Abstract
The success of transcriptome-wide association studies (TWAS) has led to substantial research toward improving the predictive accuracy of its core component of genetically regulated expression (GReX). GReX links expression information with genotype and phenotype by playing two roles simultaneously: it acts as both the outcome of the genotype-based predictive models (for predicting expressions) and the linear combination of genotypes (as the predicted expressions) for association tests. From the perspective of machine learning (considering SNPs as features), these are actually two separable steps—feature selection and feature aggregation—which can be independently conducted. In this study, we show that the single approach of GReX limits the adaptability of TWAS methodology and practice. By conducting simulations and real data analysis, we demonstrate that disentangled protocols adapting straightforward approaches for feature selection (e.g., simple marker test) and aggregation (e.g., kernel machines) outperform the standard TWAS protocols that rely on GReX. Our development provides more powerful novel tools for conducting TWAS. More importantly, our characterization of the exact nature of TWAS suggests that, instead of questionably binding two distinct steps into the same statistical form (GReX), methodological research focusing on optimal combinations of feature selection and aggregation approaches will bring higher power to TWAS protocols.
Keywords: statistical genetics, transcriptome-wide association studies, feature selection, kernel machine, statistical power
Introduction
Pioneered by several researchers in 2015 (Gamazon et al. 2015; Gusev et al. 2016), transcriptome-wide association studies (TWAS) have successfully identified many associations between genes and complex traits (Gusev et al. 2018, 2019; Mancuso et al. 2018; Theriault et al. 2018; Wu et al. 2018; Ratnapriya et al. 2019; Chen et al. 2021) and have triggered extensive methodological research (Gamazon et al. 2015; Gusev et al. 2016; Mancuso et al. 2019; Nagpal et al. 2019; Shi et al. 2020; Bhattacharya et al. 2021; Cao et al. 2021b; Tang et al. 2021). The key concept behind TWAS is genetically regulated expression (GReX), which is the component of gene expression attributed to genetic regulators. A typical TWAS procedure involves training a linear model, such as ElasticNet (Gamazon et al. 2015) or Bayesian regression (Zhou et al. 2013; Gusev et al. 2016; Nagpal et al. 2019), to estimate GReX as a weighted linear combination of regulatory DNA elements. The predicted GReX is then associated to phenotype in a separate association mapping dataset in which expression data is unavailable. The importance of GReX is evidenced by the number of publications which either seek to improve its prediction accuracy or expand its applications (Zeng et al. 2021). Researchers have refined the original ElasticNet- and Bayesian-based models of PrediXcan (Gamazon et al. 2015) and BSLMM (Zhou et al. 2013) by integrating multiple tissues (Barbeira et al. 2019; Hu et al. 2019; Zhou et al. 2020; Liu et al. 2021), adding trans-eQTLs (Luningham et al. 2020; Bhattacharya et al. 2021), and incorporating improved Bayesian methods (Nagpal et al. 2019). GReX counterparts have also been developed for LD-score (Siewert-Rocks et al. 2021), polygenic risk score (Liang et al. 2020), and fine-mapping (Mancuso et al. 2019).
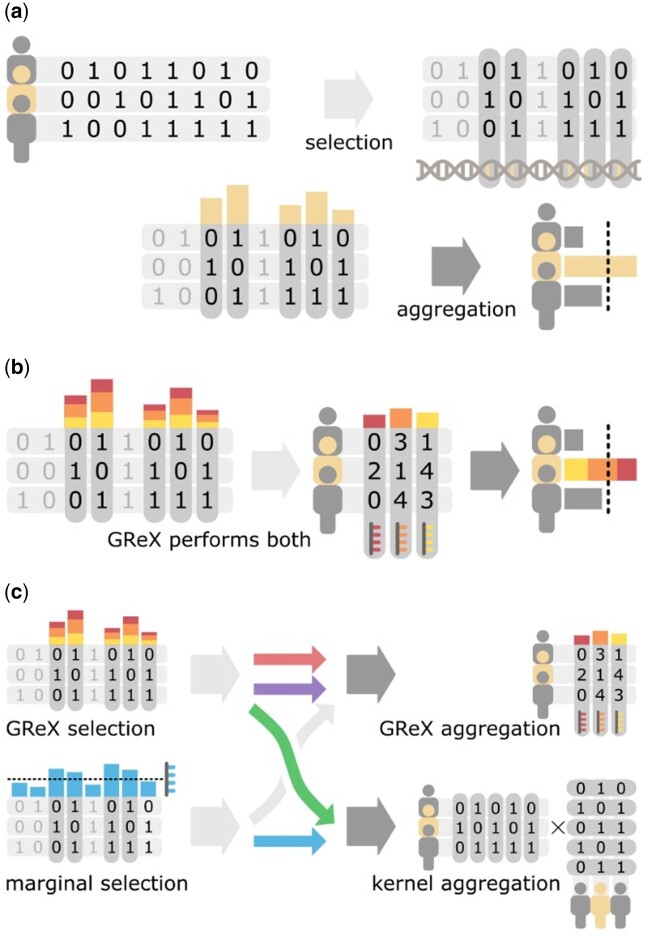
However, recent efforts in improving GReX may have overlooked its primary purpose, which is not to predict expressions but to gather relevant genetic variants for association mapping (Gamazon et al. 2015). This viewpoint is supported in practice by the low predictive accuracy of GReX models, which generally have an R2 value of 5–10% for the topmost candidates due to low expression heritability (Gamazon et al. 2015; Li et al. 2018; Mancuso et al. 2018; Bhattacharya et al. 2020). Properly speaking, GReX is a weighted linear combination of genotypes which are selected via the objective of predicting expression data, and as such, these linear combinations do not necessarily represent true biological causes of gene expression. This subtle but important distinction suggests that understanding the statistical roles underlying GReX may yield greater benefits over simply optimizing its prediction accuracy. From the perspective of machine learning on high-dimensional data, GReX combines feature selection, which reduces dimensionality by removing irrelevant features (genetic variants), and feature aggregation, which calculates aggregate statistics from selected features (variants) to maximize statistical power (Figure 1A). The process of training the expression model is equivalent to using a linear model to select variants, and the process of associating predicted expressions to phenotype is equivalent to using the same linear model to aggregate variants (Figure 1B). Given this interpretation, we investigate whether feature selection and feature aggregation can be separated into two different methods, and if so, are there methods that perform better than the GReX linear model in either step (Figure 1C)? As our analyses of real and simulated data show, the answer to both questions is yes: it is not only possible to conduct feature selection and aggregation separately, we also propose a simple combination of methods that can outperform the use of GReX alone.
Figure 1.
Function of GReX in terms of feature selection and feature aggregation. (A) Feature selection and feature aggregation are two typical steps in the statistical analysis of high-dimensional data. (B) The current practice of TWAS combines feature selection and feature aggregation into a single multiple linear regression model for estimating genetically regulated expression (GReX). (C) Separating these two fundamentally different steps allows a larger combination of methods to be applied to TWAS, providing greater flexibility in practice and potentially increased power. This study quantifies the performance of the four combinations illustrated with colored arrows.
This work extends the findings of two recent publications which replace GReX-based feature aggregation in TWAS with kernel-based methods. In our recent paper (Cao et al. 2021b), we developed a protocol called kTWAS (kernel-TWAS) using the ElasticNet model from PrediXcan to select variants and the well-known Sequence Kernel Association Test (SKAT) to associate variants to phenotype (Wu et al. 2010). We demonstrated that kTWAS outperforms TWAS in real data and simulations under different genetic architectures (Cao et al. 2021b). Independently, another group at Emory University has released VC-TWAS (Variance Component TWAS) (Tang et al. 2021), which utilizes a Bayesian linear model adapted from Tigar (Zeng and Zhou 2017; Nagpal et al. 2019) to select features and an equivalent kernel test for aggregation. Although these two publications use different approaches to model GReX and different parameterizations to conduct simulations, both studies conclude that kernel methods outperform GReX models in feature aggregation. In this work, we extend these findings by also replacing GReX-directed feature selection with a straightforward method for selecting variants based on their marginal effects on expression. By thoroughly comparing two GReX-based protocols and two protocols that disentangle feature selection from aggregation, we show that separating feature selection and aggregation into different statistical models significantly improves the power of TWAS in many conditions. This clearly shows that GReX models are not always optimal for either feature selection or aggregation, and future research should consider GReX as just one of several components that can be chosen to maximize the power of TWAS in a two-step framework.
Although TWAS is most often conducted on summary statistics (i.e., meta-analysis) rather than subject-level genotypes (Gusev et al. 2016, 2018; Theriault et al. 2018), our previous results show that the relative power between protocols utilizing summary statistics is consistent with the relative power of their counterparts utilizing subject-level genotype data (Cao et al. 2021b). We therefore chose to analyze subject-level data in order to simplify the comparison between GReX-based and disentangled TWAS protocols. For the same reason, we also restricted our comparisons to cis-genetic elements and single-tissue analyses to avoid possible complications introduced by the integration of more advanced models.
Materials and methods
Overview of novel TWAS protocol disentangling feature selection and aggregation
We implement the novel protocol called Marginal + Kernel TWAS (abbreviated mkTWAS), which replaces GReX with marginal effect-based feature selection and kernel-based feature aggregation [implemented with FastQTL (Ongen et al. 2016) and SKAT (Wu et al. 2010), respectively]. For a given focal gene, we first select genetic variants by associating individual variants with the gene’s expression level (Ongen et al. 2016), retaining significant variants as potential expression quantitative trait loci (eQTLs) for downstream association mapping. We then aggregate potential eQTLs using SKAT’s kernel-based score test to determine gene to phenotype associations (Wu et al. 2010). We include a relatively large number of potential eQTLs (with nominal P-values less than 0.05 before multiple-test correction) as input to SKAT, since our previous work found that the performance of SKAT favors a large number of weakly correlated variants over a small number of highly significant variants. We hypothesize that this is because kernel methods are more robust to noise and therefore extract weaker signals, allowing statistical power to scale with an increasing number of features (Belkin et al. 2018).
Overview of protocols under comparison
To evaluate the effectiveness of disentangled feature selection and aggregation, we compare a total of four protocols (Table 1). Protocols (1) and (2) use GReX to bind together feature selection and aggregation. Protocol (1), referred to as GReX (ElasticNet), adopts the ElasticNet linear model from PrediXcan (Gamazon et al. 2015) to estimate GReX. Protocol (2), referred to as GReX (BSLMM), adopts the Bayesian sparse linear mixed model (BSLMM) to estimate GReX. As the existing BSLMM tool Fusion (Gusev et al. 2016) operates on summary statistics, we instead incorporate the weights of the BSLMM model into PrediXcan for association mapping. Protocols (3) and (4) separate feature selection and aggregation into different models. Protocol (3), referred to as ElasticNet + Kernel, uses the ElasticNet model from PrediXcan (Gamazon et al. 2015) for feature selection and SKAT (Lee et al. 2013) for feature aggregation as implemented in our previous method kTWAS (Cao et al. 2021b). Protocol (4), referred to as Marginal + Kernel, uses marginal genotype-expression effects for feature selection and SKAT for aggregation, as described above in our novel method mkTWAS. Type-I error is experimentally quantified by simulating under the null hypothesis for each protocol. Details of each protocol and the type-I error simulations are detailed below.
Table 1.
Design of compared protocols
| Protocol no. and name | Feature selection | Feature aggregation | Implementation |
|---|---|---|---|
| GReX (ElasticNet) | GReX: ElasticNet | GReX: ElasticNet | PrediXcan |
| GReX (BSLMM) | GReX: Bayesian model | GReX: Bayesian model | PrediXcan/BSLMM |
| ElasticNet + Kernel | GReX: ElasticNet | Kernel | PrediXcan + SKAT |
| Marginal + Kernel | Marginal effects | Kernel | FastQTL + SKAT |
Notations
In this section, we use to denote a matrix of genotypes over individuals and genetic variants, and , , and to denote vectors of genetic variants, phenotypes, and gene expressions respectively. We use to denote vectors of coefficients for genetic markers and for residuals. Vectors corresponding to a particular variant site are indexed by the subscript .
Details of analytic protocols
Four protocols were applied in simulations and real data analysis, with two GReX-based protocols chosen to represent flagship TWAS methods in practical use. (1) Uses ElasticNet (Friedman et al. 2010; Simon et al. 2011) implemented by PrediXcan (Gamazon et al. 2015), as a representative of regularized GReX models, which is applied to both feature selection and aggregation. (2) Uses BSLMM (Zhou et al. 2013) as a representative of Bayesian GReX models, also applied to both feature selection and aggregation. As the widely used BSLMM tool Fusion operates on summary statistics (Gusev et al. 2016), we instead use PrediXcan to conduct subject-level association mapping from the weights estimated by BSLMM.
Two additional protocols were chosen to represent methods separating feature selection and aggregation. (3) Combines ElasticNet feature selection with Kernel-based feature aggregation. Expression data is used to train an ElasticNet model such that , where the objective function minimizes . Training is conducted using the R package glmnet (Friedman et al. 2010; Simon et al. 2011) in simulations, while pre-trained coefficients from the PrediXcan website are used in real data analysis (http://predictdb.org). Unlike the standard TWAS protocol however, the predicted expressions are not used directly to conduct association mapping. Instead, the weighted genetic variants are formed into a kernel , where is a matrix of the selected variants, is a diagonal matrix of variant weights, and is the number of genetic variants. Using SKAT, we conduct a score-test , where is the kernel described above. Complete details are in our recent publication (Cao et al. 2021b), and the code is available on GitHub (https://github.com/theLongLab/kTWAS). (4) Marginal + Kernel: This protocol uses FastQTL (Ongen et al. 2016) to carry out eQTL analyses on each gene and select a large number of genetic variants from potential eQTLs (variants with a nominal P-value lower than 0.05, without multiple-test correction). Note that the marginal effect of each variant is computed individually from the eQTLs. Selected variants are formed into the same kernel and SKAT score test as described in protocol (3), except the diagonal matrix contains the log-base-10 P-values from eQTL mapping. The code is available on GitHub (https://github.com/theLongLab/mkTWAS).
Type-I error estimation
Although both GReX-based protocols (1) and (2) are well-established and the type-I error of protocol (3), ElasticNet + Kernel, was recently assessed (Cao et al. 2021b), the type-I error may still vary depending on the simulations and implementations. We therefore generated random phenotypes using individual data from the 1000 Genomes Project (Auton et al. 2015) to measure the type-I error for each protocols. The type-I error is estimated using the top 5% cutoff for the most significant P-values obtained by the null hypothesis simulation. More specifically, we calculate corresponding statistics using real genotype and randomly simulated phenotype and take the P-value ranked at the top 5% (among all simulated rounds under the null) as the cutoff for power simulations. This ensures that the type-I error of all protocols under comparison is exactly 5%. As shown in Supplementary Table S7, all protocols have type-I errors comparable to their theoretical values.
Real data analysis
For all four protocols, feature selection was performed on GTEx whole blood data (GTEx Consortium 2015). Association tests were conducted on genotype data for seven diseases in WTCCC (Wellcome Trust Case Control Consortium 2007). Out of 393,273 features (SNPs) recorded in WTCCC, 363,217 are shared with GTEx and utilized for feature selection. The sample size for each of the seven WTCCC diseases is listed in Supplementary Table S8.
Success rate analysis
To assess the relevance of the genes identified by each protocol, we examine the proportion of discovered genes which have annotations in the DisGeNET database (Pinero et al. 2015, 2017). Specifically, for any pair of protocols and yielding corresponding sets of associated genes, we take the difference of the sets (genes only in and not ), and (genes only in but not ), and find the proportion of genes in each set difference which are annotated in DisGeNET.
Simulations
Gene expressions are simulated using genotype information from GTEx (GTEx Consortium 2015), and phenotypes are simulated using information from the 1000 Genomes Project (Auton et al. 2015).
Causal scenarios
We simulate the two commonly assumed scenarios pleiotropy and causality (Supplementary Figure S1). All variants are sampled from a region including the relevant gene body and 1 Mb of flanking sequences at both sides. Under pleiotropy, the phenotype and expression are independently caused by the same genetic variants, so that and . Under causality, phenotype is caused by genotype directly and also via the intermediary effect of expression, so that and . Note that the function which maps genotype to simulated expressions is identical in both scenarios, but the functions and for simulating phenotype differ.
Genetic architecture models
The functions , and are defined differently depending on the specific genetic architecture. In the additive model, given genetic variants in a genotype matrix where , the expression model is defined as . We set as 2, 5, and 10 in our simulations. The effect size is drawn from the standard normal distribution . In the interaction model, two genetic variants are chosen to affect gene expression or phenotype through one of three definitions. The “heterogeneous” model is equivalent to the logical operation “OR,” in which the presence of a mutant allele in either or both variant sites causes a phenotypic change. The “epistatic” model is equivalent to the logical operation “AND,” in which phenotypic change occurs only when a mutation is present at both variant sites. Finally, the “compensatory” is equivalent to the logical operation “XOR,” in which a mutant allele can cause phenotypic change at either site, but if mutations occur at both sites their effect is negated. In all of the above models, the genetic component contributing to expression or phenotype is simulated as a value between and , which is later rescaled based on expression or trait heritability. Under pleiotropy, is defined identically to , except that the variance component is rescaled by expression heritability instead of local trait heritability. Under causality, the additive genetic architecture defines . In the interaction architectures, letting denote the median of the gene expression we define:
.
Illustrated examples on the evaluation of the above formulas are provided in Supplementary Table S9.
Variance component
The residual is randomly drawn from a normal distribution where the parameter is a scaling parameter which ensures that the expression heritability or trait heritability, denoted , is maintained at a pre-specified value. Specifically, let denote the genetic component of expression or phenotype which is calculated from , , or . Then is derived using the equation , where is pre-specified for a particular simulation. This ensures that the simulated expressions or phenotypes have the desired level of heritability.
Results
Simulations
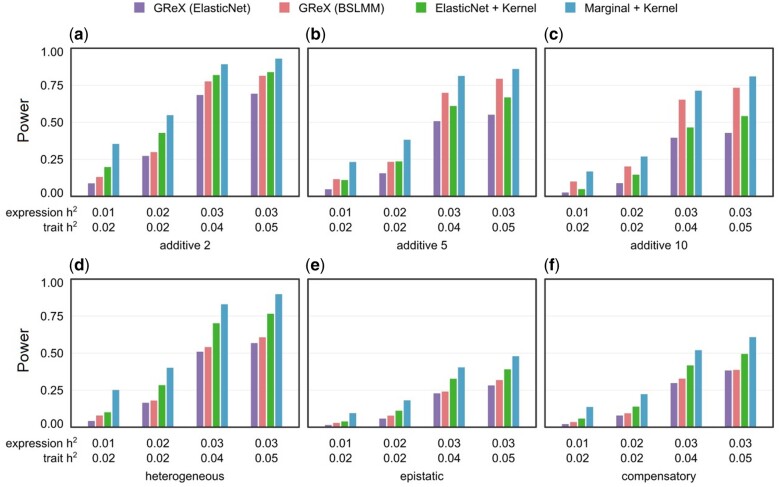
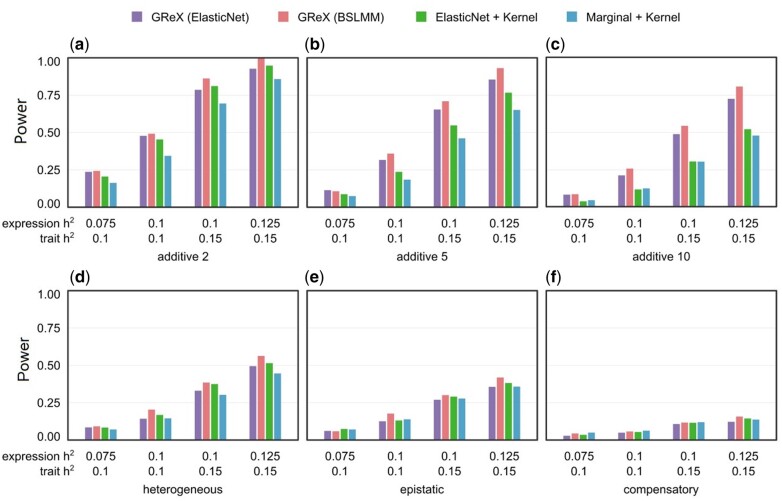
To thoroughly investigate the conditions under which disentangling feature selection and aggregation outperforms GReX alone, we conduct simulations comparing the four protocols in two scenarios: causality, where genotype causes phenotype via the intermediary of expression, and pleiotropy, where genotype causes phenotype and expression independently (Supplementary Figure S1). As previous publications show that TWAS enjoys higher power in pleiotropy than causality (Veturi and Ritchie 2018; Cao et al. 2021a; Tang et al. 2021), the pleiotropic simulations have reduced heritability to better distinguish power differences between each protocol (see Materials and Methods). In both scenarios, we simulate expression and phenotype with an additive genetic architecture and three interactive architectures labeled epistatic, compensatory, and heterogeneous (Figures 2 and 3). Under pleiotropy, the two disentangled methods (Marginal + Kernel and ElasticNet + Kernel) significantly outperform the GReX-based protocols (Figure 2). Marginal + Kernel also outperforms ElasticNet + Kernel, showing that a simple marginal effect-based model can outperform a regularized ElasticNet model in feature selection (Figure 2). Under causality, the GReX-based protocols have higher power in the additive case, with GReX (BSLMM) leading (Figure 3, A–C). This is unsurprising since the additive architecture consists of the same causal relations assumed by GReX (genotype causes expression and expression causes phenotype). In the interaction architectures under causality, all protocols have similar power with BSLMM leading slightly (Figure 3, D–F). Detailed mathematical formulas and parameterizations of our simulations are available in Materials and Methods.
Figure 2.
Power comparison of protocols in simulated pleiotropy scenario. The pleiotropic scenario simulates independent associations from genotype to phenotype and expressions (see Supplementary Figure S1 and Online Methods). Power is indicated on the y-axis. (A–C) are results under an additive genetic architecture, with differing expression heritability and local trait heritability denoted below each panel. The total number of contributing genetic variants is 2, 5, and 10 in each panel (left to right). (D–F) are results under interaction architectures, with expression heritability and local trait heritability denoted below. From left to right, the specific interactions are heterogeneous (logical “OR”), epistatic (logical “AND”), and compensatory (logical “XOR”).
Figure 3.
Power comparison of protocols in simulated causality scenario. The causality scenario simulates dependence of phenotype on genotype via gene expression (see Supplementary Figure S1 and Online Methods). Panels (A–F) have the same layout as Figure 4.
Real data analysis
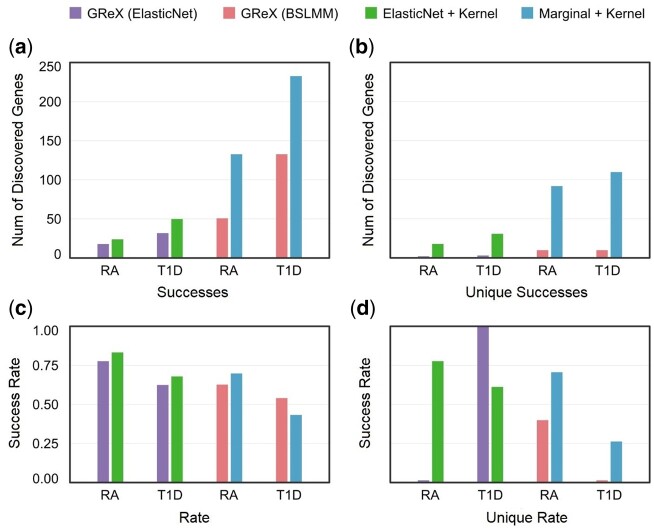
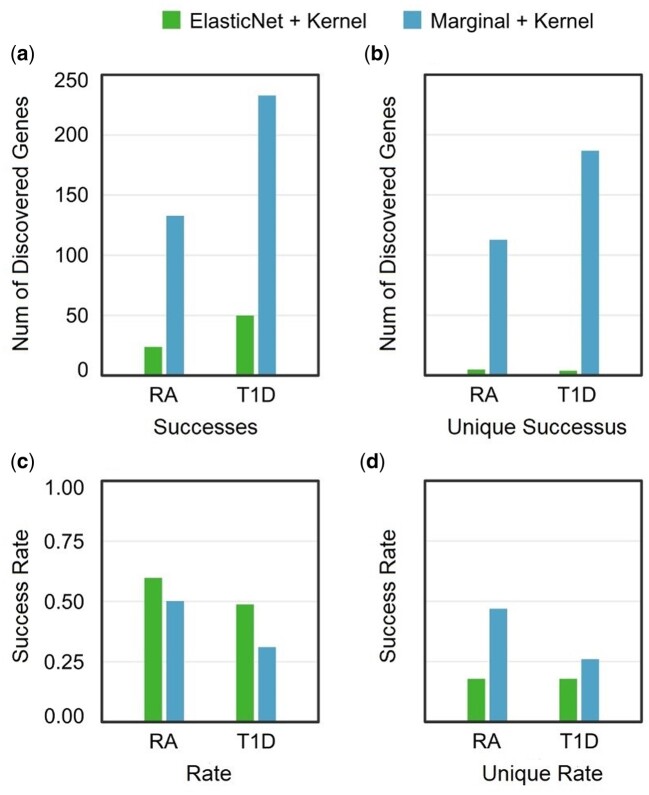
We first compare the four protocols above by analyzing WTCCC genotype data (Wellcome Trust Case Control 2007), with complete outcomes listed in Supplementary Tables S1–S3. For quantitative evaluation, we chose type 1 diabetes (T1D) and rheumatoid arthritis (RA) out of seven possible WTCCC diseases, as all four protocols discovered a large number of candidate genes in these diseases (P-value less than 0.05 after Bonferroni correction). For both diseases, Marginal + Kernel identifies the largest number of significant genes out of all protocols (Supplementary Tables S1 and S2). To validate the functional relevance of the identified genes, we refer to the DisGeNET database of human gene-disease associations (Pinero et al. 2015, 2017). We assess each protocol on the number of their discovered genes which are reported as disease-associated in DisGeNET (successes), as well as the proportion (success ratio) of these validated genes among all of the significant genes identified by the given protocol (Supplementary Table S4). Due to implementation differences, Marginal + Kernel and GReX (BSLMM) can assess all 19,696 genes in DisGeNET, whereas ElasticNet + Kernel and GReX (ElasticNet) only assess 7252 genes for which corresponding ElasticNet models are available from the PrediXcan website (Gamazon et al. 2015). As such, we only compare between the pairs Marginal + Kernel vs GReX (BSLMM) and ElasticNet + Kernel vs GReX (ElasticNet), giving a total of four comparisons over two diseases. In three of these four comparisons, the protocols disentangling feature selection and feature aggregation (Marginal + Kernel, ElasticNet + Kernel) outperform GReX-based protocols in both number and proportion of successfully identified genes (Figure 4, A and C). The only exception is between Marginal + Kernel and GReX (BSLMM) in T1D, where Marginal + Kernel has a slightly lower success ratio but a much higher number of successes (Figure 4, A and C). Since protocols which identify fewer genes tend to find a higher proportion of known disease-associated genes, we also compare the number and proportion of genes which are exclusively identified by only one of the two protocols under comparison (see Materials and Methods). Complete outcomes are listed in Supplementary Table S5. Again, in three out of four comparisons the disentangled protocols outperform GReX-based protocols (Figure 4, B and D). The only exception is between ElasticNet + Kernel and GReX (ElasticNet) in T1D, where ElasticNet + Kernel has a lower success ratio but identifies a much larger number genes (Figure 4, B and D). We perform an additional comparison between the two disentangled protocols Marginal + Kernel and ElasticNet + Kernel to evaluate the effectiveness of GReX on feature selection alone. We find that the simple marginal effects model used in Marginal + Kernel outperforms the more complex ElasticNet model used in ElasticNet + Kernel in total number of successes, but has a lower success ratio (Figure 5, A and C). However, when omitting genes identified by both protocols, Marginal + Kernel substantially outperforms ElasticNet + Kernel in both the total number and the ratio of exclusive successes (Figure 5, B and D).
Figure 4.
Comparison of GReX-based vs disentangled protocols in WTCCC data. Each figure compares two pairs of protocols in which the same number of genes are assessed over two WTCCC diseases (T1D and RA): GReX (ElasticNet) vs ElasticNet + Kernel (left), and GReX (BSLMM) vs Marginal + Kernel (right). (A) Total number of discovered genes (successes) which are reported as disease-associated in DisGeNET. (B) Number of discovered genes (successes) discovered exclusively by one of the two protocols under comparison. (C) Proportion (success rate) of all discovered genes which are validated by DisGeNET. (D) Proportion (success rate) of genes discovered exclusively by each protocol which are validated by DisGeNET.
Figure 5.
Comparison of marginal effect- and ElasticNet-based feature selection in WTCCC data. The two disentangled protocols ElasticNet + Kernel (left) and Marginal + Kernel (right) are compared on two WTCCC diseases (T1D and RA). Both protocols use kernel-based feature aggregation, but have different feature selection methods. (A) Number of discovered genes (successes) which are reported as disease-associated in DisGeNET. (B) Number of discovered genes (successes) discovered exclusively by one of the two protocols under comparison. (C) Proportion (success rate) of all discovered genes which are validated by DisGeNET. (D) Proportion (success rate) of genes discovered exclusively by each protocol which are validated by DisGeNET.
In addition to the above comparisons, for each disease we investigate whether the protocols can identify the top three genes with the highest gene-disease association scores in DisGeNET. The DisGeNET score takes into account the number of sources that report an association, the type of curation for each source, animal models where the association was studied, and the number of supporting publications discovered via text mining. This evidence is combined to score each gene by the confidence of its gene-disease association. The top three genes for RA are TNF, PTPN22, and SLC22A4, of which Marginal + Kernel is able to detect TNF and PTPN22, whereas none of the other protocols can identify any of the three genes. For T1D, the top three genes are PTPN22, INS, and HNF1A, of which Marginal + Kernel identifies PTPN22, whereas the remaining protocols do not identify any of the three genes.
Following standard practice in methodological works (Gamazon et al. 2015; Gusev et al. 2016; Hu et al. 2019; Nagpal et al. 2019; Wainberg et al. 2019; Li et al. 2020; Yuan et al. 2020), we searched the literature for additional evidence that the identified genes are relevant to disease. As discussed above, the only protocol which associates PTPN22 with T1D and RA is Marginal + Kernel. PTPN22 is highly scored in DisGeNET and has extensive literature support (Bottini et al. 2006). Among the five WTCCC diseases with an insufficient number of identifiable genes for quantitative comparison, Marginal + Kernel is the only protocol which associates TCF7L2 with type 2 diabetes and IRGM with Crohn’s disease. Among the five WTCCC diseases with an insufficient number of identifiable genes for quantitative comparison, Marginal + Kernel is the only protocol which associates TCF7L2 with type 2 diabetes and IRGM with Crohn’s disease. Both genes are well-supported by literature (Hattersley 2007; Prescott et al. 2010; Villareal et al. 2010; Baskaran et al. 2014). Based on our literature search, the top 5 significant genes for all four protocols are generally well supported. A comprehensive listing and discussion of the relevant literature for each gene is included in Supplementary Notes and Table S6.
Discussion
Our results show that in most cases, decoupling feature selection and aggregation allows even a simple feature selection method, based on the individual effects of genetic variants, to outperform a complex regularized model, which utilizes the combined linear effects of all potential eQTLs. Combined with the two preceding publications which apply kernel-based feature aggregation to TWAS (Cao et al. 2021b; Tang et al. 2021), this clearly demonstrates that GReX is not an optimal choice for all conditions. Although GReX has been a successful approach for leveraging expression data in GWAS, it is inherently limited by its use of a single linear model to solve two high-dimensional machine learning problems. Current TWAS development has largely treated GReX as a monolithic component, perhaps because the underlying statistical understanding of GReX as a genotype (not expression) model has been overlooked. By separating feature selection and feature aggregation into independent procedures, we show that many potential combinations of methods for conducting TWAS have been overlooked, some of which can yield improved power and specificity in commonly seen genetic architectures.
Another branch of TWAS method development is from the perspective of instrumental variables in causal inference. Among several tools toward this line, we chose a recently published tool, PTWAS (Zhang et al. 2020) for a comparison against mkTWAS by looking at the overlap between their outcomes and DisGeNET. Based on our procedure, it is verified that PTWAS also has a well-controled type-I-error (Supplementary Table S7) and identified lots of sensible genes (Supplementary Tables S1–S3). However, as a comparison, we found that mkTWAS discovered more genes than PTWAS (Supplementary Figure S2). The rates of success are similar however the success rates among genes uniquely identified by mkTWAS significantly outperform PTWAS (Supplementary Figure S2).
The simplicity of our marginal effects model suggests that feature selection plays an underappreciated role in TWAS and deserves further investigation. We have not thoroughly investigated the theoretical trade-offs between single marginal-effect-based approach and the advanced ElasticNet-based approach yet. One interpretation of our marginal method’s effectiveness is that it is more lenient in selecting variants, which allows a wider number of genes with poorly predicted expressions to be analyzed. For instance, PrediXcan can only analyze around 1/3 of the genes which have well-predicted expressions, whereas Marginal + Kernel can analyze all available genes in the transcriptome. We also propose that the larger number of variants selected by marginal effects pairs better with kernel-based aggregation, due to the previously discussed robustness of the kernel test to noise. These findings suggest that while feature selection and aggregation methods can be independently developed, it is also necessary to consider their compatibility when integrated in a two-step TWAS framework.
In order to simplify the design and interpretation of the protocols in this study, we did not consider trans-eQTLs and multiple tissue-based methods. As a future work, we will examine whether the conclusions of this study remain valid when potential trans-eQTLs are included in the protocols and simulations. Our findings can also apply to other types of middle-omic directed association mapping studies such as PWAS on proteins (Okada et al. 2016; Brandes et al. 2020) and IWAS (Xu et al. 2017) on brain images. This opens many additional opportunities for applying new or existing combinations of tools to various datasets.
Data availability
mkTWAS (https://github.com/theLongLab/mkTWAS), kTWAS (https://github.com/theLongLab/kTWAS), BSLMM (http://www.xzlab.org/software.html), PrediXcan (https://github.com/hakyim/PrediXcan), PTWAS (https://github.com/xqwen/ptwas), 1000 Genomes Project (https://www.internationalgenome.org/), GTEx genes expression and genotype are available at https://gtexportal.org/home/ and https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000424.v8.p2, WTCCC (https://www.wtccc.org.uk/).
Supplementary material is available at GENETICS online.
Funding
Q.L. is supported by an Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant (RGPIN-2017-04860), a Canada Foundation for Innovation JELF grant (36605), a New Frontiers in Research Fund (NFRFE-2018-00748), and an HBI Pilot grant. Q.Z. is supported by an NSERC Discovery Grant (RGPIN-2018-05147) and a University of Calgary VPR Catalyst grant. C.C. is supported by an Alberta Children's Hospital Research Institute (ACHRI) scholarship. D.K. is supported by an NSERC USRA award.
Conflicts of interest
The authors declare that there is no conflict of interest.
Supplementary Material
Literature cited
- Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. ; 1000 Genomes Project Consortium. 2015. A global reference for human genetic variation. Nature. 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbeira AN, Pividori M, Zheng J, Wheeler HE, Nicolae DL, et al. 2019. Integrating predicted transcriptome from multiple tissues improves association detection. PLoS Genet. 15:e1007889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskaran K, Pugazhendhi S, Ramakrishna BS.. 2014. Association of IRGM gene mutations with inflammatory bowel disease in the Indian population. PLoS One. 9:e106863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkin M, Ma S, Mandal S.. 2018. To understand deep learning we need to understand kernel learning. In: Jennifer D, Andreas K, editors. Proceedings of the 35th International Conference on Machine Learning. PMLR, Proceedings of Machine Learning Research, Stockholm, Sweden. p. 541–549.
- Bhattacharya A, Garcia-Closas M, Olshan AF, Perou CM, Troester MA, et al. 2020. A framework for transcriptome-wide association studies in breast cancer in diverse study populations. Genome Biol. 21:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Li Y, Love MI.. 2021. MOSTWAS: multi-omic strategies for transcriptome-wide association studies. PLoS Genet. 17:e1009398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottini N, Vang T, Cucca F, Mustelin T.. 2006. Role of PTPN22 in type 1 diabetes and other autoimmune diseases. Semin Immunol. 18:207–213. [DOI] [PubMed] [Google Scholar]
- Brandes N, Linial N, Linial M.. 2020. PWAS: Proteome-Wide Association Study. Cham: Springer International Publishing. p. 237–239. [Google Scholar]
- Cao C, Ding B, Li Q, Kwok D, Wu J, et al. 2021a. Power analysis of transcriptome-wide association study: implications for practical protocol choice. PLoS Genet. 17:e1009405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao C, Kwok D, Edie S, Li Q, Ding B, et al. 2021b. kTWAS: integrating kernel machine with transcriptome-wide association studies improves statistical power and reveals novel genes. Brief Bioinform. 22:bbaa270. [DOI] [PubMed] [Google Scholar]
- Chen H, Wang T, Huang S, Zeng P.. 2021. New novel non-MHC genes were identified for cervical cancer with an integrative analysis approach of transcriptome-wide association study. J Cancer. 12:840–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R.. 2010. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 33:1–22. [PMC free article] [PubMed] [Google Scholar]
- Gamazon ER, Wheeler HE, Shah KP, Mozaffari SV, Aquino-Michaels K, et al. ; GTEx Consortium. 2015. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet. 47:1091–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium. 2015. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 348:648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A, Ko A, Shi H, Bhatia G, Chung W, et al. 2016. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet. 48:245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A, Lawrenson K, Lin X, Lyra PC Jr, Kar S, et al. ; Ovarian Cancer Association Consortium. 2019. A transcriptome-wide association study of high-grade serous epithelial ovarian cancer identifies new susceptibility genes and splice variants. Nat Genet. 51:815–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A, Mancuso N, Won H, Kousi M, Finucane HK, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium. 2018. Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat Genet. 50:538–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattersley AT. 2007. Prime suspect: the TCF7L2 gene and type 2 diabetes risk. J Clin Invest. 117:2077–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Li M, Lu Q, Weng H, Wang J, et al. ; Alzheimer’s Disease Genetics Consortium. 2019. A statistical framework for cross-tissue transcriptome-wide association analysis. Nat Genet. 51:568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Teslovich TM, Boehnke M, Lin X.. 2013. General framework for meta-analysis of rare variants in sequencing association studies. Am J Hum Genet. 93:42–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Lappalainen T, Manichaikul A, Palmer AA, Wheeler H, et al. 2020. Predicted expression risk scores improve portability of trans-ethnic portability of polygenic risk scores. Biol Genomes. [Google Scholar]
- Li B, Verma SS, Veturi YC, Verma A, Bradford Y, et al. 2018. Evaluation of PrediXcan for prioritizing GWAS associations and predicting gene expression. Pac Symp Biocomput. 23:448–459. [PMC free article] [PubMed] [Google Scholar]
- Li Q, Cao C, Perera D, He J, Chen X, et al. 2020. Statistical model integrating interactions into genotype-phenotype association mapping: An application to reveal 3D-genetic basis underlying Autism. bioRXiv. doi:10.1101/2020.07.27.222364.
- Liu L, Zeng P, Xue F, Yuan Z, Zhou X.. 2021. Multi-trait transcriptome-wide association studies with probabilistic Mendelian randomization. Am J Hum Genet. 108:240–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luningham JM, Chen J, Tang S, De Jager PL, Bennett DA, et al. 2020. Bayesian genome-wide TWAS method to leverage both cis- and trans-eQTL information through summary statistics. Am J Hum Genet. 107:714–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso N, Freund MK, Johnson R, Shi H, Kichaev G, et al. 2019. Probabilistic fine-mapping of transcriptome-wide association studies. Nat Genet. 51:675–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso N, Gayther S, Gusev A, Zheng W, Penney KL, et al. ; PRACTICAL consortium. 2018. Large-scale transcriptome-wide association study identifies new prostate cancer risk regions. Nat Commun. 9:4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal S, Meng X, Epstein MP, Tsoi LC, Patrick M, et al. 2019. TIGAR: an improved Bayesian tool for transcriptomic data imputation enhances gene mapping of complex traits. Am J Hum Genet. 105:258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H, Ebhardt HA, Vonesch SC, Aebersold R, Hafen E.. 2016. Proteome-wide association studies identify biochemical modules associated with a wing-size phenotype in Drosophila melanogaster. Nat Commun. 7:12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongen H, Buil A, Brown AA, Dermitzakis ET, Delaneau O.. 2016. Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics. 32:1479–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinero J, Bravo A, Queralt-Rosinach N, Gutierrez-Sacristan A, Deu-Pons J, et al. 2017. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 45:D833–D839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinero J, Queralt-Rosinach N, Bravo A, Deu-Pons J, Bauer-Mehren A, et al. 2015. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database (Oxford). 2015:bav028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott NJ, Dominy KM, Kubo M, Lewis CM, Fisher SA, et al. 2010. Independent and population-specific association of risk variants at the IRGM locus with Crohn's disease. Hum Mol Genet. 19:1828–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnapriya R, Sosina OA, Starostik MR, Kwicklis M, Kapphahn RJ, et al. 2019. Retinal transcriptome and eQTL analyses identify genes associated with age-related macular degeneration. Nat Genet. 51:606–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Chai X, Yang Y, Cheng Q, Jiao Y, et al. 2020. A tissue-specific collaborative mixed model for jointly analyzing multiple tissues in transcriptome-wide association studies. Nucleic Acids Res. 48:e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siewert-Rocks KM, Kim SS, Yao DW, Shi H, Price AL. 2021. Leveraging gene co-expression to identify gene sets enriched for disease heritability. Biol Genomes. doi:10.1101/2021.07.22.453442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon N, Friedman J, Hastie T, Tibshirani R.. 2011. Regularization paths for Cox's proportional Hazards model via coordinate descent. J Stat Softw. 39:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Buchman AS, De Jager PL, Bennett DA, Epstein MP, et al. 2021. Novel Variance-Component TWAS method for studying complex human diseases with applications to Alzheimer's dementia. PLoS Genet. 17:e1009482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriault S, Gaudreault N, Lamontagne M, Rosa M, Boulanger MC, et al. 2018. A transcriptome-wide association study identifies PALMD as a susceptibility gene for calcific aortic valve stenosis. Nat Commun. 9:988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veturi Y, Ritchie MD.. 2018. How powerful are summary-based methods for identifying expression-trait associations under different genetic architectures? Pac Symp Biocomput. 23:228–239. [PMC free article] [PubMed] [Google Scholar]
- Villareal DT, Robertson H, Bell GI, Patterson BW, Tran H, et al. 2010. TCF7L2 variant rs7903146 affects the risk of type 2 diabetes by modulating incretin action. Diabetes. 59:479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainberg M, Sinnott-Armstrong N, Mancuso N, Barbeira AN, Knowles DA, et al. 2019. Opportunities and challenges for transcriptome-wide association studies. Nat Genet. 51:592–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium. 2007. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 447:661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Shi W, Long J, Guo X, Michailidou K, et al. ; kConFab/AOCS Investigators. 2018. A transcriptome-wide association study of 229,000 women identifies new candidate susceptibility genes for breast cancer. Nat Genet. 50:968–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Kraft P, Epstein MP, Taylor DM, Chanock SJ, et al. 2010. Powerful SNP-set analysis for case-control genome-wide association studies. Am J Hum Genet. 86:929–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wu C, Pan W; Alzheimer's Disease Neuroimaging Initiative. 2017. Imaging-wide association study: integrating imaging endophenotypes in GWAS. Neuroimage. 159:159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Zhu H, Zeng P, Yang S, Sun S, et al. 2020. Testing and controlling for horizontal pleiotropy with probabilistic Mendelian randomization in transcriptome-wide association studies. Nat Commun. 11:3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng P, Dai J, Jin S, Zhou X.. 2021. Aggregating multiple expression prediction models improves the power of transcriptome-wide association studies. Hum Mol Genet. 30:939–951. [DOI] [PubMed] [Google Scholar]
- Zeng P, Zhou X.. 2017. Non-parametric genetic prediction of complex traits with latent Dirichlet process regression models. Nat Commun. 8:456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Quick C, Yu K, Barbeira A, Luca F, et al. ; GTEx Consortium. 2020. PTWAS: investigating tissue-relevant causal molecular mechanisms of complex traits using probabilistic TWAS analysis. Genome Biol. 21:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Jiang Y, Zhong X, Cox NJ, Liu C, et al. 2020. A unified framework for joint-tissue transcriptome-wide association and Mendelian randomization analysis. Nat Genet. 52:1239–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Carbonetto P, Stephens M.. 2013. Polygenic modeling with bayesian sparse linear mixed models. PLoS Genet. 9:e1003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
mkTWAS (https://github.com/theLongLab/mkTWAS), kTWAS (https://github.com/theLongLab/kTWAS), BSLMM (http://www.xzlab.org/software.html), PrediXcan (https://github.com/hakyim/PrediXcan), PTWAS (https://github.com/xqwen/ptwas), 1000 Genomes Project (https://www.internationalgenome.org/), GTEx genes expression and genotype are available at https://gtexportal.org/home/ and https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000424.v8.p2, WTCCC (https://www.wtccc.org.uk/).
Supplementary material is available at GENETICS online.