ABSTRACT
Promoting sustainable agriculture and improving nutrition are the main United Nation’s sustainable development goals by 2030. New technologies are required to achieve zero hunger, and genome editing technology is the most promising one. In the last decade, genome editing (GE) using the CRISPR/Cas system has attracted researchers as a safer and easy tool for genome editing in several living organisms. GE has revolutionized the field of agriculture by improving biotic and abiotic stresses and yield improvement. GE technologies were developed fast lately to avoid the obstacles that face GM crops. GE technology, depending on site directed nuclease (SDN), is divided into three categories according to the modification methods. Developing transgenic-free edited plants without introducing foreign DNA meet the acceptance and regulatory ratification of several countries. There are several ongoing efforts from different countries that are rapidly expanding to adopt the current technological innovations. This review summarizes the different GE technologies and their application as a way to help in ending hunger.
KEYWORDS: CRISPR/Cas, base editing, prime editing, food security, crop improvenet
Introduction
Eradicating hunger and malnutrition are great significant challenges that many countries work to solve. Global climatic changes and economic downturns due to the COVID-19 pandemic are hindering the world from achieving the Sustainable Development Goals (SDGs), with a final goal of eliminating hunger by 2030. In general, the main factors affecting food security include limited availability of land and water resources, high rates of population growth, and limitation of the domestic food systems to respond to demand (availability, nutrition, access). According to the World Food Programme (www.wfp.org), more than 135 million people are suffering from acute hunger and the number is expected to surpass 840 million by 2030. In addition, unpredictable global climatic changes, and the emergence of resistant pathogens are severe threats to global food security. To ensure food security, all stakeholders should work together to establish a stringent program by improving the uptake of agricultural and food security research into policy and practice to solve food insecurity. To eliminate poverty, we need to increase food production sustainably and ensure that food is well distributed. Several challenges face agricultural production, including sustainably and maintaining yield, grain quality, improving the nutritional value, and acquiring resistance against biotic and abiotic stresses. Scientists have the great responsibility toward achieving zero hunger by the year 2030, improving nutrition, and promoting sustainable agriculture.
One way to ensure food security is through developing sustainable crops that are adopted to changing environments. Crops were developed during the Green Revolution (started in the mid-1940s and attributed to Norman Borlaug, the Nobel Peace Prize winner, 1970) with high-yielding varieties that produced an increased grain per area planted. However, conventional breeding techniques are not sufficient to meet these challenges and achieve food security. The new precision biotechnologies, involving genetically modified (GM) and genome editing (GE) technology, could speed up breeding, and the integration of ‘omics’ technology makes that possible. New Breeding Techniques (NBT) need exhaustive knowledge of the genome of the target species to enable the development of a new crop carrying the target trait(s). The pangenome, the entire gene set of all strains of a species, provides useful informations concerning the genomic variations in the gene pool for a given cultivated species. Pangenome represents genes present in all strains (core genome) and that which is only present in some strains (variable or accessory genome). Super-pangenome offers good opportunities for crop improvement by studying the complete genomic variation range of a given genus. Super-pangenome includes wild species and their application for crop improvement.1 Structural variations in the given genomes play an essential role in plant genetics. They include phenotyping-based selection, marker-based breeding, genomics-assisted breeding (GAB), genetically modified organisms (GMO), and genome editing (GE). Moreover, developing superior varieties requires prerequisite identifications of markers/loci/genes that are connected with the trait of interest.2 It is worth to mention that GE technology emerged in 2003 to improve various crop characteristics and does not involve using genes derived from different organisms other than the species of interest.
Recently, foods improved using GE technology have received considerable attention concerning its safety. GE is a precise modification that has no or minor changes to traditional crops modified breeding. In the coming decade, it is expected that GE will replace GM as it improves various crop characteristics through a higher success rate and lack of external gene insertion. Instead, the target genes are identified, cut, and modified in very precise ways.
GE could be achieved through enzymes that are collectively called site-directed nucleases (SDN) directed by DNA binding domain or by RNA molecules to bind to the genome’s target site. Thus, they affect only specific endogenous sequences. Protein-binding systems include meganucleases (MegaNs), zinc finger nuclease (ZFN), transcription activator-like effector nucleases (TALENs), while the RNA binding molecules have clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein system (CAS). CRISPR/Cas9 system requires short guide sequence RNA (sgRNA) to direct Cas9 nuclease to cleave the double-stranded DNA target site complementary to the sgRNA. CRISPR-Cas system is the most commonly used system of eukaryotic genomes, as it has several advantages, including precise manipulation, effectiveness, ease of use, inexpensiveness, and allowing multiple genome manipulation.3–5
Evolution of CRISPR-Cas Systems
First Generation of CRISPR-Cas Systems
CRISPR/Cas9 System
The CRISPR system was first identified as an adaptive defensive mechanism that confers resistance to foreign genetic elements in bacteria and archaea, and the CRISPR-Cas system was first used for eukaryote genome editing 2013. The programmed Cas9 binds to a small guide RNA (sgRNA), the CRISPR/Cas9 endonuclease first scans the genomic DNA and binds upstream of the G-rich protospacer adjacent motif (NGG-PAM) sequences (Fig. 1a). Cas9 generate a double-strand breakage (DSBs) in the targeted DNA and sgRNA guides Cas9 to the target site to form the RNA–DNA duplex followed by an adjacent protospacer motif (PAM) of the genome.6–8 Moreover, CRISPR/Cas9 genome editing was extensively implemented for trait improvement of different economically important crops.9–12 Cas9 nickase (nCas9) is to be mutated in one of the catalytic residues of the nuclease domains (H840A in HNH and D10A in RuvC) in a way that can only generate single-strand breaks (SSBs), a process that can reduce off-target effects of CRISPR/Cas system13 In addition, nCas9 increase editing specificity and improve targeting ranges for gene, epigenetic, and base editing.14,15
Figure 1.
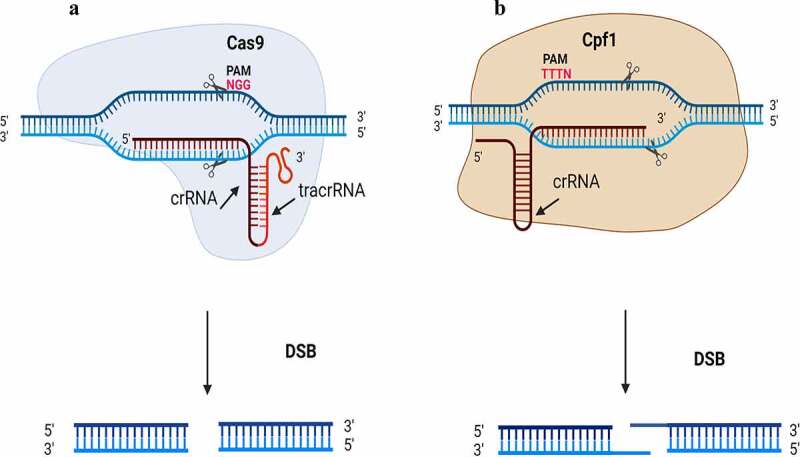
CRISPR-Cas9 versus CRISPR-Cpf1; Cas9, crRNA and tracrRNA represent a fused single-guide RNA, while Cpf1 needs crRNA only. The PAM sequence is trinucleotide 5′-NGG-3′for Cas9 while is 5ʹ-TTN for Cpf1. Cas9 cleavage dsDNA with blunt ends ends 3 nt upstream of the PAM site, while Cpf1 cleaves in a 5ʹ overhang sticky ends 18–23 bases apart from the PAM. Complementary sequence to the target DNA is linked to 5′ crRNA end of Cas9 and to 3′ ends of crRNA.
CRISPR/ Cpf1 (Cas12a) System
The Cpf1 (previously named Cas12a) is a class-II, Type V CRISPR derived from Prevotella and Francisella, and it is significantly more accurate and efficient in genome manipulation. Cpf1 is a small size monomeric protein, processes pre-crRNA into mature crRNA without a tracrRNA with more working efficiency. It recognizes T-rich PAM sequence, either 5ʹ -TTTN-3ʹ or 5ʹ -TTTV-3ʹ sequence (V = A, C or G), bind downstream of the motif and form struggled ends of DNA with 4–8 nucleotide-long15–17 (Fig. 1b). The overhangs with sequence complementarity into a genome would provide more efficient genomic insertions and offer more flexibility in base editing and epigenetic modulation.18
New Generation of CRISPR-Cas Systems
Base Editing System
Base editing (BE) is the precise modification of a single-nucleotide variants at a certain loci in the target DNA (nuclear or organellar) or RNA of a living cell using a catalytically impaired Cas nuclease that is fused to a nucleotide deaminase. The recently developed DNA BEs include adenine BEs (ABEs), cytosine BEs (CBEs), dual-base editors, and organellar BEs.
CBEs were the first developed DNA BEs to enable C.G to T.A transitions using Cas9 nickase fused to cytidine deaminase and uracil glycosylase inhibitor (UGI). The engineered sgRNA–CBE binds to the target DNA generating a ssDNA R-loop. Thus, the non-target ssDNA becomes accessible to CBE cytidine deaminase causing hydrolytic deamination of an exposed cytosine (C). The cellular mismatch of the deaminated base repair results in changing C-to-T. Uracil (U) base excision repair (BER) either keeps the original base pair or gives rise to an indel. UGI subverts uracil base excision repair and increases the probability of switching C-to-T (Fig. 2a). Interestingly, CBEs have been applied in different plant species, including Arabidopsis,19 rice,20 wheat,21 maize,22 tomato and potato,23 cotton,24 soybean,25 strawberry,26 apple and pear.27–29 While the activity window within the single-stranded DNA in the R loop that has access to the cytidine deaminase is ~4–10 nucleotides long for the majority of the Cas9-based Bes, it ranges from 8 to 13 next to the T-rich PAM motif (counted as 1) for Cas12a-based CBE. Nevertheless, developing Cas12a nickase is complicated and was only designed for base editing mammalian cells 28, 29.
Figure 2.
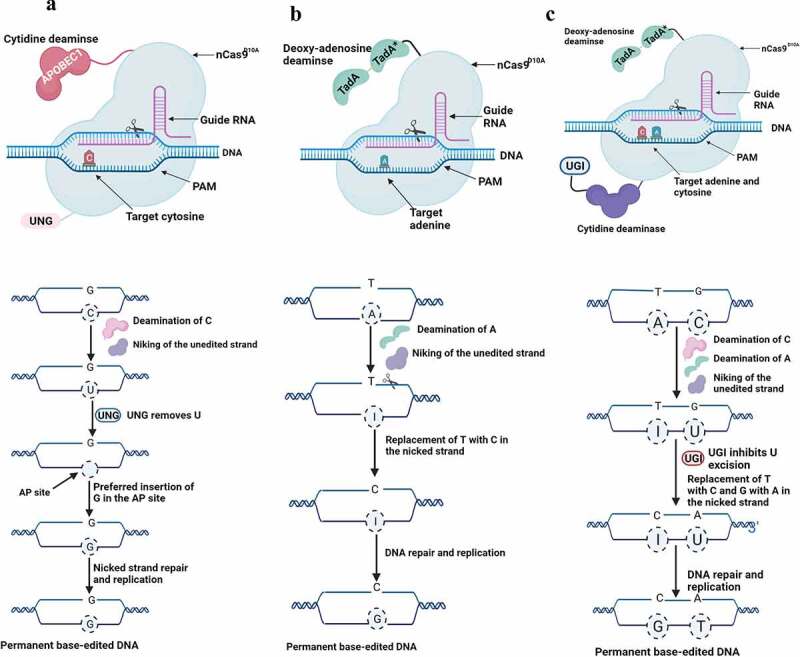
Nuclear base editing mechanism: a) Cytosine base editing (CBE) consisting of cytidine deaminase, nCas9 and UGI. The cytosine deaminase converts the desired “C” to “U,” the resulted mismatch can be corrected by base editing repair or cellular mismatch repair. The nick produced by nCas9 in the guanidine “G”-containing unedited DNA strand remove the “G” by cellular mismatch repair using uracil as a template for repair leading to the targeted “T•A.” Uracil is removed from the DNA by uracil DNA N-glycosylase, thus, reverting to the original “C•G” binding. The rate of obtaining “T•A” is enhanced through the increase of UGI protein b) Adenine base editing (ABE) contains a heterodimeric deaminase linked to nCas9. ABE is composed of wild-type TadA monomer and engineered TadA (TadA*) monomer. The target base “A” is deaminated to inosine (i) leading to converting “A•T” pair to an “I•T” bp. Inosine pairs with “C” during the replication. c) Dual-base editor converts “C-to-T” and “A-to-G.” The complex nucleoprotein is composed of nCas9, adenosine deaminase, cytidine deaminase and UGI. In the dual-base editing, deamination of “C” and “A” is performed by cytidine and adenosine deaminase, respectively. The dual base editors has the same mode of action of that of both CBE and ABE.
ABEs were developed through combining adenine deaminase and nCas9 and were used to convert an A-T base pair to a G-C base pair using tRNA adenosine deaminase (TadA) variant, which works on a ssDNA substrate.30 A TadA works as a dimer to catalyze deamination, a heterodimeric protein consisting of the wild-type TadA non-catalytic monomer. An engineered catalytic monomer (TadA*) was developed and fused with nCas9 to convert A to G (Fig. 2b). ABEs were applied to develop rice,31–35 wheat,33 Arabidopsis and Brassica napus,36 Nicotiana benthamiana,37 poplar,38 and moss.39
Dual-base editors were developed through fusing cytidine and adenosine deaminases into Cas protein (Fig. 2c) to manage C-to-T and A-to-G substitutions into a genomic region of interest. Nevertheless, individual CBE and ABE operations and dual-base editors share the same mode of action; deamination of C and A which is carried out by cytidine and adenosine deaminase, respectively.
Genome Editing for Plant Organelles
The genome of the mitochondria and chloroplasts encode essential genes for cellular respiration and photosynthesis, respectively. Gene editing in plant organelles was developed to efficiently promote point mutagenesis in chloroplasts and mitochondria using a protein-based system such as ZFN and TALEN. Although the CRISPR–Cas-derived techniques are highly effective for nuclear genome editing, they are not applicable for editing organellar DNA. It is difficult to deliver or to express both gRNA and the Cas enzyme to organelles. Mitochondria-Targeted genome editing was used in plants to disrupt the cytoplasmic male sterility (CMS)-associated genes using mitochondria-TALENs.40,41 Also, genome editing could be used to induce point mutation in the 16S rRNA gene in the chloroplast genome to enhance antibiotic resistance.42,43
Organelle base editing-based systems were developed based on mitochondrial targeting signal (MTS) or Chloroplast transit peptide (CTP), a TALE array, a DddA cytidine deaminase (DdCBE) as well as a UGI to catalyze cytosine deamination, inducing C-to-T conversions (Fig. 3). The mitochondrial genome editing system depends on the introduced genes’ nuclear gene expression, followed by transporting the expressed proteins to the mitochondria. While, in chloroplast base editing, a biolistic DNA gun is used for direct delivery. DdCBE with DNA binding domains of TALEN system was recently used for editing chloroplast and mitochondrial genome of lettuce, rapeseed, Arabidopsis, and rice.43–45
Figure 3.
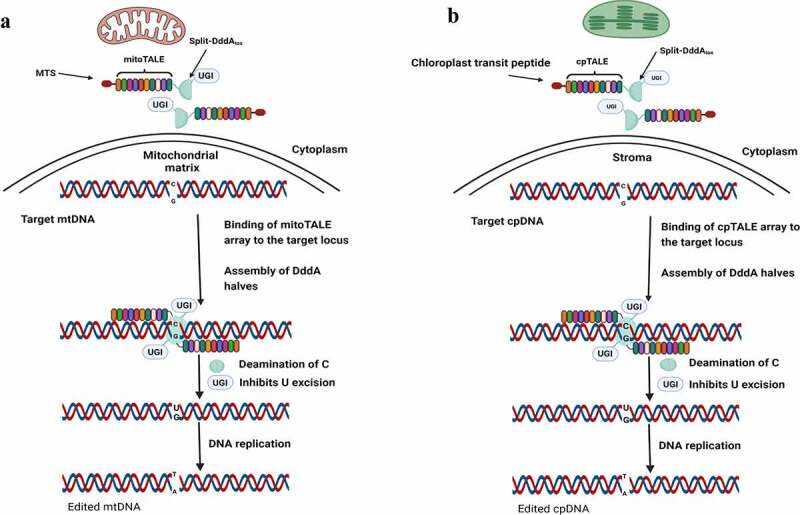
Mechanism of Organelle base editing involves TALE array, a DddA cytidine deaminase (DdCBE), a UGI to catalyze cytosine deamination, inducing “C-to-T” conversions and mitochondrial targeting signal (MTS) for editing mitochondrial DNA (a) or Chloroplast transit peptide (CTP) for editing Chloroplast DNA (b). The MTS transports the two halves of TALE into the mitochondrial matrix. The chloroplast transit peptide target the TALE array to chloroplast matrix. Two TALE arrays bind to the desired DNA sequence bringing the two DssA inactive halves into proximity. After reconstitution of active DddAtox, it deaminate “C” in the double-stranded DNA.
CRISPR/Cas13
The Cas13a was first identified in 201646 as an RNA-targeting CRISPR enzyme. Cas13b is class 2 – type VI, identified as RNA-guided RNA-targeting CRISPR–Cas system. It is used for precise target and cleavage ssRNA47 without PAM requirements.48 Cas13 was modified to adapt to RNA base editing (RBE) by using dead Cas13b (dCas13b) for either “A-to-I” editing or “C-to-U” editing. Fusing the adenosine deaminases to dCas13b deaminates A, the target A induced “A–C” mismatch in the mRNA–gRNA duplex result in editing “A-to-I.”49 While fusing engineered ADAR2 (acting as cytosine deaminase) to dCas13b results in editing “C-to-U” by an induced “C–C” or “C–U” mismatch in the mRNA–gRNA duplex (Fig. 4).
Figure 4.
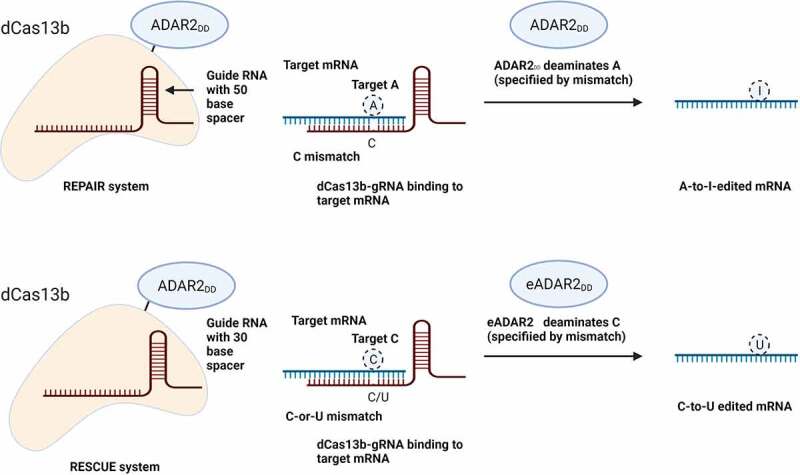
Mechanism of base editing in RNA. A) In the REPAIR system, “A-to-I” editing is using dCas13 fused to ADAR2. REPAIR use 50-nucleotide RNA with a 50-nucleotide mRNA-gRNA duplex. “A–C” mismatch in the RNA–gRNA duplex determines the target A. RESCUE system editing “C-to-U.” The optimum results are to be achieved with a gRNA of 30-nucleotide spacer. The target “C” is specified by an induced “C–C” or “C–U” mismatch in the mRNA–gRNA duplex.
Prime Editing System (PE)
PEs are multipurpose and precise genome editing tools that introduce all possible transition and transversion mutations and small indels. They use nCas9 that is fused to an engineered reverse transcriptase (RT) to write new genetic information into a specified DNA site. The RT is programmed with a prime editing gRNA (pegRNA) that specifies the target site and encodes the desired edit.50,51 PegRNA consists of an RT template, gRNA (crRNA), and a primer-binding site (PBS) (Fig. 5). The nCas9 nick the editing strand of the double-stranded DNA, and it is used for priming the reverse transcription of PBS on the pegRNA.50 Incorporating the edited DNA strand into the target DNA results in a heteroduplex DNA that contains only one edited strand. To resolve the heteroduplex, DNA repair machinery uses the edited strand as a template to copy the information from the edited strand to the complementary one. PEs led to a permanent incorporation of the preferred edit into the targeted region of the double-stranded DNA (Fig. 6).
Figure 5.
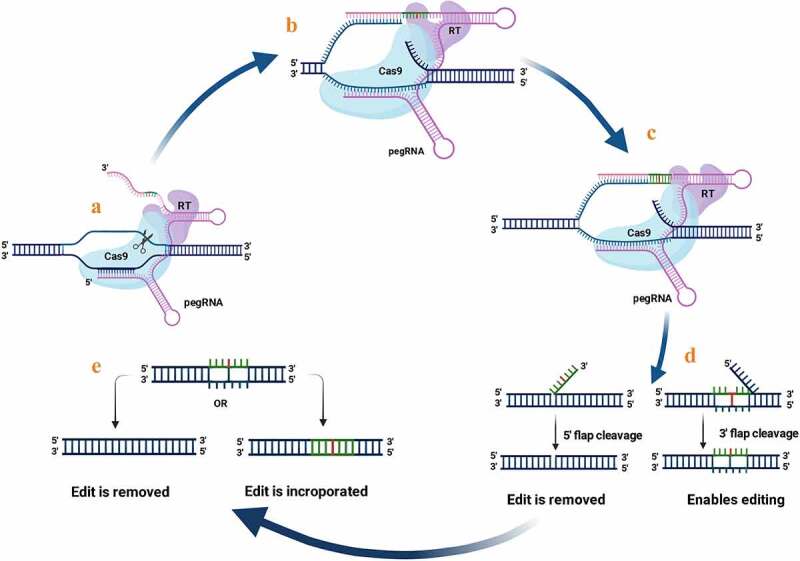
Prime editing mechanism: a) Nicking the desired DNA sequence at the PAM strand by the fusion protein, b) the exposed 3ʹ-hydroxyl group prime the reverse transcription of the RT template of the pegRNA, c) reverse transcription, d) the branched intermediate form containing two flaps of DNA: a 3ʹ flap (containing the edited sequence), and a 5ʹ flap (containing the dispensable, unedited DNA sequence) followed by flap cleavage, and e) ligation and mismatch repair; either the incorporating edite strand or remove it.
Figure 6.
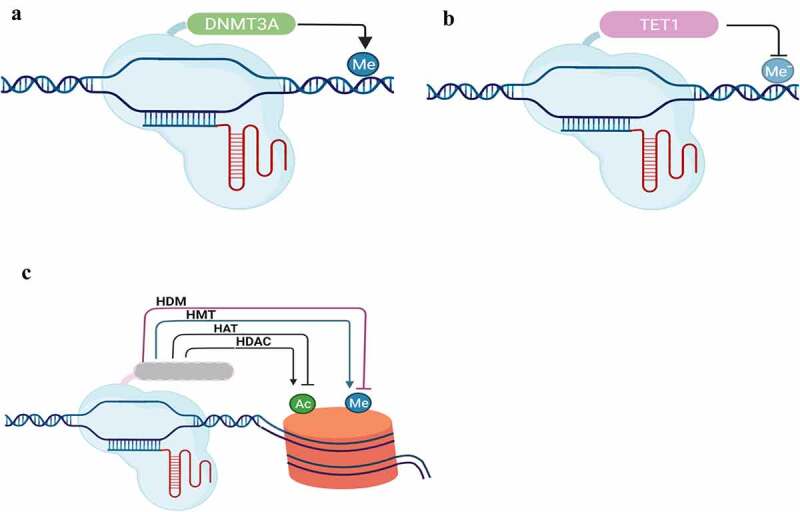
CRISPR/dCas9-based engineering of the epigenome. The techniques are based on using inactive-Cas (dCas) to allocate the desired protein to the target sequence. For DNA methylation, cytosine methylation could be used by linking dCas with DNMT3A or MQ1 (a), while demethylation of cytosine could be edited by linking Tet1 with dCas (b). Chromatin modifiers could be edited either by acetylation/deacetylation using HDAC/HAT or methylation/demethylation using HMT/HDM with dCas9.
Epigenome Editing System
Recent (CRISPR)/Cas-based epigenome editing (epi-editing) technologies site-specifically perform epigenetic modifications (methylases/demethylases) to endogenous DNA and histones, altering the chromatin structure and changing the accessibility of the transcriptional machinery, causing changes in gene expression. Histone methylation takes place at lysine and arginine residues.
The modifications of Epi-editing systems include histone acetylation using p300, histone demethylation using LSD1, cytosine methylation using MQ1 or DNMT3A, and cytosine demethylation using Tet1. In this system, dead Cas9 (dCas9) is fused with epigenetic effector domains to precise targeting promoter and enhancer regions to alter gene activation or repression. Epi-editing changes gene expression or cellular phenotype without alteration of DNA sequences and is inheritable in plants. They are used to de-methylate promoter to activate gene or to methylate promoter to deactivate it.
Applications of Genome Editing in Agriculture
CRISPR-based genome editing approach participated in different crop improvements, including abiotic (Table S1), and biotic stress management (Table S2) and breeding improvement (Table S3). The last decade witnessed increase in publications for the applications of this technology in different plants (Fig. 7). GE has been used to improve more than 40 crops, mainly rice, tomato, maize, wheat, and potato (Fig. 8).
Figure 7.
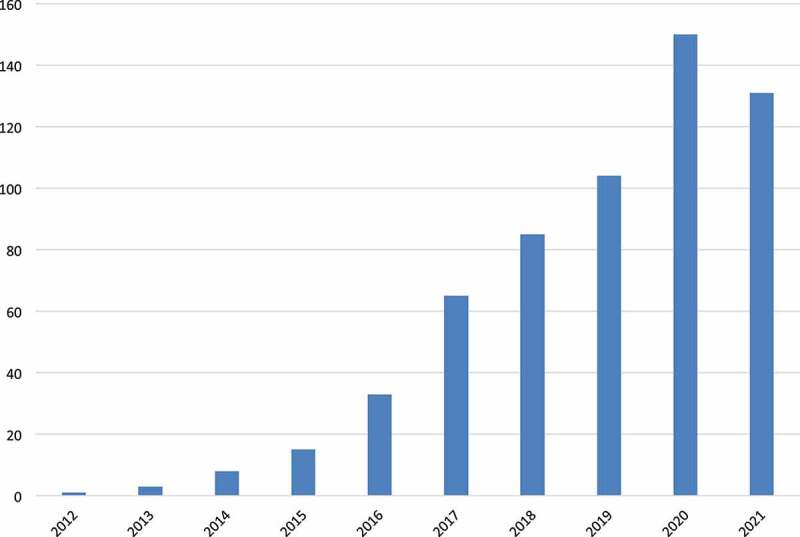
Number of CRISPR-based plant genome-editing publications over the last 10 years.
Figure 8.
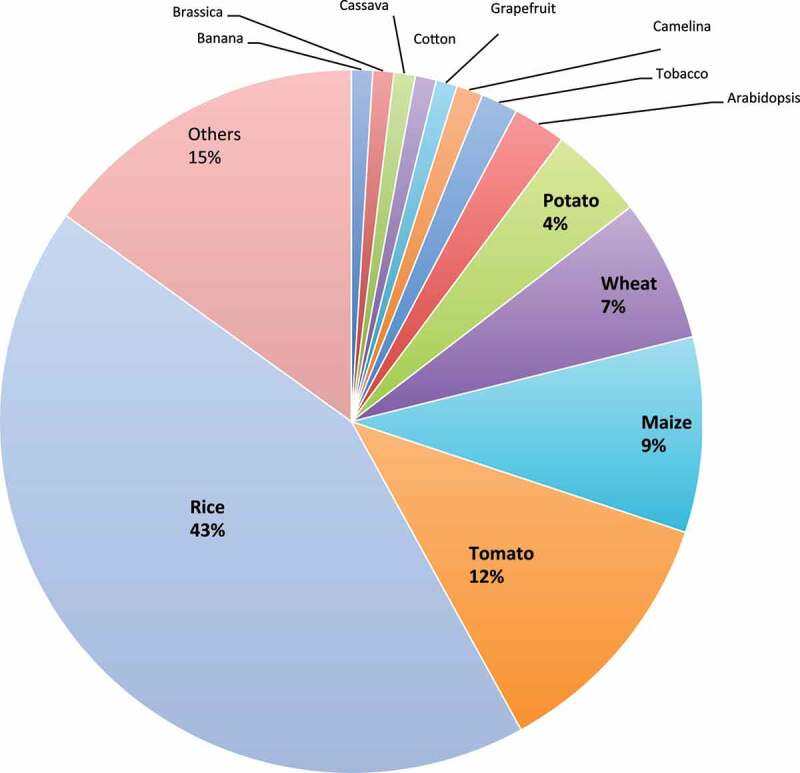
Percentage of major crops modified by CRISPR system with the aim of crop improvement.
Abiotic Stress Tolerance
Abiotic stresses are major threats to agricultural productivity and are anticipated to become more threatening in agricultural systems due to climate change. Recently, researchers targeted genome editing techniques for broadening resistance of crop tolerance, including drought, salinity, high temperature, and other climate change factors.
Salinity Tolerance
The area of salinization globally is increasing 10% annually, and it is expected by the year 2050 that more than 50% of arable lands worldwide will be salinized.51 In rice, the CRISPR/ Cas9 system was used to knock down the gene OsRR22 (encoding the rice type-B response).21 Moreover, CRISPR-Cas9 was used to mutate the salt and drought tolerance (DST) gene in indica rice cv. MTU1010. The mutated dst with 366 bp deletion showed enhanced leaf water retention due to its broadened-width leaves and the reduced stomatal density. Thus, dst mutant alleles could improve salt and drought tolerance as well as enhancing grain yield in rice.52
Moreover, in tomatoes, CRISPR/Cas9 was used to induce mutation to the SlARF4 gene. Down-regulation of SlARF4 promoted root development leading to higher soluble sugars and chlorophyll content under stress conditions. Thus, Mutated tomato plants successfully increased leaf relative water content and Abscisic acid (ABA) content and reduced stomatal conductance under normal and stressful conditions to tolerate osmotic and salt stresses53.
Drought Tolerance
Precise genomic DNA modification CRISPR‐Cas was used to introduce the native maize GOS2 promoter into the 5′‐untranslated region of the native ARGOS8 gene. ARGOS8 variants enhanced grain yield by five bushels per acre under flowering stress conditions. Nevertheless, it showed no yield loss under well‐watered conditions.54 Also, in tomato, the CRISPR/Cas9 system was utilized to generate slmapk3 mutants by knocking out SlMAPK3. Mutants showed up- or down-regulated expressions of drought stress-responsive genes (SlLOX, SlGST, and SlDREB), suggesting that knocking out SlMAPK3 protected cell membranes from oxidative damage and modulated transcription of stress-related genes.55
Thermotolerance Enhancement
The CRISPR-Cas9-mediated genome editing for heat tolerance was achieved by targeting the tomato’s SlAGAMOUS-LIKE 6 (SIAGL6) gene. Knocking out the SIAGL6 gene improved the fruit setting of tomatoes under heat stress.56 Moreover, CRISPR-Cas was used to disrupt OsMYB30 that belongs to B-Amylase (BMY) genes that regulate starch degradation and the accumulation of maltose. Thus cold-tolerant rice lines that are protecting against cold stress were generated57.
Disease Resistance
Plant pathogens, viruses, fungi, and bacteria cause various damages to crop health, and reduce yield significantly. CRISPR/Cas system has been adapted to generate varieties that can resist pathogen attack based on alteration of plant immunity. CRISPR-Cas targets plant susceptibility (Su) genes that encode factors that support pathogen infection and decrease disease susceptibility. For controlling bacterial infection, knocking out Xa13, OsSWEET13, OsSWEET13 by CRISPR/Cas9 enhanced resistance to bacterial blight in rice. While knocking down TaMLO and TaEDR1 genes in wheat control the spread of powdery mildew.58,59 Moreover, CRISPR-Cas system was used to develop rice tungro virus resistance by disrupting the Su eIF4G in rice.60
Also, the CRISPR-Cas system was used to modify genes that facilitate pathogen growth in the host. In rice, mutating the fungal genes ALB1and RSY1 with CRISPR/Cas9 was used to control the spread of rice blast, while knocking out USTA and UvSLT2 has improved was used to prevent smut disease.61 While targeting viral genome mediated by CRISPR-Cas system was implemented to control geminivirus by disrupting viral replication, including Tomato Yellow Leaf Curl Virus (TYLCV) and Wheat Dwarf Virus (WDV) infection in tomato and barely targeting viral genome region directly (MP, CP, Rep, IR).62,63
Increasing Yield and Quality
Grain yield is controlled through quantitative trait loci (QTLs) that affect grain number, weight, and size. CRISPR/Cas system was used to knockout plant genes that negatively regulate crop yields. In the rice, CRISPR/Cas9 was used to silence the expression of OsGS3, which has a negative impact on grain size and consequently on grain yield. While editing OSGW genes resulted in an increase in grain weight in rice, knockout TaCKX2-D1 increased grain number64. Moreover, fruit size in tomato was improved by mutating SICLV3 promoter and SIENO gene using the CRISPR/Cas9 system.65
Nutritional Enhancement
Gene editing plays an important role in facing the malnourishment pandemic by increasing desirable nutritional metabolites, reducing anti-nutrients and altering the composition of macronutrients. Starch quality was improved by disrupting the two genes; IbGBSSI and IbSBEII in sweet potato66. Nutrient quality in bananas was improved by knocking down LCYe gene leading to increasing beta-carotene content.67 Disrupting the alpha-gliadin gene in wheat was used to lower gluten contents to reduce allergenicity68 Moreover, the oleic acid content was increased by editing FAD2 gene in Brassica napus and peanut.69
Knockouts of the SBEIIb gene in rice decreased levels of amylopectin in favor of amylose in the rice endosperm70 In cassava, reduced starchy content was performed by knocking out two genes involved in amylose biosynthesis.71
CRISPR-Cas9 has also used to modify the starch contents in potato through knocking out of the GBSS gene in potato.72 Also, in strawberry, CRISPR/Cas9 was used to alter its sugar content through editing the gene FvebZIPs1.1.26
Manipulating Plant Breeding
CRISPR/Cas system was applied to develop breeding materials through several methods such as controlling the development of male sterility, hybrid vigor, and self-incompatibility, and hybrid seed production is sometimes challenging to restrict self-pollination. Knocking down fertility genes such as TMS5 in rice and TaNP1 in wheat, using CRISPR/Cas9, developed male sterility.69
Knocking down Male sterility gene can be classified as either cytoplasmic male sterility (CMS) and genic male sterility (GMS), depending on the fertility gene source.70
Self-fertilization is important to enhance plant breeding. In potato, knocking out the S-RNase locus resulted in RNA degradation of the pollen tube and developed self-fertile lines.71
Haploid induction is essential for the breeder to develop double haploid and stabilize the genetic architecture of inbred lines. The CRISPR-Cas system participated in haploid induction in several plants such as wheat, maize, rice, and Arabidopsis. Knocking down the pollen-specific phospholipase gene in rice (OsMTL) resulted in spermatid chromatin fragmentation and haploid induction.72
Regulatory Approaches to Genome Editing
GE could generate genetic variants at specific particular target sites that are indistinguishable from naturally evolved ones. The legislation and regulation of genome-edited plants in many countries are still evolving. Several countries have adapted their biosafety regulations based on this classification of variations induced by site-directed nuclease. Depending on the editing approach, three types of alterations could be distinguished.73 In the SDN-1, the DNA breakage introduces base-pair changes or insertions/deletions. Spontaneous repair is unguided repair and leads to gene silencing. SDN-2 requires a small DNA donor to target and repair the DNA break and generate a specific change. The donor carries the designed mutation(s) and flanking sequences complementary to both ends of the break. Homologous recombination between both the donor and the target cause DNA swap and allow introduction of the mutations. SDN-3 requires a prominent DNA donor of foreign origin to repair the damaged target sequence. The repair mechanism is also performed through the homologous recombination between the target and donor sequences. The donor DNA could contain part or complete gene and is considered transgenic.
The debate on the regulation of gene-edited crops is still going on. Certain countries regulate gene edited plants as GMOs based on the processed used for developing them, including EU union, Switzerland, and New Zealand. Other countries focus on the end-product and regulate them based on case-by-case rather than the process used for developing GE plants. This group includes the U.S., Canada, Argentina, Colombia, Brazil, Argentina, Chile Canada, and Japan. They authorize the registration of gene-edited crops if the end-product is free from any foreign DNA. While the discussion is ongoing for the third group of countries to follow the group to authorization authorize, such as the United Kingdom, Norway, Kenya, Nigeria, Paraguay, Uruguay, Norway, India, and Philippines.72,74
Argentina was the first to declare that GE crops are not be regulated under biosafety legislation if the plant do not contain foreign DNA, followed by Chile, Brazil, and Colombia. The decision taken by the biosafety authorities is based on case-by-case basis; plants containing new genetic materials will be regulated as a GMO.75–79 In contrast, USDA in the USA decided not to impose regulation on most plants produced by SDN-1 or SDN-2 once the CRISPR gene has been crossed out. GE via loss-of-function mutation only narrows the scope of improving plant yield and quality. It is worth to mention that the intragenics (within gene)/ cisgenics (whole gene) approaches are obviously more effective in leveraging genetic diversity and could be addressed to maximize the potential of agricultural improvements.
Conclusion
Precise, efficient, and rapid genome editing technique is revolutionizing crop improvement by avoiding the genetic modification, gene disruption, and introduction unwanted genes (such as selectable marker genes). Genome editing can provide unprecedented solutions to food insecurity and malnutrition by developing higher-yielding, more nutritious crops and resilient to the impacts of biotic stresses and climate change.
GE mainly depends on the identification of target genes, delivery of CRISPR machinery to the right cells, selecting, and regenerating the right cells to crops. With the wealth of information obtained from genome studies and identifying genes related to crop improvement, GE techniques are providing innovative solution that could help address global food security by developing climate-smart crops with improved yield.
GE techniques that resulted in altering DNA/chromatin confirmation (epigenome editing), minimum but precise change in the genome (base editors), or precise insertion of short DNA fragments (prime editing) are promising candidates to bring about global regulation-overhaul, changes in the policy frameworks, and improved consumer acceptance.
Supplementary Material
Funding Statement
The author(s) reported there is no funding associated with the work featured in this article.
Disclosure Statement
The authors declare that they have no conflict of interest.
Supplementary Material
Supplemental data for this article can be accessed here.
References
- 1.Khan AW, Garg V, Roorkiwal M, Golicz AA, Edwards D, Varshney RK.. Super-pangenome by integrating the wild side of a species for accelerated crop improvement. Trends Plant Sci. 2020;25:148–58. doi: 10.1016/j.tplants.2019.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh RK, Prasad A, Muthamilarasan M, Parida SK, Prasad M. Breeding and biotechnological interventions for trait improvement: status and prospects. Planta. 2020;252:1–18. doi: 10.1007/s00425-020-03465-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voytas DF, Gao C. Precision genome engineering and agriculture: opportunities and regulatory challenges. PLoS Biol. 2014;12(6):e1001877. doi: 10.1371/journal.pbio.1001877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant. 2015;8:1274–84. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Wang J, Meng X, Hu X, Sun T, Li J, Wang K, Yu H. xCas9 expands the scope of genome editing with reduced efficiency in rice. Plant Biotechnol J. 2019;17(6):709–11. doi: 10.1111/pbi.13053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10:957–63. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. science. 2012;337:816–21. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brooks C, Nekrasov V, Lippman ZB, van Eck J. Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 2014;166:1292–97. doi: 10.1104/pp.114.247577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao C. The future of CRISPR technologies in agriculture. Nat Rev Mol Cell Biol. 2018;19:275–76. doi: 10.1038/nrm.2018.2. [DOI] [PubMed] [Google Scholar]
- 11.Castel B, Tomlinson L, Locci F, Yang Y, Jones JDG, Lai E-M. Optimization of T-DNA architecture for Cas9-mediated mutagenesis in Arabidopsis. PloS One. 2019;14(e0204778):e0204778. doi: 10.1371/journal.pone.0204778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolabu TW, Park -J-J, Chen M, Cong L, Ge Y, Jiang Q, Debnath S, Li G, Wen J, Wang Z. Improving the genome editing efficiency of CRISPR/Cas9 in Arabidopsis and Medicago truncatula. Planta. 2020;252:1–14. doi: 10.1007/s00425-020-03415-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han K, Pierce SE, Li A, Spees K, Anderson GR, Seoane JA, Lo Y-H, Dubreuil M, Olivas M, Kamber RA, et al. CRISPR screens in cancer spheroids identify 3D growth-specific vulnerabilities. Nature. 2020;580:136–41. doi: 10.1038/s41586-020-2099-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kleinstiver BP, Sousa AA, Walton RT, Tak YE, Hsu JY, Clement K, Welch MM, Horng JE, Malagon-Lopez J, Scarfò I, et al. Engineered CRISPR–Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat Biotechnol. 2019;37:276–82. doi: 10.1038/s41587-018-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu H, Ding Y, Zhou Y, Jin W, Xie K, Chen -L-L. CRISPR-P 2.0: an improved CRISPR-Cas9 tool for genome editing in plants. Mol Plant. 2017;10:530–32. doi: 10.1016/j.molp.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 16.Moon SB, Feo T, Harvey TA, Prum RO. Highly efficient genome editing by CRISPR-Cpf1 using CRISPR RNA with a uridinylate-rich 3′-overhang. Nat Commun. 2018;9:1–11. doi: 10.1038/s41467-017-02088-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zetsche B, Gootenberg J, Abudayyeh O, Slaymaker I, Makarova K, Essletzbichler P, Volz S, Joung J, Van der oost J, Regev A. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–71. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arora L, Narula A. Gene editing and crop improvement using CRISPR-Cas9 system. Front Plant Sci. 2017;8:1932. doi: 10.3389/fpls.2017.01932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, Xiong X, Wang F, Liang J, Li J. Gene disruption through base editing‐induced messenger RNA missplicing in plants. New Phytol. 2019;222:1139–48. doi: 10.1111/nph.15647. [DOI] [PubMed] [Google Scholar]
- 20.Huang X, Su F, Huang S, Mei F, Niu X, Ma C, Zhang H, Zhu X, Zhu J-K, Zhang J. Novel Wx alleles generated by base editing for improvement of rice grain quality. J Integr Plant Biol. 2021;63:1632–38. doi: 10.1111/jipb.13098. [DOI] [PubMed] [Google Scholar]
- 21.Zhang A, Liu Y, Wang F, Li T, Chen Z, Kong D, Bi J, Zhang F, Luo X, Wang J, et al. Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol Breed. 2019;39:1–10. doi: 10.1007/s11032-019-0954-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zong, Y., Wang, Y., Li, C., Zhang, R., Chen, K., Ran, Y., Qiu, J.L., Wang, D. and Gao, C. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat Biotechnol. 2017;35:438–40. doi: 10.1038/nbt.3811. [DOI] [PubMed] [Google Scholar]
- 23.Veillet, F., Perrot, L., Chauvin, L., Kermarrec, M.P., Guyon-Debast, A., Chauvin, J.E., Nogué, F. and Mazier, M. Transgene-free genome editing in tomato and potato plants using agrobacterium-mediated delivery of a CRISPR/Cas9 cytidine base editor. Int J Mol Sci. 2019;20:402. doi: 10.3390/ijms20020402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin, L., Li, J., Wang, Q., Xu, Z., Sun, L., Alariqi, M., Manghwar, H., Wang, G., Li, B., Ding, X. and Rui, H. High‐efficient and precise base editing of C• G to T• A in the allotetraploid cotton (Gossypium hirsutum) genome using a modified CRISPR/Cas9 system. Plant Biotechnol J. 2020;18:45–56. doi: 10.1111/pbi.13168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai, Y., Chen, L., Zhang, Y., Yuan, S., Su, Q., Sun, S., Wu, C., Yao, W., Han, T., Hou, W. Target base editing in soybean using a modified CRISPR/Cas9 system. Plant Biotechnol J. 2020;18:1996. doi: 10.1111/pbi.13386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xing, S., Chen, K., Zhu, H., Zhang, R., Zhang, H., Li, B., Gao, C. Fine-tuning sugar content in strawberry. Genome Biol. 2020;21:1–14. doi: 10.1186/s13059-020-02146-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malabarba J, Chevreau E, Dousset N, Veillet F, Moizan J, Vergne E. New strategies to overcome present CRISPR/Cas9 limitations in apple and pear: efficient dechimerization and base editing. Int J Mol Sci. 2021;22:319. doi: 10.3390/ijms22010319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L, Hu S, Chen X. Non-viral delivery systems for CRISPR/Cas9-based genome editing: challenges and opportunities. Biomaterials. 2018;171:207–18. doi: 10.1016/j.biomaterials.2018.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Ding C, Yu W, Wang Y, He S, Yang B, Xiong Y-C, Wei J, Li J, Liang J, et al. Cas12a base editors induce efficient and specific editing with low DNA damage response. Cell Rep. 2020;31:107723. doi: 10.1016/j.celrep.2020.107723. [DOI] [PubMed] [Google Scholar]
- 30.Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR. Programmable base editing of A• T to G• C in genomic DNA without DNA cleavage. Nature. 2017;551:464–71. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molla KA, Shih J, Yang Y. Single-nucleotide editing for zebra3 and wsl5 phenotypes in rice using CRISPR/Cas9-mediated adenine base editors. aBIOTECH. 2020;1:106–18. doi: 10.1007/s42994-020-00018-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hua K, Tao X, Yuan F, Wang D, Zhu J-K. Precise A·T to G·C base editing in the rice genome. Mol Plant. 2018;11:627–30. [DOI] [PubMed] [Google Scholar]
- 33.Li C, Zong Y, Wang Y, Jin S, Zhang D, Song Q, Zhang R, Gao C. Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 2018;19:1–9. doi: 10.1186/s13059-018-1443-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan F, Kuang Y, Ren B, Wang J, Zhang D, Lin H, Yang B, Zhou X, Zhou H. Highly efficient A· T to G· C base editing by Cas9n-guided tRNA adenosine deaminase in rice. Mol Plant. 2018;11:631–34. doi: 10.1016/j.molp.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 35.Hao LI, Ruiying Q, Xiaoshuang L, Shengxiang L, Rongfang X, Jianbo Y, Pengcheng W. CRISPR/Cas9-mediated adenine base editing in rice genome. Rice Sci. 2019;26:125–28. doi: 10.1016/j.rsci.2018.07.002. [DOI] [Google Scholar]
- 36.Kang B-C, Yun J-Y, Kim S-T, Shin Y, Ryu J, Choi M, Woo JW, Kim J-S. Precision genome engineering through adenine base editing in plants. Nature Plants. 2018;4:427–31. doi: 10.1038/s41477-018-0178-x. [DOI] [PubMed] [Google Scholar]
- 37.Wang Z, Liu X, Xie X, Deng L, Zheng H, Pan H, Li D, Li L, Zhong C. ABE8e with polycistronic tRNA-gRNA expression cassette significantly improves adenine base editing efficiency in nicotiana benthamiana. Int J Mol Sci. 2021;22:5663. doi: 10.3390/ijms22115663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li G, Sretenovic S, Eisenstein E, Coleman G, Qi Y. Highly efficient C‐to‐T and A‐to‐G base editing in a Populus hybrid. Plant Biotechnol J. 2021;19:1086–88. doi: 10.1111/pbi.13581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guyon‐Debast A, Alboresi A, Terret Z, Charlot F, Berthier F, Vendrell‐Mir P, Casacuberta JM, Veillet F, Morosinotto T, Gallois J-L, et al. A blueprint for gene function analysis through Base Editing in the model plant Physcomitrium (Physcomitrella) patens. New Phytol. 2021;230:1258–72. doi: 10.1111/nph.17171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arimura S, Yanase S, Tsutsumi N, Koizuka N. The mitochondrial genome of an asymmetrically cell-fused rapeseed, Brassica napus, containing a radish-derived cytoplasmic male sterility-associated gene. Genes Genet Syst. 2018;93:43–148. [DOI] [PubMed] [Google Scholar]
- 41.Kazama, T., Okuno, M., Watari, Y, Yanase, S., Koizuka, C., Tsuruta, Y., Sugaya, H., Toyoda, A., Itoh, T., Tsutsumi, N., Toriyama, K. Curing cytoplasmic male sterility via TALEN-mediated mitochondrial genome editing. Nature Plants. 2019;5:722–30. doi: 10.1038/s41477-019-0459-z. [DOI] [PubMed] [Google Scholar]
- 42.Fromm H, Galun E, Edelman M. A novel site for streptomycin resistance in the “530 loop” of chloroplast 16S ribosomal RNA. Plant Mol Biol. 1989;12:499–505. doi: 10.1007/BF00036964. [DOI] [PubMed] [Google Scholar]
- 43.Kang B-C, Bae S-J, Lee S, Lee JS, Kim A, Lee H, Baek G, Seo H, Kim J, Kim J-S, et al. Chloroplast and mitochondrial DNA editing in plants. Nature Plants. 2021;7:899–905. doi: 10.1038/s41477-021-00943-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li S, Chang L, Zhang J. Advancing organelle genome transformation and editing for crop improvement. Plant Commun. 2021;2:100141. doi: 10.1016/j.xplc.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakazato I, Okuno M, Yamamoto H, Tamura Y, Itoh T, Shikanai T, Takanashi H, Tsutsumi N, Arimura S-I. Targeted base editing in the plastid genome of Arabidopsis thaliana. Nature Plants. 2021;7:906–13. doi: 10.1038/s41477-021-00954-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DBT, Shmakov S, Makarova KS, Semenova E, Minakhin L, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353. doi: 10.1126/science.aaf5573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cox, DB., Gootenberg, JS., Abudayyeh, OO., Franklin, B., Kellner, MJ., Joung, J., Zhang, F. RNA editing with CRISPR-Cas13. Science. 2017;358:1019–27. doi: 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palaz F, Kalkan AK, Can Ö, Demir AN, Tozluyurt A, Özcan A, Ozsoz M. CRISPR-Cas13 system as a promising and versatile tool for cancer diagnosis, therapy, and research. ACS Synth Biol. 2021;10:1245–67. doi: 10.1021/acssynbio.1c00107. [DOI] [PubMed] [Google Scholar]
- 49.Molla, KA., Sretenovic, S., Bansal, KC., Qi, Y. Precise plant genome editing using base editors and prime editors. Nature Plants. 2021;1–22. [DOI] [PubMed] [Google Scholar]
- 50.Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–57. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jamil A, Riaz S, Ashraf M, Foolad MR. Gene expression profiling of plants under salt stress. CRC Crit Rev Plant Sci. 2011;30:435–58. [Google Scholar]
- 52.Kumar VVS, Verma RK, Yadav SK, Yadav P, Watts A, Rao MV, Chinnusamy V. CRISPR-Cas9 mediated genome editing of drought and salt tolerance (OsDST) gene in indica mega rice cultivar MTU1010. Physiol Mol Biol Plants. 2020;26:1099. doi: 10.1007/s12298-020-00819-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bouzroud S, Gasparini K, Hu G, Barbosa MAM, Rosa BL, Fahr M, Bendaou N, Bouzayen M, Zsögön A, Smouni A, et al. Down regulation and loss of auxin response factor 4 function using CRISPR/Cas9 alters plant growth, stomatal function and improves tomato tolerance to salinity and osmotic stress. Genes. 2020;11:272. doi: 10.3390/genes11030272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi J, Gao H, Wang H, Lafitte HR, Archibald RL, Yang M, Hakimi SM, Mo H, Habben JE. ARGOS 8 variants generated by CRISPR‐Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol J. 2017;15:207–16. doi: 10.1111/pbi.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, L., Chen, L., Li, R., Zhao, R., Yang, M., Sheng, J., Shen, L. Reduced drought tolerance by CRISPR/Cas9-mediated SlMAPK3 mutagenesis in tomato plants. J Agric Food Chem. 2017;65:8674–82. doi: 10.1021/acs.jafc.7b02745. [DOI] [PubMed] [Google Scholar]
- 56.Klap C, Yeshayahou E, Bolger AM, Arazi T, Gupta SK, Shabtai S, Usadel B, Salts Y, Barg R. Tomato facultative parthenocarpy results from Sl AGAMOUS‐LIKE 6 loss of function. Plant Biotechnol J. 2017;15:634–47. doi: 10.1111/pbi.12662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeng Y, Wen J, Zhao W, Wang Q, Huang W. Rational improvement of Rice yield and cold tolerance by editing the three genes OsPIN5b, GS3, and OsMYB30 with the CRISPR–Cas9 system. Front Plant Sci. 2020;10:1663. doi: 10.3389/fpls.2019.01663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y, Bai Y, Wu G, Zou S, Chen Y, Gao C, Tang D. Simultaneous modification of three homoeologs of Ta EDR 1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 2017;91:714–24. doi: 10.1111/tpj.13599. [DOI] [PubMed] [Google Scholar]
- 59.Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu J-L. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol. 2014;32:947–51. doi: 10.1038/nbt.2969. [DOI] [PubMed] [Google Scholar]
- 60.Macovei, A., Sevilla, NR., Cantos, C., Jonson, GB., Slamet‐Loedin, I., Čermák, T., Voytas, DF., Choi, IR. Chadha‐Mohanty, P. Novel alleles of rice eIF4G generated by CRISPR/Cas9‐targeted mutagenesis confer resistance to Rice tungro spherical virus. Plant Biotechnol J. 2018;16:1918–27. doi: 10.1111/pbi.12927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harishchandra, DL., Chethana, KWT., Zhang, W., Xing, QK., Peng, JB., Brooks, S., Hyde, KD., Li, XH. CRISPR/Cas9: contemporary designer nucleases for efficient genome editing in phytopathogenic fungi. Curr Res Environ Appl Mycology (J Fungal Biol). 2021;11:341–63. [Google Scholar]
- 62.Tashkandi M, Ali Z, Aljedaani F, Shami A, Mahfouz MM. Engineering resistance against Tomato yellow leaf curl virus via the CRISPR/Cas9 system in tomato. Plant Signal Behav. 2018;13:e1525996. doi: 10.1080/15592324.2018.1525996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kis A, Hamar É, Tholt G, Bán R, Havelda Z. Creating highly efficient resistance against wheat dwarf virus in barley by employing CRISPR/Cas9 system. Plant Biotechnol J. 2019;17:1004. doi: 10.1111/pbi.13077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou J, Xin X, He Y, Chen H, Li Q, Tang X, Zhong Z, Deng K, Zheng X, Akher SA, et al. Multiplex QTL editing of grain-related genes improves yield in elite rice varieties. Plant Cell Rep. 2019;38:475–85. doi: 10.1007/s00299-018-2340-3. [DOI] [PubMed] [Google Scholar]
- 65.Rodríguez-Leal D, Lemmon ZH, Man J, Bartlett ME, Lippman ZB. Engineering quantitative trait variation for crop improvement by genome editing. Cell. 2017;171:470–80. doi: 10.1016/j.cell.2017.08.030. [DOI] [PubMed] [Google Scholar]
- 66.Wang H, Wu Y, Zhang Y, Yang J, Fan W, Zhang H, Zhao S, Yuan L, Zhang P. CRISPR/Cas9-based mutagenesis of starch biosynthetic genes in sweet potato (Ipomoea Batatas) for the improvement of starch quality. Int J Mol Sci. 2019;20:4702. doi: 10.3390/ijms20194702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Waltz E. Vitamin A super banana in human trials. Nat Biotechnol. 2014;32:857. doi: 10.1038/nbt0914-857. [DOI] [PubMed] [Google Scholar]
- 68.Altenbach SB, Chang H-C, Rowe MH, Yu XB, Simon-Buss A, Seabourn BW, Green PH, Alaedini A. Reducing the immunogenic potential of wheat flour: silencing of alpha gliadin genes in a US wheat cultivar. Front Plant Sci. 2020;11:20. doi: 10.3389/fpls.2020.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang H, Cui T, Zhang L, Yang Q, Yang Y, Xie K, Fan C, Zhou Y. Modifications of fatty acid profile through targeted mutation at BnaFAD2 gene with CRISPR/Cas9-mediated gene editing in Brassica napus. Theor Appl Genet. 2020;133:2401–11. doi: 10.1007/s00122-020-03607-y. [DOI] [PubMed] [Google Scholar]
- 70.Sun, Y., Jiao, G., Liu, Z., Zhang, X., Li, J., Guo, X., Du, W., Du, J., Francis, F., Zhao, Y., Xia, L. Generation of high-amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes. Front Plant Sci. 2017;8:298. doi: 10.3389/fpls.2017.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bull SE, Seung D, Chanez C, Mehta D, Kuon J-E, Truernit E, Hochmuth A, Zurkirchen I, Zeeman SC, Gruissem W, et al. Accelerated ex situ breeding of GBSS-and PTST1-edited cassava for modified starch. Sci Adv. 2018;4:eaat6086. doi: 10.1126/sciadv.aat6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Andersson, M., Turesson, H., Olsson, N., Fält, AS., Ohlsson, P., Gonzalez, MN., Samuelsson, M., Hofvander, P. Genome editing in potato via CRISPR‐Cas9 ribonucleoprotein delivery. Physiol Plant. 2018;164:378–84. doi: 10.1111/ppl.12731. [DOI] [PubMed] [Google Scholar]
- 73.Barman, HN., Sheng, Z., Fiaz, S., Zhong, M., Wu, Y., Cai, Y., Wang, W., Jiao, G., Tang, S., Wei, X., Hu, P. Generation of a new thermo-sensitive genic male sterile rice line by targeted mutagenesis of TMS5 gene through CRISPR/Cas9 system. BMC Plant Biol. 2019;19:1–9. doi: 10.1186/s12870-019-1715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Horner HT, Palmer RG. Mechanisms of genic male sterility. Crop Sci. 1995;35:1527. doi: 10.2135/cropsci1995.0011183X003500060002x. [DOI] [Google Scholar]
- 75.Eggers E-J, van der Burgt A, van Heusden SAW, de Vries ME, Visser RGF, Bachem CWB, Lindhout P. Neofunctionalisation of the Sli gene leads to self-compatibility and facilitates precision breeding in potato. Nat Commun. 2021;12:1–9. doi: 10.1038/s41467-021-24267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ahmad S, Tang L, Shahzad R, Mawia AM, Rao GS, Jamil S, Wei C, Sheng Z, Shao G, Wei X, et al. CRISPR-based crop improvements: a way forward to achieve zero hunger. J Agric Food Chem. 2021;69:8307–23. doi: 10.1021/acs.jafc.1c02653. [DOI] [PubMed] [Google Scholar]
- 77.Schmidt SM, Belisle M, Frommer WB. The evolving landscape around genome editing in agriculture: many countries have exempted or move to exempt forms of genome editing from GMO regulation of crop plants. EMBO Rep. 2020;21:e50680. doi: 10.15252/embr.202050680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kuppu, S., Ron, M., Marimuthu, MP., Li, G., Huddleson, A., Siddeek, MH., Terry, J., Buchner, R., Shabek, N., Comai, L., Britt, AB. A variety of changes, including CRISPR/Cas9‐mediated deletions, in CENH3 lead to haploid induction on outcrossing. Plant Biotechnol J. 2020;18:2068–80. doi: 10.1111/pbi.13365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Metje-Sprink J, Menz J, Modrzejewski D, Sprink T. DNA-free genome editing: past, present and future. Front Plant Sci. 2019;9:1957. doi: 10.3389/fpls.2018.01957. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


