence lifetime than was measured in solution. This is because AT-ROL in solution degrades within minutes under atmospheric oxygen conditions, due to oxidation reactions that form AT-RAL. As such, it is conceivable that the AT-ROL signal measured in cells is purer than the one measured in solution.
Regarding sensitivity of the FLIM technique to small light stimuli, it must be noted that the fluorescent signal intensity is proportional to fluorophore concentrations. Therefore, we used intense white light photobleaching to maximize the release of retinoids and thus the fluorescent signal available for identification of these fluorophores. However, milder light exposures such as the multiphoton excitation light itself can also result in detectable changes in fluorescence intensity and lifetime (see supplementary Fig. S4), thereby supporting the sensitivity of this technique.
Finally, while the presented imaging approach does not directly measure hyperpolarization and neural signaling of photoreceptors in response to small light stimuli, it can be used to assay visual cycle function, which is affected in diseases like Leber congenital amaurosis and retinitis pigmentosa [45], or used to determine presence of endogenous photopigments in lab-grown photoreceptors and whole retina explants to optimize their culture. Overall, multiphoton FLIM of photoresponse allows for noninvasive imaging of thick mouse retinal explants and whole live organoids with minimal handling at multiple timepoints in a higher-throughput format than traditional photoactivation (e.g., patch clamp) or analytical chemistry measurements (e.g., high-performance liquid chromatography, mass spectrometry).
Acknowledgments
The authors thank Matthew Stefely for the help with figure illustrations, and Maria A. Fernandez Zepeda for her technical assistance.
Funding
National Eye Institute10.13039/100000053 (U01EY032333, R01 EY021218); National Institute of Biomedical Imaging and Bioengineering10.13039/100000070 (U01EB029371); National Heart, Lung, and Blood Institute10.13039/100000050 (U01HL145792); The Somatic Cell Genome Editing (SCGE) Consortium of the NIH Common Fund; Morgridge Institute for Research (Interdisciplinary Postdoctoral Fellowship); Retina Research Foundation10.13039/100013779 (Daniel M. Albert Chair, Emmett A. Humble Chair, Kathryn and Latimer Murfee Chair, M. D. Matthews Research Professorship); Muskingum County Community Foundation10.13039/100002239 (Sarah E. Slack Prevention of Blindness fund); The McPherson Eye Research Institute (Sandra Lemke Trout Chair in Eye Research); Research to Prevent Blindness10.13039/100001818; Carl Marshall and Mildred Almen Reeves Foundation10.13039/100010657; Eunice Kennedy Shriver National Institute of Child Health and Human Development10.13039/100009633 (U54HD090256).
Disclosures
D.M.G. has an ownership interest in Opsis Therapeutics LLC, which has licensed the technology to generate retinal organoids from pluripotent stem cell sources referenced in this publication. D.M.G. also declared intellectual rights through the Wisconsin Alumni Research Foundation and a consultant role with FUJIFILM Cellular Dynamics International. All other authors declared no conflicts of interest.
Data availability
Data underlying the results presented in this paper are not publicly available at this time but may be obtained from the authors upon reasonable request.
Supplemental document
See Supplement 1 (7.8MB, pdf) for supporting content.
References
- 1.Meyer J. S., Shearer R. L., Capowski E. E., Wright L. S., Wallace K. A., McMillan E. L., Zhang S. C., Gamm D. M., “Modeling early retinal development with human embryonic and induced pluripotent stem cells,” Proc. Natl. Acad. Sci. 106(39), 16698–16703 (2009). 10.1073/pnas.0905245106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jin Z.-B., Okamoto S., Xiang P., Takahashi M., “Integration-Free Induced Pluripotent Stem Cells Derived from Retinitis Pigmentosa Patient for Disease Modeling,” Stem Cells Trans. Medicine 1(6), 503–509 (2012). 10.5966/sctm.2012-0005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamba D. A., Gust J., Reh T. A., “Transplantation of Human Embryonic Stem Cell-Derived Photoreceptors Restores Some Visual Function in Crx-Deficient Mice,” Cell Stem Cell 4(1), 73–79 (2009). 10.1016/j.stem.2008.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonzalez-Cordero A., Kruczek K., Naeem A., Fernando M., Kloc M., Ribeiro J., Goh D., Duran Y., Blackford S. J. I., Abelleira-Hervas L., Sampson R. D., Shum I. O., Branch M. J., Gardner P. J., Sowden J. C., Bainbridge J. W. B., Smith A. J., West E. L., Pearson R. A., Ali R. R., “Recapitulation of Human Retinal Development from Human Pluripotent Stem Cells Generates Transplantable Populations of Cone Photoreceptors,” Stem Cell Rep. 9(3), 820–837 (2017). 10.1016/j.stemcr.2017.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li L., Zhao H., Xie H., Akhtar T., Yao Y., Cai Y., Dong K., Gu Y., Bao J., Chen J., Zhang M., Zhong K., Xu W., Xue T., “Electrophysiological characterization of photoreceptor-like cells in human inducible pluripotent stem cell-derived retinal organoids during in vitro maturation,” Stem Cells 39(7), 959–974 (2021). 10.1002/stem.3363 [DOI] [PubMed] [Google Scholar]
- 6.Ingram N. T., Sampath A. P., Fain G. L., “Voltage-clamp recordings of light responses from wild-type and mutant mouse cone photoreceptors,” J. Gen. Physiol. 151(11), 1287–1299 (2019). 10.1085/jgp.201912419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woodruff M. L., Wang Z., Chung H. Y., Redmond T. M., Fain G. L., Lem J., “Spontaneous activity of opsin apoprotein is a cause of Leber congenital amaurosis,” Nat. Genet. 35(2), 158–164 (2003). 10.1038/ng1246 [DOI] [PubMed] [Google Scholar]
- 8.Cowan C. S., Renner M., de Gennaro M., Gross-Scherf B., Goldblum D., Hou Y., Munz M., Rodrigues T. M., Krol J., Szikra T., Cuttat R., Waldt A., Papasaikas P., Diggelmann R., Patino-Alvarez C. P., Galliker P., Spirig S. E., Pavlinic D., Gerber-Hollbach N., Schuierer S., Srdanovic A., Balogh M., Panero R., Kusnyerik A., Szabo A., Stadler M. B., Orgül S., Picelli S., Hasler P. W., Hierlemann A., Scholl H. P. N., Roma G., Nigsch F., Roska B., “Cell Types of the Human Retina and Its Organoids at Single-Cell Resolution,” Cell 182(6), 1623–1640.e34 (2020). 10.1016/j.cell.2020.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hillmann D., Spahr H., Pfäffle C., Sudkamp H., Franke G., Hüttmann G., “In vivo optical imaging of physiological responses to photostimulation in human photoreceptors,” Proc. Natl. Acad. Sci. U. S. A. 113(46), 13138–13143 (2016). 10.1073/pnas.1606428113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang F., Kurokawa K., Lassoued A., Crowell J. A., Miller D. T., “Cone photoreceptor classification in the living human eye from photostimulation-induced phase dynamics,” Proc. Natl. Acad. Sci. U. S. A. 116(16), 7951–7956 (2019). 10.1073/pnas.1816360116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lassoued A., Zhang F., Kurokawa K., Liu Y., Bernucci M. T., Crowell J. A., Miller D. T., “Cone photoreceptor dysfunction in retinitis pigmentosa revealed by optoretinography,” Proc. Natl. Acad. Sci. U. S. A. 118(47), e2107444118 (2021). 10.1073/pnas.2107444118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pandiyan V. P., Maloney-Bertelli A., Kuchenbecker J. A., Boyle K. C., Ling T., Chen Z. C., Park B. H., Roorda A., Palanker D., Sabesan R., “The optoretinogram reveals the primary steps of phototransduction in the living human eye,” Sci. Adv. 6(37), 1 (2020). 10.1126/sciadv.abc1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Redmond T. M., Yu S., Lee E., Bok D., Hamasaki D., Chen N., Goletz P., Ma J. X., Crouch R. K., Pfeifer K., “Rpe65 is necessary for production of 11-cis-vitamin A in the retinal visual cycle,” Nat. Genet. 20(4), 344–351 (1998). 10.1038/3813 [DOI] [PubMed] [Google Scholar]
- 14.Imanishi Y., Batten M. L., Piston D. W., Baehr W., Palczewski K., “Noninvasive two-photon imaging reveals retinyl ester storage structures in the eye,” J. Cell Biol. 164(3), 373–383 (2004). 10.1083/jcb.200311079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuksa V., Imanishi Y., Batten M., Palczewski K., Moise A. R., “Retinoid cycle in the vertebrate retina: experimental approaches and mechanisms of isomerization,” Vision Res. 43(28), 2959–2981 (2003). 10.1016/S0042-6989(03)00482-6 [DOI] [PubMed] [Google Scholar]
- 16.Wu Q., Chen C., Koutalos Y., “All-Trans Retinol in Rod Photoreceptor Outer Segments Moves Unrestrictedly by Passive Diffusion,” Biophys. J. 91(12), 4678–4689 (2006). 10.1529/biophysj.106.086728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palczewska G., Boguslawski J., Stremplewski P., Kornaszewski L., Zhang J., Dong Z., Liang X. X., Gratton E., Vogel A., Wojtkowski M., Palczewski K., “Noninvasive two-photon optical biopsy of retinal fluorophores,” Proc. Natl. Acad. Sci. U. S. A. 117(36), 22532–22543 (2020). 10.1073/pnas.2007527117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sparrow J. R., Gregory-Roberts E., Yamamoto K., Blonska A., Ghosh S. K., Ueda K., Zhou J., “The bisretinoids of retinal pigment epithelium,” Prog. Retinal Eye Res. 31(2), 121–135 (2012). 10.1016/j.preteyeres.2011.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takeuchi S., Tahara T., “Ultrafast Fluorescence Study on the Excited Singlet-State Dynamics of all-trans-Retinal,” J. Phys. Chem. A 101(17), 3052–3060 (1997). 10.1021/jp962765s [DOI] [Google Scholar]
- 20.Kono M., Goletz P. W., Crouch R. K., “11-cis- and All-trans-Retinols Can Activate Rod Opsin: Rational Design of the Visual Cycle†,” Biochemistry 47(28), 7567–7571 (2008). 10.1021/bi800357b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krzysztof Palczewski P., Jager S., Buczylko J., Crouch R. K., Lucille Bredberg D., Peter Hofmann K., Ann Asson-Batres M., Saari J. C., Stein Professor D., is J., “Rod Outer Segment Retinol Dehydrogenase: Substrate Specificity and Role in,” Biochemistry 33(46), 13741–13750 (1994). 10.1021/bi00250a027 [DOI] [PubMed] [Google Scholar]
- 22.Parker R. O., Crouch R. K., “Retinol dehydrogenases (RDHs) in the visual cycle,” Exp. Eye Res. 91(6), 788–792 (2010). 10.1016/j.exer.2010.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen C., Tsina E., Cornwall M. C., Crouch R. K., Vijayaraghavan S., Koutalos Y., “Reduction of All-trans Retinal to All-trans Retinol in the Outer Segments of Frog and Mouse Rod Photoreceptors,” Biophys. J. 88(3), 2278–2287 (2005). 10.1529/biophysj.104.054254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen C., Thompson D. A., Koutalos Y., “Reduction of all-trans-retinal in vertebrate rod photoreceptors requires the combined action of RDH8 and RDH12,” J. Biol. Chem. 287(29), 24662–24670 (2012). 10.1074/jbc.M112.354514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen C., Blakeley L. R., Koutalos Y., “Formation of all-trans Retinol after Visual Pigment Bleaching in Mouse Photoreceptors,” Invest. Ophthalmol. Visual Sci. 50(8), 3589–3595 (2009). 10.1167/iovs.08-3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erez Y., Presiado I., Gepshtein R., Simkovitch R., Huppert D., “Time-resolved fluorescence study of all-trans-retinal,” J. Mod. Opt. 61(19), 1589–1604 (2014). 10.1080/09500340.2014.948508 [DOI] [Google Scholar]
- 27.Bachilo S. M., Gillbro T., “Fluorescence of Retinal Schiff Base in Alcohols,” J. Phys. Chem. A 103(15), 2481–2488 (1999). 10.1021/jp983646a [DOI] [Google Scholar]
- 28.Chihara K., Takemura T., Yamaoka T., Yamamoto N., Schaffer A., Becker R. S., “Visual pigments—10. Spectroscopy and photophysical dynamics of retinol and retinyl ether,” Photochem. Photobiol. 29(5), 1001–1008 (1979). 10.1111/j.1751-1097.1979.tb07804.x [DOI] [Google Scholar]
- 29.Chmykh Y. G., Nadeau J. L., “Characterization of Retinol Stabilized in Phosphatidylcholine Vesicles with and without Antioxidants,” ACS Omega 5(29), 18367–18375 (2020). 10.1021/acsomega.0c02102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Georghiou S., Churchich J. E., “Nanosecond spectroscopy of retinol,” J. Biol. Chem. 250(3), 1149–1151 (1975). 10.1016/S0021-9258(19)41902-9 [DOI] [PubMed] [Google Scholar]
- 31.Kane M. A., Napoli J. L., “Quantification of Endogenous Retinoids,” Methods Mol. Biol. 652, 1–54 (2010). 10.1007/978-1-60327-325-1_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sparrow J. R., Wu Y., Nagasaki T., Yoon K. D., Yamamoto K., Zhou J., “Fundus autofluorescence and the bisretinoids of retina,” Photochem. Photobiol. Sci. 9(11), 1480–1489 (2010). 10.1039/c0pp00207k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Müller B., “Organotypic Culture of Adult Mouse Retina,” Methods Mol. Biol. 1940, 181–191 (2019). 10.1007/978-1-4939-9086-3_13 [DOI] [PubMed] [Google Scholar]
- 34.Ames A., Nesbett F. B., “In Vitro Retina as an Experimental Model of the Central Nervous System,” J. Neurochem. 37(4), 867–877 (1981). 10.1111/j.1471-4159.1981.tb04473.x [DOI] [PubMed] [Google Scholar]
- 35.Capowski E. E., Samimi K., Mayerl S. J., Phillips M. J., Pinilla I., Howden S. E., Saha J., Jansen A. D., Edwards K. L., Jager L. D., Barlow K., Valiauga R., Erlichman Z., Hagstrom A., Sinha D., Sluch V. M., Chamling X., Zack D. J., Skala M. C., Gamm D. M., “Reproducibility and staging of 3D human retinal organoids across multiple pluripotent stem cell lines,” Development 146(1), dev171686 (2018). 10.1242/dev.171686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Digman M. A., Caiolfa V. R., Zamai M., Gratton E., “The Phasor Approach to Fluorescence Lifetime Imaging Analysis,” Biophys. J. 94(2), L14–L16 (2008). 10.1529/biophysj.107.120154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mata N. L., Radu R. A., Clemmons R. S., Travis G. H., “Isomerization and Oxidation of Vitamin A in Cone-Dominant Retinas: A Novel Pathway for Visual-Pigment Regeneration in Daylight,” Neuron 36(1), 69–80 (2002). 10.1016/S0896-6273(02)00912-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J. S., Estevez M. E., Cornwall M. C., Kefalov V. J., “Intra-retinal visual cycle required for rapid and complete cone dark adaptation,” Nat. Neurosci. 12(3), 295–302 (2009). 10.1038/nn.2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kiser P. D., Palczewski K., “Retinoids and Retinal Diseases,” Annu. Rev. Vis. Sci. 2(1), 197–234 (2016). 10.1146/annurev-vision-111815-114407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saha K., Sontheimer E. J., Brooks P. J., Dwinell M. R., Gersbach C. A., Liu D. R., Murray S. A., Tsai S. Q., Wilson R. C., Anderson D. G., Asokan A., Banfield J. F., Bankiewicz K. S., Bao G., Bulte J. W. M., Bursac N., Campbell J. M., Carlson D. F., Chaikof E. L., Chen Z. Y., Cheng R. H., Clark K. J., Curiel D. T., Dahlman J. E., Deverman B. E., Dickinson M. E., Doudna J. A., Ekker S. C., Emborg M. E., Feng G., Freedman B. S., Gamm D. M., Gao G., Ghiran I. C., Glazer P. M., Gong S., Heaney J. D., Hennebold J. D., Hinson J. T., Khvorova A., Kiani S., Lagor W. R., Lam K. S., Leong K. W., Levine J. E., Lewis J. A., Lutz C. M., Ly D. H., Maragh S., McCray P. B., McDevitt T. C., Mirochnitchenko O., Morizane R., Murthy N., Prather R. S., Ronald J. A., Roy S., Roy S., Sabbisetti V., Saltzman W. M., Santangelo P. J., Segal D. J., Shimoyama M., Skala M. C., Tarantal A. F., Tilton J. C., Truskey G. A., Vandsburger M., Watts J. K., Wells K. D., Wolfe S. A., Xu Q., Xue W., Yi G., Zhou J., “The NIH Somatic Cell Genome Editing program,” Nature 592(7853), 195–204 (2021). 10.1038/s41586-021-03191-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maeder M. L., Stefanidakis M., Wilson C. J., Baral R., Barrera L. A., Bounoutas G. S., Bumcrot D., Chao H., Ciulla D. M., DaSilva J. A., Dass A., Dhanapal V., Fennell T. J., Friedland A. E., Giannoukos G., Gloskowski S. W., Glucksmann A., Gotta G. M., Jayaram H., Haskett S. J., Hopkins B., Horng J. E., Joshi S., Marco E., Mepani R., Reyon D., Ta T., Tabbaa D. G., Samuelsson S. J., Shen S., Skor M. N., Stetkiewicz P., Wang T., Yudkoff C., Myer V. E., Albright C. F., Jiang H., “Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10,” Nat. Med. 25(2), 229–233 (2019). 10.1038/s41591-018-0327-9 [DOI] [PubMed] [Google Scholar]
- 42.Sinha D., Steyer B., Shahi P. K., Mueller K. P., Valiauga R., Edwards K. L., Bacig C., Steltzer S. S., Srinivasan S., Abdeen A., Cory E., Periyasamy V., Siahpirani A. F., Stone E. M., Tucker B. A., Roy S., Pattnaik B. R., Saha K., Gamm D. M., “Human iPSC Modeling Reveals Mutation-Specific Responses to Gene Therapy in a Genotypically Diverse Dominant Maculopathy,” Am. J. Hum. Genet. 107(2), 278–292 (2020). 10.1016/j.ajhg.2020.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carter-Dawson L. D., Lavail M. M., “Rods and cones in the mouse retina. I. Structural analysis using light and electron microscopy,” J. Comp. Neurol. 188(2), 245–262 (1979). 10.1002/cne.901880204 [DOI] [PubMed] [Google Scholar]
- 44.Palczewska G., Maeda T., Imanishi Y., Sun W., Chen Y., Williams D. R., Piston D. W., Maeda A., Palczewski K., “Noninvasive multi–photon fluorescence microscopy resolves retinol and retinal–condensation products in mouse eyes,” Nat. Med. 16(12), 1444–1449 (2010). 10.1038/nm.2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarkar H., Toms M., Moosajee M., “Involvement of Oxidative and Endoplasmic Reticulum Stress in RDH12-Related Retinopathies,” Int. J. Mol. Sci. 22(16), 8863 (2021). 10.3390/ijms22168863 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data underlying the results presented in this paper are not publicly available at this time but may be obtained from the authors upon reasonable request.