Abstract
Targeted protein degradation using the auxin-inducible degron (AID) system is garnering attention in the research field of Caenorhabditis elegans, because of the rapid and efficient target depletion it affords, which can be controlled by treating the animals with the phytohormone auxin. However, the current AID system has drawbacks, i.e., leaky degradation in the absence of auxin and the requirement for high auxin doses. Furthermore, it is challenging to deplete degron-fused proteins in embryos because of their eggshell, which blocks auxin permeability. Here, we apply an improved AID2 system utilizing AtTIR1(F79G) and 5-phenyl-indole-3-acetic acid (5-Ph-IAA) to C. elegans and demonstrated that it confers better degradation control vs the previous system by suppressing leaky degradation and inducing sharp degradation using 1,300-fold lower 5-Ph-IAA doses. We successfully degraded the endogenous histone H2A.Z protein fused to an mAID degron and disclosed its requirement in larval growth and reproduction, regardless of the presence of maternally inherited H2A.Z molecules. Moreover, we developed an eggshell-permeable 5-Ph-IAA analog, 5-Ph-IAA-AM, that affords an enhanced degradation in laid embryos. Our improved system will contribute to the disclosure of the roles of proteins in C. elegans, in particular those that are involved in embryogenesis and development, through temporally controlled protein degradation.
Keywords: Caenorhabditis elegans, AID, degron, targeted protein degradation, protein knockdown, auxin, 5-Ph-IAA, 5-Ph-IAA-AM, histone H2A.Z
Introduction
Targeted protein degradation induced by a degron tag allows conditional loss-of-function analyses to be performed (Natsume and Kanemaki 2017; Wu et al. 2020). We pioneered the development of the auxin-inducible degron (AID) system, which has been widely applied in studies using yeast and mammalian cells (Nishimura et al. 2009). Plants have a unique degradation pathway that is controlled by the phytohormone auxin (also known as indole-3-acetic acid or IAA). Auxin binds to and activates an F-box protein, transport inhibitor response 1 (TIR1), which is incorporated into the SCF E3 ligase complex (Dharmasiri et al. 2005; Kepinski and Leyser 2005). The SCF–TIR1 E3 ligase bound to auxin recognizes the AUX/IAA-family transcriptional inhibitors for degradation via the ubiquitin-proteasome pathway, resulting in the transcription of the auxin-response genes (Chapman and Estelle 2009). The expression of TIR1 in nonplant cells results in the formation of a functional SCF E3 ligase complex with the endogenous SCF components because they are highly conserved among eukaryotes. In these cells, a protein fused with a degron tag derived from IAA17 of Arabidopsis thaliana is recognized by the SCF–TIR1 E3 ligase in the presence of IAA and is targeted for polyubiquitylation and subsequent degradation by the proteasome (Nishimura et al. 2009). One advantage of the AID system is that target depletion is very rapid (typically within a few hours), thus allowing the observation of the primary phenotype caused by the loss of expression of the protein of interest before it is complicated or compromised by secondary effects.
Caenorhabditis elegans is a model multicellular organism that is used in various research fields, including developmental biology, neurobiology, and ethology (Wood 1988). In turn, RNA interference has been used for many years to perform loss-of-function analyses (Fire et al. 1998). For the purpose of improving conditional control, the AID system was successfully applied to C. elegans (Zhang et al. 2015), and genetic tools related to this methodology have been developed (Martinez et al. 2020; Ashley et al. 2021). The combination of a 44-amino acid degron tag (namely AID*, shown in Supplementary Figure S1A) and TIR1 derived from A. thaliana (AtTIR1) encompassing the D170E and M473L mutations has been used in C. elegans studies (Morawska and Ulrich 2013; Zhang et al. 2015). Time- and tissue-specific protein depletion can be achieved by the controlled expression of AtTIR1 and the administration of IAA. Moreover, we recently reported that an IAA analog carrying an acetoxymethyl group, IAA-AM, yielded better degradation in embryos surrounded by an eggshell (Negishi et al. 2019).
Although the AID system is gaining popularity, we and others have noted leaky degradation (also called basal degradation) of degron-fused proteins in the absence of auxin (Natsume et al. 2016; Martinez et al. 2020; Schiksnis et al. 2020). Moreover, high doses of auxin, typically more than 500 µM IAA, are required for inducing degradation. In fact, the presence of IAA affects lifespan and progeny number (Loose and Ghazi 2021) (this article). These are major drawbacks that preclude sharp degradation control using a low dose of the inducing ligand. We recently overcame these problems by developing an improved version of the system called AID2, for use in yeast and mammalian cells (Yesbolatova et al. 2020). In the AID2 system, we used a TIR1 mutant derived from rice [OsTIR1(F74G)] and 5-phenyl-IAA (5-Ph-IAA). OsTIR1(F74G) has a hole in the ligand-binding pocket, so that it does not stably associate with IAA; in turn, 5Ph-IAA has a phenyl moiety that fits into the hole. Importantly, OsTIR1(F74G) yielded a negligible level of leaky degradation and afforded a higher sensitivity to the new ligand in mammalian cells. Hence, the AID2 system achieves sharp degradation control of AID-fused proteins using 5-Ph-IAA at a concentration of a few µM.
Here, we applied AID2 to the nematode C. elegans (Figure 1A). For this purpose, we introduced a transgene encoding AtTIR1(F79G) into the C. elegans genome. We found that, compared with AtTIR1, the expression of AtTIR1(F79G) caused an undetectable level of basal degradation. Furthermore, we demonstrated that significantly low concentrations of 5-Ph-IAA induced efficient degradation in nematodes expressing AtTIR1(F79G). We successfully degraded the endogenous HTZ-1 protein (the C. elegans homolog of the histone variant H2A.Z) fused with an mAID degron (Supplementary Figure S1A) and revealed that it is required for larval growth and development, which had not been demonstrated by conventional gene knock-out. By changing the timing of 5-Ph-IAA treatment, we temporally dissected the roles of HTZ-1 during larval development. Moreover, we synthesized a 5-Ph-IAA analog carrying an acetoxymethyl group, 5-Ph-IAA-AM, that afforded better target degradation in embryos surrounded by an eggshell. Our study clearly indicates that the AID2 system is a better option for future studies using C. elegans.
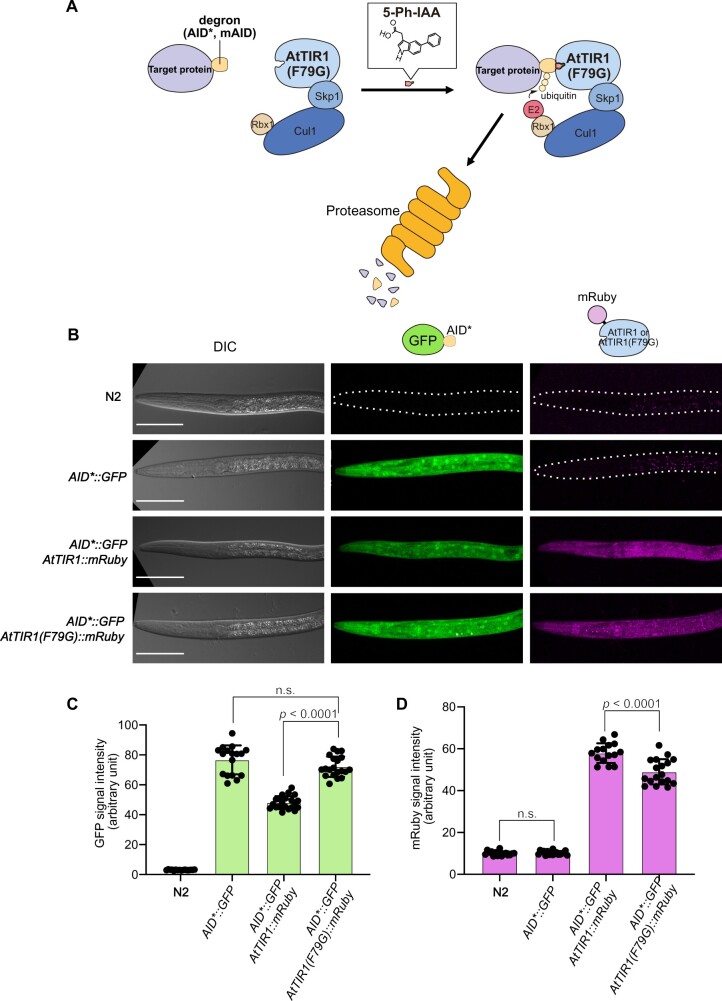
Figure 1.
Generation of C. elegans strains for testing the AID and AID2 systems. (A) Schematic illustration of the AID2 system. The AID* and mAID tags are recognized by AtTIR1(F79G) bound with 5-Ph-IAA. (B) Representative images of the strains used in the evaluations. The genotype of each strain is shown on the left side. All scale bars, 50 µm. (C, D) Quantification of the mean intensity of the GFP and mRuby signals, respectively. Error bars, standard deviation. The statistical analysis was performed using the unpaired t test.
Materials and methods
Plasmid construction
All plasmids used in this study are listed in Supplementary Table S1. The F79G mutation in the AtTIR1(D170E, M473L)::mRuby gene was introduced by PCR-mediated mutagenesis with the 5′-GGAAAGCCACACggCGCCGA-3′ and 5′-GCCGTGTGGCTTTCCCTTGAGCT-3′′ primer pair and using pLZ31 (Addgene no. 71720) as a template. The PCR product was circularized using NEBuilder. Subsequently, a dual-marker selection cassette (Norris et al. 2015) was added as follows. AtTIR1(F79G)::mRuby was amplified from the construct described above. The GFP-NeoR dual-marker selection cassette was amplified from the loxp_myo2_neor plasmid (a gift from Dr John Calarco), and a fragment containing the homology arms of the Chr. II MosSCI site (ttTi5605) was amplified from pOB4 [(Obinata et al. 2018), a gift from Dr Shinsuke Niwa]. These fragments were assembled using NEBuilder, to construct a donor plasmid, pTN4 (Peft-3::AtTIR1(F79G)::mRuby, Addgene no. 174212).
The plasmids used for tagging htz-1 with GFP::mAID were constructed as follows. For Cas9 and sgRNA expression, a guide RNA sequence (5′-AGCTTCAATCTAGTTCGCAA-3′′) targeting the N-terminal coding region of the htz-1 gene was predicted by the CRISPR direct tool (https://crispr.dbcls.jp/), and pTK73_htz-1 was constructed (Addgene no. 174546). A plasmid containing GFP::mAID, pGFPmAID (Addgene no. 174213), was constructed for the purpose of generating donor plasmids by cloning two homology arms. To facilitate the addition of homology arms, GFP-mAID is flanked by two ccdB negative-selection markers. Each ccdB is flanked by Spe1 sites, thus allowing the replacement of the ccdB markers with two homology arms. pGFPmAID_htz-1 (Addgene 174214), for tagging at the endogenous htz-1 locus, was constructed using the Gibson assembly with pGFPmAID digested with Spe1 and two PCR-amplified homology arms (519- and 586-bp fragments derived from the upstream and downstream regions of the htz-1 start codon, respectively), according to Dickinson et al. (2015).
Caenorhabditis elegans culture and strain construction
The C. elegans strains used in this study are listed in Supplementary Table S2. They were cultured by standard methods (Brenner 1974). To generate the strain containing the transgene Peft-3::AtTIR1(F79G)::mRuby, CRISPR–Cas9-mediated single-copy insertion was performed. According to a previously reported method (Obinata et al. 2018), 10 ng/µl of pTN4, 50 ng/µl of pDD162 (Peft-3::Cas9+sgRNA, Addgene no. 47549) and 50 ng/µl of pTK73_pOB4 [(Obinata et al. 2018); a gift from Dr Shinsuke Niwa] were injected into the wildtype N2 with the injection markers pCFJ90 (Pmyo-2::mCherry, Addgene no. 8984) and pCFJ104 (Pmyo-3::mCherry, Addgene no. 19328). After injection, dual selection was performed as previously described (Norris et al. 2015). First, F1 worms were treated with G418 for NeoR selection. Then, the survivors were further selected based on the expression of GFP in the pharynx. Subsequently, the selection cassette was excised by injection of pDD104 (Peft-3::Cre, Addgene no. 47551). To confirm the proper insertion of the transgene, genomic PCR was performed with the following primers (Supplementary Figure S1B): P1: 5′-TGCTGAATGGAGTACAACTGCAATG-3′′; P2: 5′-CGAGGGAGGTGTAGGTGTCTG-3′′; P3: 5′-GTCCAATTACTCTTCAACATCCC-3′′; and P4: 5′-TGGCAAAAATCCTAAAAACGAC-3′′. To extract genome DNA, 10 adult worms were lysed in a solution containing 10 mM Tris–HCl, pH 8.5, 50 mM KCl, 1 mM EDTA, 0.5% Tween 20, and 0.5 mg/ml of Proteinase K at 60°C for 1 h. After inactivation of Proteinase K at 95°C for 15 min, the lysate was directly used for PCR. The confirmed strain was crossed with the HS3280 strain (AID*::GFP), to generate the HS3545 strain harboring AtTIR1(F79G)::mRuby; AID*::GFP.
For generating the SA1532 strain expressing GFP::mAID::HTZ-1, CRISPR–Cas9-mediated tagging with GFP-mAID was performed by microinjection using 50 ng/µl of pGFPmAID_htz-1, 40 ng/µl of pTK73-htz-1, 50 ng/µl of pDD162, 40 ng/µl of pRF4, and injection markers (Figure 3A). Following the injection, the heterozygote progenies possessing GFP::mAID::htz-1 were identified by PCR using the following primers (Supplementary Figure S4A): P5: 5′-GTAGCTTGGAATCTTTTTCGGACG-3′′; P6: 5′-GTTGCATCACCTTCACCCTCTCC-3′′; and P7: 5′-TATAGGGCGAATTGGGGCGGTGAGATATTCGAGG-3′′. The homozygote animal derived from the heterozygote one was backcrossed with N2 at least twice.
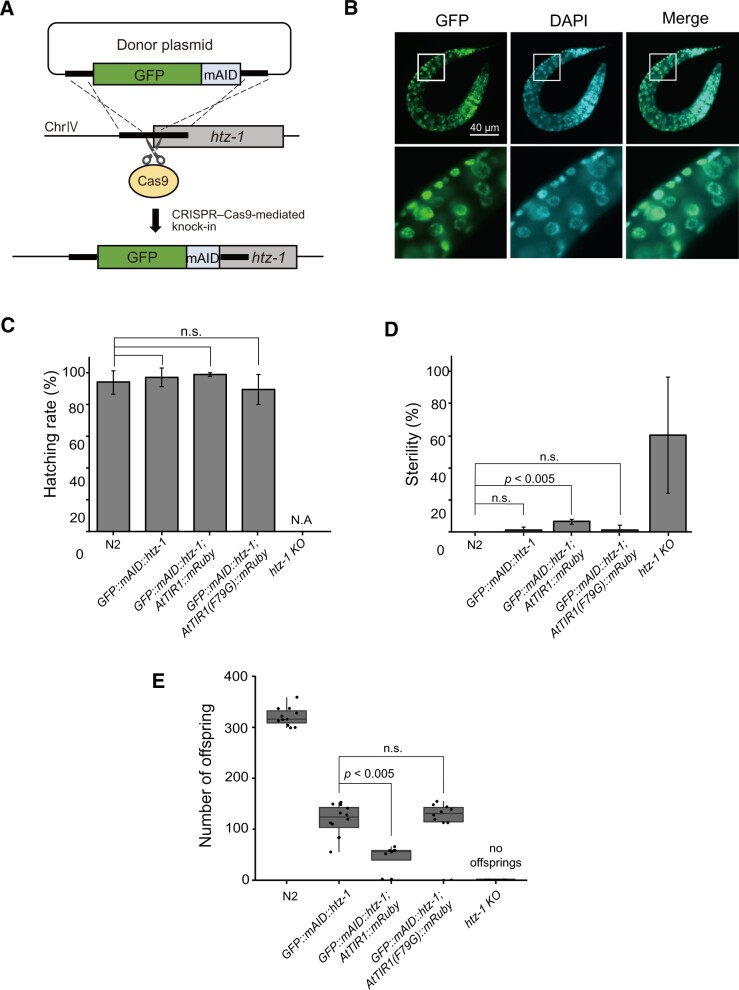
Figure 3.
The AID2 system suppresses leaky phenotypes of a mutant strain expressing the GFP::mAID-fused histone variant H2A.Z (HTZ-1), which are observed with an analogous mutant strain generated by using the original AID system. (A) Schematic illustration showing tagging of the endogenous htz-1 gene with GFP::mAID. (B) Microscopic observation of GFP::mAID::HTZ-1 in L1 larvae after DAPI staining. Scale bar, 40 µm. (C) The hatching efficiency of the indicated strains was tested (n = 3, more than 20 animals per experiment). (D) The sterile percentage of the indicated strains. Statistical significance was tested using the t-test (n = 3 or 4, more than 20 animals per experiment). (E) The offspring number of the indicated strains was tested (n ≥ 8). The statistical analysis was performed using the unpaired t test.
For htz-1 KO strain (SA1463), htz-1 (tm2469) was obtained from CGC and balanced over tmC25 (tmIs1241). GFP-negative animals were used as homozygous htz-1 KO animals for the experiments.
Synthesis of 5-Ph-IAA AM ester
1H and 13C NMR spectra were recorded on a JEOL ECZ400 NMR spectrometer (JEOL, Japan). Chemical shifts are shown as δ values from TMS as the internal reference. Peak multiplicities are quoted in Hz. MS spectram was measured with autoflex MALDI-TOF-MS (BURUKER, Japan). Column chromatography was carried out on Merck silica gel 60 (230–400 mesh, Merck, Japan). All chemicals were purchased from Tokyo Chemical Industry Japan (Tokyo, Japan).
Bromomethyl acetate (300 mg, 2.0 mmol) was added dropwise to the solution of 5-Ph-IAA (500 mg, 2.0 mmol) and trimethylamine (402 mg, 4.0 mmol) in DMF (10 ml), followed by stirring for 18 h at room temperature. The reaction mixture was added to water (100 ml) and extracted with EtOAc (50 ml × 3). The organic layer was washed with 1 M HCl, 1 M aqueous Na2CO3 and brine, and then dried over Na2SO4. The residue was purified by a silica gel column chromatography (hexane:EtOAc = 7:3), to yield a 5-phenylindole 3-acetic acid acetoxymethyl ester as a crystal (442 mg; yield, 69%). 1H-NMR (400 MHz, CDCl3) δ 8.18 (s, 1H), 7.77 (s, 1H), 7.64 (d, J = 8.2 Hz, 2H), 7.44–7.41 (m, 3H), 7.32 (t, J = 9.1 Hz, 2H), 7.09 (s, 1H), 5.76 (s, 2H), 3.84 (s, 2H), 1.97 (s, 3H), 13C-NMR (100 MHz, CDCl3) δ 170.7, 169.7, 142.4, 135.6, 133.3, 128.6, 127.5, 127.3, 126.4, 124.0, 122.1, 117.2, 111.5, 107.6, 79.4, 31.0, 20.5, MALDI-TOF-MS m/z 346.1 [M+Na]+.
Ligand treatment
IAA purchased from Nacalai Tesque (no. 19119-61) was dissolved in EtOH at a concentration of 400 mM. 5-Ph-IAA was synthesized as described previously (Yesbolatova et al. 2020). The stock of 5-Ph-IAA was prepared in DMSO at a concentration of 100 mM.
The young larvae were placed on an NGM plate containing the indicated concentrations of IAA or 5-Ph-IAA. The laid embryos by adult animals were picked and subsequently treated with M9 buffer containing a concentration of 50 or 100 µM 5-Ph-IAA or a concentration of 50 µM 5-Ph-IAA-AM. The buffer also contained polystyrene beads with a diameter of 20 µm (Polyscience no. 18329), for embryo imaging (Bao and Murray 2011). Both 5-Ph-IAA and 5-Ph-IAA-AM are commercially available from BioAcademia.
Observation and data processing
The images included in Figures 1, 2, 6 and Supplementary Figures S1, S3, and S6 were acquired with a Zeiss LSM700 confocal microscope using a 63× N.A. 1.40 oil-immersion objective. The images shown in Figures 3, 4 and Supplementary Figures S4 and S5 were obtained with an IX-83 wide-field microscope (Olympus) using a SPlan10PL10 × 0.30 objective (Olympus) or a UPlanSApo 40 × 0.95 objective (Olympus). The image-acquisition setting was identical in each experiment. Acquired images were processed by Fiji (Schindelin et al. 2012). GraphPad Prism 6, R (R Core Team 2020) and RStudio (R Studio Team 2020) were used for drawing graphs and performing statistical analyses. The T1/2 and DC50 values reported in Figure 2 were calculated using a nonlinear fitting, one-phase decay model.
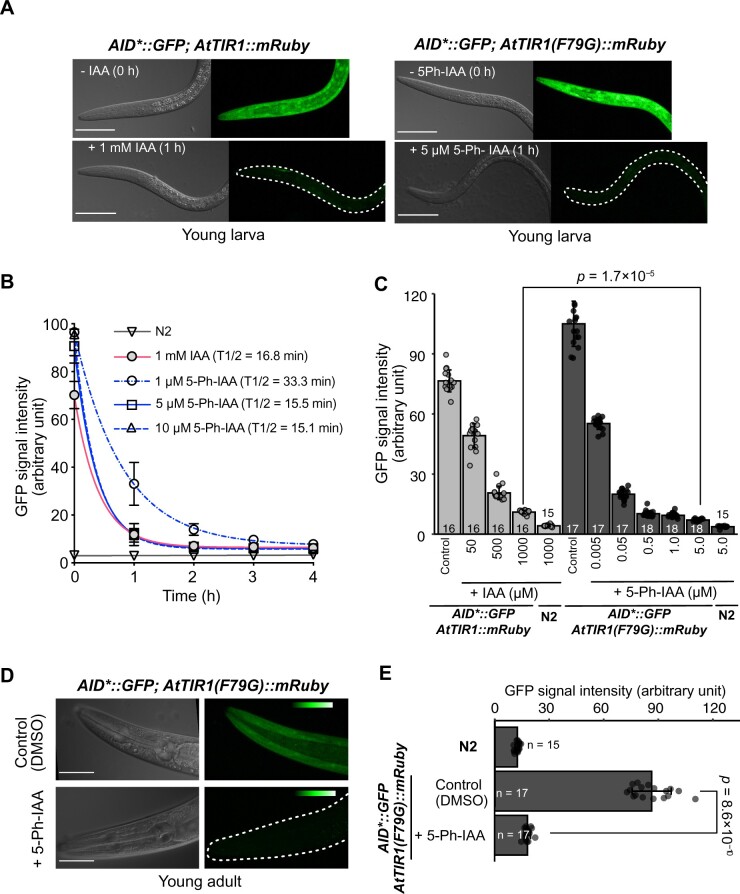
Figure 2.
Induced degradation of the AID*::GFP reporter by the AID and AID2 systems. (A) Representative images acquired before and after ligand treatment. The indicated larva was treated with 1 mM IAA (left) or 5 µM 5-Ph-IAA for 1 h. All scale bars, 50 µm. (B) Time-course evaluation of the mean GFP intensity in the larvae treated with the ligands. The red line shows the GFP level in the larvae expressing AtTIR1::mRuby treated with 1 mM IAA (n > 7). The other lines indicate the GFP level in the larvae expressing AtTIR1(F79G)::mRuby treated with 1, 5, and 10 µM 5-Ph-IAA (n = 7, 12, and 7, respectively). Error bars, standard deviation. The T1/2 for each condition is indicated in the graph. (C) Quantification of the GFP level in the indicated larval strains treated over one generation with different concentrations of IAA or 5-Ph-IAA. The numbers of treated animals are shown at the bottom of each column. Error bars, standard deviation. The statistical analysis was performed between the treatment with 1000 µM IAA and the treatment with 5 µM 5-Ph-IAA (Wilcoxon rank-sum test, P = 1.7 × 10−5). (D) Representative images of reporter protein expression in a young adult expressing AtTIR1(F79G)::mRuby after treatment with DMSO (upper panels) or 5 µM 5-Ph-IAA (lower panels) for 4 h. The color table indicates the intensity of the fluorescent signal. Scale bars, 50 µm. (E) Quantification of mean GFP intensity in the adults treated with 5-Ph-IAA for 4 h. The numbers of treated animals are shown on the left side of each column. Error bars, standard deviation. The statistical analysis was performed between the treatment with DMSO and the treatment with 5 µM 5-Ph-IAA (Wilcoxon rank-sum test, P = 8.6 × 10−10).
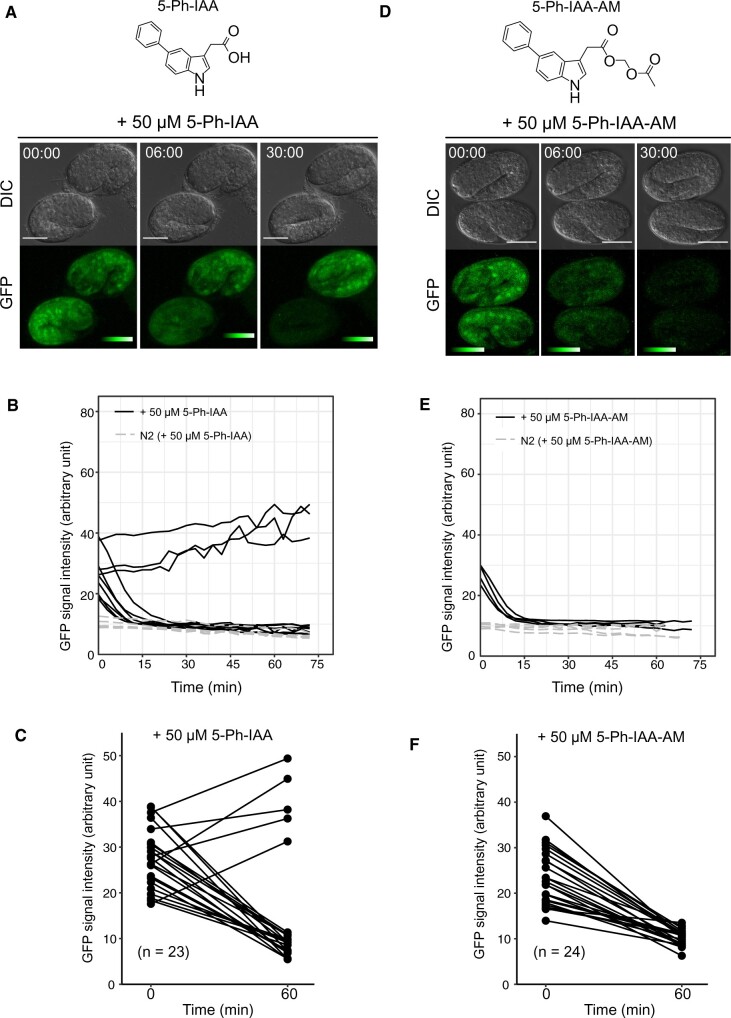
Figure 6.
5-Ph-IAA-AM triggers better degradation in the C. elegans embryos. (A) Chemical structure of 5-Ph-IAA (top). Time-lapse images of the embryos (AID*::GFP; AtTIR1(F79G)::mRuby) treated with 50 µM 5-Ph-IAA (bottom). Scale bars, 20 µm. The color table indicates the intensity of the fluorescent signal. (B) GFP intensity in the 10 and 5 embryos [AID*::GFP; AtTIR1(F79G)::mRuby and wildtype N2, respectively] treated with 50 µM 5-Ph-IAA. Images were captured at 3 min intervals. (C) Efficacy of triggering reporter degradation using 50 µM 5-Ph-IAA. The GFP intensity was measured at 0 and 60 min (n = 23). (D) Chemical structure of 5-Ph-IAA-AM (top). Time-lapse images of the embryos [AID*::GFP; AtTIR1(F79G)::mRuby] treated with 50 µM 5-Ph-IAA-AM (bottom). Scale bars, 20 µm. The color table indicates the intensity of the fluorescent signal. (E) GFP intensity of the 4 and 5 embryos [AID*::GFP; AtTIR1(F79G)::mRuby and wildtype N2, respectively] treated with 50 µM 5-Ph-IAA-AM. Images were captured at 3 min intervals. (F) Efficacy of triggering reporter degradation using 50 µM 5-Ph-IAA-AM. GFP intensity was measured at 0 and 60 min (n = 24).
Figure 4.
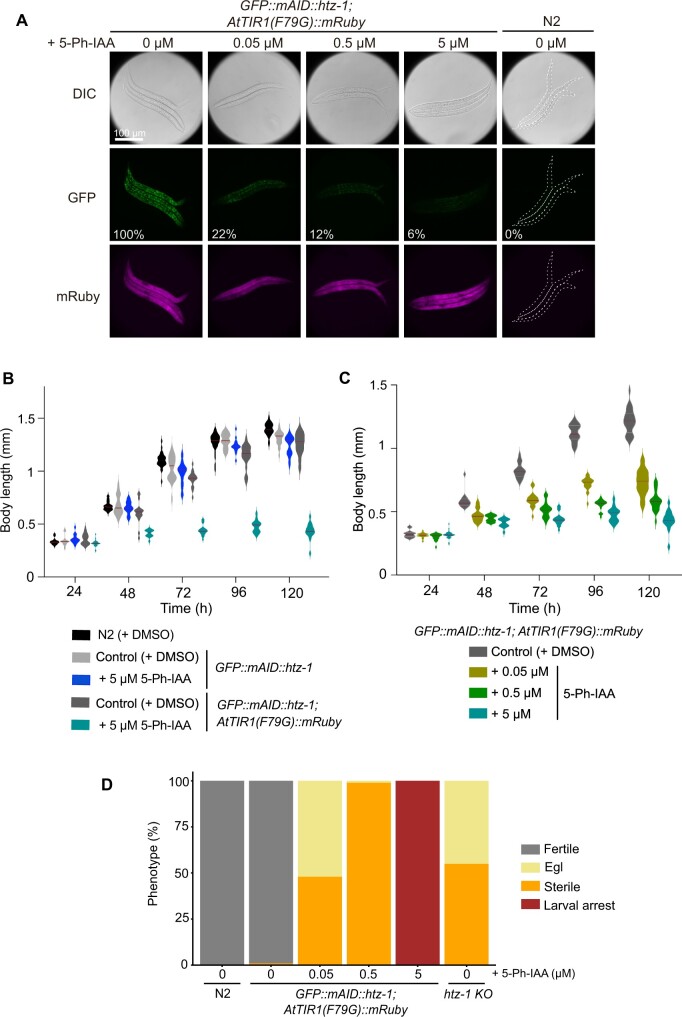
Phenotypic analyses after targeted depletion of HTZ-1. (A) Targeted depletion of GFP::mAID::HTZ-1 was observed in the presence of the indicated concentrations of 5-Ph-IAA. Images of differential interference contrast (DIC, upper), GFP (middle), and mRuby (bottom) are shown. The numbers in the middle panels indicate the relative intensity of the GFP signal (0% in the control N2 strain and 100% in the GFP::mAID::htz-1; AtTIR1(F79G)::mRuby strain without 5-Ph-IAA treatment). Scale bar, 100 µm. (B) The growth of wildtype N2 and the strain expressing GFP::mAID::HTZ-1 with or without AtTIR1(F79G)::mRuby were tested in the presence or absence of 5 µM 5-Ph-IAA. The body length was measured at the indicated time points. More than 11 animals were counted for each time point. (C) Embryos expressing GFP::mAID::HTZ-1 and AtTIR1(F79G)::mRuby were treated with 0.05, 0.5, or 5 µM 5-Ph-IAA for testing the body length at the indicated time points. More than 11 animals were counted for each point. (D) Phenotypes of the GFP::mAID::htz-1; AtTIR1(F79G)::mRuby strain treated with the indicated concentration of 5-Ph-IAA (n = 92, 96, 95, 97, and 37 for N2, 0, 0.05, 0.5, and 5 µM 5-Ph-IAA, respectively). The phenotypes of the htz-1-KO strain were also tested (n = 77). The statistical analysis was performed using the unpaired t test.
Analyses of worm phenotypes
Hatching rate and sterility
Adult worms were placed on an NGM or auxin-containing plate for egg-laying for 2 h. Subsequently, adult larvae were removed, and the number of eggs was counted. Hatched animals were counted 24 h later.
Phenotypic definitions for viable larvae
Phenotypes were scored according to the following four categories. Arrested larvae up to L2 were scored as “Larval arrest”; worms possessing embryos that were capable of laying on a plate were classified as “Fertile”; worms possessing embryos that failed to lay on a plate were scored as “Egg-laying defect (Egl)”; and worms that did not have any embryos were classified as “Sterile.”
Progeny size
In the investigation of the effect of the ligands (Supplementary Figure S2A), L1 larvae were placed on plates containing 1 mM IAA, 5 µM 5-Ph-IAA, or the respective solvents (0.25% ethanol or 0.1% DMSO) and were subsequently transferred to new plates daily over 4 days. After confirming that hermaphrodite animals finished laying the eggs, the sum of the hatched progeny was counted for each treatment. In the HTZ-1-depletion experiments (Figure 5), young adults treated with the indicated concentration of 5-Ph-IAA were plated singly. They were transferred to a fresh plate every 24 h until no progeny was detected. The number of hatched animals on each plate was counted.
Figure 5.
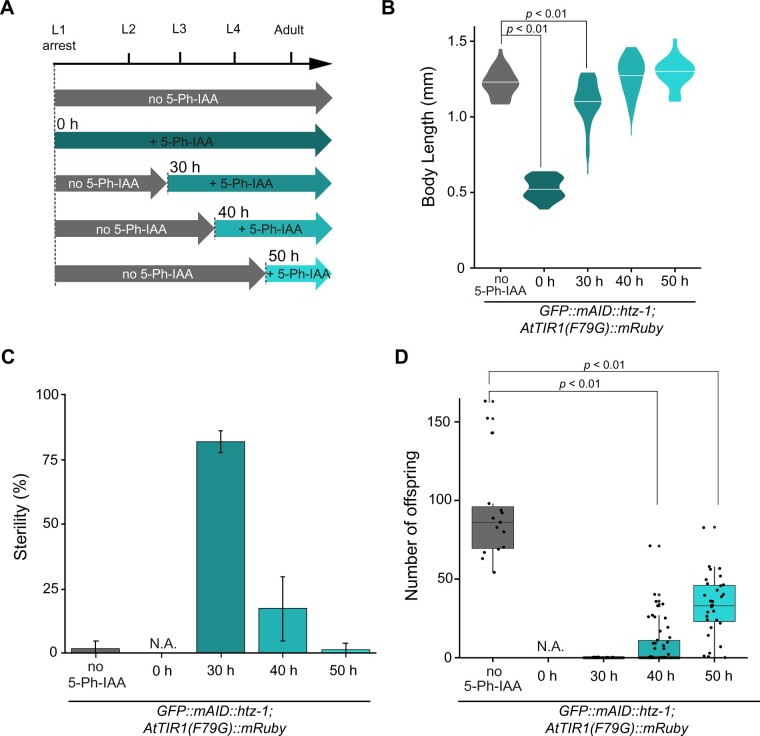
Stage-specific degradation of HTZ-1 yields differential phenotypes regarding larval growth, sterility and progeny number. (A) Experimental scheme showing the timing of 5 µM 5-Ph-IAA addition. (B) Body length of the GFP::mAID::htz-1;AtTIR1(F79G)::mRuby strain and htz-1-KO strain at 120 h in each experiment (n ≥ 19). (C) Sterility of the GFP::mAID::htz-1; AtTIR1(F79G)::mRuby strain in each experiment (n = 3, more than 20 animals per experiment). (D) Number of viable progeny from the GFP::mAID::htz-1; AtTIR1(F79G)::mRuby strain treated with DMSO (n = 15) or 5 µM 5-Ph-IAA at 30 (n = 37), 40 (n =50), and 50 h (n = 32).
Body length
Body length was evaluated at each time point until 120 h after egg laying or L1 diapause. At each time point, cultured animals were mounted and paralyzed on a 2% agar pad containing 0.4% sodium azide. They were imaged using differential interference contrast with an IX-83 wide-field microscope (Olympus) equipped with a 10× objective (Olympus, SPlan10PL, numerical aperture, 0.30). Body length was measured using the ImageJ software (Mörck and Pilon 2006).
Results
AtTIR1(F79G) shows a negligible level of leaky degradation in C. elegans
When the AID system was originally established in C. elegans, Zhang et al. used a codon-optimized AtTIR1 gene containing the D170E and M473L mutations, which were reported to enhance auxin affinity in A. thaliana (Yu et al. 2013; Zhang et al. 2015). To employ the AID2 system in C. elegans (Figure 1A), we introduced an additional F79G mutation in AtTIR1(D170E and M473L), which corresponds to an F74G mutation in OsTIR1 [hereafter called AtTIR1 and AtTIR1(F79G) for the original and F79G mutant AtTIR1 proteins, respectively] (Zhang et al. 2015; Yesbolatova et al. 2020). We inserted the transgene encoding AtTIR1(F79G) fused with a red fluorescent protein (mRuby) into the ttTi5605 locus (chromosome II: position 8,420,204) using a CRISPR–Cas9-mediated knock-in approach (Supplementary Figure S1, B and C) (Zhang et al. 2015; Obinata et al. 2018). The expression of AtTIR1(F79G)::mRuby was driven by the eft-3 promoter, to achieve ubiquitous expression in all somatic and germ cells (Zhang et al. 2015). Consistently, we detected the expression of AtTIR1(F79G)::mRuby in all organs including the germline (Supplementary Figure S1D). We crossed the strain expressing AtTIR1(F79G)::mRuby with a strain expressing an AID*-fused GFP reporter (AID*::GFP) (Supplementary Figure S1A). To monitor the level of basal degradation, we measured the intensity of AID*::GFP in larvae expressing AtTIR1::mRuby or AtTIR1(F79G)::mRuby (Zhang et al. 2015). In the strain that expressed AtTIR1::mRuby, the reporter level was decreased down to 73% compared with that detected in the parental strain without the transgene, indicating the presence of basal degradation in the absence of the ligand, consistent with previous reports (Figure 1, B and C) (Martinez et al. 2020; Schiksnis et al. 2020). In contrast, the expression level of the reporter was unaffected in the larvae expressing AtTIR1(F79G)::mRuby (Figure 1, B and C), suggesting that AtTIR1(F79G) yielded a lower level of basal degradation than did AtTIR1 in C. elegans, as observed in cultured cells (Yesbolatova et al. 2020). We also noted that the expression level of AtTIR1 was higher than that of AtTIR1(F79G), possibly because AtTIR1 was introduced into the ttTi5605 locus using a transposon-based MosSCI system and the genotype of the two strains was slightly different (Figure 1D) (Frøkjaer-Jensen et al. 2008; Zhang et al. 2015). This observation also suggests that the suppression of basal degradation observed in this study might not be attributed entirely to the ligand-independent activity of AtTIR1(F79G); rather, it may be partly caused by the lower expression level of AtTIR1(F79G). Nevertheless, as described below, this level of AtTIR1(F79G) was able to induce sharp degradation using significantly lower concentrations of the ligand.
The AID2 system enables sharp target depletion using low ligand doses in C. elegans
Initially, we wished to determine whether 5-Ph-IAA is safer than IAA in C. elegans. It has recently been reported that a 1-mM IAA treatment increased the lifespan of C. elegans (Loose and Ghazi 2021). That study suggested that IAA potentially causes side-effects in C. elegans. To clarify this issue, we grew the L1 larvae expressing AID*::GFP with AtTIR1 or AtTIR1(F79G) in the presence of 1 mM IAA or 5 µM 5-Ph-IAA, respectively. We found that the worms treated with 1 mM IAA produced a reduced number of offspring (Supplementary Figure S2A). In contrast, this effect was not observed in those treated with 5 µM 5-Ph-IAA. These results indicate that treating C. elegans with 1 mM IAA affects egg production or embryogenesis. We concluded that treating C. elegans with 5 µM 5-Ph-IAA is safer than treating them with 1 mM IAA.
To test whether the combination of AtTIR1(F79G) and 5-Ph-IAA induces the degradation of the AID*::GFP reporter, we treated the larvae harboring AtTIR1 or AtTIR1(F79G) on plates containing 1 mM IAA or 5 µM 5-Ph-IAA, respectively. Before the treatment, the fluorescence intensity of the reporter was higher in the larvae expressing AtTIR1(F79G) than it was in those expressing AtTIR1 (Figure 2, A and B). Consistent with the previous report, treatment with 1 mM IAA reduced the level of the reporter in the larvae expressing AtTIR1 with a half-life of 16.8 min (Zhang et al. 2015). We found that treatment with 5 µM 5-Ph-IAA similarly reduced the reporter level with a half-life of 15.5 min in the larvae expressing AtTIR1(F79G), although the ligand concentration was 200 times lower (Figure 2, A and B). We further tested treatment with 1 and 10 µM 5-Ph-IAA, which resulted in a half-life of 33.6 and 15.1 min, respectively (Figure 2B). These results showed that the depletion kinetics reached nearly a maximum after treatment with 5 µM 5-Ph-IAA. We achieved sharper degradation with the combination of AtTIR1(F79G) and 5 µM 5-Ph-IAA compared with the conventional AID condition, because the initial expression level of the reporter was higher. Taking the background fluorescent level of the wildtype N2 strain, 95% of the AID*::GFP reporter was degraded by treating with 5 µM 5-Ph-IAA for 4 h.
Next, we grew the L1 larvae on plates containing different concentrations of the ligands. We then monitored the intensity of the AID*::GFP reporter in young larvae (second generation) born from adult worms. We observed that the reporter expression was lowest in the larvae treated with 5 µM 5-Ph-IAA (Figure 2C). Moreover, this expression level was lower than that observed in the AtTIR1-expressing larvae treated with 1 mM IAA (P = 1.7 × 10−5). The estimated DC50 value (the ligand concentration required for 50% degradation) was 4.5 nM and 59 µM for 5-Ph-IAA combined with AtTIR1(F79G) and IAA combined with AtTIR1, respectively. This result indicates that the AID2 system using 5-Ph-IAA requires ∼1,300-fold lower ligand concentrations than does the conventional AID system. Furthermore, we tested the efficacy of the AID2 system in adult worms. Treatment of these worms with 5 µM 5-Ph-IAA for 4 h led to loss of the reporter (Figure 2, D and E). These results clearly indicate that the combination of AtTIR1(F79G) and 5-Ph-IAA improves ligand sensitivity and depletes the target protein even more sharply in C. elegans larvae and adult individuals.
Subsequently, we investigated the cross-reactivity of IAA and 5-Ph-IAA (Supplementary Figure S2B). The combination 5-Ph-IAA and AtTIR1(F79G) worked the best. However, a high dose of 1 mM IAA combined with AtTIR1(F79G) reduced the reporter level to 62.4%, suggesting weak cross-reactivity of IAA to AtTIR1(F79G). Consistent with this notion, 5 µM IAA did not show significant cross-reactivity to AtTIR1(F79G). Therefore, we concluded that IAA cross-reacted to AtTIR1(F79G) only at high doses.
In all experiments shown above, we used AtTIR1(F79G)-fused with mRuby [AtTIR1(F79G)::mRuby]. We also generated a strain expressing AtTIR1(F79G) without mRuby (Supplementary Figure S3, A and B) and tested the degradation of the AID*::GFP reporter. As expected, we confirmed that AtTIR1(F79G) without mRuby achieved depletion of the reporter similarly to the case with AtTIR1(F79G)::mRuby (Supplementary Figure S3C).
Application of the AID2 system for degrading the histone variant H2A.Z
To evaluate the targeted depletion of an endogenous protein by the AID2 system in C. elegans, we employed H2A.Z (namely HTZ-1) as a target for evaluation. The histone variant H2A.Z is involved in multiple fundamental chromatin transactions, including transcriptional regulation, DNA repair, DNA replication, and chromatin modification (Giaimo et al. 2019; Long et al. 2020). The mAID degron fused with GFP (GFP::mAID, Figure 3; Supplementary Figure S1A) was introduced into the genomic htz-1 locus using CRISPR–Cas9-mediated genome editing, as described in Materials and methods, and the integration of the GFP::mAID tag was confirmed (Supplementary Figure S4, A and B). Expression of GFP::mAID::HTZ-1 in the L1 larvae was observed in the whole body (Figure 3B, upper panels), and the HTZ-1 fusion protein colocalized with genomic DNA (Figure 3B, lower panels). These observations are consistent with the characteristics of the endogenous HTZ-1 protein without a tag, which is ubiquitously expressed and incorporated into chromatin (Whittle et al. 2008). The hatching efficiency and sterility of the animals expressing GFP::mAID::HTZ-1 with AtTIR1(F79G)::mRuby were undiscriminated from those of the control N2 strain (Figure 3, C and D). In contrast, homozygous htz-1 mutant derived from heterozygous mothers (htz-1-KO) exhibited a sterile phenotype, as reported previously (Figure 3D) (Whittle et al. 2008). Interestingly, the animals expressing GFP::mAID::HTZ-1 with AtTIR1::mRuby showed a significantly higher level of sterility compared with the N2 control, suggesting that AtTIR1 caused problems leading to sterility possibly due to its leaky degradation. The number of offspring of the animals expressing GFP::mAID::HTZ-1 was reduced when compared with N2 animals with and without the expression of AtTIR1(F79G)::mRuby to the same extent (Figure 3E). This observation suggests that the GFP::mAID tag, not AtTIR1(F79G), affected the number of offspring. Notably, the combination of GFP::mAID::HTZ-1 with AtTIR1::mRuby, but not AtTIR1(F79G)::mRuby, further reduced the number of offspring (Figure 3E), consistent with the notion that AtTIR1 caused increased sterility (Figure 3D). Therefore, we concluded that the use of the AID2 system is advantageous for suppressing leaky phenotypes, which were expressed when using the original AID system for C. elegans.
Next, we tested the targeted depletion of GFP::mAID::HTZ-1. By treating the animals expressing both GFP::mAID::HTZ-1 and AtTIR1(F79G)::mRuby with 5-Ph-IAA, we observed that GFP::mAID::HTZ-1 was depleted in a concentration-dependent manner (Figure 4A). Conversely, the expression of AtTIR1(F79G)::mRuby alone did not affect the level of GFP::mAID::HTZ-1 in the absence of the ligand (Supplementary Figure S4C and D), supporting the notion that AtTIR1(F79G) yields a negligible level of leaky degradation (Figure 1). When we observed an individual animal, the expression of AtTIR1(F79G)::mRuby is observed in the whole body including the germline with and without the ligand (Supplementary Figure S5, mRuby). The expression of GFP::mAID::HTZ-1 is observed in the whole body without 5-Ph-IAA, and the signal is reduced by adding 5-Ph-IAA in the body, including the germline (Supplementary Figure S5, GFP).
Developmental defects in response to the targeted depletion of HTZ-1
The consequence of GFP::mAID::HTZ-1 depletion was evaluated through observation of the phenotypes that appeared after the ligand treatment. Embryos expressing GFP::mAID::HTZ-1 or both GFP::mAID::HTZ-1 and AtTIR1(F79G)::mRuby were treated with or without 5 µM 5-Ph-IAA (Figure 4B). The GFP::mAID::htz-1 larvae treated with 5-Ph-IAA grew normally, as did those treated with the control; this showed that 5-Ph-IAA did not cause side effects, consistent with the notion that a 5-µM 5-Ph-IAA treatment did not affect the offspring number reported in Supplementary Figure S2A. In sharp contrast, the GFP::mAID::htz-1; AtTIR1(F79G)::mRuby larvae treated with 5 µM 5-Ph-IAA showed a significant growth defect, which was not observed in the control-treated larvae (Figure 4B). To evaluate the growth defect upon HTZ-1 depletion, we treated GFP::mAID::htz-1; AtTIR1(F79G)::mRuby larvae with different concentrations of 5-Ph-IAA (Figure 4C). After depletion of GFP::mAID::HTZ-1, as shown in Figure 4A, we observed growth defects in a dose-dependent manner. These results clearly indicate that HTZ-1 is essential for the normal growth of C. elegans larvae.
Next, we tested the reproduction of the animals in the presence of different concentrations of the ligand. It has been reported that htz-1-KO animals show defects in progeny production, as expressed by the sterile (no egg production) and egg-laying defective (Egl) phenotypes (Figure 4D) (Whittle et al. 2008). We recapitulated similar phenotypes when the larvae expressing GFP::mAID::HTZ-1 with AtTIR1(F79G)::mRuby were grown in the presence of 0.05 µM 5-Ph-IAA (Figure 4D). Treatment of these larvae with 0.5 µM 5-Ph-IAA caused a sterile phenotype in all worms. Larval growth was severely affected after treatment with the highest dose of 5-Ph-IAA (5 µM) (Figure 4, B and C), and the larvae did not reach the L3 stage (defined as larval arrest in Figure 4D). This observation supports the previously proposed hypothesis that maternal HTZ-1 is inherited to homo-knock-out daughters and rescues growth defects during larval developments (Whittle et al. 2008). The larval arrest observed at 5 µM 5-Ph-IAA is a new phenotype that was not observed with htz-1-KO, demonstrating that the AID2 system enables the depletion of maternally inherited proteins during development. Since some embryos taken out from the Egl htz-1-KO animals show larval arrest (Whittle et al. 2008), this new phenotype is a disclosure of roles of HTZ-1, but not an artificial effect of the AID2 system. These results also indicate that the differential phenotypes observed here were caused by the different levels of HTZ-1 depletion.
Stage-specific degradation of HTZ-1 during larval growth
To narrow down the time frame within which loss of HTZ-1 causes defects in growth and reproduction, we treated the larvae with 5 µM 5-Ph-IAA in a stage-specific manner. Figure 5A shows the experimental scheme indicating the timing of the 5-Ph-IAA addition after the release from the L1 larval arrest. When the ligand treatment was started at 0 h after the release (Figure 5A, second row), the treated larvae arrested their growth (Figures 4 and 5B). When the ligand treatment was started at 30 h (Figure 5A, third row), the growth defect was mostly rescued (Figure 5B), and the worm exhibited the sterile phenotype (Figure 5C). When the ligand was applied at 40 or 50 h (Figure 5A, fourth and fifth rows, respectively), we observed no growth defect and a weak effect on sterility (egg-production deficiency) (Figure 5, B and C). The appearance of the weak phenotype was not triggered by inefficient depletion of GFP::mAID::HTZ-1 in these stages, because significant depletion was observed in the whole animal body even including the germline after the ligand treatment (Supplementary Figure S5). This observation suggests that HTZ-1 in the L2 to L3 stages (between 30 and 40 h) plays a critical role in egg production. Even though most of the worms treated with the ligand at 40 or 50 h were able to produce eggs (Figure 5C), the number of viable progeny from the worms was significantly decreased (Figure 5D), suggesting that HTZ-1 in the L3 to L4 stages (between 40 and 50 h) plays roles in the completion of egg maturation and/or embryogenesis. Importantly, the htz-1-KO animals are not appropriate for the analysis of these stage-specific roles because of the presence of maternal HTZ-1, and we could not dissect these phenotypes using these animals (Figure 5, B–D). Thus, these observations underscore the advantages of the AID2 system in the analysis of the stage-specific functions of HTZ-1 during larval development.
5-Ph-IAA-AM induces better degradation in embryos than does 5-Ph-IAA
Rapid target depletion in the laid embryos will be useful for dissecting the role of proteins in embryogenesis. Initially, we tested the efficacy of protein depletion in embryos using 5-Ph-IAA. We placed embryos expressing AtTIR1(F79G)::mRuby and AID*::GFP into drops of M9 buffer containing 5 or 50 µM 5-Ph-IAA, and subsequently monitored the reporter intensity (Figure 6, A and B; Supplementary Figure S6A). We found that the treatment with 50 µM 5-Ph-IAA caused a rapid decrease in fluorescence intensity in many embryos (78%, 18/23 embryos), but 5 µM 5-Ph-IAA did not (Figure 6, B and C; Supplementary Figure S6A). We tested a higher concentration, 100 µM 5-Ph-IAA, and obtained a similar result to 50 µM 5-Ph-IAA (80%, 16/20) (Supplementary Figure S6B). The degradation of the reporter reached a maximum value within 15–30 min after degradation was induced (Figure 6, A and B; treatment with 50 µM 5-Ph-IAA). However, some embryos did not respond to the treatment (Figure 6A, upper embryo). These results suggest that 5-Ph-IAA can be used for loss-of-function experiments in C. elegans embryos, although treatment with 5-Ph-IAA is not always effective, possibly because the C. elegans embryo develops within the eggshell, which blocks its permeability to many compounds (Carvalho et al. 2011).
We have previously reported that the modification of auxin by attaching an acetoxymethyl group improved the efficiency of protein degradation in embryos (Negishi et al. 2019). Therefore, we wished to develop a more efficient 5-Ph-IAA analog for use in embryos. We synthesized 5-Ph-IAA-AM, which contains an acetoxymethyl (AM) group (Figure 6D). We found that 50 µM 5-Ph-IAA-AM effectively triggered rapid depletion of the AID*::GFP reporter, in contrast with the DMSO- and the 5 µM 5-Ph-IAA-AM-treated embryos (Figure 6E;Supplementary Figure S6, C and D). In sharp contrast with the unmodified 5-Ph-IAA, 5-Ph-IAA-AM caused rapid degradation in all embryos (Figure 6F; 24/24 embryos). We confirmed that treating the embryos with 50 µM 5-Ph-IAA-AM did not cause developmental abnormality (Supplementary Figure S6E). These results indicate that treating with 50 µM 5-Ph-IAA-AM is a better condition for targeting proteins in embryos.
Discussion
In this article, we overcame the limitations of the original AID system (which is currently used in C. elegans studies), i.e., basal degradation and the requirement for high doses of IAA, by establishing AID2 via a combination of AtTIR1(F79G), the AID*/mAID degron and 5-Ph-IAA. As presented in Figure 1C, the expression level of AID*::GFP in larvae with or without AtTIR1(F79G)::mRuby was comparable. Similarly, the GFP::mAID::HTZ-1 level in larvae with or without AtTIR1(F79G)::mRuby was quantitatively indistinguishable (Supplementary Figure S4, C and D). Therefore, we did not observe basal degradation in our experimental condition when AtTIR1(F79G)::mRuby was expressed under the control of the eft-3 promoter. We expect that the use of AtTIR1(F79G) will overcome the problems associated with leaky degradation and will promote the functional characterization of proteins fused with AID* or mAID. Please note that the AID* and mAID tags contain the domain II that is recognized by AtTIR1 (Supplementary Figure S1A). Both degron tags are compatible with AtTIR1(F79G) and work efficiently in the presence of 5-Ph-IAA.
Researchers have typically been using 1 mM IAA for inducing degradation in C. elegans using the original AID system (Zhang et al. 2015). Recently, 1-naphthaleneacetic acid (NAA), a synthetic auxin, has been proposed as an alternative to IAA (Martinez et al. 2020). However, NAA has also been used at 1 mM and is not compatible with AtTIR1(F79G). We and others showed that treating nematodes with 1 mM IAA affected the number of progeny (Supplementary Figure S2A) and lifespan (Loose and Ghazi 2021), which might be attributed to the inhibition of the target of rapamycin complex 1 (TORC1) recently reported in yeast (Nicastro et al. 2021). In fact, the inhibition of TORC1 in C. elegans causes a reduction in the number of germline progenitors and extends the lifespan of the animals (Korta et al. 2012; Blackwell et al. 2019). In contrast, treating nematodes with 5 µM 5-Ph-IAA did not change the number of progeny (Supplementary Figure S2A) or larval growth (Figure 4B), which implies that 5-Ph-IAA is a safer degradation inducer compared with IAA. Importantly, we showed that 5-Ph-IAA induced sharper degradation (Figure 2C) and worked at 1,300-fold lower concentrations than did IAA (Figure 2C). We suggest that users should test 5 µM 5-Ph-IAA for inducing degradation in larvae and adults.
The observation that 5-Ph-IAA worked at lower concentrations can be explained in part by the fact that 5-Ph-IAA shows higher affinity to AtTIR1(F79G) (Uchida et al. 2018). In addition, 5-Ph-IAA is likely to be more cell permeable, because the lipophilicity (logP) of 5-Ph-IAA and IAA is 2.8 and 1.1, respectively. We assume that the combination of these two features of 5-Ph-IAA confers higher sensitivity and faster degradation in C. elegans. We also speculate that the leaky degradation by AtTIR1 was mainly caused by IAA or IAA-like compounds in medium or C. elegans. Because AtTIR1(F79G) is less reactive to IAA (Supplementary Figure S2B), AtTIR1(F79G) may not show the leaky degradation.
Moreover, we showed that treating laid embryos with 5-Ph-IAA-AM induced better degradation than did treatment with 5-Ph-IAA (Figure 6). The AM modification is widely used to enhance cell permeability and to prepare a prodrug (Schultz 2003). Thus, it is likely that 5-Ph-IAA-AM shows better eggshell permeability than does 5-Ph-IAA. In the embryos, 5-Ph-IAA-AM is converted to 5-Ph-IAA by cytoplasmic esterases, leading to the accumulation of 5-Ph-IAA. Note that the AM cleavage reaction also produces formaldehyde as a by-product (Schultz 2003). Although we did not observe any problems when the embryos were treated with 50 µM 5-Ph-IAA-AM (Supplementary Figure S6E), researchers should consider this issue when using 5-Ph-IAA-AM at high concentrations.
To evaluate the targeted degradation of an endogenous protein by the AID2 system in C. elegans, we focused on the histone variant H2A.Z (HTZ-1), which is widely conserved among metazoans and is involved in multiple chromatin transactions (Giaimo et al. 2019). Because HTZ-1 is maternally inherited by the embryos and has multiple roles, it has been challenging to dissect its roles during larval development. In fact, it was only possible to observe sterile and egg-laying defect phenotypes in the postembryonic stage using genetic null homozygous individuals (htz-1-KO) (Whittle et al. 2008). Using the AID2 system, we observed differential phenotypes after the depletion of GFP::mAID::HTZ-1 (Figure 4). Importantly, we detected a severe growth defect when the larvae were treated with 5 µM 5-Ph-IAA from the L1 stage (Figure 4, B and C). It is likely that this growth defect was not observed in the case of htz-1-KO because the maternal HTZ-1 inherited by daughters rescues the defect (Figure 4D) (Whittle et al. 2008). These observations indicate that the AID2 system will be particularly useful to study the roles of essential proteins in embryogenesis and development.
Loss of HTZ-1 caused the larval arrest, sterile and egg-laying defect phenotypes after the depletion timing during larval growth (Figure 5). These results clearly indicate that HTZ-1 is involved in multiple developmental processes, each of which can be potentially dissected by changing the timing of 5-Ph-IAA addition. Further functional dissection of HTZ-1 and other proteins will be possible by expressing AtTIR1(F79G)::mRuby under a tissue-specific promoter, instead of the ubiquitous eft-3 promoter used in this study, to allow temporally and spatially controlled degradation simultaneously.
In conclusion, this study demonstrated that the AID2 system employing AtTIR1(F79G) and the AID*/mAID degron is a more effective tool than the original AID system and enables the depletion of proteins in all stages of C. elegans development. Furthermore, it enables the examination of the role of proteins during embryogenesis and development, as well as the temporal dissection of the roles of proteins involved in multiple processes. We believe that the AID2 system presented in this paper will be useful for future C. elegans studies.
Data availability
All plasmids are listed in Supplementary Table S1 with the availability information. All strains are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplementary material is available at GENETICS online.
Supplementary Material
Acknowledgments
We thank Drs Shinsuke Niwa (Tohoku University) and John Calarco (University of Toronto) for sharing plasmids. We also thank Drs David Matus (Stony Brook University) and Christopher Hamell (Cold Spring Harbor Laboratory) for sharing unpublished results. We also thank A. Konno, Y. Oma and M. Aida (Tohoku University) for supporting the construction of htz-1 KO strains. Some strains listed in Supplementary Table S2 were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Funding
This research was supported by JSPS KAKENHI grants (JP20K06616 for N.H.; JP17KT0013 for A.S.; JP16KT0078 for H.S.; JP19H03253 for K.H.; JP20H05378, JP21H02151, JP20K21261, and JP21H04746 for M.H.; JP20H05396 and JP21H04719 for M.T.K.) and a JST CREST grant (JPMJCR21E6 for M.T.K.). M.H. was also supported by the JSPS Core-to-Core Program (Advanced Research Networks) entitled “Establishment of International Agricultural Immunology Research-core for a Quantum Improvement in Food Safety” and the Japan–Czech Republic Research Cooperative Program between JSPS and CAS grant number JPJSBP120202501. S.K. was supported by the Division for Interdisciplinary Advanced Research and Education, Tohoku University.
Conflicts of interest
The authors declare that there is no conflict of interest.
Literature cited
- Ashley GE, Duong T, Levenson MT, Martinez MAQ, Johnson LC, et al. 2021. An expanded auxin-inducible degron toolkit for Caenorhabditis elegans. Genetics. 217:7250–7257. doi:10.1093/genetics/iyab006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Z, Murray JI.. 2011. Protocol mounting Caenorhabditis elegans embryos for live imaging of embryogenesis. Cold Spring Harb Protoc. 2011:pdb.prot065599.doi: 10.1101/pdb.prot065599. [DOI] [PubMed] [Google Scholar]
- Blackwell TK, Sewell AK, Wu Z, Han M.. 2019. TOR signaling in Caenorhabditis elegans development, metabolism, and aging. Genetics. 213:329–360. doi: 10.1534/genetics.119.302504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho A, Olson SK, Gutierrez E, Zhang K, Noble LB, et al. 2011. Acute drug treatment in the early C. elegans embryo. PLoS One. 6:e24656. doi: 10.1371/journal.pone.0024656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman EJ, Estelle M.. 2009. Mechanism of auxin-regulated gene expression in plants. Annu Rev Genet. 43:265–285. doi: 10.1146/annurev-genet-102108-134148. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M.. 2005. The F-box protein TIR1 is an auxin receptor. Nature. 435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- Dickinson DJ, Pani AM, Heppert JK, Higgins CD, Goldstein B.. 2015. Streamlined genome engineering with a self-excising drug selection cassette. Genetics. 200:1035–1049. doi: 10.1534/genetics.115.178335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, et al. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 391:806. [DOI] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, et al. 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaimo BD, Ferrante F, Herchenröther A, Hake SB, Borggrefe T.. 2019. The histone variant H2A.Z in gene regulation. Epigenetics Chromatin. 12:37.doi: 10.1186/s13072-019-0274-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O.. 2005. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- Korta DZ, Tuck S, Hubbard EJA.. 2012. S6K links cell fate, cell cycle and nutrient response in C. elegans germline stem/progenitor cells. Development. 139:859–870. doi: 10.1242/dev.074047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long H, Zhang L, Lv M, Wen Z, Zhang W, et al. 2020. H2A.Z facilitates licensing and activation of early replication origins. Nature. 577:576–581. doi: 10.1038/s41586-019-1877-9. [DOI] [PubMed] [Google Scholar]
- Loose JA, Ghazi A.. 2021. Auxin treatment increases lifespan in Caenorhabditis elegans. Biol Open. 10:bio058703. doi:10.1242/bio.058703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez MAQ, Kinney BA, Medwig-Kinney TN, Ashley G, Ragle JM, et al. 2020. Rapid degradation of Caenorhabditis elegans proteins at single-cell resolution with a synthetic auxin. G3 (Bethesda). 10:267–280. doi: 10.1534/g3.119.400781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawska M, Ulrich HD.. 2013. An expanded tool kit for the auxin‐inducible degron system in budding yeast. Yeast. 30:341–351. doi: 10.1002/yea.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mörck C, Pilon M.. 2006. C. elegans feeding defective mutants have shorter body lengths and increased autophagy. BMC Dev Biol. 6:39.doi: 10.1186/1471-213X-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume T, Kiyomitsu T, Saga Y, Kanemaki MT.. 2016. Rapid protein depletion in human cells by auxin-inducible degron tagging with short homology donors. Cell Rep. 15:210–218. doi: 10.1016/j.celrep.2016.03.001. [DOI] [PubMed] [Google Scholar]
- Natsume T, Kanemaki MT.. 2017. Conditional degrons for controlling protein expression at the protein level. Annu Rev Genet. 51:83–102. doi: 10.1146/annurev-genet-120116-024656. [DOI] [PubMed] [Google Scholar]
- Negishi T, Asakawa M, Kanemaki M, Sawa H.. 2019. Modified auxin improves the auxin-inducible degradation (AID) system for laid C. elegans embryos. microPubl Biol. 2019:10–13. doi: 10.17912/micropub.biology.000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro R, Raucci S, Michel AH, Stumpe M, Osuna GMG, et al. 2021. Indole-3-acetic acid is a physiological inhibitor of TORC1 in yeast. PLoS Genet. 17:e1009414.doi: 10.1371/journal.pgen.1009414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M.. 2009. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- Norris AD, Kim HM, Colaiácovo MP, Calarco JA.. 2015. Efficient genome editing in Caenorhabditis elegans with a toolkit of dual-marker selection cassettes. Genetics. 201:449–458. doi: 10.1534/genetics.115.180679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obinata H, Sugimoto A, Niwa S.. 2018. Streptothricin acetyl transferase 2 (Sat2): a dominant selection marker for Caenorhabditis elegans genome editing. PLoS One. 13:e0197128.doi:10.1371/journal.pone.0197128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2020. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- R Studio Team. 2020. RStudio: Integrated Development Environment for R. Boston, MA: R Studio, PBC. [Google Scholar]
- Schiksnis E, Nicholson A, Modena M, Pule M, Arribere J, et al. 2020. Auxin-independent depletion of degron-tagged proteins by TIR1. microPubl Biol. 2020:30–32. doi:10.17912/micropub.biology.000213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods. 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz C. 2003. Prodrugs of biologically active phosphate esters. Bioorg Med Chem. 11:885–898. doi:10.1016/S0968-0896(02)00552-7. [DOI] [PubMed] [Google Scholar]
- Uchida N, Takahashi K, Iwasaki R, Yamada R, Yoshimura M, et al. 2018. Chemical hijacking of auxin signaling with an engineered auxin-TIR1 pair. Nat Chem Biol. 14:299–305. doi: 10.1038/nchembio.2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle CM, McClinic KN, Ercan S, Zhang X, Green RD, et al. 2008. The genomic distribution and function of histone variant HTZ-1 during C. elegans embryogenesis. PLoS Genet. 4:e1000187.doi: 10.1371/journal.pgen.1000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood WB. 1988. The Nematode Caenorhabditis elegans. NY, USA: Cold Spring Harbor Laboratory. [Google Scholar]
- Wu T, Yoon H, Xiong Y, Dixon-Clarke SE, Nowak RP, et al. 2020. Targeted protein degradation as a powerful research tool in basic biology and drug target discovery. Nat Struct Mol Biol. 27:605–614. doi: 10.1038/s41594-020-0438-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- , Yesbolatova A, Saito Y, Kitamoto N, Makino-Itou H, Ajima R, et al. 2020. The auxin-inducible degron 2 technology provides sharp degradation control in yeast, mammalian cells, and mice. Nat Commun. 11. 5701. doi: 10.1038/s41467-020-19532-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Moss BL, Jang SS, Prigge M, Klavins E, et al. 2013. Mutations in the TIR1 auxin receptor that increase affinity for auxin/Indole-3-acetic acid proteins result in auxin hypersensitivity. Plant Physiol. 162:295–303. doi: 10.1104/pp.113.215582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ward JD, Cheng Z, Dernburg AF.. 2015. The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development. 142:4374–4384. doi: 10.1242/dev.129635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All plasmids are listed in Supplementary Table S1 with the availability information. All strains are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplementary material is available at GENETICS online.