Abstract
2,3-Dichloro-1-propanol is more chemically stable than its isomer, 1,3-dichloro-2-propanol, and is therefore more difficult to degrade. The isolation of bacteria capable of complete mineralization of 2,3-dichloro-1-propanol was successful only from enrichments at high pH. The bacteria thus isolated were found to be members of the α division of the Proteobacteria in the Rhizobium subdivision, most likely Agrobacterium sp. They could utilize both dihaloalcohol substrates and 2-chloropropionic acid. The growth of these strains in the presence of 2,3-dichloro-1-propanol was strongly affected by the pH and buffer strength of the medium. Under certain conditions, a ladder of four active dehalogenase bands could be visualized from this strain in activity gels. The enzyme involved in the complete mineralization of 2,3-dichloro-1-propanol was shown to have a native molecular weight of 114,000 and consisted of four subunits of similar molecular weights.
Epichlorohydrin (1-chloro-2,3-epoxypropane) and its precursors (1,3-dichloro-2-propanol [1,3-DCP], 2,3-dichloro-1-propanol [2,3-DCP], and 3-chloro-1,2-propanediol [3-CPD]) are halohydrins used widely as solvents and as starting materials for resins, polymers, agrochemicals, and pharmaceuticals. 2,3-DCP, 1,3-DCP, and epichlorohydrin are carcinogenic, mutagenic, and genotoxic. According to a U.S. Environmental Protection Agency assessment (available at http://www.epa.gov/ngispgm3/iris/), 2,3-DCP showed significant effects on rats dosed with 35 mg of 2,3-DCP/kg of body weight/day. The resulting mortality is attributed to myocardial degeneration and kidney and liver malfunction. 2,3-DCP has been shown to be more toxic than 1,3-DCP to the testes and kidneys, though it is less hepatotoxic (7, 16). The Environmental Protection Agency Prioritized Chemical List showed that the overall score (as the sum of the persistence, bioaccumulation, and toxicity scores for human health risk potential added to the corresponding scores for ecological risk) for 1,3-DCP and epichlorohydrin were 11 out of 18 each. Both epichlorohydrin and 1,3-DCP have a high risk factor for animal and human toxicity with regards to the environment. No information was available for 2,3-DCP. However, due to its greater stability, 2,3-DCP is likely to be more persistent than 1,3-DCP and may pose a substantial environmental threat.
To date, few bacteria capable of the complete degradation of 2,3-DCP have been isolated. A single Pseudomonas strain capable of growth on 2,3-DCP was isolated from 300 samples of contaminated soil (9). The same group later isolated 13 isolates from a further 1,000 similar samples, suggesting 2,3-DCP-degrading bacteria to be quite rare (10). These bacteria were shown to degrade only the S enantiomer of 2,3-DCP and were of use in enantiospecific preparation of (R)-2,3-DCP and (S)-epichlorohydrin. Haloalcohol dehalogenases with activity against 1,3-DCP have either no activity or only fractional activity (<50%) against racemic 2,3-DCP (2, 10, 15, 21, 22).
The initial aim of this study was to isolate and characterize pure bacterial cultures or consortia capable of completely degrading chlorinated aliphatic compounds based on enrichments using 2,3-DCP as the sole carbon and energy source. Microbes that produce dehalogenases are widely distributed in nature, apparently having evolved to degrade naturally occurring halogenated compounds in order either to exploit them as a carbon source for growth or as a means of protection against the toxicity of these compounds (18).
MATERIALS AND METHODS
Microbe isolation.
Enrichments were carried out using soil samples from 10 different locations in Cardiff, United Kingdom, suspected to have been exposed to chlorinated compounds (i.e., from parks, gardens, riverbank, estuary bank, and beach sand). The horticultural soils are likely to have been exposed to chlorinated agrochemicals, and the river is polluted with a wide range of chlorinated industrial intermediates and products. Ten-gram soil samples were each mixed with 100 ml of standard basal salts (SBS) medium (20) containing filter-sterilized 2,3-DCP (0.22-μm-pore-size Millipore filter) (0.5 g of C per liter) and incubated in an orbital shaker (150 rpm) at 30°C overnight. Of these soil suspensions, 1 to 10 ml was inoculated into a series of flasks containing 100 ml of SBS medium supplemented with filter-sterilized 2,3-DCP (0.5 g of C per liter). The pH values of the medium were varied from 7 to 9 by addition of either NaOH or NaHCO3, and the cultures were shaken as before. Chloride release from enrichments was measured daily using a Corning 926 chloride analyzer (Corning Ltd., Halstead, Essex, England). Once the chloride release reached 50% of the theoretical maximum, 1% aliquots of the culture were subcultured into fresh SBS–2,3-DCP medium. After subculturing several times and after the soil particles had been completely eliminated, serial dilutions using 100-μl aliquots were plated onto SBS plates supplemented with 2,3-DCP (0.5 g of C per liter). Suspected isolates were picked and inoculated onto fresh supplemented SBS plates. Once the isolates showed significant growth on the plates, single colonies were inoculated into liquid SBS medium containing various carbon sources and the chloride release as a percentage of the total available and the turbidity (600 nm) of the cultures were measured.
Preparation of CFEs and broken-cell preparations.
Preliminary experiments showed that maximum chloride release could be obtained by growth in supplemented high-phosphate (HP) medium. The mineral base of this medium was as follows: 12.5 g of K2HPO4, 3.8 g of KH2PO4, 1.0 g of (NH4)2SO4, and 0.1 g of MgSO4 · 7H2O per liter and 10 ml of trace element solution (20). This was supplemented with an appropriate, filter-sterilized carbon source (0.5 g of C per liter). These media were also used for preparation of cell extracts (CFEs). Cultures in mid-exponential phase were harvested by centrifugation at 10,000 × g for 15 min. The cells were washed with and then resuspended in 50 mM Tris-H2SO4 (pH 8.0). They were disrupted by passage through a French pressure cell (Apex Construction Ltd., London, United Kingdom) at least twice. Cell debris and intact cells were separated by centrifugation at 45,000 × g for 45 min. Supernatant solutions were decanted, stored at −20°C, and used as CFEs. The debris and intact cells were used for assays of cell-bound material after resuspension in 50 mM Tris-H2SO4 (pH 8.0). All preparations were carried out at 4°C.
Identification of bacterial isolates.
The gram-negative status of the isolates was determined by Gram staining. The isolates were also identified by API 20 NE kits (bioMerieux, Basingstoke, United Kingdom) for gram-negative isolates and by sequencing PCR products from 16S rRNA genes using universal bacterial primers (63f and 1387r) (14). As previous isolates (9, 10) were Pseudomonas spp., growth on Pseudomonas Selective Agar medium (products CM559 and SR102; Oxoid, Basingstoke, United Kingdom) at 30°C was also tested. Protein profile comparisons of the isolates used CFEs and were carried out under denaturing conditions by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (13).
Dehalogenase activity assays.
Qualitative assays of dehalogenase activity were performed using electrophoretic zymograms under native conditions based on the methods of Laemmli (13) and Hardman and Slater (6). Gels were soaked in 50 mM organochloride substrate for 1 h at 30°C. After washing several times, the gels were developed in 0.1 M AgNO3 until brown bands appeared and then fixed in 5% (vol/vol) acetic acid and dried. Quantitative assays of whole cells were detected by measurement of the chloride release from 10-ml HP medium cultures containing substrate (0.5 g of C/liter) grown aerobically at 30°C. Dehalogenase activities of the protein extracts were determined by assay at 30°C of the CFEs or broken cells in 5 ml of 0.2 M Tris-H2SO4 (pH 8.0) containing 10 mM (each) substrate. Resultant chloride release was detected as described above. One unit of enzyme activity was defined as the activity that catalyzed the formation of 1 μmol of halide/min/mg of protein.
Protein determination.
Protein concentrations of the CFEs were determined by the Sedmak and Grossberg method (17), and the soluble protein concentration of the cell membranes was determined by the biuret assay (4).
Enzyme purification.
CFEs were fractionated by the stepwise addition of (NH4)2SO4 (low heavy metal; Fisher Scientific) from 45 to 80% of saturation at 0°C. The precipitate was dissolved and dialyzed against 10 mM Tris-H2SO4–1 mM dithiothreitol (DTT) (pH 8.0). The dialyzed (NH4)2SO4 fractions were absorbed onto an ion-exchange chromatography DEAE Sepharose CL-6B column. The column was washed with 10 mM Tris-H2SO4 (pH 8.0) overnight to remove unbound protein. Elution was carried out in a total of 500 ml of 10 mM Tris-H2SO4–1 mM DTT (pH 8.0) with a linear gradient of 0 to 0.5 M (NH4)2SO4 and a flow rate of 15 ml/h. Eighty fractions were collected using a 2211 Superrac fraction collector (LKB Pharmacia, Milton Keynes, United Kingdom) set to change every 30 min. During elution, the protein concentration in the eluent was continuously measured at 280 nm via a 2138 UVICORD S detector and a 2210 one-channel recorder (LKB Pharmacia). Each fraction was tested for activity using a microtiter plate assay; each well contained 100 μl of the ion-exchange eluate, 100 μl of 200 mM Tris-H2SO4 (pH 8.0), and 50 μl of substrate to a final concentration of 10 mM with substrate being added last to initiate the reaction. The plate was incubated at 30°C for 30 min. Active fractions were observed by the addition of 50 μl of 0.1 M AgNO3 to each well and exposure to UV light for 2 min. The brown wells which resulted from the reaction of free chloride with AgNO3 indicated the active fractions. The pooled active fractions were placed in a dialysis sac and then concentrated with polyethylene glycol (molecular weight [MW], 12,000). The concentrated fractions were applied to a gel filtration Sephacryl S-200-HR column equilibrated with 10 mM Tris-H2SO4–1 mM DTT (pH 8.0) overnight at a flow rate of 10 ml/h. The fraction collector was set to give fractions every 15 min, and 80 fractions were taken. Active fractions were dialyzed against 1.0 liter of 10 mM Tris-H2SO4 (pH 8.0) overnight at 4°C and finally concentrated with polyethylene glycol 12000.
MW determination.
Relative MW determinations under denaturing conditions were performed by SDS-PAGE (13) and under nondenaturing conditions by gel filtration on a Sephacryl S-200-HR column.
Chemicals and materials.
All halogenated compounds were purchased from Sigma (Poole, United Kingdom) or Fisher/Acros Chimica (Loughborough, United Kingdom). Unless otherwise stated, other chemicals and media were from BDH or Oxoid. DEAE-Sepharose CL-6B and Sephacryl S-200-HR were purchased from LKB Pharmacia. A culture of Agrobacterium tumefaciens strain HK7 (degrades 1,3-DCP but not 2,3-DCP) was obtained from M. Lewis (3, 13a).
Nucleotide sequence accession numbers.
The DNA sequences of the PCR product from the amplification of a 16S rRNA gene from both strains using 63f and 1387r primers (14) are registered at the EMBL nucleotide sequence database as accession no. AJ276434 (NHC2) and AJ276433 (NHG3).
RESULTS AND DISCUSSION
Microbe isolation.
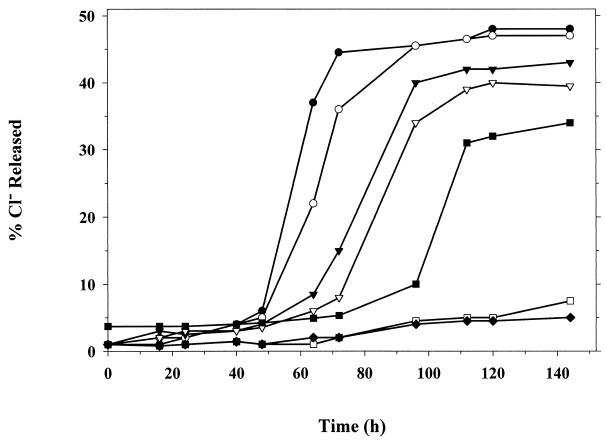
In initial experiments with medium adjusted to pH 7.0, no microbes that grew on 2,3-DCP were isolated and little dechlorination was observed above background decomposition. Various devices such as marble chips to encourage biofilm development and activated charcoal had no effect. However, if the pH of the medium was raised, dechlorination took place. After a few subcultures in medium with initial pH set between 7.5 and 9.0, the higher was the pH, the earlier was the commencement of dechlorination. Approximately 50% dechlorination was observed in most enrichments. The 2,3-DCP itself was stable at high pH, as shown by uninoculated controls (Fig. 1).
FIG. 1.
The Cl− released from 2,3-DCP batch culture enrichment at various starting pH values. ●, pH 9 culture; ○, pH 8.5 culture; ▾, pH 8.0 culture; ▿, pH 7.5 culture; ■, pH 7.0 culture; □, uninoculated control (pH 9.0); ⧫, uninoculated control (pH 8.5). Times for cultures were measured from the time of 1% subculture.
Two strains that grew with 2,3-DCP as their sole carbon and energy source were isolated. These strains were isolated from the pH 8.5 and 9.0 enrichments and named NHC2 and NHG3, respectively. No strains were isolated from pH 7.0, 7.5, and 8.0 cultures. The alteration of pH helped the destabilization of 2,3-DCP. Both acid and alkali enhance abiotic breakdown of halogenated organic compounds wherein a nucleophilic substitution or elimination is the major reaction mechanism or pathway (5, 12). With haloalcohols, susceptibility to pH shifts arises as a direct result of the arrangement of both halide and hydroxyl groups.
The growth of the isolated bacteria on 2,3-DCP was strongly affected by the pH and buffer capacity of the medium. The susceptibility to pH change was such that the release of Cl− ceased when the pH of medium dropped below 4.6. However, when a highly buffered medium (HP medium) was used, the pH could be maintained at approximately 7, under which conditions the Cl− released could reach 100%. This indicated that the bacteria are capable of dehalogenating both stereoisomers of 2,3-DCP.
Preliminary characterization of strains NHC2 and NHG3 showed them to be gram-negative rods with a circular and smooth-surfaced colony morphology. Both strains failed to grow on Pseudomonas Selective Agar medium. Results from API 20 NE kits yielded the classification number 1467745 for both strains, which suggests a low discrimination of Agrobacterium radiobacter, Pseudomonas paucimobilis, or Chryseomonas luteola. Using the further tests suggested in the API documentation, a lack of a yellow colony color and positive reaction on MacConkey agar suggested the strains to be Agrobacterium sp. (99%).
Both strains were studied further by 16S rRNA gene characterization. The Ribosome Database Project similarity matrix package (www.cme.msu.edu/RDP/html/analyses.html) suggested the strains to be in the Rhizobium-Agrobacterium group. For strain NHC2, the closest 12 matches were Agrobacterium spp. (0.993 to 0.986 match); for strain NHG3, high-similarity matches (>0.950) were with a range of Rhizobium-Agrobacterium α-subdivision Proteobacteria including Ochrobacterium anthropi, Brucella melitensis, A. tumefaciens, and Sinorhizobium fredii.
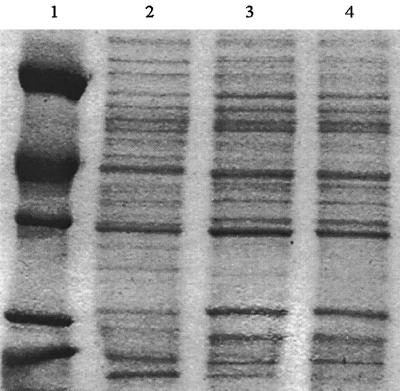
As bacteria derived from soil able to grow on ordinary laboratory media, a tentative classification of the isolates as Agrobacterium seemed appropriate. In addition, SDS-PAGE protein profile comparisons between these strains showed NHC2 and NHG3 to be very similar (Fig. 2). Identically prepared extracts from A. tumefaciens strain HK7 (a 1,3-DCP-degrading strain) (3) were clearly related though not identical.
FIG. 2.
SDS-PAGE protein profile comparisons of A. tumefaciens HK7 and strains NHC2 and NHG3. Lanes: 1, protein marker; 2, strain HK7; 3, strain NHC2; 4, strain NHG3. Protein markers were (MW) bovine serum albumin (66,000); egg albumin (45,000), glycerol dehydrogenase (36,000), carbonic anhydrase (29,000), and trypsinogen (24,000).
Substrate range of activity in whole cells.
In order to investigate the ability of the strains to degrade other halogenated substrates, strains NHC2 and NHG3 were grown on various halogenated compounds. Maximum Cl− release from the cultures, after 2 days of incubation at 30°C, is given in Table 1. Both strains almost completely dechlorinated 2,3-DCP, 1,3-DCP, and 3-CPD. There was less activity on the brominated analogues (2,3-dibromopropanol and 1,3-dibromopropanol). Most other similar substrates were not metabolized, with the exception of 2-chloropropionic acid (2-MCPA), which was almost fully dechlorinated and provided a good growth substrate. This latter finding was very significant. The presence of two different groups of dehalogenase was also observed in a haloalkane utilizer, Xanthobacter autotrophicus GJ10 (8, 11), which had both a haloalkane and a haloacid dehalogenase. This is the first report of haloacid utilizers being able to also degrade haloalcohol compounds. The pattern of activity was similar for the two strains, and strain NHG3 alone was selected for further investigations.
TABLE 1.
Halide release by strains NHC2 and NHG3 on various substrates at 0.5 g of C/litera
| Substrate | Halide release by strain on culture type
|
|||
|---|---|---|---|---|
| NHC2
|
NHG3
|
|||
| Liquid | Solid | Liquid | Solid | |
| 2-MCPA | 95 | +++ | 88 | +++ |
| Dichloroacetic acid | 2 | − | 4 | − |
| 2-Chloropropionamide | 10 | − | 9 | − |
| 1,3-Dichloropropane | 0 | − | 0 | − |
| 1,6-Dichlorohexane | 0 | − | 0 | − |
| 1-Chloropentane | 0 | − | 0 | − |
| 2-Chlorobutane | 0 | − | 0 | − |
| 1,3-Dibromopropanol | 24 | ++ | 25 | ++ |
| 2,3-Dibromopropanol | 29 | ++ | 31 | +++ |
| 6-Chlorohexanol | 9 | − | 13 | − |
| 1-Chloropropanol | 10 | + | 25 | + |
| 3-CPD | 100 | +++ | 100 | +++ |
| 1,3-Dichloropropanol | 89 | +++ | 95 | +++ |
Symbols: +++, very significant growth; ++, significant growth; +, sparse growth; −, no growth. The halide released in liquid cultures was determined as the percentage of the total halide content of the appropriate substrates in stationary-phase cultures.
Enzyme expression.
Preliminary experiments showed haloalcohol dehalogenase activity to be evident in 2,3-DCP cultures in resting cells, whole cells, and CFEs. No extracellular activity was detected. Cells grown with succinate (0.5 g of C/liter) as sole carbon source also exhibited enzyme activity. Further investigations showed that the cell membranes (broken cells) of strain NHG3 also exhibited activity against 1,3-DCP and 2,3-DCP. Table 2 shows the specific activity of the haloalcohol dehalogenases present in strain NHG3 toward haloalcohol substrates.
TABLE 2.
Specific activities of haloalcohol dehalogenases in the cellsa
| Substrate | Sp act (U)
|
||
|---|---|---|---|
| Resting cells | CFEs | Membrane | |
| 1,3-DCP | 0.720 | 0.314 | 0.256 |
| 2,3-DCP | 0.256 | 0.054 | 0.032 |
| CPD | 0.254 | 0.017 | 0.021 |
The cells were grown with 2,3-DCP (0.5 g of C/liter) as sole carbon and energy source.
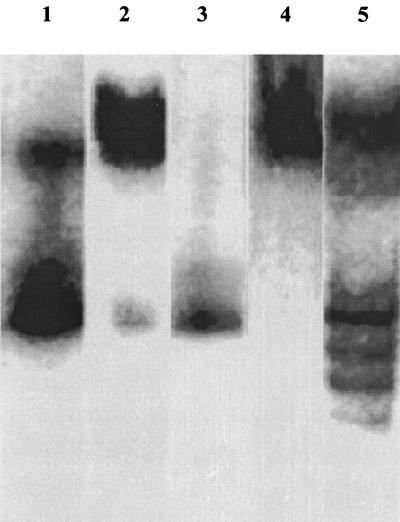
The strain was found to express in the soluble fraction at least two haloalcohol dehalogenases and two 2-MCPA dehalogenases as shown from native PAGE zymographic analysis. In CFEs from cells grown on 3-CPD, two 3-CPD dehalogenases were observed (Fig. 3, lane 1), whereas those cultures grown on 2-MCPA showed two 2-MCPA dehalogenases (Fig. 3, lane 2, upper broad band). The smaller band appeared only when the gel was developed in the presence of 2,3-DCP. Interestingly, extracts from 2-MCPA-grown cells developed with 2,3-DCP showed only the smaller band (Fig. 3, lane 3), and extracts from 3-CPD-grown cells developed with 2-MCPA showed only the two 2-MCPA dehalogenases (Fig. 3, lane 4). CFEs derived from cells grown with 2,3-DCP, succinate, or any other growth substrate and developed with 2,3-DCP showed only a single band like that shown in Fig. 3, lane 3. These observations make it likely that 2,3-DCP dehalogenase is constitutive while the other bands derive from enzymes inducible under various conditions.
FIG. 3.
Dehalogenase zymograms of CFEs of strain NHG3 under various substrate conditions. CFEs from cells grown on various substrates as sole carbon source were electrophoresed under native conditions and developed with a variety of substrates. Lane 1, grown and developed on 50 mM 3-CPD; lane 2, grown and developed on 50 mM 2-MCPA–1 mM 2,3-DCP; lane 3, grown on 2-MCPA and developed on 50 mM 2,3-DCP; lane 4, grown on 3-CPD and developed on 50 mM 2-MCPA; lane 5, grown on 2-MCPA–2,3-DCP and developed on 2-MCPA–1,3-DCP.
When the strains were grown on a mixture of 2,3-DCP and 2-MCPA and developed in 1,3-DCP, four active bands were shown, at least one of which coincided with the 2,3-DCP dehalogenase band (Fig. 3, lane 5). These might have been created as a consequence of the formation of homo- and heterotetramers from different combinations of the two subunits (15). For 1,3-DCP-degrading enzymes, four active bands of the Ib dehalogenase from Corynebacterium sp. strain N-1074 formed from various combinations of 32,000- and 35,000-MW subunits (15). Similarly, five active bands of dehalogenase A from Arthrobacter erithii H10a consisting of 31,500- and 34,000-MW subunits (2) were also observed. However, neither Ib nor dehalogenase A had activity against 2,3-DCP. The reason for the appearance of the four and/or five active bands that comigrated with the two subunits is still unclear. It is likely to have occurred either within cells or as a consequence of conditions imposed during the preparation of the crude extract (2). With respect to strain NHG3, it was most likely that the creation of four active bands was a result of the growth conditions. The ladder of four active bands was observed only when the strain was grown on the mixture of 2-MCPA and 2,3-DCP and developed on 1,3-DCP.
There was also evidence of membrane-associated haloalcohol dehalogenase activity in strain NHG3 (Table 2). This raises the possibility of a novel pathway for a haloalcohol dehalogenation. To date, the information regarding haloalcohol dehalogenases has been limited to hydrolytic haloalcohol dehalogenases (19). It was possible that the degradation of haloalcohol by strain NHG3 might involve an oxygenase type of haloalcohol dehalogenase. The oxidation route would be via 2-MCPA, which could account for the presence of dehalogenases of this compound in this strain. Rhodococcus erythropolis Y2 was shown to carry a second haloalkane dehalogenase of the oxygenase type which was found to be a membrane-associated enzyme (1).
Characteristics of the 2,3-DCP dehalogenase.
None of the various inhibitors used inhibited 2,3-DCP dehalogenase activity completely. The thiol reagent iodoacetamide, at a concentration of up to 1 mM, had no effect on enzyme activity. The activity of the enzyme decreased by 38% when 20 mM substrate analogue 2-MCPA was incorporated and by 77% when 3-MCPA was incorporated at the same concentration. Also, the divalent ions Cu2+ and Zn2+ were found to inhibit activity. When at 1 mM, Cu2+ led to a 76% loss and Zn2+ led to a 61% loss of 2,3-DCP dehalogenase activity, whereas the presence of Ca2+ at a 1 mM concentration increased the enzyme activity by 25%.
With increasing temperature, 2,3-DCP dehalogenase activity increased markedly up to 40°C. Above this temperature, a broad maximum activity up to 60°C was observed, after which it declined sharply. The enzyme also demonstrated a broad pH optimum, ranging from 8.8 to 9.5. The enzyme activity decreased markedly below pH 8.5 and above pH 9.8.
The 2,3-DCP dehalogenase was found to be relatively heat labile. Thirty-two percent of the enzyme activity was lost after incubation at 50°C for 1 min. However, the relative activity remained unchanged, being around 68% compared to the untreated activity after 15 min. After incubation at 50°C for 120 min, only 20% of the activity remained. Evidence for the instability of the enzyme was strengthened by the effect of storage of purified enzyme at 4°C. Three days of storage had no effect on the activity against 2,3-DCP. However, 20% of the activity of the enzyme was lost after 7 days of storage, and after 30 days of storage, only 30% of the activity remained. However, storage of both CFEs and purified enzyme at −20 and −80°C, respectively, for 1 month had no effect on enzyme activity.
Enzyme purification.
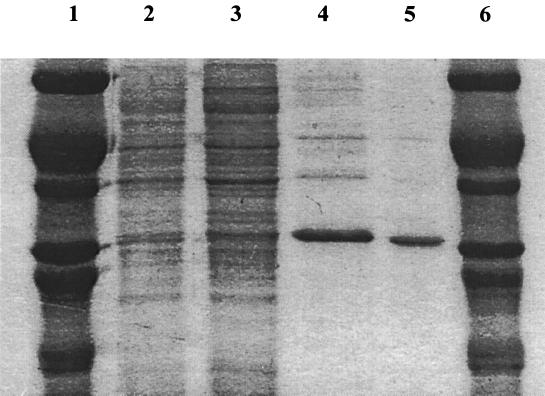
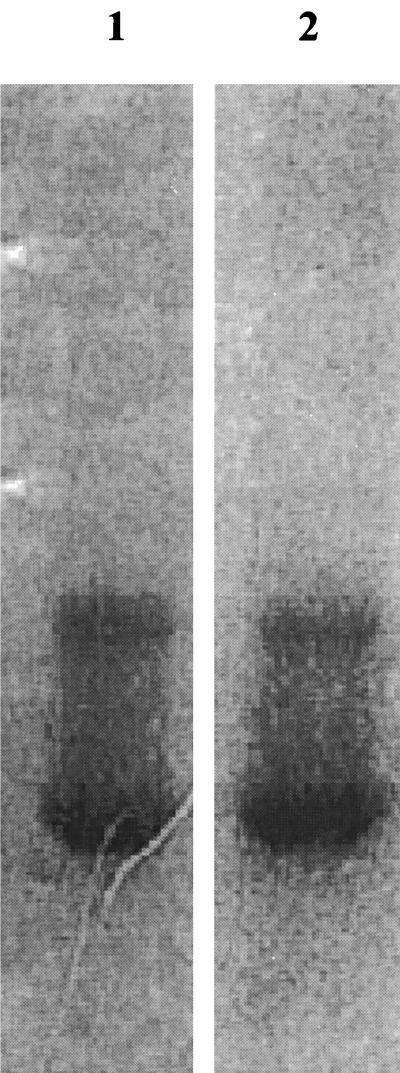
The 2,3-DCP dehalogenase from strain NHG3 was partially purified. Three purification steps were employed: (NH4)2SO4 precipitation, ion-exchange chromatography on a DEAE Sepharose CL-6B column, and gel filtration chromatography on a Sephacryl S-200-HR column. The 2,3-DCP dehalogenase was separated from the 2-MCPA dehalogenases by ion exchange and further purified on a gel filtration Sephacryl S-200-HR column. These three purification steps resulted in a 10-fold increase in activity with a 0.3% yield. The purification scheme is presented in Table 3 and shown in Fig. 4 on SDS-polyacrylamide gels to demonstrate the purity of the enzyme. The purified enzyme was visualized on an activity gel (native PAGE) as shown in Fig. 5. After development in either 50 mM 1,3-DCP or 50 mM 2,3-DCP, two bands of the 2,3-DCP dehalogenase were visible. This suggested that the 2,3-DCP dehalogenase may have two different conformations. The upper band was shown only in purified preparations using buffer containing DTT.
TABLE 3.
The scheme of purification of 2,3-DCP dehalogenase
| Stage | Vol (ml) | Total protein (mg) | Total activity (μmol of Cl− min−1) | Sp act (U) | Yield (%) | Fold increase in sp act |
|---|---|---|---|---|---|---|
| CFE | 40 | 722.80 | 44.24 | 0.06 | 100 | 1.00 |
| (NH4)2SO4 preparation | 10 | 425.50 | 42.25 | 0.10 | 59 | 1.62 |
| Ion exchange | 2 | 4.32 | 2.33 | 0.54 | 0.7 | 8.81 |
| Gel filtration | 1 | 1.86 | 1.18 | 0.64 | 0.3 | 10.37 |
FIG. 4.
SDS-PAGE analysis during the purification of 2,3-DCP dehalogenase. Lanes 1 and 6, protein markers; lane 2, CFE; lane 3, (NH4)2SO4 precipitation; lane 4, ion exchange; lane 5, gel filtration fraction. Protein markers (MW) were bovine serum albumin (66,000), egg albumin (45,000), glyceraldehyde dehydrogenase (36,000), carbonic anhydrase (29,000), trypsinogen (24,000), and trypsin inhibitor (20,100).
FIG. 5.
Native PAGE zymograms of purified 2,3-DCP dehalogenase. Lane 1, developed on 50 mM 1,3-DCP; lane 2, developed on 50 mM 2,3-DCP.
The MW under native and denatured conditions revealed the 2,3-DCP dehalogenase of strain NHG3 to be a tetrameric protein with an MW of 114,000 (data not shown), and its four subunits showed similar MWs (28,500) (Fig. 4).
The activity of the purified 2,3-DCP dehalogenase (gel filtration fraction) against a range of haloalcohols, haloalkanoic acids, and haloalkanes was tested. The activity of the 2,3-DCP dehalogenase showed substrate specificity limited to chloropropanols (1,3-DCP, 2,3-DCP, and 3-CPD) and their brominated analogues. Setting the specific activity of the purified enzymes against 2,3-DCP as 1, specific activity against other substrates was much higher (1,3-DCP, 2.82-fold; CPD, 1.65-fold; 3-chloropropanol, 10.13-fold; 1,3-DCP, 6.31-fold; 2,3-DBP, 5.71-fold). Also, as expected, it was found that the enzyme had activity against both (R)- and (S)-3-CPD, indicating that the 2,3-DCP dehalogenase was not a stereoselective enzyme. No activity was found on either haloalkanoic acids or haloalkanes. Using double-reciprocal plots, it was found that the Vmax and Km of purified 2,3-DCP dehalogenase were 0.761 ± 0.109 U and 5.643 ± 0.279 mM, respectively.
Other published reports of 2,3-DCP dehalogenase activity are cited with reference to 1,3-DCP activity. Some 1,3-DCP dehalogenases show no activity against 2,3-DCP (2, 22). Others, like the NHG3 enzyme reported here, show a marked preference for 1,3-DCP. Enzyme from Pseudomonas sp. strain OS-K-29 showed 10-fold-greater specific activity on 1,3-DCP than on 2,3-DCP (10); for Alcaligenes sp. strain DS-K-S-38, the value was 2.1-fold (21). The stereospecific enzyme from Corynebacterium sp. strain N-1074 showed 833-fold-greater activity on 1,3-DCP than on 2,3-DCP.
From native PAGE analysis, the activity and mobility of the haloalcohol dehalogenases from strain NHG3 were found to be similar to those of the haloalcohol dehalogenases of A. tumefaciens strain HK7 (3) and Pseudomonas sp. strain AD1 (22). However, the latter strains showed activity against 1,3-DCP and 3-CPD only, not 2,3-DCP (or 2-MCPA).
ACKNOWLEDGMENTS
The work described in this paper was carried out within the UK-Indonesia Biodiversity for Biotechnology Development Project (1994–1999) funded by the UK Department for International Development (DFID).
We thank the British Council and the International Institute for Biotechnology (www.bio.ukc.ac.uk/IIBMIRCEN/) for facilitating the collaboration established during this project.
REFERENCES
- 1.Armfield S J, Sallis P J, Baker P B, Bull A T, Hardman D J. Biodegradation of haloalkanes by Rhodococcus erythropolis Y2: the presence of an oxygenase-type dehalogenase complements a halidohydrolase activity. Biodegradation. 1995;6:237–246. doi: 10.1007/BF00700463. [DOI] [PubMed] [Google Scholar]
- 2.Assis H M S, Sallis P J, Bull A T, Hardman D J. Biochemical characterisation of a haloalcohol dehalogenase from Arthrobacter erithii H10a. Enzyme Microb Technol. 1998;22:568–574. doi: 10.1016/s0141-0229(97)00254-8. [DOI] [PubMed] [Google Scholar]
- 3.Bull, A. T., D. J. Hardman, P. J. Sallis, and B. M. Stubbs. October 1992. Dehalogenation of organohalogen-containing compounds. European patent 0 510 987 A1 and U.S. patent 5,470,742 (November 1995).
- 4.Gornall A G, Bardawill C J, David M M. Determination of serum proteins by means of the biuret reaction. J Biol Chem. 1949;177:751–766. [PubMed] [Google Scholar]
- 5.Gu B, Siegrist R J. Dehalogenation of chlorinated organic compounds by strong alkalis. ASCE J Environ Eng. 1997;123:982–987. [Google Scholar]
- 6.Hardman D J, Slater J H. Dehalogenase in soil bacteria. J Gen Microbiol. 1981;123:117–128. [Google Scholar]
- 7.Hirata M, Tanaka A, Omura M, Inoue N. Acute toxicity of dichloropropanols in mice: comparison between toxicities of two isomers. Jpn J Ind Health. 1993;35:S400. [Google Scholar]
- 8.Janssen D B, Scheper A, Dijkhuizen L, Witholt B. Degradation of halogenated aliphatic compounds by Xanthobacter autotrophicus GJ10. Appl Environ Microbiol. 1985;49:673–677. doi: 10.1128/aem.49.3.673-677.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kasai N, Tsujimura K, Unoura K, Suzuki T. Degradation of 2,3-dichloro-1-propanol by a Pseudomonas species. Agric Biol Chem. 1990;45:29–34. [Google Scholar]
- 10.Kasai N, Tsujimura K, Unoura K, Suzuki T. Isolation of (S)-2,3-dichloro-1-propanol assimilating bacterium, its characterization, and its use in preparation of (R)-2,3-dichloro-1-propanol and (S)-epichlorohydrin. J Ind Microbiol. 1992;10:37–43. [Google Scholar]
- 11.Keuning S, Janssen D B, Witholt B A. Purification and characterization of hydrolytic haloalkane dehalogenase from Xanthobacter autotrophicus GJ10. J Bacteriol. 1985;163:635–639. doi: 10.1128/jb.163.2.635-639.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krijgsheld K R, van der Gen A. Assessment of the impact of the emission of certain organochlorine compounds on the aquatic environment. Part III: epichlorohydrin. Chemosphere. 1986;15:881–893. [Google Scholar]
- 13.Laemmli U K. Cleavage of structural protein during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 13a.Lewis, M., and S. D. Greenaway. Biodegradation, in press.
- 14.Marchesi J R, Sato T, Weightman A J, Martin T A, Fry J C, Hiom S J, Wade W G. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol. 1998;64:795–799. doi: 10.1128/aem.64.2.795-799.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura T, Nagasawa T, Yu F, Watanabe I, Yamada H. Resolution and some properties of enzyme involved in enantioselective transformation of 1,3-dichloro-2-propanol to (R)-3-chloro-1,2-propanediol by Corynebacterium sp. strain N-1074. J Bacteriol. 1992;174:7613–7619. doi: 10.1128/jb.174.23.7613-7619.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Omura M, Hirata M, Zhao M, Tanaka A, Inoue N. Comparative testicular toxicities of two isomers of dichloropropanol, 2,3-dichloro-1-propanol, and 1,3-dichloro-2-propanol, and their metabolites alpha-chlorohydrin and epichlorohydrin, and the potent testicular toxicant 1,2-dibromo-3-chloropropane. Bull Environ Contam Toxicol. 1995;55:1–7. doi: 10.1007/BF00212381. [DOI] [PubMed] [Google Scholar]
- 17.Sedmak J J, Grossberg S E. A rapid, sensitive and versatile assay for protein using coomassie blue G250. Anal Biochem. 1977;79:544–552. doi: 10.1016/0003-2697(77)90428-6. [DOI] [PubMed] [Google Scholar]
- 18.Slater J H. Microbial dehalogenation of haloaliphatic compounds. In: Ratledge C, editor. Biochemistry of microbial degradation. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 379–421. [Google Scholar]
- 19.Slater J H, Bull A T, Hardman D J. Microbial dehalogenation of halogenated alcanoic acids, alcohols and alkanes. Adv Microb Physiol. 1997;38:133–176. doi: 10.1016/s0065-2911(08)60157-5. [DOI] [PubMed] [Google Scholar]
- 20.Slater J H, Lovatt D, Weightman A J, Senior E, Bull A T. The growth of Pseudomonas putida on chlorinated aliphatic acids and its dehalogenase activity. J Gen Microbiol. 1979;114:125–136. [Google Scholar]
- 21.Suzuki T, Kasai N, Yamamoto R, Minamiura N. Isolation of a bacterium assimilating (R)-3-chloro-1,2-propanediol and production of (S)-3-chloro-1,2-propanediol using microbial resolution. J Ferment Bioeng. 1992;73:443–448. [Google Scholar]
- 22.van den Wijngaard A J, Janssen D, Witholt B. Degradation of epichlorohydrin and halohydrins by bacterial cultures isolated from freshwater sediments. J Gen Microbiol. 1989;135:2199–2208. [Google Scholar]