Abstract
Ammonia-oxidizing bacteria were detected by PCR amplification of DNA extracted from filtered water samples throughout the water column of Mono Lake, California. Ammonia-oxidizing members of the β subdivision of the division Proteobacteria (β-subdivision Proteobacteria) were detected using previously characterized PCR primers; target sequences were detected by direct amplification in both surface water and below the chemocline. Denaturing gradient gel electrophoresis analysis indicated the presence of at least four different β-subdivision ammonia oxidizers in some samples. Subsequent sequencing of amplified 16S rDNA fragments verified the presence of sequences very similar to those of cultured Nitrosomonas strains. Two separate analyses, carried out under different conditions (different reagents, locations, PCR machines, sequencers, etc.), 2 years apart, detected similar ranges of sequence diversity in these samples. It seems likely that the physiological diversity of nitrifiers exceeds the diversity of their ribosomal sequences and that these sequences represent members of the Nitrosomonas europaea group that are acclimated to alkaline, high-salinity environments. Primers specific for Nitrosococcus oceanus, a marine ammonia-oxidizing bacterium in the γ subdivision of the Proteobacteria, did not amplify target from any samples.
Mono Lake is an alkaline, hypersaline, closed-basin lake in central California just east of the Sierra Nevada Mountains (38°N, 119°W). Exceptionally heavy rainfall in 1982 and 1983 caused a freshening of the surface layer; the resulting large-density difference between surface and deep waters caused the lake to become ectogenically meromictic (15, 18). The lake became stratified with a chemocline in the vicinity of 15 m (total depth, approximately 30 m) and did not turn over or mix thoroughly until 1988. In the interval, the difference in salinity between the deep water and surface layer gradually decreased (15), due to evaporation and removal of surface water to supply drinking water to southern California. After 1988, the lake resumed its normal pattern of annual winter holomixis (15).
The seasonal density stratification leads to stratification in oxygen and other parameters, such as nitrogenous nutrients (16), and this stratification is expected to be reflected in the depth distribution of microbial activities. Both ammonium and methane accumulate in the deep layer (25); the major biological sinks for these compounds are thought to be oxidation by obligately aerobic chemolithotrophic bacteria in the surface layer and, for methane, anaerobic oxidation in the deep layer. The depth distributions of methane and ammonia oxidation, measured by radiotracer experiments, were previously reported (19).
Ammonia-oxidizing bacteria are responsible for the oxidation of ammonia to nitrite, the first step in nitrification, in terrestrial and aquatic habitats of all kinds and are found in the β and γ subdivisions of the division Proteobacteria. The number of species of ammonia oxidizers that have been described is relatively small (21), although a greater diversity of uncultured strains has been detected in various environments (10, 12, 22, 23, 36, 37, 41, 43).
Considering Mono Lake's recent history of salinity variation, it was not obvious what kinds of ammonia oxidizers might be responsible for the nitrification rates observed in the lake (19). While the lake is in a “normal” temperate phase, the usual freshwater bacteria, such as Nitrosomonas and Nitrosospira strains in the β subdivision, might be favored, while Nitrosococcus oceanus strains, representatives of the γ-subdivision ammonia oxidizers, might be favored in the lake's current saline state. This question was investigated by using PCR and sequence analysis focusing on the 16S rRNA gene in nitrifying bacteria.
MATERIALS AND METHODS
Water samples.
Water column samples were collected near a permanently moored buoy at a station near the middle of the lake (28-m depth) in April and July 1995. Water samples from discrete depths were collected with a 5-liter polyvinyl chloride sampling bottle (25) and stored on ice in 1-liter brown polyethylene bottles prior to filtration. A peristaltic pump was used to circulate water samples (400 to 700 ml) through Sterivex (0.22-μm-pore-size) filter heads (Millipore). Filters were stored frozen in 1.8 ml of lysis buffer (40 mM EDTA, 50 mM Tris, 0.74 M sucrose [pH 8.3]) prior to DNA extraction.
Bacterial strains and DNA extraction.
Bacterial strains were maintained in a pure culture as previously described (45), using freshwater (33) or seawater (48) medium as appropriate. High-molecular-weight, bacterial genomic DNA from various strains in the culture collection (required for controls and for standardization in the denaturing gradient gel electrophoresis [DGGE] analysis) was obtained from cells harvested from 1-liter cultures and purified as previously described (41).
DNA was released from the cells collected on the filters by gentle enzymatic and detergent lysis with a slight modification of standard methods (3). Nucleic acids were purified by chloroform extraction and visualized on 1% (wt/vol) horizontal agarose minigels run in 1× TAE buffer (40 mM Tris, 5 mM sodium acetate, 1 mM EDTA). High-molecular-weight, good-quality DNA was obtained from all samples and was used in PCR without further purification. The DNA samples are referred to by month and sample number (e.g., 4.18 = sample number 18 collected in April) (see Table 3).
TABLE 3.
Results of PCR experiments with Mono Lake DNA extractsa
| Sample
|
Expt
|
rRNA sequence obtained
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Direct amplification of genomic DNA extracts with:
|
Two-stage amplification: reamplification of EUB amplification product with:
|
P2P3 amplification: reamplification of NitA-NitB amplification product with:
|
Laboratory A | Laboratory B | |||||||
| No. | Depth (m) | EUB | NitA-NitB | NitD-NitB | NitA-NitB | NitD-NitB | NOC1-NOC2 | P2P3 | N.euro* | ||
| 4.2 | 1 | + | + | − | + | + | − | + | + | ||
| 4.7 | 1 | + | − | − | + | + | − | + | + | ||
| 4.5 | 5 | + | + | + | + | + | − | + | + | 4.5PCR | 4.5-A–4.5-E |
| 4.8 | 5 | + | + | + | + | + | − | + | + | 4.8PCR | 4.8-A–4.8-E |
| 4.3 | 10 | + | + | − | + | + | − | + | + | ||
| 4.9 | 10 | + | − | − | − | + | − | + | + | ||
| 4.10 | 11 | + | − | − | + | − | − | − | |||
| 4.11 | 12 | + | − | − | + | + | − | + | + | ||
| 4.12 | 12 | + | − | ||||||||
| 4.13 | 13 | + | + | − | + | + | − | + | + | ||
| 4.14 | 15 | + | + | − | + | + | − | + | |||
| 4.15 | 15 | + | + | − | + | + | − | + | + | ||
| 4.16 | 17 | + | − | − | − | + | − | + | + | ||
| 4.17 | 20 | + | − | − | − | − | − | − | |||
| 4.20 | 20 | + | − | − | + | − | − | − | |||
| 4.18 | 23 | + | + | + | + | + | − | + | + | 4.18.4, 4.18.8 | 4.18-A–4.18-J |
| 4.19 | 25 | + | − | − | − | − | − | − | |||
| 7.6 | 1 | + | − | − | − | ND | − | ND | |||
| 7.15 | 1 | + | + | − | + | + | − | + | + | 7.15PCR | 7.15-A–7.15-J |
| 7.2 | 5 | + | + | ND | − | ND | − | ND | |||
| 7.5 | 5 | + | − | − | + | + | − | + | |||
| 7.4 | 7 | + | − | − | + | − | − | − | |||
| 7.7 | 10 | + | − | − | − | ND | − | ND | |||
| 7.13 | 10 | + | + | − | + | + | − | + | + | ||
| 7.19 | 10 | + | − | − | + | + | − | + | + | ||
| 7.3 | 12 | + | + | ND | − | ND | − | ND | |||
| 7.12 | 13 | + | − | − | + | + | − | + | |||
| 7.11 | 14 | + | − | − | + | + | − | + | |||
| 7.1 | 15 | + | + | − | − | − | − | − | |||
| 7.1 | 17 | + | − | − | + | + | − | + | |||
| 7.18 | 20 | + | + | + | + | + | − | + | + | 7.18.5, 7.18.6 | 7.18-B–7.18-H |
| 7.14 | 23 | + | + | − | + | + | − | + | + | ||
| 7.17 | 25 | + | + | − | + | + | − | + | + | ||
See Table 1 for primer abbreviations. The names of the rDNA sequences reflect derivation as sequenced directly from the PCR fragment (e.g., 4.5PCR) or from a cloned PCR fragment (e.g., 4.18.4). +, detection of expected size PCR fragment by agarose gel electrophoresis; −, denotes no PCR fragments detected; ND, no data due to insufficient sample to perform this analysis; N. euro*, detection of a band corresponding to the position of the N. europaea standard band in DGGE.
PCR amplification, cloning, and sequencing.
The PCR primers used here have been described previously, and their positions relative to that of the E. coli 16 S ribosomal DNA (rDNA) are listed in Table 1. General bacterial primers (EUB1 and EUB2 [24]) were used to verify that the sample DNA was of PCR amplification quality. The NitA and NitB primers (41) were designed to be specific for all nine described ammonia oxidizers within the β subdivision of the Proteobacteria (11, 50), and the NitD primer is 100% specific for Nitrosomonas europaea (47). The NOC1 and NOC2 primers (42) are specific for N. oceanus. Amplification conditions for all of these primer sets were optimized according to the method of Cobb and Clarkson (7). The primer sequences and optimal assay conditions (slightly different from previously published conditions) are shown in Table 1. In addition to those reagents listed in the table, the reaction mixture contained DNA template (1 μl of purified DNA, from either a culture or an extract of natural sample) and 2.5 U of Taq polymerase in a total reaction volume of 100 μl. When two-stage amplification was performed, 1 μl of the initial (first-stage) reaction mixture was used as the template in the second amplification without further purification.
TABLE 1.
Primer sequences and optimal amplification reaction conditions
| Primer sequence (5′→3′) (location relative to Escherichia coli 16S rRNA) | Optimal reaction condition
|
|||||
|---|---|---|---|---|---|---|
| Annealing temp | Primers (nM) | Mg (mM) | Deoxynucleoside triphosphates (μM) | No. of cycles | Reference | |
| EUB1 (9–27) GAGTTTGATCCTGGCTCAG | ||||||
| EUB2 (1525–1542) AGAAAGGAGGTGATCCAGCC | 60 | 200 | 1.5 | 50 | 25 | 24 |
| NitA (137–160) CTTAAGTGGGGAATAACGCATCG | ||||||
| NitB (1213–1234) TTACGTGTGAAGCCCTACCCA | 60 | 150 | 1.5 | 100 | 25 | 41 |
| NitD (423–445) TAGTCGGAAAGAAAGAGTTGCAA | 60 | 150 | 1.5 | 200 | 25 | 47 |
| NOC1 (25–45) CGTGGGAATCTGGCCTCTAGA | ||||||
| NOC2 (1168–1188) AGATTAGCTCCGCATCAGCT | 60 | 100 | 3.0 | 400 | 25 | 42 |
| P2 (540–557) ATTACCGCGGCTGCTGG | ||||||
| P3 (339–346) CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGGCCTACGGGAGGCAGCAGb | 55a | 200 | 1.5 | 200 | 25 | 27 |
| GM5F (341–358) CCTACGGGAGGCAGCAG | ||||||
| DS907R (907–928) CCCCGTCAATTCCTTTGAGTTT | 55 | 200 | 1.5 | 200 | 25 | 39 |
A touchdown program was used for this primer set, which involved several annealing temperatures, with 55°C being the maximum temperature used.
Includes a 40-bp GC clamp.
Fragments (194 bp each) for DGGE analysis (see below) were produced with a touchdown amplification procedure (8) with the P2 and P3 primers of Muyzer et al. (27). The NitA-NitB fragment (1 μl of the reaction mixture) which had been amplified from the EUB1-EUB2 fragment was used as the template for amplification with the P2 and P3 primers. The primers used to sequence an internal fragment of the products generated by amplification with the NitA and NitB primers were the GM5F and DS907R primers reported by Teske et al. (39).
For every PCR experiment, both positive and negative controls were included. The positive control consisted of purified N. europaea (or Rhodocyclus purpureas as a substitute for N. europaea) or N. oceanus DNA, and the negative controls were PCRs containing all reagents and no DNA. Each sample for which a result is reported yielded consistent results in at least two separate PCR experiments. PCR-amplified fragments were resolved by electrophoresis on 1% (wt/vol) horizontal agarose minigels run in 1× TAE buffer.
PCR products from five samples were subjected to sequence analysis in Laboratory A (University of California, Santa Cruz). The NitD-NitB amplification product was sequenced from three samples which amplified directly with the NitD and NitB primers (sequences identified as 4.5PCR, 4.8PCR, and 7.15PCR). The PCR products were purified with Wizard spin columns (Promega) and were sequenced directly with the NitB and NitD primers and the internal eubacterial primers 795r and 773f with a DyeDeoxy Terminator Cycle Sequencing kit (Perkin-Elmer) and a 373 automated DNA sequencer (Applied Biosystems) by following the manufacturers' recommendations.
The direct NitA-NitB amplification products of the two other samples (4.18 and 7.18) were cloned into PGem Vector (Promega) before sequencing. Plasmid DNA from two clones from each of the two original samples (clones 7.18.5 and 7.18.6 from July 1995 and clones 4.18.4 and 4.18.8 from April 1995) was isolated by standard protocols (3), purified with Wizard spin columns (Promega), and sequenced as described above. A total of seven NitD-NitB sequences were obtained from the Laboratory A sequence analysis.
Two years later, PCR products from the same five samples (4.5, 4.8, 4.18, 7.15, and 7.18) were reanalyzed by using updated procedures and instrumentation in Laboratory B (Princeton University). For these samples, PCR products from NitA-NitB, NitD-NitB, and GM5F-DS907R reactions were cloned into the pCR2.1 vector according to the manufacturer's instructions (Invitrogen Corp., San Diego, Calif.). Plasmid DNAs containing inserts were isolated for sequencing by alkaline lysis procedures (QIAprep spin columns; Qiagen, Inc., Chatsworth, Calif.) or by direct PCR of recombinant clones with vector-specific primers (Invitrogen Corp.). Plasmid templates containing NitA-NitB or GM5F-DS907R fragments were sequenced with forward and reverse vector primers with an ABI 310 DNA sequencer (Big Dye-Terminator Cycle Sequencing Ready Reaction FS kit; Perkin-Elmer Applied Biosystems, Foster City, Calif.) according to the manufacturer's instructions. The Laboratory B analysis yielded 34 new sequences of the GM5F-DS907R fragment.
Hybridization.
To verify that the amplified sequences represented rRNA genes, the amplified fragments were analyzed by hybridization with the EUB amplification product of N. europaea DNA, which was previously verified as being of rDNA origin (41). The PCR amplified DNA was purified (Geneclean; BIO 101, San Diego, Calif.), labeled with digoxigenin by random priming according to the Genius protocol (Boehringer Mannheim), purified again, and used at a final concentration of 100 ng ml−1. The test DNAs were bound to a nylon membrane via a slot blotting procedure (46). The hybridization conditions and procedures for posthybridization washes and visualization of the probe-target conjugate were those recommended by the manufacturer (Boehringer Mannheim).
DGGE.
DGGE was performed using a Bio-Rad mini-Protean II gel system modified for buffer recirculation as described by Myers et al. (28) with a linear gradient with 8% (wt/vol) acrylamide stock solutions (acrylamide-N,N′-methylenebisacrylamide; 37:1) which contained 20% (or 35%) and 70% denaturant (100% denaturant = 7 M urea [Sigma] and 40% formamide [Fisher] deionized with AG501-X8 mixed-bed resin [Bio-Rad]). Electrophoresis was performed at a constant voltage of 105 V and at 60°C for 3 to 4 h. After electrophoresis, the gels were stained in ethidium bromide and visualized with UV transillumination.
Phylogenetic analyses.
Sequences were initially compared to the available databases by using the BLAST network service (1) to determine phylogenetic affiliation. Sequences were then compiled in Sequence Navigator (Perkin-Elmer Applied Biosystems) and aligned with representative nitrifier sequences from the Ribosomal Database Project. Evolutionary distance, parsimony, and maximum-likelihood analyses were performed on the aligned sequences with programs from the Phylogenetic Inference package (PHYLIP, version 3.5 [9]).
Nucleotide sequence accession numbers.
The rDNA sequences of the 41 unique Mono Lake clones have been deposited in GenBank (accession numbers AF266805 to AF266845).
RESULTS AND DISCUSSION
The density and chemical distributions of Mono Lake were determined at the time of both sample collections. The oxic-anoxic interface was at a 12 to 13 m depth in April 1995 and at 13 to 17 m in July 1995. The characteristics of the water column above and below the oxycline are summarized in Table 2 and described in more detail in the work of Joye et al. (19). Salinity was not measured at the time of sampling but can be estimated from subsequent measurements (17). It ranged from approximately 74 g kg−1 in the surface water to 88 g kg−1 in the bottom water. This gradient coincided with a strong thermal gradient, which together were sufficient to maintain stratification beyond the normal winter mixing season (17). Ammonia oxidation was detected in the surface layer down to 12 m in both July and April and also at 15 and 17 m in July (19).
TABLE 2.
Chemical concentrations illustrating the stratified condition of Mono Lake at time of samplinga
| Characteristic | Surface (0–10 m) | Oxycline (10–15 m) | Deep (15–25 m) |
|---|---|---|---|
| [O2] (μM) | 250 | 0 | |
| Salinity (g liter−1)b | 75 | 80–86 | 88 |
| [CH4] (μM) | 0 | 2 | 5 |
| [NH4+] (μM) | 1 | 2–10 | 25 |
| Temp (°C) | 20 | 8 | 4 |
Parameter values are representative of both April and July 1995 sampling dates (only ammonium concentration differed significantly between April and July, being higher in July). The oxycline interval is defined by the region of rapid change in oxygen concentration from nearly 250 to 0 μM in the space of a few meters, so no independent value is provided for that variable in the oxycline region.
Reference 17
Detection of β-subdivision ammonia-oxidizing sequences.
DNA was successfully amplified with the EUB primers from all samples, yielding a single band of the correct size (Table 3). Extracts from 6 of the 11 depths from April and from 8 of the 12 depths from July yielded the correct (1,080-bp) fragment upon direct amplification with the NitA and NitB primers. Target sequences were detected both above and below the chemocline in both months, indicating the presence of nitrifiers, even in anoxic water. In several cases (at depths of 1 and 10 m in April and 1, 5, and 10 m in July), only one or two of the two or three samples collected from each depth amplified directly with the NitA and NitB primers, and these results were completely reproducible. Only three and one sample(s) from April and July, respectively, amplified directly with the NitD and NitB primers, suggesting that the N. europaea target was relatively more abundant in those samples.
When the NitA and NitB primers were used in a two-stage amplification, samples from most depths yielded a fragment of the correct size (Table 3). Similarly, many more samples yielded the correct-size amplification product in a two-stage amplification with the NitD and NitB primers than when direct amplification with the same primers was used.
A double-nested approach was used because previous research had shown it to be more sensitive at detecting rare sequences (41). While the selectivity of PCR is enhanced by this procedure, detection of the target is more sensitive and perhaps more specific by this procedure than by direct amplification (2, 5). Thus, detection of the NitA-NitB target by two-stage amplification should be a reliable indicator of the presence or absence of the target sequence, but no quantitative information about the relative abundance of nitrifiers or individual nitrifier targets can be derived from these data. Based on the two-stage amplification results, β-subdivision ammonia-oxidizer target sequences were present at all depths in the lake, and N. europaea-like sequences were present at most of those depths. In one case, two-stage amplification with NitD and NitB was positive, but neither single-stage nor two-stage amplification with NitA and NitB was positive. This NitD-NitB fragment was not studied further; it may reflect the greater selectivity of the specific NitD primer for a rare target that was enriched by EUB amplification sufficiently to detect with NitD but not with NitA.
The NitA and NitB primers were among the first to be developed for the β-subdivision ammonia oxidizers (41), and a few of the more recently developed primers have slightly improved specificity (40). The NitA and NitB primers contain slight mismatches in order to amplify all nine known strains. Because of the potential ambiguity resulting from nonspecific amplification of the possible targets for the NitA and NitB primers, successful amplification with these primers was interpreted as preliminary evidence that β-subdivision ammonia oxidizers were present in the samples.
The NitD primer has 100% homology with N. europaea; the next-closest sequence in the database, at 83% similarity, is Nitrosomonas eutropha. The analysis by Utaker and Nes (40) concluded that NitD is absolutely specific for N. europaea, based on the published sequences. Therefore, both single- and two-stage amplification with the NitD and NitB primers is strong evidence that β-subdivision ammonia oxidizers are present, specifically N. europaea.
NitA and NitB PCR products that were analyzed by hybridization reacted strongly with the rRNA probe (results not shown), indicating that the amplified sequences represented ribosomal sequences from the environment. These fragments were then further characterized by more-specific techniques. Because it is possible that nonnitrifier fragments of the right size could be amplified from the complex milieux of the natural samples, PCR amplification alone is not proof that β-subdivision nitrifiers were present at all depths where amplification was obtained. Because it seemed unlikely that the familiar freshwater-terrestrial nitrifier, N. europaea, would be present in the highly alkaline Mono Lake environment, the NitD-NitB amplification seemed most suspect. Therefore, we focused on the depths where NitD-NitB amplification was obtained to evaluate more specifically whether nitrifier sequences were indeed present. Validation of these sequences as belonging to nitrifiers would tend to validate the NitA-NitB amplifications, whereas the opposite was not true.
Characterization of nitrifier sequences.
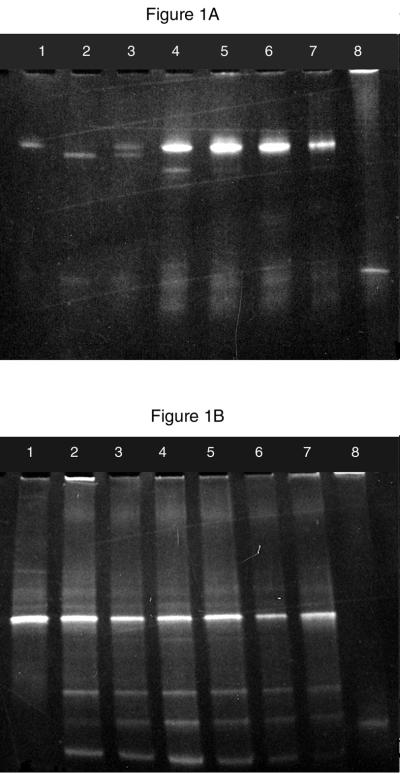
DGGE results represent another line of evidence strongly indicating the presence of N. europaea-like sequences among the Mono Lake ammonia oxidizers. The nine strains of β-subdivision ammonia oxidizers each yielded unique DGGE fragments in gradient gels run under the conditions described here (22). Bands obtained from Mono Lake samples appeared most similar to bands obtained from N. europaea and Nitrosomonas marina; therefore, a mixed standard containing both N. europaea and N. marina DNA was routinely run on DGGE gels containing Mono Lake samples (Fig. 1A, lane 3). In most samples, a band that migrated essentially identically with the band from pure cultures of N. europaea was observed (Fig. 1). Several samples also yielded additional bands, indicating the presence of multiple species of β-subdivision ammonia oxidizers in the sample (Fig. 1B).
FIG. 1.
DGGE of 16S rDNA fragments from cultured cells and from Mono Lake DNA extracts from the following standard DNAs (A) and sample numbers (B). (A) Lanes: 1, N. europaea; 2, N. marina; 3, an equal mixture of N. europaea and N. marina; 4, sample 4.15; 5, sample 7.14; 6, sample 7.15; 7, sample 7.18; 8, Nitrosospira briensis. (B) Lanes: 1, N. europaea; 2, sample 4.2; 3, sample 4.3; 4, sample 4.5; 5, sample 4.8; 6, 4.11; 7, sample 4.7; 8, N. briensis.
The number of bands observed in a gradient pattern from a mixed sample represents the minimum number of different sequences present. The band pattern of the Mono Lake samples indicates the presence of at least four distinct sequences derived from amplification with the NitA-NitB primers, which is a small number compared to what would be expected from DGGE analysis of the total EUB amplification (26). These bands result from intense preselection (two previous PCRs, one of which is highly selective for the ammonia-oxidizer group) and reflect diversity among the β-subdivision ammonia oxidizers only. Sequencing of amplification products indicated the presence of only 1 NitD-NitB sequence per band.
Six of the seven Mono Lake sequences from the Laboratory A analysis (all except 4.18.8, obtained at 23 m in April 1995) were very similar to each other and to the β-subdivision ammonia-oxidizer sequences in the database, especially those of N. europaea and N. eutropha (Table 4). Sequence 4.18.8, which was derived from the same NitA-NitB amplification reaction as 4.18.4, was an outlier; its genetic distance from the other Mono Lake sequences and from the known N. europaea sequences was about 0.10. Nevertheless, 4.18.8 is clearly a nitrifier sequence, as it is more similar to the nitrifiers than to any other sequence in the database.
TABLE 4.
Genetic distance among seven aligned rDNA sequences, comparing clones or direct amplification products from Laboratory A analysis of Mono Lake DNA extracts (boldface)
| Extract | Genetic distance
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N. europaea | N. europaea 2 | N. eutropha | N. mobilis | N. marina | N. multiformis | N. tenuis | N. briensis | 4.5PCR | 4.8PCR | 4.18.4 | 4.18.8 | 7.15PCR | 7.18.6 | 7.18.5 | |
| N. europaea | 0.0000 | 0.0138 | 0.0103 | 0.0404 | 0.0799 | 0.0703 | 0.0664 | 0.0801 | 0.0069 | 0.0069 | 0.0071 | 0.1013 | 0.0103 | 0.0191 | 0.0310 |
| N. europaea 2 | 0.0000 | 0.0244 | 0.0552 | 0.0958 | 0.0819 | 0.0779 | 0.0919 | 0.0173 | 0.0156 | 0.0215 | 0.1050 | 0.0244 | 0.0335 | 0.0446 | |
| N. eutropha | 0.0000 | 0.0477 | 0.0800 | 0.0779 | 0.0740 | 0.0840 | 0.0173 | 0.0173 | 0.0304 | 0.1211 | 0.0208 | 0.0298 | 0.0527 | ||
| Nitrosococcus mobilis | 0.0000 | 0.0779 | 0.0478 | 0.0477 | 0.0610 | 0.0477 | 0.0477 | 0.0533 | 0.1504 | 0.0515 | 0.0591 | 0.0855 | |||
| N. marina | 0.0000 | 0.0837 | 0.0797 | 0.0802 | 0.0876 | 0.0878 | 0.0993 | 0.1895 | 0.0918 | 0.1000 | 0.1251 | ||||
| Nitrosolobus multiformis | 0.0000 | 0.0086 | 0.0316 | 0.0779 | 0.0780 | 0.1043 | 0.1923 | 0.0799 | 0.0900 | 0.1309 | |||||
| Nitrosovibrio tenuis | 0.0000 | 0.0262 | 0.0740 | 0.0741 | 0.0091 | 0.1919 | 0.0760 | 0.0860 | 0.1273 | ||||||
| Nitrosospira briensis | 0.0000 | 0.0878 | 0.0879 | 0.1138 | 0.2010 | 0.0858 | 0.1002 | 0.1336 | |||||||
| 4.5PCR | 0.0000 | 0.0034 | 0.0134 | 0.0863 | 0.0103 | 0.0157 | 0.0226 | ||||||||
| 4.8PCR | 0.0000 | 0.0156 | 0.0913 | 0.0072 | 0.0157 | 0.0226 | |||||||||
| 4.18.4 | 0.0000 | 0.0939 | 0.0205 | 0.0317 | 0.0388 | ||||||||||
| 4.18.8 | 0.0000 | 0.1082 | 0.0993 | 0.1047 | |||||||||||
| 7.15PCR | 0.0000 | 0.0203 | 0.0272 | ||||||||||||
| 7.18.6 | 0.0000 | 0.0273 | |||||||||||||
| 7.18.5 | 0.0000 | ||||||||||||||
The genetic distance between the seven Laboratory A sequences and that of the positive control, N. europaea, was more than that expected to result from detection of a true N. europaea strain in our samples or from the inadvertent sequencing of our positive control. However, when the opportunity arose to work in Laboratory B, which had never been used to analyze N. europaea DNA, we repeated the sequence analyses of the same samples. The NitA-NitB fragments from the Laboratory A PCR experiments, which amplified directly with the NitD and NitB primers, were reamplified with the GM5F and DS907R primers, using DNA from Rhodocyclus purpureas as the positive control. (An R. purpureas sequence was never retrieved from analysis of the lake samples, so contamination with the positive control is unlikely to have occurred.) If contamination was responsible for our Laboratory A results, these products should have contained solely N. europaea sequences, rather than a diverse group of sequences. The genetic distances reported here are much greater than would be expected from introduction of error by the use of Taq polymerase (6). Assuming an error rate of 10−4 for Taq polymerase, the expected error rate (or difference) per base in the amplified 582-bp fragment is computed to be 0.00117 per base. The smallest genetic distance between our environmental sequences (4.5PCR and 4.8PCR) (Table 4) and the published N. europaea sequences was about 0.069 per base. Thus, these differences likely represent real variability of naturally occurring sequences.
In the Laboratory B analysis, 10 clones each were sequenced from samples 4.5 and 4.8 and 15 each from samples 4.18, 7.15, and 7.18, to yield a total of 65 sequences. Of those clones, 5, 4, 9, 10, and 6 sequences, respectively (a total of 34), were unique. Multiple clones of the same sequence were obtained from a single sample, and the same sequences were obtained multiple times from various depths, indicating the presence of the same strains at many depths. Although 10 or 15 clones is not an exhaustive number, the fact that the same sequences were obtained more than once indicates that the degree of diversity in these samples is well represented by the unique sequences reported.
The 34 sequences obtained from the Laboratory B analysis are included in the phylogenetic analyses using 582 bp (an approximately 550-bp internal fragment plus primers) that resulted from the GM5F-DS907R amplification. This fragment includes the V6 and V7 variable regions (4), and most of the variability lies in the V6 region. The V6 region was not included in the NitD-NitB fragment, on which the Laboratory A sequence analysis was based. The seven Laboratory A sequences could not be included in the phylogenetic analysis because they do not include the 180 bp upstream of the NitD site. A combined analysis of the NitD-NitB fragments and GM5F-DS907R fragments would have reduced the common region to only 400 bp and would have included only the V7 region, which is the least variable of the variable regions. Thus, we considered that analysis was not informative and do not present results from it here.
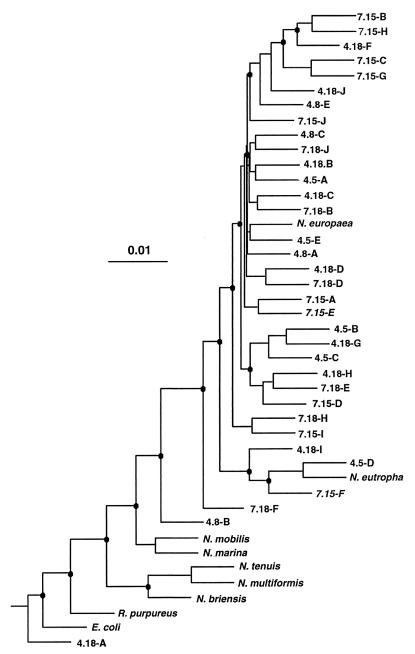
The distance tree (Fig. 2) shows that the greatest similarities among Laboratory B sequences were found between sequences from the same depth (e.g., 7.15-B and 7.15-H), but sequences from different depths were interspersed among clusters in the tree. This implies that the assemblage at 5 m was very similar to that at 20 or 23 m and that the assemblage present in April was very similar to that present in July. Greater differences might have been expected between the shallow oxic habitat and the deep anoxic habitat, but viability and persistence under these conditions complicate the comparison.
FIG. 2.
Evolutionary distance dendogram of the β-subdivision proteobacteria nitrifiers and associated Mono Lake clones, based on 582 bp of a 16S rRNA sequence (GM5F-DS907R fragment). DNA sequences from the Mono Lake samples are identified by month and DNA sample number, as listed in Table 3. Branch points supported (bootstrap values ≥75%) by parsimony and distance analyses are indicated by solid circles.
Stackebrandt and Goebel (34) compared DNA-DNA homology data with 16S rRNA sequence data and suggested that a sequence difference of greater than 2.5% is sufficient to differentiate bacterial species. By this criterion, five of the seven Laboratory A NitB-NitD sequences lie within the sequence range of N. europaea, and three of those seven are indistinguishable from sequences of N. eutropha. These sequences were derived from NitD-NitB amplifications, so they were strongly preselected to be similar to those of N. europaea and N. eutropha. This comparison is made for 750 bp of the 780-bp NitD-NitB fragment, which includes variable regions V3, V4, V7, V8, and V9 (4). Most of the variability lies within the V3 and V8 regions. None of the 34 unique sequences derived from NitA-NitB amplifications performed in Laboratory B lie within 2.5% similarity with the sequence of N. europaea (although 32 of them are within 5% of it), and 5 of these 34 are not differentiable (less than 2.5% different) from the sequence of N. eutropha.
Slightly different levels of sequence distance might be observed if the complete 16S rRNA gene could be sequenced, but it is likely that the total 16S rRNA sequences differ to the same degree, on average, as those observed in the portion (about one-third for the 34 Laboratory B sequences and one-half for the Laboratory A sequences) of the 16S rRNA gene considered here.
Both the Laboratory A and Laboratory B analyses yielded one sequence that was different than all the others. In the Laboratory A analysis, this sequence, 4.18.8, was aligned most closely with the β-subdivision Nitrosomonas-type nitrifiers and was nearly identical in an overlapping 400 bp with one of the sequences (4.8-B) obtained in the Laboratory B analysis. The outlier from the Laboratory B analysis (4.18-A, obtained at 23 m), based on 582 bp of unambiguous sequence, is not derived from a nitrifier, being most similar (95%) to a Thiomicrospira thyasirae strain. The environmental sequence was not a perfect match for the Thiomicrospira in the database, and the NitA and NitB primer sequences with which it was amplified are not present in the published sequence.
Significance of N. europaea and N. eutropha sequences in a hypersaline environment.
The finding that organisms like N. europaea and N. eutropha are present in the ammonia-oxidizer assemblage in Mono Lake has ecological significance, firstly because N. europaea is the most commonly isolated terrestrial ammonia oxidizer; for that reason it has been the subject of intense physiological and biochemical study of the pathways and enzymes of nitrification. If this strain is present in the environment, then it may be valid to extrapolate from laboratory studies to the environment in terms of the kinetics, biochemistry, and ecology of nitrification. Secondly, finding N. europaea in the environment suggests that at least some organisms in culture are in fact representative of those in nature, although the quantitative contribution of N. europaea to the natural assemblage cannot be determined.
One important caveat prevents this interpretation, however. This analysis is based entirely on ribosomal genes, which are the genes most commonly used to distinguish phylogenetic groups. However, the alkaline and saline conditions of Mono Lake are not particularly conducive to the growth of the type strains of N. europaea, which do not have a salt requirement, grow in distilled-water medium, and are inhibited by high-salt and -ammonium concentrations (20). The highest salt tolerance attributed to various genospecies of N. europaea by Koops et al. (20) was less than 500 mM NaCl (29 g liter−1). Hunik et al. (14) found that N. europaea activity (in terms of oxygen consumption) was 90% inhibited by the presence of 500 mM NaCl but that several different ions cause a similar degree of inhibition. Stehr et al. (35) obtained N. eutropha and N. europaea isolates from the Elbe River estuary that exhibited higher levels of salt tolerances than the Nitrosospira isolates from the same location. The highest level of salt tolerance tested was 500 mM NaCl (29 g liter−1). The total salt content of Mono Lake at present is on the order of 74 to 88 g liter−1, varying with depth and stratification (17); sodium and chloride are the major ions. The surface water at 74 g liter−1 is less salty than the deep water but has still more than twice the salinity of seawater. The presence of ammonia oxidizers in the deep water cannot therefore result simply from inoculation of the deep layer with normal, i.e., freshwater, Nitrosomonas strains from the upper layer. It also therefore seems unlikely that the nitrifiers represented by these ribosomal sequences are physiologically identical to the cultured N. europaea or N. eutropha. Koops et al. (20) identified a subgroup of the genus Nitrosomonas which was composed of halotolerant “genospecies,” including some N. europaea strains and Nitrosomonas halophila. These strains had salt requirements but had optimal salt concentrations of 300 mM NaCl, much less than the salt concentration of Mono Lake waters. The sequences described here may represent a new more-halophilic group of Nitrosomonas species.
The genus Nitrosomonas includes marine members (e.g., N. marina and N. halophila) with documented salt requirements. However, the Mono Lake sequences presented here were more similar to N. europaea and N. eutropha than to either the N. halophila (not shown) or N. marina sequences (Table 4; Fig. 2). DGGE analysis did detect bands in some of the samples that appeared to migrate identically with bands from the N. marina standard DNA, but these bands were not analyzed further. The positive NitA-NitB amplifications at most of the depths also imply the presence of β-subdivision nitrifiers in addition to N. europaea- or N. eutropha-like organisms, and further analysis of those fragments could yield information about the broader ammonia-oxidizer assemblage. This study focused on the N. europaea-N. eutropha group because of our initial surprise at finding sequences so close to the cultured strains in this extreme environment. The sequence and DGGE results show that a wider diversity of β-subdivision ammonia oxidizers is also present.
The physiological diversity deriving from selective pressures in the environment probably exceeds the phylogenetic diversity of microorganisms reflected in their rRNA sequences. Environmental parameters such as salinity and temperature probably exert greater selection on functional genes (e.g., genes which encode enzymes with different salt tolerances or membrane proteins) than on ribosomal genes. Thus, detecting an organism identified as nearly identical to N. europaea, according to its ribosomal sequence in Mono Lake, does not necessarily imply that a nitrifier with the physiological traits of N. europaea in culture could reside in Mono Lake. Future work in this area could fruitfully focus on genes for ammonia monooxygenase, the enzyme responsible for the first step in the oxidation of ammonia, which is more variable among nitrifying strains than is the ribosomal sequence of the same strains (31).
These results suggest that N. europaea- and N. eutropha-like bacteria are present and perhaps dominant members of the ammonia-oxidizer assemblage of Mono Lake. Hiorns et al. (12) and Hastings et al. (10) obtained quite different results from English lakes: they were unable to detect N. europaea by using specific PCR amplification and oligonucleotide hybridization probes but did detect Nitrosospira-like sequences in several freshwater and sediment environments. Enrichment cultures yielded strains in the Nitrosomonas lineage, however (10). Sequences of the amplified fragments were not reported, so the actual degree of identity or lack thereof in those samples cannot be compared to the data reported here. Nevertheless, the results suggest important differences in species composition of ammonia oxidizers among different kinds of aquatic environments. Subsequently, Whitby et al. (49) used the Hastings et al. (10) primers to retrieve two groups of sequences from aquatic sediments that were very similar to N. europaea and N. eutropha, while Nitrosospira still dominated sequences retrieved from the water column of a freshwater lake.
Using a different set of nitrifier-specific primers, Stephen et al. (36, 37) detected a preponderance of Nitrosospira sequences in soils and marine sediments. Again, enrichment cultures contained Nitrosomonas-like sequences. Kowalchuk et al. (22) used the same primers and also reported detecting a preponderance of Nitrosospira-like sequences in coastal sand dune environments, and Nitrosomonas-like sequences were detected only in the seawardmost dune site of their study. Compost material and manure also contained mainly Nitrosospira sequences (23). Relatives of the most easily culturable Nitrosomonas types were detected only in enrichment cultures from these materials. Both Phillips et al. (29), working with seawater, and Prinčič et al. (30), working with wastewater, detected N. eutropha-like sequences in natural assemblages. Hovanec and DeLong (13) found that N. europaea-like targets were more common in seawater aquarium filters than were Nitrosospira-like targets, providing more evidence for salt-tolerant N. europaea strains. It appears from this summary of recent work that direct detection of strains or species by PCR amplification of extracted DNA detects different strains than does culturing of the same samples. Strains amenable to enrichment may be those which tolerate or require high-substrate concentrations, while those which predominate in the environment may be a less uniform group with substrate preferences that vary depending on the environment. That might explain the prevalence of N. europaea in culture and would predict that high-ammonium environments might select for N. europaea. While we did not attempt any isolations, our results differ from most of the reports summarized above in detecting sequences in the environment that are very similar to those of the easily cultured strains. This may be in part an artifact associated with selectivity of different PCR primers, which could be investigated by comparing results from other 16S rRNA nitrifier primers on the same samples.
Several of the β-subdivision ammonia-oxidizer sequences reported here were detected in samples (at 23 m in April 1995 and at 20 m in July 1995) collected from below the oxic-anoxic interface (12 to 13 m in April and 13 to 17 m in July) of the stratified lake. The present analysis cannot evaluate whether the target DNA derived from intact cells or whether cells were active in these samples. Joye et al. (19) estimated ammonium oxidation rates in these same samples using methyl fluoride-sensitive bicarbonate fixation rates (25). Significant rates were found at 15 and 17 m in July, and amplification with NitA-NitB and NitD-NitB primers was detected in both of these samples (Table 3). The sample obtained at 20 m in July had evidence of ammonium oxidation activity, but the time course of bicarbonate fixation was not linear. This 20-m sample was the source of the sequences identified by the 7.18 prefix. The amplification results and retrieval of nitrifier sequences, in combination with the detection of methyl fluoride-sensitive bicarbonate fixation at these depths, imply the presence of viable and even active ammonia oxidizers of the β-subdivision type in this environment. Depths below the oxycline were not sampled by Joye et al. (19) in April, so we cannot make a direct comparison with those samples. Detection of ammonia oxidizers in the anoxic hypolimnion of lakes has been reported previously, using the NitA and NitB primers in the analysis of the water column of a lake in northern Germany (47) and by immunofluorescence of lake sediments (of a lake in the U.S. midwest) and enrichment samples derived from anoxic sediments (32). In the latter case, the organisms clearly remained viable and could be retrieved by enrichment culturing, even in very unsuitable environmental conditions. Without culturing the assemblage in Mono Lake, it cannot be verified that the retrieved sequences actually represent viable organisms. However, multiple Nitrosomonas sequences were detected below the chemocline in both April and July 1995, and activity was detected in July. Thus, it is possible that these sequences do represent viable, if not actively growing, cells. The combined approach of assaying presence, diversity, and activity in the same samples has potential for understanding regulation of nitrification in natural environments.
It is also of ecological significance that N. oceanus was not detected in Mono Lake samples. None of the samples were amplified in a two-stage amplification procedure with the NOC1 and NOC2 primers. A direct amplification with these primers was not attempted because the higher-sensitivity two-stage amplification failed to obtain successful amplification. It is conceivable that reliance on the two-stage amplification procedure might have missed N. oceanus if it were rare and if it were selected against by the EUB primers in the initial amplification. The results of direct and two-stage amplification with the NitAB primers (Table 3) suggest the opposite effect, however, but we cannot rule out the presence of N. oceanus at some low level. N. oceanus is the only known ammonia oxidizer in the γ subdivision of the Proteobacteria, and it has been detected using the NOC1 and NOC2 primers in seawater from the Southern California Bight (42) and in several permanently ice-covered, saline lakes in Antarctica (43). Immunofluorescence with specific antisera (44) has also detected N. oceanus in these two environments (43, 45). That N. oceanus was not readily detectable in Mono Lake suggests that some factor other than salinity or temperature limits its distribution.
ACKNOWLEDGMENTS
T. J. Hollibaugh, C. Culbertson, L. Miller, and T. Connell assisted with the field work. Mandy Mazzotta extracted the DNA which was used in these experiments.
This research was supported by NSF grants to B.B.W. and S.B.J.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Altwegg M. General problems associated with diagnostic applications of amplification methods. J Microbiol Methods. 1995;23:21–30. [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Green Publishing Associates and Wiley-Interscience; 1989. [Google Scholar]
- 4.Barry T, Powell R, Gannon R. A general method to generate DNA probes for microorganisms. Bio/Technology. 1990;8:233–236. doi: 10.1038/nbt0390-233. [DOI] [PubMed] [Google Scholar]
- 5.Block W. A biochemical perspective of the polymerase chain reaction. Biochemistry. 1991;30:2735–2747. doi: 10.1021/bi00225a001. [DOI] [PubMed] [Google Scholar]
- 6.Cha R S, Thilly W G. PCR methods and applications. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1993. Specificity, efficiency and fidelity of PCR; pp. 518–528. [DOI] [PubMed] [Google Scholar]
- 7.Cobb B D, Clarkson J M. A simple procedure for optimising the polymerase chain reaction (PCR) using modified Taguchi methods. Nucleic Acids Res. 1994;22:3801–3805. doi: 10.1093/nar/22.18.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Don R H, Cox P T, Wainwright B, Baker K, Mattick J S. “Touchdown” PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 10.Hastings R C, Saunders J R, Hall G H, Pickup R W, McCarthy A J. Application of molecular biological techniques to a seasonal study of ammonia oxidation in a eutrophic freshwater lake. Appl Environ Microbiol. 1998;64:3674–3682. doi: 10.1128/aem.64.10.3674-3682.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Head I M, Hiorns W D, Martin T, McCarthy A J, Saunders J R. The phylogeny of autotrophic ammonia-oxidizing bacteria as determined by analysis of 16S ribosomal RNA gene sequences. J Gen Microbiol. 1993;139:1147–1153. doi: 10.1099/00221287-139-6-1147. [DOI] [PubMed] [Google Scholar]
- 12.Hiorns W D, Hastings R C, Head I M, Saunders J R, McCarthy A J, Hall G H, Pickup R W, Embley T M. Amplification of 16S ribosomal RNA genes of autotrophic ammonia-oxidizing bacteria demonstrates the ubiquity of Nitrosospiras in the environment. Microbiology. 1995;141:2793–2800. doi: 10.1099/13500872-141-11-2793. [DOI] [PubMed] [Google Scholar]
- 13.Hovanec T A, DeLong D F. Comparative analysis of nitrifying bacteria associated with freshwater and marine aquaria. Appl Environ Microbiol. 1996;62:2888–2896. doi: 10.1128/aem.62.8.2888-2896.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunik J H, Meijer H J G, Tramper J. Kinetics of Nitrosomonas europaea at extreme substrate, product and salt concentrations. Appl Microbiol Biotechnol. 1992;37:802–807. [Google Scholar]
- 15.Jellison R, Melack J M. Meromixis in hypersaline Mono Lake, California. 1. Stratification and vertical mixing during the onset, persistence, and breakdown of meromixis. Limnol Oceanogr. 1993;38:1008–1019. [Google Scholar]
- 16.Jellison R, Miller L G, Melack J M, Dana G L. Meromixis in hypersaline Mono Lake, California. 2. Nitrogen fluxes. Limnol Oceanogr. 1993;38:1020–1039. [Google Scholar]
- 17.Jellison R, Romero J, Melack J M. The onset of meromixis during restoration of Mono Lake, California: unintended consequences of reducing water diversions. Limnol Oceanogr. 1998;43:706–711. [Google Scholar]
- 18.Johannesson K H, Lyons W B. The rare earth element geochemistry of Mono Lake water and the importance of carbonate complexing. Limnol Oceanogr. 1994;39:1141–1154. [Google Scholar]
- 19.Joye S B, Connell T L, Miller L G, Oremland R S, Jellison R S. Oxidation of ammonia and methane in an alkaline, saline lake. Limnol Oceanogr. 1999;44:178–188. [Google Scholar]
- 20.Koops H-P, Bottcher B, Moller U C, Pommerening-Roser A, Stehr G. Classification of eight new species of ammonia-oxidizing bacteria: Nitrosomonas communis sp. nov., Nitrosomonas ureae sp. nov., Nitrosomonas aestuarii sp. nov., Nitrosomonas marina sp. nov., Nitrosomonas nitrosa sp. nov., Nitrosomonas eutropha sp. nov., Nitrosomonas oligotropha sp. nov. and Nitrosomonas halophila sp. nov. J Gen Microbiol. 1991;137:1689–1699. [Google Scholar]
- 21.Koops H-P, Moller U C. The lithotrophic ammonia-oxidizing bacteria. In: Balows A, et al., editors. The prokaryotes: a handbook on the biology of bacteria; ecophysiology, isolation, identification, applications. Vol. 3. New York, N.Y: Springer-Verlag; 1992. pp. 2625–2637. [Google Scholar]
- 22.Kowalchuk G A, Stephen J R, De Boer W, Prosser J I, Embley T M, Woldendorp J A. Analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl Environ Microbiol. 1997;63:1489–1497. doi: 10.1128/aem.63.4.1489-1497.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kowalchuk G A, Naoumenko Z S, Derikx P J L, Felske A, Stephen J R, Arkhipchenko I A. Molecular analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in compost and composted materials. Appl Environ Microbiol. 1999;65:396–403. doi: 10.1128/aem.65.2.396-403.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liesack W, Weyland H, Stackebrandt E. Potential risks of gene amplification by PCR as determined by 16S rDNA analysis of a mixed-culture of strict barophilic bacteria. Microb Ecol. 1991;21:191–198. doi: 10.1007/BF02539153. [DOI] [PubMed] [Google Scholar]
- 25.Miller L G, Jellison R, Oremland R S, Culbertson C W. Meromixis in hypersaline Mono Lake, California. 3. Biogeochemical response to stratification and overturn. Limnol Oceanogr. 1993;38:1040–1051. [Google Scholar]
- 26.Murray A E, Hollibaugh J T, Orrego C. Phylogenetic compositions of bacterioplankton from two California estuaries compared by denaturing gradient gel electrophoresis of 16S rDNA fragments. Appl Environ Microbiol. 1996;62:2676–2680. doi: 10.1128/aem.62.7.2676-2680.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muyzer G, De Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myers R M, Maniatis T, Lerman L S. Detection and localization of single base changes by denaturing gradient gel electrophoresis. Methods Enzymol. 1987;155:501–527. doi: 10.1016/0076-6879(87)55033-9. [DOI] [PubMed] [Google Scholar]
- 29.Phillips C J, Smith Z, Embley T M, Prosser J I. Phylogenetic differences between particle-associated and planktonic ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in the northwestern Mediterranean Sea. Appl Environ Microbiol. 1999;65:779–786. doi: 10.1128/aem.65.2.779-786.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prinčič A, Mahne I, Megušar F, Paul E A, Tiedje J M. Effects of pH and oxygen and ammonium concentrations on the community structure of nitrifying bacteria from wastewater. Appl Environ Microbiol. 1998;64:3584–3590. doi: 10.1128/aem.64.10.3584-3590.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rotthauwe J-H, de Boer W, Liesack W. Comparative analysis of gene sequences encoding ammonia monooxygenase of Nitrosospira sp. AHB1 and Nitrosolobus multiformis C-71. FEMS Microbiol Lett. 1995;133:131–135. doi: 10.1111/j.1574-6968.1995.tb07873.x. [DOI] [PubMed] [Google Scholar]
- 32.Smorczewski W T, Schmidt E L. Numbers, activities, and diversity of autotrophic ammonia-oxidizing bacteria in a freshwater, eutrophic lake sediment. Can J Microbiol. 1991;37:828–833. [Google Scholar]
- 33.Soriano S, Walker N. Isolation of ammonia oxidizing autotrophic bacteria. J Appl Bacteriol. 1968;31:493–497. doi: 10.1111/j.1365-2672.1968.tb00397.x. [DOI] [PubMed] [Google Scholar]
- 34.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-RNA reassociation and 16S rRNA sequence analysis in the present species definition of bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 35.Stehr G, Bottcher B, Dittberner R, Rath G, Koops H P. The ammonia-oxidizing nitrifying population of the River Elbe estuary. FEMS Microbiol Ecol. 1995;17:177–186. [Google Scholar]
- 36.Stephen J R, McCaig A E, Smith Z, Prosser J I, Embley T M. Molecular diversity of soil and marine 16S rRNA gene sequences related to β-subgroup ammonia-oxidizing bacteria. Appl Environ Microbiol. 1996;62:4147–4154. doi: 10.1128/aem.62.11.4147-4154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephen J R, Kowalchuk G A, Bruns M-A V, McCaig A E, Phillips C J, Embley T M, Prosser J I. Analysis of the β-subgroup proteobacterial ammonia oxidizer populations in soil by denaturing gradient gel electrophoresis analysis and hierarchical phylogenetic probing. Appl Environ Microbiol. 1998;64:2958–2965. doi: 10.1128/aem.64.8.2958-2965.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teske A, Alm E, Regan J M, Toze S, Rittmann B E, Stahl D A. Evolutionary relationships among ammonia- and nitrite-oxidizing bacteria. J Bacteriol. 1994;176:6623–6630. doi: 10.1128/jb.176.21.6623-6630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teske A, Wawer C, Muyzer G, Ramsing N B. Distribution of sulfate-reducing bacteria in a stratified fjord (Mariager Fjord, Denmark) as evaluated by most-probable-number counts and denaturing gradient gel electrophoresis of PCR-amplified ribosomal DNA fragments. Appl Environ Microbiol. 1996;62:1405–1415. doi: 10.1128/aem.62.4.1405-1415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Utaker J B, Nes I F. A qualitative evaluation of the published oligonucleotides specific for the 16s rRNA gene sequences of the ammonia-oxidizing bacteria. Syst Appl Microbiol. 1998;21:72–88. doi: 10.1016/S0723-2020(98)80010-6. [DOI] [PubMed] [Google Scholar]
- 41.Voytek M A, Ward B B. Detection of ammonium-oxidizing bacteria of the beta-subclass of the class Proteobacteria in aquatic samples with the PCR. Appl Environ Microbiol. 1995;61:1444–1450. doi: 10.1128/aem.61.4.1444-1450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Voytek M A. Detection, abundance and diversity of aquatic nitrifying bacteria. Ph.D. dissertation. Santa Cruz: University of California; 1996. [Google Scholar]
- 43.Voytek M A, Ward B B, Priscu J C. The abundance of ammonia-oxidizing bacteria in Lake Bonney, Antarctica determined by immunofluorescence, PCR and in situ hybridization. In: Priscu J C, editor. Ecosystem processes in a polar desert: the McMurdo Dry Valleys, Antarctica. Antarctica Research Series 72. Washington, D.C.: American Geophysical Union; 1998. pp. 217–228. [Google Scholar]
- 44.Ward B B, Perry M J. Immunofluorescent assay for the marine ammonium-oxidizing bacterium Nitrosococcus oceanus. Appl Environ Microbiol. 1980;39:913–918. doi: 10.1128/aem.39.4.913-918.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ward B B, Carlucci A F. Marine ammonia- and nitrite-oxidizing bacteria: serological diversity determined by immunofluorescence in culture and in the environment. Appl Environ Microbiol. 1985;50:194–201. doi: 10.1128/aem.50.2.194-201.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ward B B, Cockcroft A R, Kilpatrick K A. Antibody and DNA probes for detection of nitrite reductase in seawater. J Gen Microbiol. 1993;139:2285–2293. doi: 10.1099/00221287-139-9-2285. [DOI] [PubMed] [Google Scholar]
- 47.Ward B B, Voytek M A, Witzel K-P. Population diversity of ammonium oxidiziers investigated by specific PCR amplification. Microb Ecol. 1997;33:87–96. doi: 10.1007/s002489900011. [DOI] [PubMed] [Google Scholar]
- 48.Watson S W. Characteristics of a marine nitrifying bacterium, Nitrosocystis oceanus sp. nov. Limnol Oceanogr. 1965;10:R274–R289. [Google Scholar]
- 49.Whitby C B. Phylogenetic differentiation of two closely related Nitrosomonas spp. that inhabit different sediment environments in an oligotrophic freshwater lake. Appl Environ Microbiol. 1999;65:4855–4862. doi: 10.1128/aem.65.11.4855-4862.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woese C R, Weisburg W G, Paster B J, Hahn C M, Tanner R S, Krieg N R, Koops H-P, Harms H, Stackebrandt E. The phylogeny of the purple bacteria: the β-subdivision. Syst Appl Microbiol. 1984;5:327–336. doi: 10.1016/s0723-2020(84)80034-x. [DOI] [PubMed] [Google Scholar]