Abstract
Since the discovery of eukaryotic small RNAs as the main effectors of RNA interference in the late 1990s, diverse types of endogenous small RNAs have been characterized, most notably microRNAs, small interfering RNAs (siRNAs) and PIWI-interacting RNAs (piRNAs). These small RNAs associate with Argonaute proteins and, through sequence-specific gene regulation, affect almost every major biological process. Intriguing features of small RNAs, such as their mechanisms of amplification, rapid evolution and non-cell-autonomous function, bestow upon them the capacity to function as agents of intercellular communications in development, reproduction and immunity, and even in transgenerational inheritance. Although there are many types of extracellular small RNAs, and despite decades of research, the capacity of these molecules to transmit signals between cells and between organisms is still highly controversial. In this Review, we discuss evidence from different plants and animals that small RNAs can act in a non-cell-autonomous manner and even exchange information between species. We also discuss mechanistic insights into small RNA communications, such as the nature of the mobile agents, small RNA signal amplification during transit, signal perception and small RNA activity at the destination.
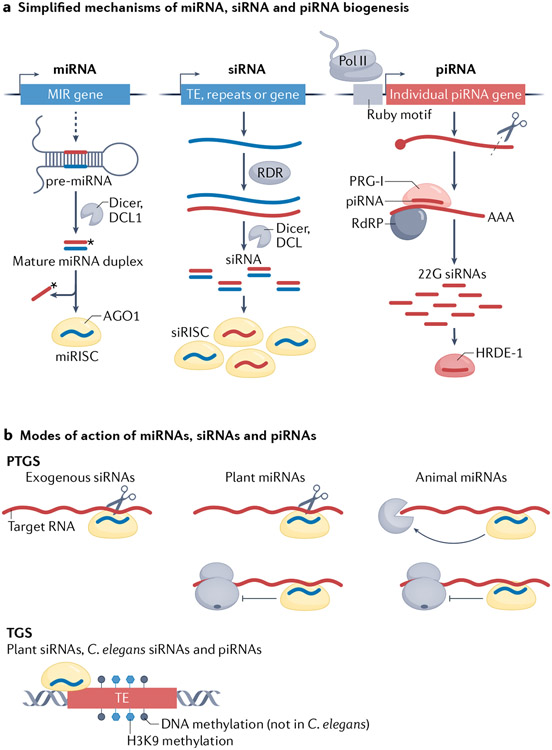
Although numerous species of short RNA may be present and functional in cells, in this Review, we discuss only small RNAs of 20–40 nucleotides (nt) in length that bind and target Argonaute proteins to genomic loci in a sequence-specific manner to regulate gene expression, such as microRNAs (miRNAs), small interfering RNAs (siRNAs) and PIWI-interacting RNAs (piRNAs). For discussion of the well-understood mechanisms of the biogenesis and cell-autonomous activities of these small RNAs, we refer the reader to BOX 1 and FIG. 1, and for discussion of various endogenous siRNAs, we refer the reader to BOX 2 and FIG. 2.
Box 1 ∣. The biogenesis and cell-autonomous activities of small RNAs in plants and animals.
Broadly speaking, small RNAs can be classified into microRNAs (miRNAs), small interfering RNAs (siRNAs) and PIWI-interacting RNAs (piRNAs)224,225 (FIG. 1a). Small RNAs associate with Argonaute proteins to form RNA-induced silencing complexes (RISCs), which either directly cleave target RNAs or recruit other proteins to the target RNAs and/or their associated chromatin to initiate post-transcriptional or transcriptional gene silencing226 (FIG. 1b). Of the different eukaryote Argonaute protein families, the Argonaute family is present in plants and the Argonaute and PIWI families are present in most animals226. Within plant and animal lineages, Argonaute genes have undergone duplication, sub-functionalization and loss, resulting in a gene family with differing members across species, probably reflecting diverse populations of small RNAs. For example, in Arabidopsis thaliana, ARGONAUTE 1 (AGO1) is the effector of almost all miRNAs, whereas the AGO4, AGO6, and AGO9 proteins associate with endogenous siRNAs produced from transposable elements227.
The biogenesis of miRNAs and siRNAs from their precursors is performed by RNase III-like enzymes such as Drosha, Dicer or DICER-LIKE (DCL) proteins, whereas the biogenesis of piRNAs is Dicer independent228 (FIG. 1a). A second distinctive feature is that piRNAs associate with PIWI effectors, which comprise a distinct clade of Argonaute proteins228. Although miRNAs and siRNAs are mostly Dicer-dependent and Argonaute-binding small RNAs, they differ in their precursors (although the distinction can be blurry in the case of evolutionarily young miRNAs)229. The former are transcribed from genes and are usually processed by Drosha and Dicer in animals and by DCL1 in plants to generate mature miRNAs224,230-232 (FIG. 1a). The precursors of siRNAs are long double-stranded RNAs (dsRNAs) or long hairpin RNAs, which are processed by Dicer in a more random or processive manner, so the resulting siRNA population is diverse and siRNAs can originate from the entire length of their precursor224,230,232 (FIG. 1a). The long dsRNA precursors can be the product of convergent transcription, viral-RNA replication or, in organisms such as plants and worms, the activity of RNA-dependent RNA polymerases233 (FIG. 1a). In plants and some animal species, Dicer or DCL proteins have diversified to produce different types or sizes of small RNAs. For example, Drosophila melanogaster Dicer 1 and Dicer 2 specialize in the biogenesis of miRNAs and siRNAs, respectively224. In A. thaliana, DCL1 is the major DCL protein for miRNA biogenesis, whereas DCL2, DCL3 and DCL4 generate siRNAs of 22 nucleotides, 24 nucleotides and 21 nucleotides, respectively234.
The cell-autonomous activities of small RNAs have been well studied and are briefly summarized here (FIG. 1b). Small RNAs can form RISCs with Argonaute or PIWI proteins, which in turn exert regulatory functions235,236. In general, cytoplasmic RISCs engage in post-transcriptional gene silencing through RNA cleavage, RNA decay or translation repression, whereas nuclear RISCs cause transcriptional gene silencing through inhibition of RNA polymerase II (Pol II)-mediated transcript elongation, and can lead to DNA or histone methylation235. For example, in worms, small RNAs guide the trimethylation of histone H3 Lys9 (REF.237), H3 Lys27 (REF.238) and H3 Lys23 (REF.239). A. thaliana nuclear AGO1 RISCs may even promote gene expression240. Animal miRNAs undergo hybridization between their ‘seed’ sequence and their target transcripts and cause translation repression or RNA decay, whereas plant miRNAs have extensive complementarity with their targets and cause RNA cleavage and translation repression. Notably, a small number of miRNAs in plants and one miRNA in C. elegans (mir-243) trigger the production of secondary siRNAs87,241 (BOX 2). siRNAs derived from exogenous genetic material such as transgenes, viruses and injected dsRNAs tend to cause target RNA cleavage, but endogenous siRNAs from plants and C. elegans differ in biogenesis, size, associated Argonaute proteins and activity (BOX 2). Through transcriptional gene silencing, piRNAs and their associated PIWI proteins are most notable for their functions in genome defence against transposable elements, although they are also known to regulate protein-coding genes242.
Fig. 1 ∣. Schematics of mechanisms of small RNA biogenesis and modes of action.
a ∣ Mechanisms of microRNA (miRNA), small interfering RNA (siRNA) and PIWI-interacting RNA (piRNA) biogenesis. For piRNAs, a Caenorhabditis elegans pathway is featured. miRNA (MIR) genes are transcribed into primary miRNAs containing stem–loop structures (not shown; dashed arrow), which are processed into shorter, similarly structured precursor miRNAs (pre-miRNAs). A pre-miRNA is processed by the RNase Dicer in animals and by DICER-LIKE 1 (DCL1) in plants into a miRNA–miRNA* duplex, which is loaded into an effector protein of the Argonaute family. The ejection of the miRNA* strand results in the formation of the miRNA-induced silencing complex (miRISC). siRNAs are produced from exogenously provided or endogenously produced long double-stranded RNA (dsRNA). Endogenous dsRNA can arise through different means, including through the activities of RNA-dependent RNA polymerases (RDRs or RdRPs). The dsRNA is processed by Dicer into many siRNA duplexes, which are loaded into an Argonaute protein to form siRNA-induced silencing complexes (siRISCs). piRNAs are transcribed as independent transcription units and most have an upstream ‘Ruby’ motif required for their expression223. PIWI-related gene 1 protein (PRG-1) is an Argonaute protein required for primary piRNA activity; heritable RNA interference (RNAi) deficient protein 1 (HRDE-1) is an Argonaute protein that carries RdRP-amplified 22-nucleotide, 5′-guanosine secondary siRNAs (22G siRNAs), which is required specifically for heritable RNAi in the germ line. b ∣ Small RNAs guide their effector proteins to specific loci to initiate either post-transcriptional gene silencing (PTGS) through transcript cleavage, RNA decay (degradation) and translation repression, or transcriptional gene silencing (TGS), which involves DNA methylation or histone H3 Lys9 (H3K9) methylation at DNA loci that are homologous to the small RNA, often transposable element (TE) loci. Different mechanisms observed in different organisms are summarized in the same scheme. Note that there is no DNA cytosine methylation in C. elegans nematodes. AGO1, ARGONAUTE 1; Pol II, RNA polymerase II.
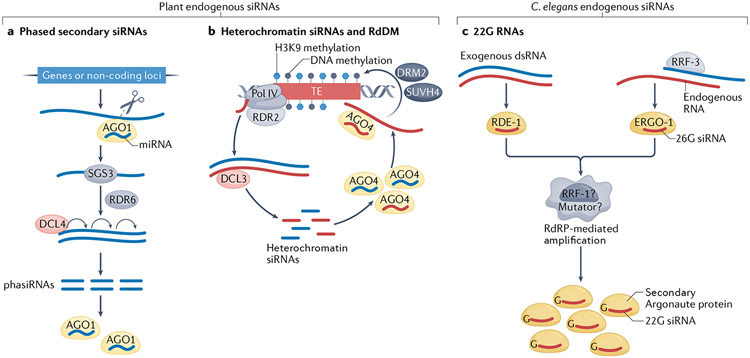
Box 2 ∣. The variety of endogenous siRNAs in Arabidopsis thaliana and Caenorhabditis elegans.
Endogenous sequences that can produce small interfering RNAs (siRNAs) in plants and worms include transposable elements, other non-coding loci and protein-coding genes. One main class of endogenous siRNAs in plants is phased secondary siRNAs (phasiRNAs), which are derived from protein-coding genes or from non-coding loci87. The biogenesis of phasiRNAs is initiated by the targeting of a transcript by one or two Argonaute protein-containing microRNA (miRNA)-induced silencing complexes (miRISCs), which leads to the production of double-stranded RNA (dsRNA) (FIG. 2a). The dsRNA is processively cleaved by DICER-LIKE 4 (DCL4) to produce 21-nucleotide (nt) secondary siRNAs; in cases where the initial targeting by miRISC causes transcript cleavage, siRNA production occurs in the phase defined by the cleavage. Some phasiRNAs act in trans to regulate target genes and are known as trans-acting siRNAs. In grasses, thousands of loci produce not only 21-nt phasiRNAs but also DCL5-dependent 24-nt phasiRNAs specifically in reproductive tissues.
A second class of plant endogenous siRNAs is 24-nt heterochromatin siRNAs, which function to silence transposable elements through a process known as RNA-directed DNA methylation243. Their biogenesis entails transcription of transposable elements or other repeats by RNA polymerase IV (Pol IV), dsRNA formation by RNA-DEPENDENT RNA POLYMERASE 2 (RDR2) and dicing of the dsRNA by DCL3 (FIG. 2b). These siRNAs are incorporated into ARGONAUTE 4 (AGO4) and are recruited to homologous chromatin loci to promote DNA cytosine methylation and gene silencing. In addition to the loci that generate phasiRNAs and heterochromatin siRNAs, protein-coding genes and ribosomal RNA genes can produce RDR-dependent and DCL-dependent siRNAs in specific conditions, such as under viral infection or when RNA decay pathways are compromised82,244,245.
In Caenorhabditis elegans, different types of ‘primary’ small RNAs target the RNA-dependent RNA polymerases (RdRPs) EGO-1 and RRF-1 to RNA molecules (such as mRNAs) to create much more abundant ‘secondary’ siRNAs that are 22 nt long and have a guanosine at their 5′ end (22G siRNAs)246 (FIG. 2c). It was recently shown that the nucleotidyltransferase RDE-3 marks RNAs as templates for siRNA production by adding non-templated stretches of alternating uridine and guanosine ribonucleotides to their 3′ termini, and that this addition facilitates RdRP recruitment54,55. The primary small RNAs that trigger the amplification (by recruiting the RdRPs to the mRNA template) can be siRNAs bound by the Argonaute protein RDE-1 and derived directly from an exogenously provided dsRNA substrate (diced by DCR-1); 26G endogenous siRNAs, which are Dicer-dependent primary small RNAs that are also generated by an RdRP, RRF-3, and which during oogenesis and embryogenesis are bound by ERGO-1; 21U PIWI-interacting RNAs (bound by PRG-1); and ribosomal RNA-derived siRNAs (the production of which depends on SUSI-1, a homologue of the exonuclease human DIS3L2)247. Unlike plant RDRs, the worm RdRPs synthesize 22G siRNAs non-processively, one-by-one (that is, the small RNAs are not cleaved from a longer precursor by Dicer). The amplified siRNAs are bound by many different worm-specific Argonaute proteins248. Amplification of siRNAs is required for transgenerational gene silencing41,249-251. Maintenance of high levels of 22G siRNAs, even transgenerationally, is often independent of the primary Argonaute protein that initiated the amplification process. Two Argonaute proteins in particular, HRDE-1 and WAGO-4, are required for maintenance of 22G RNA-induced RNA interference252-257.
Fig. 2 ∣. Endogenous siRNAs in plants and Caenorhabditis elegans.
a ∣ Phased secondary small interfering RNAs (phasiRNAs) are produced in plants following transcript targeting and cleavage by a microRNA (miRNA)–ARGONAUTE 1 (AGO1) complex, which leads to the production of double-stranded RNA (dsRNA) by RNA-DEPENDENT RNA POLYMERASE 6 (RDR6) using the transcript as a template, a process that also requires the RNA-binding protein SUPPRESSOR OF GENE SILENCING 3 (SGS3). The dsRNA is processively cleaved into phasiRNAs by DICER-LIKE 4 (DCL4), starting from the parental RNA end produced by the miRNA-guided cleavage, b ∣ A simplified model of RNA-directed DNA methylation (RdDM). Transposable elements (TEs) in plants produce 24-nucleotide small interfering RNAs (siRNAs) through a process mediated by RNA polymerase IV (Pol IV), RNA-DEPENDENT RNA POLYMERASE 2 (RDR2) and DCL3; the siRNAs are loaded into AGO4 and target this siRNA-induced silencing complex (siRISC) to nascent transcripts at homologous DNA loci. The siRISC recruits the de novo DNA methyltransferase DRM2 to perform cytosine methylation, and histone methyltransferases such as SUVH4 that methylate histone H3 Lys9 (H3K9), thereby silencing the expression of the TEs. c ∣ In Caenorhabditis elegans, ‘primary’ siRNAs derived from single-stranded RNA or from dsRNA are loaded onto the ‘primary’ Argonaute proteins ERGO-1 or RDE-1, respectively. RRF-3 is an RNA-dependent RNA polymerase (RdRP) that mediates the production of 26-nucleotide-long 5′-guanosine siRNAs (26G siRNAs). The loaded primary Argonaute protein directs the production — possibly in ‘mutator’ foci, where the RdRP RRF-1 is localized — of 22G ‘secondary’ siRNAs, which are loaded onto ‘secondary’ Argonaute proteins to direct gene silencing.
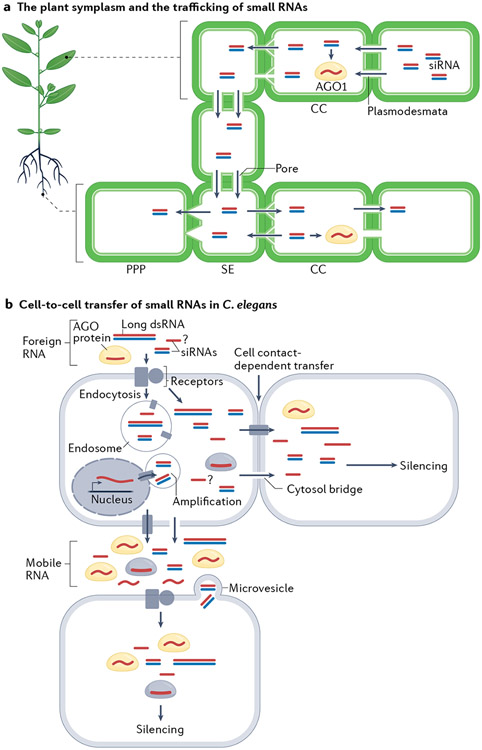
Studies performed following the discovery in the late 1980s of post-transcriptional gene silencing in plants and RNA interference (RNAi) in Caenorhabditis elegans revealed that these processes can function in a non-cell-autonomous manner1. In addition to a requirement for sequence specificity, it was realized through grafting or localized introduction of the silencing trigger that a signal travels systemically through the plant to silence homologous transgenes in post-transcriptional gene silencing2,3 (FIG. 3a). In the Nobel prize-winning study describing the requirement for double-stranded RNA (dsRNA) in RNAi, Andrew Fire, Craig Mello and colleagues found that upon injection of artificial, exogenous dsRNA into a particular worm tissue, the RNAi response spreads systemically throughout the body4 (FIG. 3b).
Fig. 3 ∣. Cell-to-cell and long-distance transfer of small RNAs.
a ∣ The cytoplasm of plant cells is connected through plasmodesmata to form the symplasm (white space in the figure), which allows the exchange of nutrients and biomolecules, including small RNAs. Sieve element (SE) cells in the phloem are connected by pores and mediate long-distance transfer of molecules. Companion cells (CCs) are connected to sieve elements through branched plasmodesmata and load mobile molecules into the sieve elements. Phloem unloading can occur through different types of cells connected to the sieve elements, depending on the cargo molecules. Phloem pole pericycle (PPP) cells are connected to sieve elements through funnel-shaped plasmodesmata and have been shown to unload proteins. How small RNAs are unloaded from phloem is unknown. Small RNAs are thought to move from cell to cell in the form of duplexes. In Arabidopsis thaliana, ARGONAUTE 1 (AGO1) is cell autonomous and consumes mobile small RNAs as they are transported from cell to cell. b ∣ In Caenorhabditis elegans, the identity of the small RNA agents that move between cells is still unknown. With regard to exogenously induced RNA interference responses, it appears that long double-stranded RNA (dsRNA) or double-stranded small interfering RNAs (siRNAs) — but not single-stranded siRNAs — can move between cells. Direct evidence for shuttling of endogenous small RNAs between cells is missing; however, manipulation of endogenous small RNAs in one tissue (neurons or gut) has non-cell-autonomous effects. Different mobile small RNAs might be secreted from cells or might move to contacting cells that are connected by membrane bridges. The small RNAs might be present in vesicles that bud out of the plasma membrane or might be contained in intraluminal vesicles that fuse with the plasma membrane. It is also possible that mobile small RNAs are released when bound by RNA-binding proteins, such as Argonaute proteins. Although not much is known about export of small RNA out of cells in worms (for example, is the RNA secreted inside microvesicles?), import involves receptors that shuttle the small RNA into the host cell. Transporters (perhaps SID-1) likely release the imported RNA from endocytosis vesicles (specialized endosomes). The imported siRNAs can then direct siRNA amplification and gene silencing.
Further studies showed that systemic RNAi can be induced by feeding the worm with RNase III-deficient bacteria that produce dsRNA corresponding to worm genes5, and that small RNAs from both exogenous and endogenous sources can serve as mobile signals in plants, and perhaps in C. elegans and other animals. Although RNA-mediated gene silencing between cells and species is increasingly recognized, the extent, mechanisms and functions of intercellular communications through small RNAs remain controversial. In this Review, we discuss recent advances in our understanding of non-cell-autonomous RNA-mediated gene silencing in plants and animals, highlight mechanistic insights and advocate caution in interpreting data in this field of research.
Trafficking of exogenous small RNAs
The non-cell-autonomous nature of RNA-mediated silencing was first recognized in studies of gene silencing triggered by exogenous sources, such as transgenes and injected long dsRNA2-5. The gene silencing mechanisms discovered using these ‘foreign’ agents help understand RNA communications in vivo. In addition, cross-species non-cell-autonomous gene silencing by such foreign RNAs is being used in agriculture to enhance plant immunity against pests and is referred to as ‘host-induced gene silencing’6,7.
Silencing by mobile siRNAs in plants
Most plant cells are connected with their neighbouring cells through channels, usually up to tens of nanometres in diameter, called ‘plasmodesmata’, thereby forming a symplasm within the apoplasm (FIG. 3a). The plasmodesmata are lined with plasma membrane that is continuous with that of the two neighbouring cells and traverses the cell wall to link the cytoplasm of adjacent cells, thereby allowing intercellular exchange of molecules, while the size-exclusion limit imposed by the plasmodesmata prevents unlimited exchange to ensure cellular identities8. Plasmodesmata and the phloem together connect cells in the entire plant to ensure systemic transport of nutrients and coordinated actions during development and in response to environmental cues. In the phloem, the non-nucleated sieve elements form a strand to conduct the long-distance transport of nutrients and macromolecules. The sieve elements are connected to their neighbouring companion cells through branched plasmodesmata9, which allows mobile molecules to be loaded into the phloem. In roots, unloading of macromolecules from the phloem occurs between sieve elements and phloem pole pericycle cells, which are connected through funnel-shaped plasmodesmata10 (FIG. 3a). The sieve elements may unload smaller molecules to any neighbouring cells10.
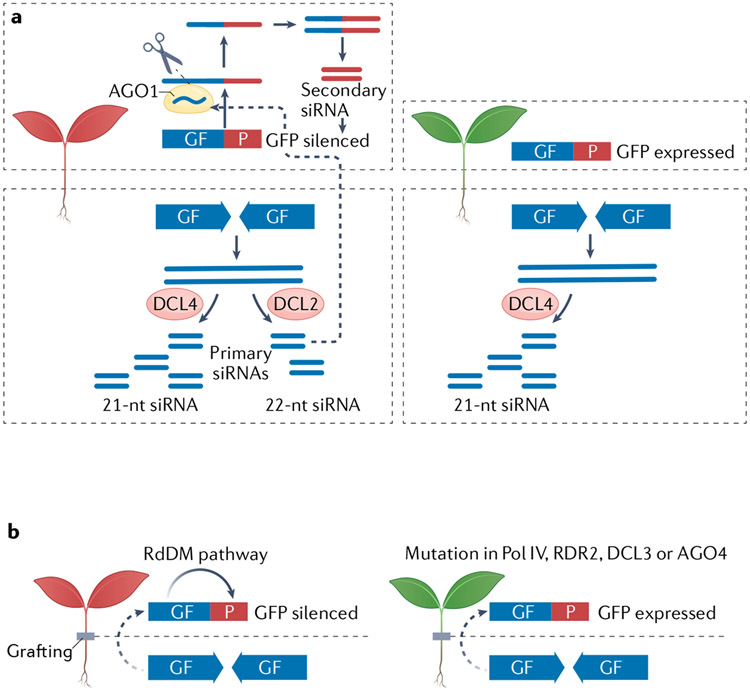
Transgenes in plants tend to trigger RNA-mediated gene silencing. Studies of cell-to-cell and systemic spread of transgene-originating silencing RNAs provided insights into the nature of the mobile-RNA signal and mechanisms underlying this cell-to-cell communication. Extensive grafting studies in mutants of small-RNA-biogenesis genes, performed mainly in Arabidopsis thaliana and sometimes in Nicotiana benthamiana, strongly implicate siRNAs as mobile signals of transgene-triggered gene silencing11-14. For example, the silencing of GFP by an inverted repeat GF (part of GFP) (IR-GF) transgene was accompanied by the production from IR-GF of siRNAs of 21, 22 and 24 nt in length13. When IR-GF plants were grafted onto wild-type roots, siRNAs of all sizes were detected in the roots. These siRNAs were also detected in grafted roots of the dicer-like 2 (dcl2) dcl3 dcl4 triple mutant, suggesting that siRNAs were mobile, although a contribution to siRNA biogenesis from DCL1 in the root could not be completely ruled out13.
Although different experimental systems have provided conflicting evidence12-18, some common themes of non-cell-autonomous RNA-mediated silencing can be gleaned. For example, signal amplification is an important component of non-cell-autonomous gene silencing, and intriguingly, factors of different ‘canonical’ small RNA pathways (BOX 2) contribute to signal amplification. The conflicting observations may be explained by the differing strengths of the silencing triggers and/or the distance of the mobile signal from the phloem or from the final destination.
A key feature of signal amplification is transitivity — a process in which a primary siRNA produced from a certain transcript triggers the production of secondary siRNAs, which are reminiscent of phased secondary siRNAs (phasiRNAs). Transitivity requires the ARGONAUTE 1 (AGO1)–SUPPRESSOR OF GENE SILENCING 3 (SGS3)–RNA-DEPENDENT RNA POLYMERASE 6 (RDR6) pathway (BOX 2; FIG. 2a). The transitivity-triggering primary siRNA tends to be 22 nt long19,20. As the enzyme that produces 22-nt siRNAs, DCL2 has a prominent role in transitivity14,21,22 and in systemic post-transcriptional gene silencing14. For example14, grafting experiments using a dcl2 mutant showed that DCL2 is required in the source and recipient tissues for systemic silencing accompanied by the production of transitive siRNAs (FIG. 4a). Similar grafting experiments using a dcl4 mutant as the source tissue led to enhanced systemic silencing, consistent with the known hierarchy of DCL4 and DCL2, whereby the former has priority of access to dsRNA23 and further highlighting the crucial roles of 22-nt siRNAs in systemic post-transcriptional gene silencing.
Fig. 4 ∣. Signal amplification in transgene silencing in plants.
a ∣ DICER-LIKE 2 (DCL2)-triggered production of secondary small interfering RNA (siRNA) is crucial for systemic RNA-mediated gene silencing. An inverted repeat transgene encoding a portion of GFP (IR-GF) expressed in the root tip leads to the silencing of GFP in the shoot, which is accompanied by the production of ’P’-specific secondary siRNAs from GFP mRNA through amplification of DCL2-dependent, 22-nucleotide (nt) primary ‘GF’-specific siRNAs. In a dcl2 mutant, GFP silencing does not occur and P-specific siRNAs are not made, although DCL4 still produces 21-nt siRNAs from the GF transgene. b ∣ siRNA amplification in systemic RNA-mediated gene silencing. In grafting experiments, IR-GF expressed in the root triggers GFP silencing in the shoot. Part of the RNA-directed DNA methylation (RdDM) pathway, comprising RNA polymerase IV (Pol IV), RNA-DEPENDENT RNA POLYMERASE 2 (RDR2), DCL3 and ARGONAUTE 4 (AGO4) is required in the recipient tissue for such systemic gene silencing. Dashed lines represent siRNA movement in the plant.
Signal amplification in systemic RNA-mediated gene silencing also entails a nuclear pathway that requires RNA polymerase IV (Pol IV), RNA-DEPENDENT RNA POLYMERASE 2 (RDR2), DCL3 and AGO4. This pathway generates 24-nt siRNAs, which are used in RNA-directed DNA methylation (BOX 2; FIG. 2b). Grafting experiments showed that mutations in the four aforementioned factors abolished or compromised GFP silencing in shoots when roots expressing an inverted repeat silencing trigger were grafted onto shoots containing one of the mutations15 (FIG. 4b). Therefore, the perception of mobile RNA signals in recipient cells requires RNA-directed DNA methylation factors. However, DNA or histone methylation is not likely involved here, as Pol V, which is crucial for siRNA-triggered DNA methylation, is largely dispensable for systemic silencing in this system15. In two other systems, inverted repeat transgenes were expressed in phloem companion cells to target the endogenous PHYTOENE DESATURASE gene (PDS) or SULFER gene (SUL) for silencing in leaf mesophyll cells16,18. Thus, the silencing of PDS or SUL requires cell-to-cell, and perhaps systemic (from older to younger leaves), signalling. In these systems, mutations in Pol IV subunits or in RDR2 compromise silencing, whereas mutations in DCL3 and AGO4 either enhance silencing or have no effects. The common observation that Pol IV and RDR2 promote non-cell-autonomous gene silencing may be explained by recent findings that Pol IV and RDR2 not only generate 24-nt siRNAs but also 21–22-nt siRNAs24,25; the 22-nt siRNAs may participate in signal amplification as discussed above. The disparate DCL3 and AGO4 observations may also be explained by the different effects of mutations in these genes on secondary siRNA production in the different systems. In the GFP transgene system15, DCL3-dependent production of 24-nt siRNAs seems to promote the production of RDR6-dependent, DCL4-dependent and/or DCL2-dependent secondary siRNAs from the target GFP RNA. In the PDS and SUL systems16,18, DCL3 probably competes with DCL4 and/or DCL2 for access to dsRNA from the inverted repeat transgenes.
Routes and regulation of silencing-RNA movement in plants
The mechanisms that regulate non-cell-autonomous RNA-mediated silencing are not well understood. Long-distance movement of silencing RNA is thought to entail cell-to-cell movement and phloem loading in source tissue, and phloem unloading followed by cell-to-cell movement in recipient tissues (FIG. 3a). The channel of cell-to-cell movement is likely the plasmodesmata, as spread of non-cell-autonomous silencing cannot access stomata, which are not connected to the symplasm through plasmodesmata26. The agents of cell-to-cell spreading of gene silencing are likely small RNA duplexes. Grafting experiments with a dcl2 dcl3 dcl4 recipient tissue strongly suggest that the mobile agents are not precursor RNAs13. Through association with the viral protein P19, siRNA–siRNA* duplexes made from transgenes expressed in the vasculature are detectable in root epidermal cells, suggesting that small RNA duplexes are mobile27. In addition, the pool of siRNAs in the epidermis exhibits depletion of the 5′ uridine feature, which is characteristic of association with AGO1, suggesting that AGO1 loading occurs as the duplexes enter a recipient cell, which reduces the pool of mobile siRNAs that can exit the cell27. As AGO1 is likely to function in a cell-autonomous manner28, it is potentially a factor that restricts the mobility of small RNAs in iterative cell-to-cell movements (FIG. 3a). AGO1 expression is thought to be ubiquitous in the plant, although local differences may exist, as in A. thaliana ovules29. Thus, association with AGO1, and with other Argonaute proteins, may be a mechanism to regulate the mobility of small RNAs. Additional regulatory mechanisms may exist. Two plasma membrane-associated and plasmodesmata-associated receptor-like kinases, BAM1 and BAM2, are redundantly required for the cell-to-cell spread of RNA-mediated silencing30. It is possible that they regulate the permeability of plasmodesmata with regard to small RNAs.
Trafficking of exogenous RNA in worms
In C. elegans, cell-specific expression of dsRNA-producing transgenes in a specific tissue (such as pharynx, gut, muscles or neurons) leads to non-cell-autonomous gene silencing responses in most other tissues, except for the nervous system (FIG. 3b). Three forward genetics screens were conducted to identify genes required specifically for such systemic RNAi31-33 (likely affecting the efficiency of RNA trafficking), but not for cell-autonomous gene silencing per se. The best characterized systemic RNAi genes are the systemic RNA interference defective-1 gene (sid-1) and sid-2.
SID-1 functions in RNA import into cells34, but is dispensable for RNA export out of cells35. SID-1 is selective for dsRNA and does not discriminate dsRNA on the basis of length36. It might be required for release of imported dsRNA from internal vesicles into the cytosol37. Different Caenorhabditis species differ dramatically in the potency of the RNAi responses that they mount, in part because of differences in non-cell-autonomous small RNA activity. However, sid-1 has functional orthologues in other Caenorhabditis species that do not exhibit systemic RNAi38. Interestingly, sid-1 does not appear to have a role in the intercellular and transgenerational small RNA responses that C. elegans can mount against some viruses39-41. It is possible that another, unidentified, RNA transporter enables such protective effects. Indeed, exogenous small RNAs can elicit systemic gene silencing in SID-1-null tissues: worm neurons, which do not express SID-1, can be sensitized to RNAi in mutants with disrupted levels of endogenous small RNAs42.
Similarly to SID-1, SID-2 affects RNA import43. It is a transmembrane protein that takes up ingested dsRNA longer than 25 nt43 and is localized to the gut apical membrane and trans-Golgi network33,44. Whereas SID-2 is required for systemic RNAi triggered by feeding on dsRNA-producing bacteria, it is dispensable if the dsRNA is introduced into the worm in other ways, such as injection or transgene-encoded dsRNA transcription, in the pharynx for example. SID-2 is conserved in other Caenorhabditis species33. Divergence in the sequence of the protein’s extracellular domain makes Caenorhabditis briggsae refractory to RNAi by feeding33,45. It could be intriguing to study whether this difference between C. elegans and C. briggsae has ecological relevance, as, for example, SID-2 was implicated recently in uptake of small RNAs from pathogenic bacteria46. However, another recent study suggested that SID-2 has other functions and no role in feeding44,47.
SID-3 is an orthologue of human activated CDC42 kinase 1 (also known as TNK2)48, which interacts with SID-4 (REF.49), and SID-5 is a late-endosome protein that is required for sid-1-independent transport of dsRNA taken up by feeding50. It is still not entirely clear whether SID-5 is involved in execution of RNAi (gene silencing) or in export of RNAs out of cells51. SID-5 is required in the gut for enabling silencing in muscles, hinting that it might be required for export of small RNAs. However, it might facilitate export to the muscles indirectly, by releasing ingested dsRNA locally to the gut’s cytoplasm51. RNA systemic defect 3 (RSD-3) also functions in RNA import (and not export) and also localizes to endosomes and to the trans-Golgi network52. Lastly, the low-density lipoprotein receptor mediated endocytosis (RME-2) is required for importing yolk and also for transfer of labelled dsRNA to oocytes — this precursor of small RNAs was suggested to ‘hitchhike’ with the yolk as it moves to the oocytes from the body cavity37. However, it was later proposed that yolk and dsRNA travel independently, and that maternal rme-2 contributes to, but is not required for, movement of small RNAs to the oocytes53.
It is still unclear which RNA species can move between cells (FIG. 3b). Tissue-specific rescue experiments involving genes required for synthesis of either primary or secondary siRNAs suggest that exogenous (dsRNA-derived) siRNA duplexes or longer (dsRNA) precursors are the molecules that move between cells. This conclusion is compatible with the selectivity of SID-1 for dsRNA (although SID-1-independent pathways might exist). The nucleotidyltransferase RDE-3 (also known as MUT-2) is required in donor cells for systemic gene silencing53. RDE-3 adds stretches of alternating non-templated uridine and guanosine ribonucleotides to the 3′ termini of its target RNAs (dubbed ‘poly(U-G) tails’)54,55. Poly(U-G) tails induce RNAi by recruiting RNA-dependent RNA polymerases, which use the poly(U-G) RNAs as templates to synthesize siRNAs55. Since RDE-3 is required in the donor tissue for systemic silencing, it is possible that the mobile RNAs are the poly(U-G)-tailed RNAs themselves, or, alternatively, that siRNAs amplified from poly(U-G)-tailed RNAs move between cells.
Non-cell-autonomous RNAs in insects
Some insects (for example, flour beetles) show a robust systemic RNAi response. In Drosophila melanogaster, injecting dsRNA into the abdomen of adult flies leads to silencing that spreads to different cell types56-58, whereas RNA injections into larvae are inefficient in inducing systemic RNAi (except in haemocytes)59. Interestingly, there appear to be both similarities and differences between insects and nematodes in the mechanisms of cell-to-cell movement of small RNAs. Accordingly, studies in insects identified genes that function in spreading of silencing responses that were not detected in C. elegans screens60.
Experiments using fluorescently labelled nucleic acids showed that long dsRNA binds to the plasma membrane of D. melanogaster S2 cells, and following internalization, the RNA can be found in large puncta inside the cell. By contrast, labelled siRNAs exhibited only low-level binding and no discernible internalization61. It is not entirely clear what underlies this length-based difference. SID-1 is insensitive to dsRNA length36; however, it is obvious that SID-1-mediated import is not the only pathway that enables RNA uptake, because D. melanogaster can naturally take up RNA despite having no known SID-1 homologue (although the uptake is not as efficient as in C. elegans). How do fly cells take up RNA in the absence of SID-1? The process depends at least in part on clathrin-mediated endocytosis and scavenger receptors62. Similar requirements were observed in other insects (for example, in red flour beetle)63. The C. elegans gene rsd-3 is a homologue of the human clathrin interactor 1 gene, and some of the genes identified in screens for RNA import in D. melanogaster are needed also for systemic gene silencing in C. elegans61. By contrast, although they have a crucial role in systemic silencing in flies, scavenger receptors have not been implicated in systemic silencing in C. elegans. Depletion of two redundant D. melanogaster scavenger receptors, SR-CI and Eater, reduces dsRNA uptake by more than 90%62. The role of scavenger receptors in RNA uptake seems to be conserved in other organisms: certain small RNAs associate with high-density lipoproteins, an association which facilitates their delivery into some mammalian cells64. A scavenger receptor, SR-BI, was found to be required for this high-density lipoprotein-assisted RNA import.
Lastly, recent evidence suggest that D. melanogaster cells use also actin-composed and tubulin-composed nanotube-like structures to transmit dsRNA and RNAi machinery to other cells, reminiscent of the plasmodesmata that enable cell-to-cell transmission of small RNAs in plants. These nanotubules were reported to transmit different factors that enable RNAi: AGO2, dsRNA and CG4572 (a putative serine carboxypeptidase required for reporter-gene silencing following soaking cells in dsRNA)65.
Physiological roles of systemic RNAi
Silencing of viral RNA in plants is well studied66, and small RNA pathways that protect C. elegans from viruses have also been explored67. The systemic amplification and activity of transgene-originating siRNAs in plants and worms may reflect similar functions of virus-originating siRNAs, which target viral RNAs for degradation. For example, it is likely that virus-originating siRNAs from virus-infected leaves are sent out to healthy leaves, where they ‘prime’ antiviral RNAi when the virus reaches these leaves. The low levels of systemic virus-originating siRNAs may trigger amplified production of virus-originating siRNAs through DCL and RDR, both of which are required for systemic antiviral responses in plants68,69.
RNAi is important for antiviral protection also in arthropods, including in D. melanogaster, where mutants defective in a dsRNA uptake pathway are especially sensitive to C virus and Sindbis virus infections70. Intriguingly, systemic antiviral RNAi in D. melanogaster depends on macrophage-like haemocytes, which import dsRNA from virus-infected cells71. The flies use a mechanism (which is analogous to the bacterial CRISPR system) in which an endogenous transposon reverse transcriptase generates virus-derived complementary DNA sequences72. These virus-derived DNA fragments are used as a template for the production of antiviral siRNAs. The DNA-encoded amplified small RNAs are released in microvesicles to other cells and provide systemic antiviral protection (FIG. 3b). Small RNAs purified from haemolymph of infected animals can transfer their protective effects to naive animals71. In mosquitoes, antiviral siRNAs are similarly generated through reverse transcription of RNA viruses73. The reverse-transcribed viral DNA exists in both linear and circular forms and its biogenesis is regulated by the DExD/H helicase domain of Dicer 2. It was suggested that through this activity, Dicer 2 functions like a pattern recognition receptor that modulates antiviral immunity in insects74. In ovaries of Aedes aegypti mosquitoes, the reverse-transcribed viral sequences regulate through small RNAs the replication of cell-fusing agent virus, potentially affecting the inheritance of the virus75.
Conceptually related studies on systemic RNAi and antiviral protection have been conducted in bees. The Israeli acute paralysis virus causes colony collapse disorder, which endangers the survival of bees, particularly in agricultural settings. RNA-mediated silencing of the virus through dsRNA feeding is efficient in preventing colony collapse disorder76. Moreover, secreted RNA can move between bees by feeding of royal jelly; the royal jelly strongly binds RNA through major royal jelly protein 3, which functions as an extracellular sequence-unspecific RNA-aggregating factor. Honeybees release this RNA-binding protein to share RNA and RNAi between them77. The shared RNA holds the potential to regulate the bees’ own genes and transposons, and corresponding sequences in bacteria, fungi and viruses. It is possible that the shared RNA has important roles in conferring social immunity78.
In addition to direct silencing of cognate targets, mobile exogenous small RNAs might affect gene expression indirectly. In A. thaliana and C. elegans, for example, it is clear that different small RNA species compete for shared resources such as RNA-processing enzymes or binding to Argonaute proteins. For example, C. elegans mutants that fail to produce certain endogenous siRNAs are especially vigorous in inducing RNAi in response to exogenously supplied dsRNA, presumably because more resources are available for the task79-81; in A. thaliana mutants that produce ribosomal RNA-derived siRNAs, the loading of miRNAs into AGO1 is impaired82, and C. elegans mutants impaired in production of piRNAs produce more ribosomal RNA-derived siRNAs83. Thus, it is possible that exposure to certain abundant exogenous small RNAs or dsRNA and their shuttling between cells will affect the utilization of other, endogenous small RNAs in different tissues, and therefore also the regulation of their target genes.
Trafficking of endogenous small RNAs
In plants, endogenous small RNAs, including siRNAs and miRNAs, can travel en masse through the phloem from shoots to roots13,84, following the movement of sugars produced by photosynthesis. However, the extent of the biological impact of systemic small RNAs is still largely unknown, as functions have been ascribed to only a handful of systemic small RNAs. Plant endogenous small RNAs can also travel locally between cells, and a number of mobile small RNAs have been shown to act as morphogenesis factors. In animals, ample endogenous extracellular small RNAs can be detected in different biological fluids, and their analysis is used for diagnosis of diseases in humans85. A number of convincing studies (described later) have ascribed functions to such circulating animal small RNAs, and similarly in C. elegans, perturbation of endogenous small RNA production generates non-cell-autonomous and transgenerational effects86.
Mobile siRNAs in plants
phasiRNAs (BOX 2) are widespread in land plants, and diverse types of protein-coding gene families or non-coding RNAs are targeted by miRNAs for phasiRNA production in different species87. In the highly conserved miR390–TAS3–AUXIN RESPONSE FACTOR (ARF) module, miR390 triggers the production of phasiRNAs from non-coding TAS3 transcripts, of which a subset comprising trans-acting siRNAs (tasiRNAs) subsequently represses the expression of ARF genes (tasiR-ARFs). It appears that the module is an evolutionary success — it is used to specify common or different developmental processes in different plants, such as leaf polarity88-90, developmental timing91-95, lateral root growth96-98, nodulation96,99, meristem organization100, female germline development101 and corolla tube formation102. The non-cell-autonomous activities of A. thaliana tasiR-ARFs are inferred from their broader domains of localization (as determined by in situ hybridization) or activity (as determined by their regulation of ARF3) in comparison with that of tasiR-ARF biogenesis, which is determined by the expression of TAS3 or phasiRNA biogenesis genes, such as SGS3 and AGO7. For example, in A. thaliana leaf primordia, AGO7 is expressed only in the top two adaxial cell layers, which implies that tasiR-ARF biogenesis occurs in these cells. However, tasiR-ARFs form a gradient of abundance emanating from the adaxial side to the abaxial (bottom) side, leading to the expression of the target gene ARF3 on the abaxial side, where it specifies abaxial identity103. In A. thaliana ovules, a central, hypodermal cell is destined to become the megaspore mother cell (MMC) that is to give rise to the female germ line. In phasiRNA biogenesis mutants, such as rdr6, sgs3 and ago7, additional hypodermal cells assume MMC characteristics, suggesting that the phasiRNA pathway is required to repress ectopic MMC formation29,101. Specific patterns of accumulation of SGS3 along the radial axis and of AGO7 along the proximal–distal axis would, in combination, limit tasiR-ARF biogenesis to the distal, epidermal layer, where both SGS3 and AGO7 are expressed. However, tasiR-ARF activity as revealed by ARF3 expression expands to the hypodermal layer as well as towards the proximal axis29,101. Expression of a tasiR-ARF-resistant ARF3 using its own promoter causes the formation of multiple MMCs101. Thus, tasiR-ARFs made in epidermal cells suppress MMC identify of hypodermal cells.
In grasses, miR2118 and miR2275 target transcripts produced from hundreds to thousands of phasiRNA-producing loci (PHAS) to generate 21-nt and 24-nt phasiRNAs, respectively, in anthers, and at a lower level in female reproductive organs104-106. Although the functions of the reproductive tissue phasiRNAs are still largely unknown, deletion of a cluster of rice MIR2118 genes results in male and female sterility107, and photoperiod-dependent or temperature-dependent male sterility in rice has been mapped to variations at specific PHAS loci108-110, implying phasiRNAs have a role in reproduction. In maize, the biogenesis of 21-nt phasiRNAs probably occurs in the anther epidermis, as the transcription factor gene ocl4, which is expressed specifically in the epidermis, is required for the accumulation of miR2118 and 21-nt phasiRNAs106. Nevertheless, 21-nt phasiRNAs are detected in all cell types in maize anthers87, and in meiocytes in rice, where they are associated with the Argonaute protein MEL1 (REF.111), suggesting that 21-nt phasiRNAs are mobile. The 24-nt reproductive phasiRNAs may also be mobile. DCL5, which is the Dicer homologue that produces 24-nt phasiRNAs in maize, is most highly expressed in the tapetum (a layer of the anther wall), and loss of the tapetum leads to severe reduction of the levels of both 24-nt phasiRNAs and their precursor PHAS RNAs106,112. Although these data suggest that 24-nt phasiRNAs are made in the tapetum, they are found in both the tapetum and the germ line106. Thus, 21-nt and 24-nt phasiRNAs may serve as agents of cell-to-cell communication. However, their molecular and biological functions are still largely unknown, although 21-nt phasiRNAs were recently reported to cause target-RNA cleavage105,113-115.
A large class of endogenous siRNAs are heterochromatin siRNAs that are derived from transposable elements and other repeats, and guide DNA methylation at their source loci or at homologous loci (BOX 1). As products of the action of DCL3, these siRNAs are primarily 24 nt long, although DCL4 and DCL2 may access the same dsRNA precursors to produce 21-nt and 22-nt siRNAs, respectively, particularly when DCL3 is absent25,116. Grafting experiments in which wild-type scions were connected to dcl2 dcl3 dcl4 mutant stocks followed by high-throughput sequencing provided strong evidence for the systemic movement of 24-nt siRNAs from shoot to root; moreover, mobile 24-nt siRNAs trigger DNA methylation and silencing in roots12,13. Therefore, mobile 24-nt siRNAs can be functional, raising the possibility that they convey specific epigenetic changes at distant tissues, including meristematic cells, which may transmit the epigenetic changes to descendent cells. Intriguingly, the levels of 24-nt siRNAs in dcl2 dcl3 dcl4 rootstock grafted onto wild-type scion were similar to those in wild-type rootstock grafted onto wild-type scion13, implying that mobile siRNAs constitute a large portion of root siRNAs. A recent study examined the species-specific 24-nt siRNAs in shoots and roots of soybean–common bean heterografts and reached a similar conclusion84. These studies suggest that roots import small RNAs on a large scale and implicate small RNAs as agents of communication. Note that the shoot-to-root transport of small RNAs in these studies may represent the direction of movement of nutrients in the phloem — from source (photosynthesizing tissue) to sink (tissue that imports nutrients). Thus, small RNAs are expected to also move through the phloem to other sink tissues, such as the shoot apical meristem.
Bulk movement of endogenous, transposable element-derived siRNAs may also occur during male germline development. In A. thaliana anthers, expression of RDR2 or DCL3, genes required for the biogenesis of 24-nt, transposable element-derived siRNAs, specifically in nurse cells known as tapetal cells, was sufficient to confer DNA methylation of homologous genetic elements in meiocytes117. In male gametophytes, the DNA of the vegetative nucleus is hypomethylated due to the expression of DNA demethylases and lack of expression of DDM1 (a gene required for DNA methylation), which leads to the expression of retrotransposons118. Transcripts from the retroelements are processed into 21-nt siRNAs, which accumulate in sperm cells118,119. Together, these studies suggest that transposable element-derived siRNAs move from nurse cells to the germ line and have regulatory functions.
Mobile miRNAs in plants
Plant miRNAs have key roles in various aspects of plant development and stress responses. Early studies concluded that miRNAs act cell autonomously, but subsequent studies documented cell-to-cell or even systemic trafficking of some miRNAs, leading to the proposal that miRNAs may serve as signals in developmental patterning or stress responses120. In soybean–common bean heterografts, root miRNAs are largely shoot-derived84, suggesting the existence of large-scale systemic trafficking of miRNAs following the source–sink nutrient trafficking route. The biological functions of most mobile miRNAs remain to be examined, and here we describe mobile miRNAs with established biological functions.
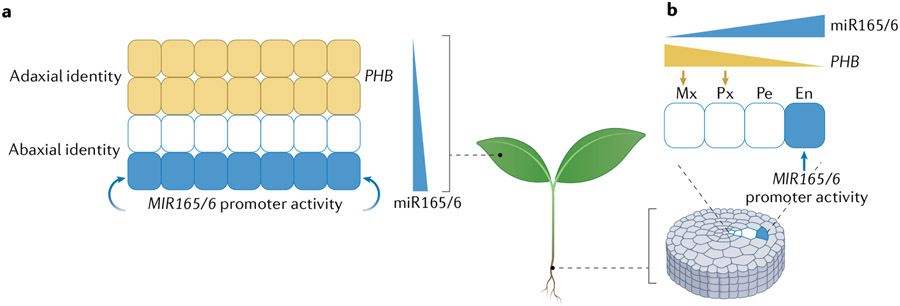
Cell-to-cell trafficking of at least two miRNAs has been shown to specify cell fates. In leaf primordia, the MIR165 and MIR166 genes are strictly expressed in the abaxial epidermis, but the mature miRNA, collectively known as miR165/6, forms a gradient of abundance, diminishing towards the adaxial side89,121 (FIG. 5a). This gradient restricts to the adaxial side the expression of the miR165/6 target genes encoding class III homeodomain leucine zipper transcription factors, including PHABULOSA, which confer adaxial identity122-124. In A. thaliana roots, which consist of concentric layers of cells with different identities, the MIR165 and MIR166 genes are specifically expressed in the endodermis, whereas the abundance of the mature miRNA and its activity decrease from the endodermis to the innermost cells125,126 (FIG. 5b). Coincidently, the levels of the target protein PHABULOSA form an inverse gradient, and the different levels of PHABULOSA contribute to the formation of distinct identities of the cell layers, such as the metaxylem, protoxylem and pericycle125,126. miR394 is another miRNA with non-cell-autonomous activities. The MIR394B gene is expressed in the epidermal layer of the shoot apical meristem, but the mature miR394 is found in the epidermal layer as well as in two underlying cell layers, where it represses the expression of its target gene to maintain stem cells in the shoot apical meristem127. In addition to miR165/6 and miR394, spatial comparison of MIR-gene promoter activity versus the accumulation of mature miRNAs and/or of their regulatory activities implicated miR160, miR164 and miR395 in cell-to-cell mobility in A. thaliana roots and miR160 in cell-to-cell mobility in leaves28.
Fig. 5 ∣. Pattern formation by the mobile microRNA miR165/6 in leaves and roots involves distinct mechanisms.
a ∣ In leaf primordia, miR165/6 is specifically produced in the abaxial epidermis and forms a diminishing gradient towards the adaxial side. This gradient leads to the all-or-none expression pattern of the miRNA target gene PHABULOSA (PHB), which in turn specifies adaxial identity. b ∣ In roots, miR165/6 is specifically produced in the endodermis (En) and forms a diminishing gradient of expression towards the inner parts of the root. This gradient results in graded expression of PHB, with increasing levels of PHB expression leading to the formation of the protoxylem (Px) and metaxylem (Mx) cell types. Pe, pericycle.
Systemic trafficking of miRNAs has also been observed. Phloem sap contains many miRNA species, suggesting that miRNAs may travel through phloem to distant tissues128,129. This was confirmed through grafting experiments. Taking advantage of the fact that miR399 is not present in phosphate-replete conditions, scions that constitutively express a MIR399 transgene were grafted onto wild-type rootstocks in both A. thaliana and tobacco, and miR399 accumulation in roots and the corresponding downregulation of its target gene PHO2 were observed, suggesting the existence of shoot-to-root movement of the miRNA, with functional consequences130. In the hen1-1 mutant, which is defective in miRNA biogenesis, the levels of miR399 and miR395 were below detection in roots; however, hen1-1 roots grafted onto wild-type scions accumulated levels of miR399 and miR395 similar to those of wild-type roots131. Notably, MIR399 transcription is quickly induced in shoots, but not in roots, by phosphate starvation, but mature miR399 first accumulates in roots130. It is possible that shoot-to-root translocation of miR399 is a means for shoots to convey the need to take up phosphate by roots. Legumes have evolved symbiosis with nitrogen-fixing rhizobia, which live in root nodules and provide ammonium to plants in exchange for photosynthates. In the legume Lotus japonicus, miR2111 may be an agent of the shoot–root signalling that regulates nodule numbers. MIR2111 genes are expressed in the phloem in leaves, and grafting experiments show that shoot-derived miR2111 promotes nodule numbers in roots132,133.
Regulation by and of mobile small RNAs in plants.
It might be worth considering the unique regulatory functions of non-cell-autonomous small RNAs in comparison with cell-autonomous small RNAs. One function is signalling from source cells to recipient cells. In the case of the stress-inducible miR399 and miR395 and the nodulation-regulating miR2111, the shoot-to-root trafficking may be a means for shoots to regulate nutrient uptake by roots. Another function of mobile small RNAs is the generation of positional information within a developmental field of cells. The best examples are tasiR-ARFs and miR165/6, which form opposing gradients along the adaxial–abaxial axis in leaves to specify adaxial–abaxial polarity89,120. Intriguingly, small RNA gradients may be interpreted differently to produce different outcomes (FIG. 5): in leaves, tasiR-ARF and miR165/6 gradients are interpreted in a threshold-dependent manner to result in sharp boundaries of target-gene expression134; in roots, the miR165/6 gradient generates a nearly inverse gradient of target-gene expression, which then leads to the formation of different cell types125,126. Regardless, the mobile small RNAs behave as morphogens that specify cell fates120.
Another consideration is how small RNA-based signalling might differ from signalling by, for example, peptides or hormones. On the one hand, gene regulation by small RNAs has the clear advantage of target gene–small RNA sequence specificity, which limits the response in recipient cells to a small number of target genes. In the case of peptides or hormones, signalling pathways usually regulate the activities of transcription factors, which can have hundreds to thousands of target genes. On the other hand, when a host of small RNAs are mobile agents, such as in the case of tapetal transposable element-derived siRNAs that move into meiocytes117, large-scale yet sequence-specific effects are possible.
As signalling molecules, non-cell-autonomous small RNAs themselves are likely regulated. Like transgene-originating siRNAs, endogenous small RNAs probably move from cell to cell through the plasmodesmata. Callose deposition, which restricts the size-exclusion limit of plasmodesmata, reduces the non-cell-autonomous activity of miR165/6 (REF.135). Similar to siRNA–siRNA* duplexes, miRNA–miRNA* duplexes (rather than their precursor miRNAs) may be the mobile agents27,28. Small RNAs appear to move through passive diffusion, resulting in a source-to-destination gradient of abundance103,121,125. However, such movement is not necessarily unregulated, as it is directional at certain cellular interfaces, such as between lateral organs and the shoot apical meristem136. As AGO1 is cell autonomous, it can be envisioned that loading of mobile miRNAs into AGO1 would reduce the levels of these mobile miRNAs that exit the cell, as shown for siRNAs28. This mechanism may apply to other Argonaute proteins. AGO10, in particular, preferentially binds miR165/6 and is crucial for preventing the accumulation of mobile miR165/6 in the shoot apical meristem137,138. AGO10 probably also sequesters miR398 from moving into the female gametophyte in A. thaliana ovules139.
Subcellular localization of small RNA biogenesis may also affect the mobility of small RNAs. When an artificial small RNA is made from a miRNA precursor, its mobility is much reduced compared with the that of the same small RNA that is generated from a phasiRNA precursor140. The subcellular locations of miRNA biogenesis and siRNA biogenesis are different. In plants, miRNA biogenesis, including the loading of miRNA–miRNA* into AGO1, probably occurs in the nucleus, with the resulting miRNA-induced silencing complexes (miRISCs) being exported to the cytoplasm141, although some level of miRISC formation in the cytoplasm cannot be excluded. This process might be the reason why miRNAs are generally not highly mobile, as their nuclear loading would reduce the levels of the mobile species, perhaps miRNA–miRNA* duplexes, that can access the plasmodesmata in the cytoplasm. By contrast, the biogenesis of phasiRNAs probably involves cytoplasmic membranes — phasiRNA precursors and the triggering miRNAs are present on membrane-bound polysomes105,142, and the biogenesis machinery is found in siRNA bodies, which are adjacent to vesicles143. This connection to membranes may allow phasiRNAs to access the plasmodesmata at the plasma membrane more readily, resulting in better cell-to-cell trafficking.
Despite their biogenesis and RISC formation in the nucleus, some miRNAs are mobile. This may be due to incomplete loading of miRNA–miRNA* duplexes into AGO1. Indeed, a non-AGO1-bound pool of some miRNAs exists in vivo144. This finding raises the question of how mobile miRNAs load into AGO1 in recipient cells. If miRNA–miRNA* duplexes are the mobile agents, upon entry into recipient cells, they first access the cytoplasm. Do they access AGO1 in the nucleus or in the cytoplasm in recipient cells? If they are loaded into AGO1 in the cytoplasm, how do mobile miRNA–miRNA* duplexes in source cells avoid cytoplasmic loading into AGO1, while miRNA–miRNA* duplexes access cytoplasmic AGO1 in destination cells? Mechanisms that regulate the nuclear and cytoplasmic loading of miRNAs may regulate miRNA mobility.
Mobile endogenous small RNAs in worms
Whereas much knowledge has been gained regarding movement of exogenous RNAs in worms (as described in detail earlier herein), it is still unclear which types of endogenous small RNAs can move from cell to cell in C. elegans. Silencing by the miRNA lin-4 was explicitly shown to be restricted to the miRNA-expressing cell81. Nevertheless, the ability of other miRNAs to move between cells has not been extensively investigated. Although chemically tagged and exogenously supplied small RNAs can be tracked, we do not yet have the technology to stain or tag endogenous small RNAs with fluorophores in vivo, so direct evidence of their movement is hard to come by. sid-1 mutants have phenotypes that suggest that endogenous small RNAs do move between cells: SID-1 is required for gene regulation in both the soma and the germ line following entry to dauer stage145; the regulatory effects of neuronally produced small RNA on inheritance are ‘noisier’ in sid-1 mutants (that is, behavioural responses which are affected by ancestral neuronal small RNAs show higher variability between animals86); and descendants of sid-1-mutant mothers develop an abnormal number of gut cells146. It is possible, however, that in addition to RNA import, sid-1 mutants have indirect or RNAi-unrelated phenotypes as well (for example, sid-1 mutants exhibit a decrease in turnover of misfolded proteins)147.
A recent study has demonstrated that rescuing the production of endogenous siRNAs in the C. elegans nervous system through neuron-specific expression of the dsRNA-binding protein RDE-4 (which is required for synthesis of some endogenous siRNAs) results in gene expression changes in the germ line86. These non-cell-autonomous effects persisted transgenerationally and affected the progeny’s chemotaxis behaviour. In this study, small RNAs were sequenced from the brain and gonads of wild-type and sid-1 animals, and most (but not all) of the non-cell-autonomous and transgenerational effects were found to be unaffected by SID-1 deficiency86. The non-cell-autonomous gene regulation by the endogenous neuronal small RNAs might result from their movement between cells. Although that is the parsimonious explanation, other explanations exist. For example, the neuronal small RNAs might regulate the germline genes indirectly, through other neuronal signalling molecules with non-cell-autonomous functions, such as hormones or neuropeptides86.
Mobile endogenous small RNAs in mammals
Much work in mammals has focused on disease diagnosis based on extracellular small RNAs and on delivering into cells regulatory small RNAs to induce RNAi in different tissues for therapeutic purposes, and recently these efforts began to bear fruit148. Such studies are beyond the scope of this Review. Here, we discuss the evidence that mammals secrete functional endogenous small RNAs.
In mammals, the extracellular environment, and bodily fluids in particular, are rich in different endogenous small RNAs (see more later). Release of extracellular small RNAs might be regulated, or it could result from bursting of dying cells, non-specifically. The small RNAs might be ‘naked’ or bound by proteins, and could travel within or outside different vesicles, such as exosomes. In recent years we have witnessed major developments in characterizing and understanding extracellular RNA in mammals.
Mast cells were shown almost 15 years ago to secrete ample vesicles that contain miRNAs and mRNAs149. This seminal work was followed up by numerous studies, and the shear amount of literature on RNA in different vesicles is overwhelming and outside the scope of this Review. Mammalian secreted miRNAs were implicated in many processes affecting development, cell migration, immunity and cancer progression and metastasis150,151. Some secreted miRNAs remain functional in adopting cells, including in well-controlled in vivo studies. For example, in Dicer1-knockout mice, which cannot produce miRNAs, the 3′ untranslated region of a liver mRNA can be regulated by miRNAs originating from transplanted wild-type adipose tissue152.
In addition to the greatly studied secretion of miRNAs, most other small RNAs are detected in mammalian exosomes as well, including small nuclear RNAs, small nucleolar RNAs, ribosomal RNA fragments, piRNAs, tRNAs, mitochondrial RNAs, Y RNAs and vault RNAs151. The functionality of most of these RNAs outside their tissue of origin is still unknown. A recent comprehensive survey of small RNAs in vesicles from different cell types estimated that ribosomal RNA fragments account for 30–94% of the small RNA reads. We only briefly discuss this topic here, since it was reviewed recently151.
Are specific small RNAs sorted into specific extracellular vesicles to execute specific functions? Many studies in mammals linked different non-cell-autonomous phenotypes with small RNA-carrying vesicles; however, for years the definitions of what constitutes exosomes or other extracellular vesicles were confusing and inconsistent. Moreover, variability in the isolation procedures was proven to cause high heterogeneity in purified vesicles153, and many have focused only on some highly expressed miRNAs154. In different cell types, different proteins are required for secretion155, and several quantitative studies concluded that in certain cases vesicles contain only a tiny number of specific small RNAs, perhaps too few to be functional156. To deal with these problems, the extracellular vesicles research community formed the Extracellular RNA Communication Consortium, the work of which resulted in more than 500 publications and was summarized in a recent review157. One of the projects of the consortium focused on a systematic comparison of ten methods for isolation of extracellular RNA from many types of biofluids158. Despite these considerable efforts, it is clear that other classes of RNA carriers await further inquiry (for example, the recently identified non-membranous nanoparticles termed ‘exomeres’)159.
The consortium’s work clearly demonstrates that RNA sequencing reproducibility differs dramatically across isolation methods. The percentage of reads mapped to ribosomal RNA, miRNA, tRNA, piRNA and other RNAs changes depending on the biofluid type. Regardless of these technical issues, the pool of small RNAs within secreted vesicles appears to be very different from the pool of intracellular small RNAs, strongly suggesting the existence of sorting mechanisms. Studies of small RNAs that are secreted from different cell types revealed different pools of secreted RNAs and proposed different small RNA-classifying mechanisms. For example, it was shown that exosomes from HEK293T cells contain a unique set of enriched miRNAs; cell-free experiments revealed that sorting of miR-223 into such vesicles depends on the RNA-binding protein Y-box protein 1 (YBX1), and moreover that YBX1 is required for secretion of miRNAs in exosomes150. It was later revealed that many other small non-coding RNA species depend on YBX1 for sorting into exosomes160. Other sorting mechanisms in other cell types include binding by sumoylated heterogeneous nuclear ribonucleoproteins A2/B1 (REF.161), the presence of a miR-1289 binding site and the sequence motif CUGCC in the 3′ untranslated region of microvesicle mRNAs162, and non-templated 3′ uridylation163. The small GTPase ARF6 was found to target precursor miRNAs to so-called oncosomes164, and interaction of RNA-binding proteins with LC3, the autophagosome factors, determines the composition of small RNAs in secreted vesicles165. To complicate the matter further, the sorting mechanisms respond to changes in cellular states: overexpression of an oncogene drives sorting of AGO2-associated miRNAs into vesicles166, and other types of stress (oxidative stress167, inescapable electric shock168 or exercise169) change the population of miRNAs in exosomes.
In addition to secretion by vesicles, small RNAs can move between cells or in the body in association with proteins, for example, bound to AGO2 (REFS170,171). High-density lipoproteins bind certain miRNAs and enable their delivery into cells64. In response to serum starvation, miRNAs bind nucleophosmin, which is secreted from certain human cells172.
Other studies have demonstrated intercellular movement of small RNAs, which depends on cell–cell contact. For example, immune cells can acquire different non-secreted molecules from cells with which they form immunological synapses173, and during this process small RNAs from B cells are adopted by T cells174. Separation of the cells by a transmembrane significantly reduced small RNA transfer (residual molecules were likely transferred by exosomes). Several studies suggested small RNAs can move between cells through gap junctions, as connexin is required for intercellular transfer of siRNAs175 and miRNA176-178. Lastly, similarly to what was found in D. melanogaster65, several studies suggest that small RNAs can move in membranous ‘nanotubes’ that connect cells in culture179,180.
Interspecies small RNA communications
Since it became clear that C. elegans nematodes take up artificial dsRNA from Escherichia coli very efficiently, it was suspected that interspecies or cross-kingdom RNAi occurs between interacting organisms such as plants, animals and pathogens. Hosts might identify pathogens by identifying their small RNAs, or might transmit protective small RNAs that kill their pathogens or reduce their infectivity. Host-induced gene silencing, whereby dsRNA expressed in plants from transgenes causes silencing in their pathogens and pests, is effective against insects, nematodes, parasitic plants, fungi and Phytophthora (water moulds) and is actively exploited as a biocontrol tool6,7. Although possible, siRNAs have not yet been shown to be the mobile agents in transgene host-induced gene silencing. Although it is reasonable to assume that interspecies small RNAs could be used as weapons or for self-defence, they might also promote helpful, even symbiotic interactions. Different endogenous small RNAs were suggested to move between organisms, but new bioinformatics tools for distinguishing the small RNA pools of interacting organisms are still required181. The detection of small RNAs from one species in another species, even when small RNAs from one species are in an Argonaute protein of the other species, may be due to tissue contamination or spurious Argonaute loading following tissue lysis. Although the evidence for the activity of interspecies small RNAs is scarce, we discuss some compelling examples that were collected in recent years.
Interspecies small RNAs in plants
The parasitic plant Cuscuta campestris uses a feeding structure called a ‘haustorium’ to obtain water and nutrients from host plants. C. campestris produces 22-nt miRNAs in the haustoria, which are detected in host stems at a certain distance from the haustoria (meaning they are truly in the host plant and do not represent experimental contamination from the parasitic plant)182. These 22-nt miRNAs target host transcripts for cleavage and induce secondary siRNAs, leading to the downregulation of the host target genes. Another example is the transfer of miRNAs from cotton plants to the fungal pathogen Verticillium dahliae183. Although infected tissue contains both plant and fungal cells, which cannot be easily separated, one experimental setup deliberately cultured fungi in vitro from hyphae recovered from infected cotton. Tens of cotton miRNAs, including miR159 and miR166, were detected from fungi that had been cultured in vitro. This was strong evidence for the transfer of miRNAs from plants to pathogenic fungi, but it is perplexing that the plant miRNAs were still detectable in fungi after 20 days of in vitro culturing. The production of miR159 and miR166 is enhanced in cotton plants upon V. dahliae infection, and the two miRNAs target fungal genes that are essential for virulence.
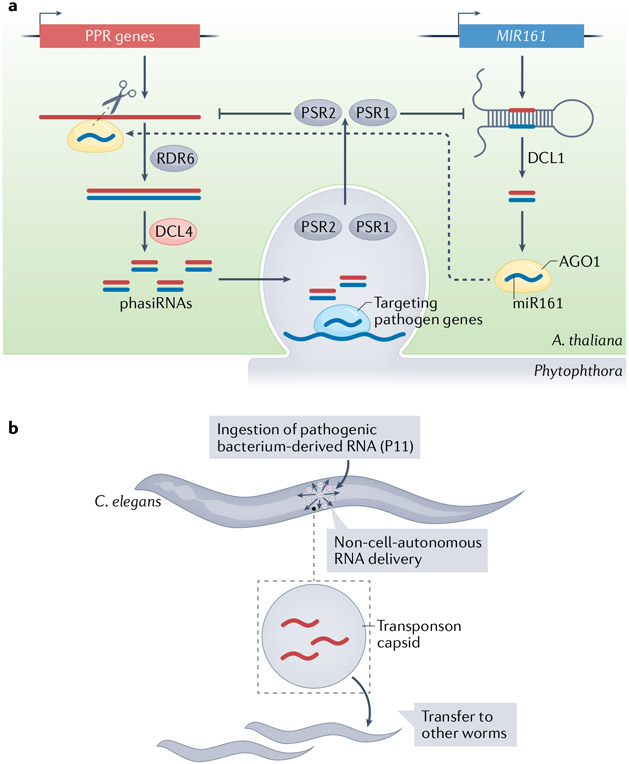
Other studies documented interspecies small RNAs between plants and fungal or Phytophthora pathogens and between plants and symbiotic bacteria184-189. In these studies, however, interspecies small RNAs were detected following crude separation of the interacting organisms, or inferred from their loading into Argonaute proteins or their regulatory activities in the partner organism, making the evidence for interspecies transfer of small RNAs not as strong as in the two examples above. Nevertheless, small-RNA biogenesis mutants and experimental target mimicry to suppress small RNA activities provide further evidence for interspecies small RNA activities. The interspecies small RNAs found in such studies include tasiRNAs from A. thaliana that target genes in the fungal pathogen Botrytis cinerea184, phasiRNAs from genes encoding pentatricopeptide repeat-containing proteins targeting the pathogen Phytophthora capsici186 (FIG. 6a), and tRNA-derived small RNAs from rhizobia that target soybean genes to regulate nodulation187. Consistent with phasiRNAs being interspecies small RNAs that move from plants to Phytophthora pathogens, Phytophthora sojae sends Phytophthora RNA silencing suppressor (PSR) proteins into plant cells to interfere with phasiRNA biogenesis190 (FIG. 6a).
Fig. 6 ∣. Interspecies small RNAs.
a ∣ Small RNA-based defence and counterdefence between plants and the oomycete (water mould) pathogen Phytophthora. In Arabidopsis thaliana, family of pentatricopeptide repeat-containing protein (PPR) genes is targeted by the microRNA miR161 to generate phased secondary small interfering RNAs (phasiRNAs), which probably enter Phytophthora capsici to silence pathogen genes as a plant defence measure. As a counterdefence, Phytophthora sojae secretes RNA silencing suppressor proteins (PSR) into plant cells to suppress the biogenesis of both microRNAs and phasiRNAs. b ∣ A bacterial (Pseudomonas aeruginosa) small RNA named ‘P11’ is taken up by Caenorhabditis elegans nematodes when the worms eat the bacteria. P11 travels from the site of ingestion to the germ line, regulates host genes and spreads to somatic tissues in a process that depends on the transporters SID-1 and SID-2 (not shown) and on encapsulation by a capsid, which is encoded by an endogenous retrotransposon. The worm uses this process to protect itself against future infections, as it induces an avoidance behaviour. AGO1, ARGONAUTE 1; DCL, DICER-LIKE; RDR6, RNA-DEPENDENT RNA POLYMERASE 6.
How small RNAs move between plants and their interacting organisms is largely unknown. Like animal cells, plant cells secret extracellular vesicles191; isolated extracellular vesicles were found to contain small RNA species184,192, suggesting that they serve as a carrier of interspecies small RNAs. However, the fungal gene-targeting trans-acting RNA species found in extracellular vesicles in one study were found in the apoplastic fluid (rather than in extracellular vesicles) in another study184,192,193, highlighting the need to establish uniform experimental standards in these studies194. The presence of apoplastic small RNAs, including select miRNAs and tasiRNAs, implies that an extracellular vesicle-independent pathway exports small RNAs from plant cells. Intriguingly, tiny RNAs (~10–17 nt long), which are probably degradation products of coding and non-coding transcripts, were the main species of small RNAs found in extracellular vesicles in one study192. Notably, 14-nt RNAs were found to enhance the endonuclease activity of human AGO3 (REF.195), raising the possibility that plant tiny RNAs are functional if in pathogens they are incorporated into an Argonaute protein with properties similar to those of human AGO3.
Natural interspecies small RNAs in worms
The existence of a molecular pathway dedicated to taking up dsRNA from the environment strongly suggests natural uptake of RNA could be happening in some circumstances in nematodes. As discussed earlier, the C. elegans protein SID-2 likely functions specifically in mediating uptake of dsRNA from the environment, since it was shown to be required for RNAi through feeding (with dsRNA-producing bacteria), but not for spreading gene silencing to different tissues (sid-2 mutants have proper systemic RNAi when the dsRNA is introduced by injection or expressed in a specific tissue under the control of tissue-specific promoters)33. Another nematode, Meloidogyne incognita, induces RNAi when it feeds on bacteria that produce dsRNA, although it has no sid-2 homologue196. A 2012 study suggested that two E. coli endogenous small RNAs, oxyS and dsrA, move to C. elegans and affect its chemosensory behaviour by downregulating the chemotaxis gene che-2 (REF.197). However, a follow-up study reported a failure to replicate these results and argued that the idea that E. coli would transmit functional RNAs to C. elegans is untenable “given that C. elegans is unlikely to encounter E. coli in nature”198. However a recent article reported that worms do learn to avoid a pathogenic bacterium, Pseudomonas aeruginosa (PA14), by taking up a P. aeruginosa-encoded non-coding RNA called ‘P11’ (FIG. 6b). The authors proposed that the bacterial RNA moves from the gut to the germ line, where it downregulates the worm gene maco-1 in a process that requires the systemic RNAi proteins SID-1 and SID-2 (REF.46). The evolution of such interspecies RNA interaction is certainly intriguing, and many new directions are open for future research47.
In addition to work in the model organism C. elegans, considerable insights into interspecies small RNAs have been gained from studies of parasitic nematodes. Heligmosomoides bakeri secretes different types of RNAs, including miRNAs and siRNAs, to manipulate its hosts. In a mouse model, these small RNAs (and also Y RNAs and nematode Argonaute proteins) are shed into the gut and modulate the immune response199. miRNAs are relatively conserved in sequence between parasites and their different hosts, and therefore it is reasonable to assume they could remain functional upon transfer. Specific exosome-carried miRNAs from the filarial nematodes Litomosoides sigmodontis and Dirofilaria immitis were detected in the serum of mouse and dog hosts200. Onchocerca species, which infect cattle and humans, similarly secrete interspecies regulatory small RNA201. Different Schistosoma parasites also secrete small RNAs in extracellular vesicles202, suggesting that interspecies small RNA secretion is conserved among parasitic nematodes, and might be used to manipulate their hosts in different ways. Whereas early studies of small RNAs secreted from parasitic nematodes focused on miRNAs, newer studies have shown that the most abundant small RNAs that the nematodes secrete are secondary (RNA-dependent RNA polymerase-amplified) siRNAs. These secreted siRNAs are enriched in siRNAs that align with transposons and newly evolved repetitive elements203.
Similarly to many parasites, parasitic nematodes often transition between multiple different hosts. For example, the lymphatic filariasis-causing nematode Brugia malayi is transmitted from mosquitoes to humans. Parasitic nematodes might use their ability to secrete small RNAs into the host to regulate genes that are required for a particular stage of their life cycle204.
Interspecies small RNAs in insects
Cases of small RNAs used by pathogens to control insect hosts have been documented in different species; they exemplify the importance of systemic RNAi in insects, but also expose this system as a liability that makes insects vulnerable to manipulation. It is likely that in some cases interspecies small RNA exchange will be used also for the benefit of the insect.
Wolbachia species, which are intracellular bacteria that infect many invertebrates and manipulate reproductive pathways, use small RNAs to control their hosts205. Two abundant Wolbachia small non-coding RNAs, WsnRNA-46 and WsnRNA-49 (not to be confused with small nuclear RNAs) were suggested to regulate both Wolbachia genes and host genes in a non-cell-autonomous manner. The authors of the study focused on regulation of the mosquito host gene Dhc, as it has perfect complementarity to the WsnRNA-46 seed region. Wolbachia-infected mosquitoes had elevated levels of Dhc, and the study authors suggested that by binding to the target mRNA, WsnRNA-46 stabilizes it205. In addition to Wolbachia, the fungal pathogen Beauveria bassiana was recently shown to use small RNAs to weaken the mosquito immune system. B. bassiana secretes a miRNA-like RNA (bba-milR1), which binds the mosquito AGO1; bba-milR1 suppresses host immunity by downregulating the Toll receptor ligand Spätzle 4 (REF.206).
Other insects can be affected by plant-derived small RNAs (host-induced gene silencing, as described earlier herein), although small RNAs have not been directly shown to be the mobile agents in these case. For example, feeding dsRNA at biologically relevant levels to the agricultural insect pest Diabrotica virgifera virgifera LeConte (western corn rootworm) induces strong RNAi responses. Such responses can be evoked by feeding the pest on corn plants expressing insect-gene-targeting dsRNA207,208. For the response to be effective, the dsRNA needs to be longer than 50–60 base pairs208, a requirement that mirrors what is required to induce RNAi in C. elegans by feeding. Also consistent with what has been found in C. elegans, plant endogenous miRNAs do not induce RNAi in the pest when it is fed on corn roots. However, whereas SID-2 is required for RNAi induction by feeding in C. elegans, a SID-2 orthologue was never discovered in the pest209. Moreover, both western corn rootworm and another coleopteran species, Colorado potato beetle (Leptinotarsa decemlineata Say), take up many endogenous long dsRNAs through feeding (from corn and tomatoes, respectively). The same effects were not observed in other insects, which are less sensitive to RNAi (Spodoptera frugiperda (J.E. Smith) and Helicoverpa zea (Boddie))210.
In bees, RNAi functions systemically, and dsRNA is transmitted in royal jelly, as described earlier herein77. Recently it was shown that plant miRNAs present in the worker bee’s food delay larval development211. Furthermore, bees can transfer ingested dsRNA to the mite Varroa destructor (an obligatory ectoparasite), and dsRNA can transfer back from the mite to parasitized bees. Reciprocal exchange of gene-silencing RNAs between the bee and its parasite enhances RNAi and decreases the mite population212.
Interspecies small RNAs in mammals
Malaria resistance in some humans correlates with secretion of some miRNAs into the parasite Plasmodium falciparum213. The authors of the study suggested that this interspecies transfer of RNA induces gene regulation in the parasite in a non-canonical way by impairing ribosome loading on mRNA and translation. It was also demonstrated that blood cells release human AGO2 bound to human miRNAs, and that AGO2–miR-451/140 silenced the expression of the essential malaria antigen, PfEMP1 (REF.214). Similarly, in both humans and mice, secretion of miRNAs in the gut affects gene regulation in bacteria (through non-canonical mechanisms). Cell-specific knockout of Dicer1, which is required for miRNA processing, revealed intestinal epithelial cells and HOPX-positive cells as the source of the secreted miRNAs215.
Can mammals take up functional small RNAs from other organisms? As discussed already, there is convincing evidence to support the claim that different parasitic nematodes secrete small RNAs that enter mammalian cells and modulate the immune response199. This interspecies exchange is facilitated by the fact that animal miRNAs are relatively conserved and nematode miRNAs share seed sites with mammalian miRNAs. In addition, worms secrete worm specific Argonaute proteins (WAGOs) preloaded with small RNAs to utilize their own amplified siRNAs and regulate host genes; this pathway is expected to be less conserved compared with the miRNAs203.
In addition to absorption of small RNAs from parasites, more debated studies proposed that functional miRNAs can be obtained from food. One study suggested that exogenous miRNAs derived from ingested plants are functional in the host and can downregulate gene expression in the liver through RNAi216. Recently, SID1 transmembrane family member 1 (SIDT1) expressed in the stomach was reported to be required for taking up dietary miRNAs, and orally administered plant-derived miR-2911 was reported to function in the liver and protect against fibrosis in a SIDT1-dependent manner217.
The existence of plant-derived exogenous miRNAs, however, is still in doubt, for a number of reasons. First, systemic RNAi is inefficient (or does not exist) in mammals, and therefore it is unclear how the foreign miRNAs would travel to the liver from the intestinal lumen. Second, it was suggested that plant miRNAs will not be functional in mammals, because unlike mammalian miRNAs, plant miRNAs are 3′ methylated and mammalian Argonaute proteins do not bind methylated small RNAs218. Third, the amounts of digested plant miRNAs were suggested to be too low to induce silencing in a non-cell-autonomous manner219. Another study220 failed to replicate the results of the study on exogenous functional plant miRNAs220, and feeding experiments with monkeys (Macaca nemestrina) did not detect xenogenic miRNAs in their blood219. The surrounding debate was comprehensively reviewed in REF.219.
Interspecies small RNAs in cephalopods
A recent study demonstrated that small RNAs from the bacterium Vibrio fischeri transfer to the squid Euprymna scolopes, where they have an important function in the symbiosis between these two organisms. The bacterial small RNA ssrA is released in membrane vesicles to the epithelial cells in the light organ of the squid. When squids are colonized with ssrA-mutant bacteria, the light organ deregulates genes encoding heightened immune function or antimicrobial activities221.
Conclusions
In this Review, we ask whether, aside from regulating genes inside their cell of origin, small RNAs have additional, perhaps different functions in other tissues or other organisms. Numerous examples exist, but for small RNAs to remain functional upon movement, at least some of the following requirements must be fulfilled51.
The small RNAs in the cell of origin need to be sorted and prepared for delivery.
These selected small RNAs must pass through the plasma membrane, the cell wall, a channel providing direct connection to another cell, a cytoplasmic bridge or a membranous nanotube.
Throughout their travel, the small RNAs need to be protected from degradation.
The mobile small RNAs need to penetrate the recipient cell and reach a cellular milieu in which they could function.
The machinery in the recipient cells must be compatible with the mobile small RNAs (this is especially important in the case of small RNAs movement between different organisms).
The small RNAs need to hybridize with target RNAs, either in the cytoplasm (to enable post-transcriptional gene silencing) or in the nucleus (to enforce transcriptional gene silencing).
It is important to note, however, that an adopted small RNA can in theory remain functional and affect gene expression or other physiological properties of the recipient cells not by inducing RNAi but through interactions with RNA-binding proteins or by influencing some other biochemical reaction indirectly. Although it is convenient to think of these six ‘requirements’, it is important to realize that not all these steps are necessarily required. For example, cell death might lead to non-specific spilling of many cellular components, including small RNAs, and some of these small RNAs might be taken up passively by other tissues, where they could have adaptive or non-adaptive effects (for example, trigger a non-sequence-specific immune response)222. Such a scenario is possible in plant–pathogen interactions, as plant cell death is often accompanied by such exchanges. Finally, it is important to note that presence (in recipient cells) does not equate to function. Very low levels of small RNAs can easily be detected using sensitive methods such as stem–loop reverse transcription–PCR or small RNA sequencing, but one needs to be mindful that certain threshold levels need to be achieved for the small RNAs to be functional.
Post-transcriptional gene silencing.
Silencing of genetic elements usually through RNA degradation or the inhibition of their translation.
Non-cell-autonomous RNA-mediated gene silencing.
RNA-mediated gene silencing in which the trigger for silencing is delivered to, or originates from, cells that are different from those that undergo gene silencing.
Symplasm.
The ‘shared’ cytoplasm of nearly all plant cells due to the presence of the plasmodesmata connecting adjacent plant cells.
Apoplasm.
The extracellular (outside the plasma membrane) environment consisting of the cell wall and other molecules therein.
Phloem.
A tissue in the plant vasculature that transmits nutrients and other molecules to distant plant parts.
siRNA–siRNA* duplexes.
Also miRNA–miRNA* duplexes, products of RNA processing by Dicer, consisting of a duplex of small RNAs with an overhang on each strand. The strand that is not loaded into an Argonaute protein is marked with an asterisk.
siRNA bodies.
In Arabidopsis thaliana, cytoplasmic foci containing factors involved in the production of endogenous siRNAs, such as ARGONAUTE 7 (AGO7), SUPPRESSOR OF GENE SiLENCiNG 3 (SGS3) and RNA-DEPENDENT RNA POLYMERASE 6 (RDR6).
Apoplastic fluid.
The fluid in the plant apoplasm that can be collected through centrifugation.
Transcriptional gene silencing.
Silencing of genetic elements at the transcriptional level, usually through the deposition of repressive histone modifications or DNA methylation.
Acknowledgements
The authors are grateful to N. Alon for help with the illustrations. Research on small RNAs in the Chen laboratory is funded by the US National Institutes of Health (GM129373). The Rechavi laboratory is funded by ERC grant no. 819151.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Fagard M & Vaucheret H (Trans)Gene silencing in plants: how many mechanisms? Annu Rev. Plant Physiol. Plant Mol. Biol 51, 167–194 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Palauqui JC, Hlmayan I, Pollien JM & Vaucheret H Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 16, 4738–4745 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voinnet O & Baulcombe DC Systemic signalling in gene silencing. Nature 389, 553 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Fire A et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Timmons L & Fire A Specific interference by ingested dsRNA. Nature 395, 854 (1998). [DOI] [PubMed] [Google Scholar]
- 6.Baulcombe DC VIGS, HIGS and FIGS: small RNA silencing in the interactions of viruses or filamentous organisms with their plant hosts. Curr. Opin. Plant Biol 26, 141–146 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Koch A & Kogel KH New wind in the sails: improving the agronomic value of crop plants through RNAi-mediated gene silencing. Plant Biotechnol. J 12, 821–831 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Petit JD, Li ZP, Nicolas WJ, Grison MS & Bayer EM Dare to change, the dynamics behind plasmodesmata-mediated cell-to-cell communication. Curr. Opin. Plant Biol 53, 80–89 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Esau K & Thorsch J Sieve plate pores and plasmodesmata, the communication channels of the symplast: ultrastructural aspects and developmental relations. Am. J. Bot 72, 1641–1653 (1985). [Google Scholar]
- 10.Ross-Elliott TJ et al. Phloem unloading in Arabidopsis roots is convective and regulated by the phloem-pole pericycle. eLife 6, e24125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen W et al. A genetic network for systemic RNA silencing in plants. Plant Physiol. 176, 2700–2719 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melnyk CW, Molnar A, Bassett A & Baulcombe, D. C. Mobile 24 nt small RNAs direct transcriptional gene silencing in the root meristems of Arabidopsis thaliana. Curr. Biol 21, 1678–1683 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Molnar A et al. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 328, 872–875 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Taochy C et al. A genetic screen for impaired systemic RNAi highlights the crucial role of DICER-LIKE 2. Plant Physiol. 175, 1424–1437 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brosnan CA et al. Nuclear gene silencing directs reception of long-distance mRNA silencing in Arabidopsis. Proc. Natl Acad. Sci. USA 104, 14741–14746 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunoyer P, Himber C, Ruiz-Ferrer V, Alioua A & Voinnet O Intra- and intercellular RNA interference in Arabidopsis thaliana requires components of the microRNA and heterochromatic silencing pathways. Nat. Genet 39, 848–856 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Schwach F, Vaistij FE, Jones L & Baulcombe DC An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 138, 1842–1852 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith LM et al. An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell 19, 1507–1521 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen HM et al. 22-nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl Acad. Sci. USA 107, 15269–15274 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cuperus JT et al. Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol 17, 997–1003 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mlotshwa S et al. DICER-LIKE2 plays a primary role in transitive silencing of transgenes in Arabidopsis. PLoS ONE 3, e1755 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parent JS, Bouteiller N, Elmayan T & Vaucheret H Respective contributions of Arabidopsis DCL2 and DCL4 to RNA silencing. Plant J. 81, 223–232 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Borges F & Martienssen RA The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol 16, 727–741 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez G et al. Paternal easiRNAs regulate parental genome dosage in Arabidopsis. Nat. Genet 50, 193–198 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Panda K, McCue AD & Slotkin RK Arabidopsis RNA polymerase IV generates 21-22 nucleotide small RNAs that can participate in RNA-directed DNA methylation and may regulate genes. Phil. Trans.R. Soc. B 375, 20190417 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voinnet O, Vain P, Angell S & Baulcombe DC Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell 95, 177–187 (1998). [DOI] [PubMed] [Google Scholar]
- 27.Devers EA et al. Movement and differential consumption of short interfering RNA duplexes underlie mobile RNA interference. Nat. Plants 6, 789–799 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Brosnan CA et al. Genome-scale, single-cell-type resolution of microRNA activities within a whole plant organ. EMBO J. 38, e100754 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su Z et al. Regulation of female germline specification via small RNA mobility in Arabidopsis. Plant Cell 32, 2842–2854 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosas-Diaz T et al. A virus-targeted plant receptor-like kinase promotes cell-to-cell spread of RNAi. Proc. Natl Acad. Sci. USA 115, 1388–1393 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tijsterman M, May RC, Simmer F, Okihara KL & Plasterk R H. Genes required for systemic RNA interference in Caenorhabditis elegans. Curr. Biol 14, 111–116 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Timmons L, Tabara H, Mello CC & Fire AZ Inducible systemic RNA silencing in Caenorhabditis elegans. Mol. Biol. Cell 14, 2972–2983 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winston WM, Sutherlin M, Wright AJ, Feinberg EH & Hunter CP Caenorhabditis elegans SID-2 is required for environmental RNA interference. Proc. Natl Acad. Sci. USA 104, 10565–10570 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winston WM, Molodowitch C & Hunter CP Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295, 2456–2459 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Jose AM, Smith JJ & Hunter CP Export of RNA silencing from C. elegans tissues does not require the RNA channel SID-1. Proc. Natl Acad. Sci. USA 106, 2283–2288 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shih JD & Hunter C P SID-1 is a dsRNA-selective dsRNA-gated channel. RNA 17, 10, 1165–57 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marré J, Traver EC & Jose AM Extracellular RNA is transported from one generation to the next in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 113, 12496–12501 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whangbo JS, Weisman AS, Chae J & Hunter CP SID-1 domains Important for dsRNA import in Caenorhabditis elegans. G3 7, 3887–3899 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Felix MA et al. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 9, e1000586 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gammon DB et al. The antiviral RNA interference response provides resistance to lethal Arbovirus infection and vertical transmission in Caenorhabditis elegans. Curr. Biol 27, 795–806 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rechavi O, Minevich G & Hobert O. Transgenerational inheritance of an acquired small RNA-based antiviral response in C. elegans. Cell 147, 1248–1256 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhuang JJ & Hunter CP Tissue specificity of Caenorhabditis elegans enhanced RNA interference mutants. Genetics 188, 235–237 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McEwan DL, Weisman AS & Hunter CP Uptake of extracellular double-stranded RNA by SID-2. Mol. Cell 47, 746–754 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Braukmann F, Jordan D, Jenkins B, Koulman A & Miska EA SID-2 negatively regulates development likely independent of nutritional dsRNA uptake.RNA Biol. 18, 888–899 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nuez I & Felix MA Evolution of susceptibility to ingested double-stranded RNAs in Caenorhabditis nematodes. PLoS ONE 7, e29811 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaletsky R et al. C. elegans interprets bacterial noncoding RNAs to learn pathogenic avoidance. Nature 586, 445–451 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rechavi O Transgenerational inheritance: that pathogen gut feeling. Curr. Biol 30, R1486–R1488 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Jose AM, Kim YA, Leal-Ekman S & Hunter CP Conserved tyrosine kinase promotes the import of silencing RNA into Caenorhabditis elegans cells. Proc. Natl Acad. Sci. USA 109, 14520–14525 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhatia S & Hunter C P SID-4/NCK-1 is important for dsRNA import in Caenorhabditis elegans. Preprint at bioRxiv 10.1101/702019 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hinas A, Wright AJ & Hunter CP SID-5 is an endosome-associated protein required for efficient systemic RNAi in C. elegans. Curr. Biol 22, 1938–1943 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jose AM Movement of regulatory RNA between animal cells. Genesis 53, 395–416 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Imae R, Dejima K, Kage-Nakadai E, Arai H & Mitani S Endomembrane-associated RSD-3 is important for RNAi induced by extracellular silencing RNA in both somatic and germ cells of Caenorhabditis elegans. Sci. Rep 6, 28198 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang E & Hunter C P SID-1 functions in multiple roles to support parental RNAi in Caenorhabditis elegans. Genetics 207, 547–557 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Preston MA et al. Unbiased screen of RNA tailing activities reveals a poly(UG) polymerase. Nat. Methods 16, 437–445 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shukla A et al. Poly(UG)-tailed RNAs in genome protection and epigenetic inheritance. Nature 582, 283–288 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dzitoyeva S, Dimitrijevic N & Manev H Intra-abdominal injection of double-stranded RNA into anesthetized adult Drosophila triggers RNA interference in the central nervous system. Mol. Psychiatry 6, 665–670 (2001). [DOI] [PubMed] [Google Scholar]
- 57.Goto A, Blandin S, Royet J, Reichhart JM & Levashina EA Silencing of Toll pathway components by direct injection of double-stranded RNA into Drosophila adult flies. Nucleic Acids Res. 31, 6619–6623 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petruk S et al. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell 127, 1209–1221 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miller SC, Brown SJ & Tomoyasu Y Larval RNAi in Drosophila? Dev. Genes Evol 218, 505–510 (2008). [DOI] [PubMed] [Google Scholar]
- 60.Tomoyasu Y et al. Exploring systemic RNA interference in insects: a genome-wide survey for RNAi genes in Tribolium. Genome Biol. 9, R10 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saleh M-C et al. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat. Cell Biol 8, 793–802 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ulvila J et al. Double-stranded RNA is internalized by scavenger receptor-mediated endocytosis in Drosophila S2 cells. J. Biol. Chem 281, 14370–14375 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Xiao D et al. Clathrin-dependent endocytosis plays a predominant role in cellular uptake of double-stranded RNA in the red flour beetle. Insect Biochem. Mol. Biol 60, 68–77 (2015). [DOI] [PubMed] [Google Scholar]
- 64.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD & Remaley AT MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol 13, 423–433 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karlikow M et al. Drosophila cells use nanotube-like structures to transfer dsRNA and RNAi machinery between cells. Sci. Rep 6, 27085 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vance V & Vaucheret H RNA silencing in plants–defense and counterdefense. Science 292, 2277–2280 (2001). [DOI] [PubMed] [Google Scholar]
- 67.Gammon DB Caenorhabditis elegans as an emerging model for virus-host interactions. J Virol. 91, e00509–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Garcia-Ruiz H et al. Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip Mosaic Virus infection. Plant Cell 22, 481–496 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang XB et al. RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 107, 484–489 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saleh M-C et al. Antiviral immunity in Drosophila requires systemic RNA interference spread. Nature 458, 346–350 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tassetto M, Kunitomi M & Andino R Circulating immune cells mediate a systemic RNAi-based adaptive antiviral response in Drosophila. Cell 169, 314–325.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Goic B et al. RNA-mediated interference and reverse transcription control the persistence of RNA viruses in the insect model Drosophila. Nat. immunol 14, 396–403 (2013). [DOI] [PubMed] [Google Scholar]
- 73.Goic B et al. Virus-derived DNA drives mosquito vector tolerance to arboviral infection. Nat. Commun 7, 12410 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Poirier EZ et al. Dicer-2-dependent generation of viral DNA from defective genomes of RNA viruses modulates antiviral immunity in insects. Cell Host Microbe 23, 353–365.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Suzuki Y et al. Non-retroviral endogenous viral element limits cognate virus replication in Aedes aegypti ovaries. Curr. Biol 30, 3495–3506.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maori E et al. IAPV, a bee-affecting virus associated with colony collapse disorder can be silenced by dsRNA ingestion. Insect Mol. Biol 18, 55–60 (2009). [DOI] [PubMed] [Google Scholar]
- 77.Maori E et al. A secreted RNA binding protein forms RNA-stabilizing granules in the honeybee royal jelly. Mol. Cell 74, 598–608.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maori E et al. A transmissible RNA pathway in honey bees. Cell Rep. 27, 1949–1959.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 79.Houri-Zeevi L & Rechavi O A matter of time: small RNAs regulate the duration of epigenetic inheritance. Trends Genet. 33, 46–57 (2017). [DOI] [PubMed] [Google Scholar]
- 80.Zhuang JJ & Hunter CP The influence of competition among C. elegans small RNA pathways on development. Genes 3, 671–685 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sarkies P, Ashe A, Le Pen J, McKie MA& Miska EA Competition between virus-derived and endogenous small RNAs regulates gene expression in Caenorhabditis elegans. Genome Res. 23, 1258–1270 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.You C et al. FIERY1 promotes microRNA accumulation by suppressing rRNA-derived small interfering RNAs in Arabidopsis. Nat. Commun 10, 4424 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wahba L, Hansen L & Fire AZ An essential role for the piRNA pathway in regulating the ribosomal RNA pool in C. elegans. Dev. Cell 56, 2295–2312.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li S et al. Unidirectional movement of small RNAs from shoots to roots in interspecific heterografts. Nat. Plants 7, 50–59 (2021). [DOI] [PubMed] [Google Scholar]
- 85.Leslie M NIH effort gambles on mysterious extracellular RNAs. Science 341, 947 (2013). [DOI] [PubMed] [Google Scholar]
- 86.Posner R et al. Neuronal small RNAs control behavior transgenerationally. Cell 177, 1814–1826.e15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu Y, Teng C, Xia R & Meyers BC PhasiRNAs in plants: their biogenesis, genic sources, and roles in stress responses, development, and reproduction. Plant Cell 32, 3059–3080 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Garcia D, Collier SA, Byrne ME & Martienssen RA Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr. Biol 16, 933–938 (2006). [DOI] [PubMed] [Google Scholar]
- 89.Nogueira FT, Madi S, Chitwood DH, Juarez MT & Timmermans MC Two small regulatory RNAs establish opposing fates of a developmental axis. Genes Dev. 21, 750–755 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xu L et al. Genetic interaction between the AS1-AS2 and RDR6-SGS3-AGO7 pathways for leaf morphogenesis. Plant Cell Physiol. 47, 853–863 (2006). [DOI] [PubMed] [Google Scholar]
- 91.Adenot X et al. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol 16, 927–932 (2006). [DOI] [PubMed] [Google Scholar]
- 92.Fahlgren N et al. Regulation of AUXIN RESPONSE FACTORS by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol 16, 939–944 (2006). [DOI] [PubMed] [Google Scholar]
- 93.Hunter C et al. Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 133, 2973–2981 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Peragine A, Yoshikawa M, Wu G, Albrecht HL& Poethig RS SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes. Dev 18, 2368–2379 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yoshikawa M, Peragine A, Park MY & Poethig RS A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes. Dev 19, 2164–2175 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hobecker KV et al. The microRNA390/TAS3 pathway mediates symbiotic nodulation and lateral root growth. Plant Physiol. 174, 2469–2486 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Marin E et al. miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 22, 1104–1117 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yoon EK et al. Auxin regulation of the microRNA390-dependent transacting small interfering RNA pathway in Arabidopsis lateral root development. Nucleic Acids Res. 38, 1382–1391 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li X et al. The REL3-mediated TAS3 ta-siRNA pathway integrates auxin and ethylene signaling to regulate nodulation in Lotus japonicus. N. Phytol 201, 531–544 (2014). [DOI] [PubMed] [Google Scholar]
- 100.Nagasaki H et al. The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proc. Natl Acad. Sci. USA 104, 14867–14871 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Su Z et al. The THO complex non-cell-autonomously represses female germline specification through the TAS3-ARF3 module. Curr. Biol 27, 1597–1609.e2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ding B et al. Developmental genetics of corolla tube formation: role of the tasiRNA-ARF pathway and a conceptual model. Plant Cell 32, 3452–3468 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chitwood DH et al. Pattern formation via small RNA mobility. Genes Dev. 23, 549–554 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Johnson C et al. Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res. 19, 1429–1440 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yang X et al. Widespread occurrence of microRNA-mediated target cleavage on membrane-bound polysomes. Genome Biol. 22, 15 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhai J et al. Spatiotemporally dynamic, cell-type-dependent premeiotic and meiotic phasiRNAs in maize anthers. Proc. Natl Acad. Sci. USA 112, 3146–3151 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Araki S et al. miR2118-dependent U-rich phasiRNA production in rice anther wall development. Nat. Commun 11, 3115 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fan Y et al. PMS1T, producing phased small-interfering RNAs, regulates photoperiod-sensitive male sterility in rice. Proc. Natl Acad. Sci. USA 113, 15144–15149 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tamim S et al. Cis-directed cleavage and nonstoichiometric abundances of 21-nucleotide reproductive phased small interfering RNAs in grasses. N. Phytol 220, 865–877 (2018). [DOI] [PubMed] [Google Scholar]
- 110.Zhou H et al. Photoperiod- and thermo-sensitive genic male sterility in rice are caused by a point mutation in a novel noncoding RNA that produces a small RNA. Cell Res. 22, 649–660 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Komiya R et al. Rice germline-specific Argonaute MEL1 protein binds to phasiRNAs generated from more than 700 lincRNAs. Plant J. 78, 385–397 (2014). [DOI] [PubMed] [Google Scholar]
- 112.Teng C et al. Dicer-like 5 deficiency confers temperature-sensitive male sterility in maize. Nat. Commun 11, 2912 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jiang P et al. 21-nt phasiRNAs direct target mRNA cleavage in rice male germ cells. Nat. Commun 11, 5191 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tian P et al. Evolution and diversification of reproductive phased small interfering RNAs in Oryza species. N. Phytol 229, 2970–2983 (2021). [DOI] [PubMed] [Google Scholar]
- 115.Zhang YC et al. Reproductive phasiRNAs regulate reprogramming of gene expression and meiotic progression in rice. Nat. Commun 11, 6031 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kasschau KD et al. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 5, e57 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Long J et al. Nurse cell–derived small RNAs define paternal epigenetic inheritance in Arabidopsis. Science 373, eabh0556 (2021). [DOI] [PubMed] [Google Scholar]
- 118.Slotkin RK et al. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136, 461–472 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Martinez G, Panda K, Kohler C & Slotkin RK Silencing in sperm cells is directed by RNA movement from the surrounding nurse cell. Nat. Plants 2, 16030 (2016). [DOI] [PubMed] [Google Scholar]
- 120.Klesen S, Hill K & Timmermans MC P Small RNAs as plant morphogens. Curr. Top. Dev. Biol 137, 455–480 (2020). [DOI] [PubMed] [Google Scholar]
- 121.Yao X et al. Two types of cis-acting elements control the abaxial epidermis-specific transcription of the MIRI65a and MIR166a genes. FEBS Lett. 583, 3711–3717 (2009). [DOI] [PubMed] [Google Scholar]
- 122.Emery JF et al. Radial patterning of Arabidopsis shoots by class III HD-ZlP and KANADI genes. Curr. Biol 13, 1768–1774 (2003). [DOI] [PubMed] [Google Scholar]
- 123.Juarez MT, Kui JS, Thomas J, Heller BA & Timmermans MC microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428, 84–88 (2004). [DOI] [PubMed] [Google Scholar]
- 124.McConnell JR et al. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713 (2001). [DOI] [PubMed] [Google Scholar]
- 125.Carlsbecker A et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465, 316–321 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Miyashima S, Koi S, Hashimoto T & Nakajima K Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development 138, 2303–2313 (2011). [DOI] [PubMed] [Google Scholar]
- 127.Knauer S et al. A protodermal miR394 signal defines a region of stem cell competence in the Arabidopsis shoot meristem. Dev. Cell 24, 125–132 (2013). [DOI] [PubMed] [Google Scholar]
- 128.Buhtz A, Springer F, Chappell L, Baulcombe DC & Kehr J Identification and characterization of small RNAs from the phloem of Brassica napus. Plant J. 53, 739–749 (2008). [DOI] [PubMed] [Google Scholar]
- 129.Yoo BC et al. A systemic small RNA signaling system in plants. Plant Cell 16, 1979–2000 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lin SI et al. Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol. 147, 732–746 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Buhtz A, Pieritz J, Springer F & Kehr J Phloem small RNAs, nutrient stress responses, and systemic mobility. BMC Plant Biol. 10, 64 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Okuma N, Soyano T, Suzaki T & Kawaguchi M MIR2111-5 locus and shoot-accumulated mature miR2111 systemically enhance nodulation depending on HARI in Lotus japonicus. Nat. Commun 11, 5192 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tsikou D et al. Systemic control of legume susceptibility to rhizobial infection by a mobile microRNA. Science 362, 233–236 (2018). [DOI] [PubMed] [Google Scholar]
- 134.Skopelitis DS, Benkovics AH, Husbands AY & Timmermans MCP Boundary formation through a direct threshold-based readout of mobile small RNA gradients. Dev. Cell 43, 265–273.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 135.Vaten A et al. Callose biosynthesis regulates symplastic trafficking during root development. Dev Cell 21, 1144–1155 (2011). [DOI] [PubMed] [Google Scholar]
- 136.Skopelitis DS et al. Gating of miRNA movement at defined cell-cell interfaces governs their impact as positional signals. Nat. Commun 9, 3107 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Liu Q et al. The ARGONAUTE10 gene modulates shoot apical meristem maintenance and establishment of leaf polarity by repressing miR165/166 in Arabidopsis. Plant J. 58, 27–40 (2009). [DOI] [PubMed] [Google Scholar]
- 138.Zhu H et al. Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell 145, 242–256 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cai H et al. Spatiotemporal control of miR398 biogenesis via chromatin remodeling and kinase signaling ensures proper ovule development. Plant Cell 33, 1530–1553 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.de Felippes FF, Ott F & Weigel D Comparative analysis of non-autonomous effects of tasiRNAs and miRNAs in Arabidopsis thaliana. Nucleic Acids Res. 39, 2880–2889 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bologna NG et al. Nucleo-cytosolic shuttling of ARGONAUTE1 prompts a revised model of the plant microRNA pathway. Mol. Cell 69, 709–719.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 142.Li S et al. Biogenesis of phased siRNAs on membrane-bound polysomes in Arabidopsis. eLife 5, e22750 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Jouannet V et al. Cytoplasmic Arabidopsis AGO7 accumulates in membrane-associated siRNA bodies and is required for ta-siRNA biogenesis. EMBO J. 31, 1704–1713 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dalmadi A, Gyula P, Balint J, Szittya G &Havelda Z AGO-unbound cytosolic pool of mature miRNAs in plant cells reveals a novel regulatory step at AGO1 loading. Nucleic Acids Res. 47, 9803–9817 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ow MC, Borziak K, Nichitean AM, Dorus S& Hall SE Early experiences mediate distinct adult gene expression and reproductive programs in Caenorhabditis elegans. PLOS Genet. 14, e1007219 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ohno H & Bao Z Small RNAs couple embryonic developmental programs to gut microbes. Preprint at bioRxiv 10.1101/2020.11.13.381830 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Long OS et al. A C. elegans model of human α1-antitrypsin deficiency links components of the RNAi pathway to misfolded protein turnover. Hum. Mol. Genet 23, 5109–5122 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Setten RL, Rossi JJ & Han S-P The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov 18, 421–446 (2019). [DOI] [PubMed] [Google Scholar]
- 149.Valadi H et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol 9, 654–659 (2007). [DOI] [PubMed] [Google Scholar]
- 150.Shurtleff MJ, Temoche-Diaz MM, Karfilis KV,Ri S & Schekman R Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. eLife 5, e19276 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.O’Brien K, Breyne K, Ughetto S, Laurent LC & Breakefield XO RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol 21, 585–606 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Thomou T et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 542, 450–455 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Witwer KW et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles 2, 20360 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kim Y-K, Yeo J, Kim B, Ha M & Kim VN Short structured RNAs with low GC content are selectively lost during extraction from a small number of cells. Mol. Cell 46, 893–895 (2012). [DOI] [PubMed] [Google Scholar]
- 155.Ostrowski M et al. Rab27a and Rab27b control different steps of the exosome secretion pathway.Nat. Cell Biol 12, 19–30 (2010). [DOI] [PubMed] [Google Scholar]
- 156.Chevillet JR et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes.Proc. Natl Acad. Sci. USA 111, 14888–14893 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Das S et al. The Extracellular RNA Communication Consortium: establishing foundational knowledge and technologies for extracellular RNA research. Cell 177, 231–242 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Srinivasan S et al. Small RNA sequencing across diverse biofluids identifies optimal methods for exRNA isolation. Cell 177, 446–462.e16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhang H et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol 20, 332–343 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Shurtleff MJ et al. Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc. Natl Acad. Sci. USA 114, E8987–E8995 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Villarroya-Beltri C et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun 4, 2980 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bolukbasi MF et al. miR-1289 and “zipcode”-like sequence enrich mRNAs in microvesicles. Mol. Ther. Nucleic Acids 1, e10 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Koppers-Lalic D et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 8, 1649–1658 (2014). [DOI] [PubMed] [Google Scholar]
- 164.Clancy JW, Zhang Y, Sheehan C & D’Souza-Schorey C An ARF6–exportin-5 axis delivers pre-miRNA cargo to tumour microvesicles. Nat. Cell Biol 21, 856–866 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Leidal AM et al. The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat. Cell Biol 22, 187–199 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.McKenzie AJ et al. KRAS-MEK signaling controls Ago2 sorting into exosomes. Cell Rep. 15, 978–987 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Matsuzaki J & Ochiya T Extracellular microRNAs and oxidative stress in liver injury: a systematic mini review. J. Clin. Biochem. Nutr 63, 6–11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Beninson LA et al. Acute stressor exposure modifies plasma exosome-associated heat shock protein 72 (Hsp72) and microRNA (miR-142-5p and miR-203). PLoS ONE 9, e108748 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ma C et al. Moderate exercise enhances endothelial progenitor cell exosomes release and function. Med. Sci. Sports Exerc 50, 2024–2032 (2018). [DOI] [PubMed] [Google Scholar]
- 170.Arroyo JD et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl Acad. Sci. USA 108, 5003–5008 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Turchinovich A, Weiz L, Langheinz A & Burwinkel B Characterization of extracellular circulating microRNA. Nucleic Acids Res. 39, 7223–7233 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wang K, Zhang S, Weber J, Baxter D &Galas DJ Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 38, 7248–7259 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Rechavi O et al. Trans-SILAC: sorting out the non-cell-autonomous proteome. Nat. Methods 7, 923–927 (2010). [DOI] [PubMed] [Google Scholar]
- 174.Rechavi O et al. Cell contact-dependent acquisition of cellular and viral nonautonomously encoded small RNAs. Genes Dev. 23, 1971–1979 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Valiunas V et al. Connexin-specific cell-to-cell transfer of short interfering RNA by gap junctions: oligonucleotide and siRNA transfer through gap junctions. J. Physiol 568, 459–468 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kizana E, Cingolani E & Marbán E Non-cell-autonomous effects of vector-expressed regulatory RNAs in mammalian heart cells. Gene Ther. 16, 1163–1168 (2009). [DOI] [PubMed] [Google Scholar]
- 177.Katakowski M, Buller B, Wang X, Rogers T & Chopp M Functional microRNA is transferred between glioma cells. Cancer Res. 70, 8259–8263 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lim PK et al. Gap junction-mediated import of microRNA from bone marrow stromal cells can elicit cell cycle quiescence in breast cancer cells. Cancer Res. 71, 1550–1560 (2011). [DOI] [PubMed] [Google Scholar]
- 179.Lu JJ, Yang WM, Li F, Zhu W & Chen Z Tunneling nanotubes mediated microRNA-155 intercellular transportation promotes bladder cancer cells’ invasive and proliferative capacity. Int. J. Nanomed 14, 9731–9743 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Morales CR et al. A TB-RBP and Ter ATPase complex accompanies specific mRNAs from nuclei through the nuclear pores and into intercellular bridges in mouse male germ cells. Dev. Biol 246, 480–494 (2002). [DOI] [PubMed] [Google Scholar]
- 181.Bermúdez-Barrientos JR, Ramírez-Sánchez O, Chow FW-N, Buck AH & Abreu-Goodger C Disentangling sRNA-Seq data to study RNA communication between species. Nucleic Acids Res. 48, e21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Shahid S et al. MicroRNAs from the parasitic plant Cuscuta campestris target host messenger RNAs. Nature 553, 82–85 (2018). [DOI] [PubMed] [Google Scholar]
- 183.Zhang T et al. Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat. Plants 2, 16153 (2016). [DOI] [PubMed] [Google Scholar]
- 184.Cai Q et al. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 360, 1126–1129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Dunker F et al. Oomycete small RNAs bind to the plant RNA-induced silencing complex for virulence. eLife 9, e56096 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Hou Y et al. A Phytophthora effector suppresses trans-kingdom RNAi to promote disease susceptibility. Cell Host Microbe 25, 153–165.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Ren B, Wang X, Duan J & Ma J Rhizobial tRNA-derived small RNAs are signal molecules regulating plant nodulation. Science 365, 919–922 (2019). [DOI] [PubMed] [Google Scholar]
- 188.Wang B et al. Puccinia striiformis f. sp. tritici microRNA-like RNA 1 (Pst-milR1), an important pathogenicity factor of Pst, impairs wheat resistance to Pst by suppressing the wheat pathogenesis-related 2 gene. N. Phytol 215, 338–350 (2017). [DOI] [PubMed] [Google Scholar]
- 189.Weiberg A et al. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Qiao Y et al. Oomycete pathogens encode RNA silencing suppressors. Nat. Genet 45, 330–333 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Rutter BD & Innes RW Extracellular vesicles isolated from the leaf apoplast carry stress-response proteins. Plant Physiol. 173, 728–741 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Baldrich P et al. Plant extracellular vesicles contain diverse small RNA species and are enriched in 10- to 17-nucleotide “tiny” RNAs. Plant Cell 31, 315–324 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.He B et al. RNA-binding proteins contribute to small RNA loading in plant extracellular vesicles. Nat. Plants 7, 342–352 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Rutter BD & Innes RW Growing pains: addressing the pitfalls of plant extracellular vesicle research. N. Phytol 228, 1505–1510 (2020). [DOI] [PubMed] [Google Scholar]
- 195.Park MS, Sim G, Kehling AC & Nakanishi K Human Argonaute2 and Argonaute3 are catalytically activated by different lengths of guide RNA. Proc. Natl Acad. Sci. USA 117, 28576–28578 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Maule AG et al. An eye on RNAi in nematode parasites. Trends Parasitol. 27, 505–513 (2011). [DOI] [PubMed] [Google Scholar]
- 197.Liu H et al. Escherichia coli noncoding RNAs can affect gene expression and physiology of Caenorhabditis elegans. Nat. Commun 3, 1073 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Akay A, Sarkies P & Miska EA E.coli OxyS non-coding RNA does not trigger RNAi in C. elegans. Sci. Rep 5, 9597 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Buck AH et al. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat. Commun 5, 5488 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Quintana JF et al. Comparative analysis of small RNAs released by the filarial nematode Litomosoides sigmodontis in vitro and in vivo. PLoS Neg. Trop. Dis 13, e0007811 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Quintana JF et al. Extracellular Onchocerca-derived small RNAs in host nodules and blood. Parasites Vectors 8, 58 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Nowacki FC et al. Protein and small non-coding RNA-enriched extracellular vesicles are released by the pathogenic blood fluke Schistosoma mansoni.J. Extracell. Vesicles 4, 28665 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Chow FW-N et al. Secretion of an Argonaute protein by a parasitic nematode and the evolution of its siRNA guides. Nucleic Acids Res. 47, 3594–3606 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Sarkies P & Miska EA Molecular biology. Is there social RNA? Science 341, 467–468 (2013). [DOI] [PubMed] [Google Scholar]
- 205.Mayoral JG et al. Wolbachia small noncoding RNAs and their role in cross-kingdom communications. Proc. Natl Acad. Sci. USA 111, 18721–18726 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Cui C et al. A fungal pathogen deploys a small silencing RNA that attenuates mosquito immunity and facilitates infection. Nat. Commun 10, 4298 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Baum JA et al. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol 25, 1322–1326 (2007). [DOI] [PubMed] [Google Scholar]
- 208.Bolognesi R et al. Characterizing the mechanism of action of double-stranded RNA activity against western corn rootworm (Diabrotica virgifera virgifera LeConte). PLoS ONE 7, e47534 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Miyata K et al. Establishing an in vivo assay system to identify components involved in environmental RNA interference in the western corn rootworm. PLoS ONE 9, e101661 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Chow YS et al. in Encyclopedia of Entomology (ed. Capinera JL) 4205–4207 (Springer, 2008). [Google Scholar]
- 211.Zhu K et al. Plant microRNAs in larval food regulate honeybee caste development. PLoS Genet. 13, e1006946 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Garbian Y, Maori E, Kalev H, Shafir S & Sela I Bidirectional transfer of RNAi between honey bee and Varroa destructor: Varroa gene silencing reduces Varroa population. PLoS Pathog. 8, e1003035 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.LaMonte G et al. Translocation of sickle cell erythrocyte microRNAs into Plasmodium falciparum inhibits parasite translation and contributes to malaria resistance. Cell Host Microbe 12, 187–199 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Wang Z et al. Red blood cells release microparticles containing human argonaute 2 and miRNAs to target genes of Plasmodium falciparum: MPs and hAgo2-miRNAs targeting P. falciparum. Emerg. Microbes Infect 6, 1–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Liu S et al. The host shapes the gut microbiota via fecal microRNA. Cell Host Microbe 19, 32–43 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Zhang L et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 22, 107–126 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Chen Q et al. SIDT1-dependent absorption in the stomach mediates host uptake of dietary and orally administered microRNAs. Cell Res. 31, 247–258 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Tian Y, Simanshu DK, Ma J-B & Patel DJ Structural basis for piRNA 2’-O-methylated 3’-end recognition by Piwi PAZ (Piwi/Argonaute/Zwille) domains. Proc. Natl Acad. Sci. USA 108, 903–910 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Svoboda P in Introduction to RNAi and miRNA pathways 1st edn 313–328 (Karolinum, 2020). [Google Scholar]
- 220.Dickinson B et al. Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat. Biotechnol 31, 965–967 (2013). [DOI] [PubMed] [Google Scholar]
- 221.Moriano-Gutierrez S et al. The noncoding small RNA SsrA is released by Vibrio fischeri and modulates critical host responses. PLoS Biol. 18, e3000934 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Tavora B et al. Tumoural activation of TLR3–SLIT2 axis in endothelium drives metastasis. Nature 586, 299–304 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Ruby JG et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127, 1193–1207 (2006). [DOI] [PubMed] [Google Scholar]
- 224.Carthew RW & Sontheimer EJ Origins and mechanisms of miRNAs and siRNAs. Cell 136, 642–655 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Kobayashi H & Tomari Y RISC assembly: coordination between small RNAs and Argonaute proteins. Biochim. Biophys. Acta 1859, 71–81 (2016). [DOI] [PubMed] [Google Scholar]
- 226.Swarts DC et al. The evolutionary journey of Argonaute proteins. Nat. Struct. Mol. Biol 21, 743–753 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Fang X & Qi Y RNAi in plants: an Argonaute-centered view. Plant Cell 28, 272–285 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Czech B et al. piRNA-guided genome defense: from biogenesis to silencing. Annu. Rev. Genet 52, 131–157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Rajagopalan R, Vaucheret H, Trejo J & Bartel DP A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20, 3407–3425 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Axtell MJ & Meyers BC Revisiting criteria for plant microRNA annotation in the era of big data. Plant Cell 30, 272–284 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Kim VN MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol 6, 376–385 (2005). [DOI] [PubMed] [Google Scholar]
- 232.Meyers BC et al. Criteria for annotation of plant microRNAs. Plant Cell 20, 3186–3190 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Chapman EJ & Carrington JC Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet 8, 884–896 (2007). [DOI] [PubMed] [Google Scholar]
- 234.Fukudome A & Fukuhara T Plant dicer-like proteins: double-stranded RNA-cleaving enzymes for small RNA biogenesis. J. Plant Res 130, 33–44 (2017). [DOI] [PubMed] [Google Scholar]
- 235.Castel SE & Martienssen RA RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet 14, 100–112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ghildiyal M & Zamore PD Small silencing RNAs: an expanding universe. Nat. Rev. Genet 10, 94–108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Gu SG et al. Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nat. Genet 44, 157–164 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Mao H et al. The Nrde pathway mediates small-RNA-directed histone H3 lysine 27 trimethylation in Caenorhabditis elegans. Curr. Biol 25, 2398–2403 (2015). [DOI] [PubMed] [Google Scholar]
- 239.Schwartz-Orbach L et al. Caenorhabditis elegans nuclear RNAi factor SET-32 deposits the transgenerational histone modification, H3K23me3. eLife 9, e54309 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Liu C et al. Arabidopsis ARGONAUTE 1 binds chromatin to promote gene transcription in response to hormones and stresses. Dev. Cell 44, 348–361.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 241.Correa RL, Steiner FA, Berezikov E & Ketting RF MicroRNA-directed siRNA biogenesis in Caenorhabditis elegans. PLoS Genet. 6, e1000903 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Tang W et al. A sex chromosome piRNA promotes robust dosage compensation and sex determination in C. elegans. Dev. Cell 44, 762–770.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Matzke MA & Mosher RA RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet 15, 394–408 (2014). [DOI] [PubMed] [Google Scholar]
- 244.Cao M et al. Virus infection triggers widespread silencing of host genes by a distinct class of endogenous siRNAs in Arabidopsis. Proc. Natl Acad. Sci. USA 111, 14613–14618 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Liu L & Chen X RNA quality control as a key to suppressing RNA silencing of endogenous genes in plants. Mol. Plant 9, 826–836 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Almeida MV, Andrade-Navarro MA & Ketting RF Function and evolution of nematode RNAi pathways. Noncoding RNA 5, 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Zhou X et al. RdRP-synthesized antisense ribosomal siRNAs silence pre-rRNA via the nuclear RNAi pathway. Nat. Struct. Mol. Biol 24, 258–269 (2017). [DOI] [PubMed] [Google Scholar]
- 248.Ketting RF & Cochella L Concepts and functions of small RNA pathways in C. elegans. Curr. Top. Dev. Biol 144, 45–89 (2021). [DOI] [PubMed] [Google Scholar]
- 249.Lev I et al. Germ granules govern small RNA inheritance. Curr. Biol 29, 2880–2891.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Rechavi O & Lev I Principles of transgenerational small RNA inheritance in Caenorhabditis elegans.Curr. Biol 27, R720–R730 (2017). [DOI] [PubMed] [Google Scholar]
- 251.Sapetschnig A, Sarkies P, Lehrbach NJ &Miska EA Tertiary siRNAs mediate paramutation in C. elegans. PLoS Genet. 11, e1005078 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Ashe A et al. PiRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 150, 88–99 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Buckley BA et al. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 489, 447–451 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Luteijn MJ et al. Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans: extremely stable Piwi-induced gene silencing. EMBO J. 31, 3422–3430 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Shirayama M et al. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 150, 65–77 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Wan G et al. Spatiotemporal regulation of liquid-like condensates in epigenetic inheritance. Nature 557, 679–683 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Xu F et al. A cytoplasmic argonaute protein promotes the inheritance of RNAi. Cell Rep. 23, 2482–2494 (2018). [DOI] [PubMed] [Google Scholar]