ABSTRACT
Deubiquitination, a type of post-translational modification, cleaves ubiquitin from target proteins, thereby regulating their stability or activity. Deubiquitination enzymes, ubiquitin-specific proteases (UBP/USP), have been reported to be involved in numerous cellular processes in plants, including meristem development, circadian clock regulation, and immunity. In contrast to model plants, however, the functions of UBP in other higher plants remain poorly understood. Here, we isolated a deubiquitination enzyme, ubiquitin-specific protease 12 (NbUBP12), from Nicotiana benthamiana, which shows high sequence homology with the core enzyme regions of UBP12 from other plants. Quantitative reverse-transcription PCR analysis revealed that NbUBP12 gene expression was significantly induced after drought treatment, and its level was higher in seed than in other tissues. Using a virus-induced gene silencing technique, we generated NbUBP12-silenced tobacco plants to analyze NbUBP12 gene function in response to drought stress and found that compared with control plants, NbUBP12-silenced plants exhibited a lower survival rate after exposure to drought stress. In addition, they were characterized by lower leaf surface temperatures and larger stomatal pore size following abscisic acid (ABA) treatment. On the basis of these observations, we suggest that NbUBP12 is involved in modulating drought resistance in N. benthamiana, which is associated with ABA-mediated stomatal closure.
KEYWORDS: Ubiquitin-specific protease 12, deubiquitination, dehydration stress, abscisic acid, Nicotiana benthamiana
Text
As sessile organisms, plants have evolved a range of mechanisms designed to conserve and efficiently utilize water, thereby enabling them to survive in adverse environments, especially water deficit. In this regard, numerous proteins known to be involved in the adaptation to changing environments are regulated by post-translational modification mechanisms (PTMs), which contribute to controlling enzymatic activity by modifying protein structure or stability, regulating entire plant life cycles, or promoting rapid responses to the surrounding environment.1,2 Among these PTMs, ubiquitination is one of the most conserved processes in eukaryotic cells. It occurs in three sequential steps, catalyzed by E1-activating, E2-conjugating, and E3 ligase enzymes,3 which contribute to modifications that trigger the degradation of the target proteins. In contrast to ubiquitination, deubiquitination is mediated by deubiquitination enzymes (DUBs) and entails the removal of the attached ubiquitin from the target proteins.4 The quality control of proteins via the ubiquitination–deubiquitination regulatory system in plants conceivably represents an effective mechanism enabling the adaptation to a changeable environment.
DUBs are classified into two groups, namely, metalloproteases and cysteine proteases, the latter of which comprise the majority of DUBs.5,6 Cysteine proteases can be further divided into four families based on their functional domains and structures: ubiquitin-specific proteases (UBPs/USPs), ubiquitin C-terminal hydrolases, ovarian tumor proteases, and Machado–Joseph disease protein domain proteases.5–9 Among these, UBPs form the largest subfamily of DUBs in eukaryotes, which in plants function via diverse pathways. In Arabidopsis, 27 genes have been found to encode UBPs,10,11 which are involved in a range of biological processes associated with responses to different environmental stimuli as well as in normal growth and development.10–12 For example, AtUBP12 and its homolog AtUBP13 function in plant immunity, circadian clock regulation, flowering, seed development, jasmonate signaling, and leaf senescence,13–18 in which their target proteins such as MYC215 and ROOT GROWTH FACTOR RECEPTOR 1 and ORESARA 116 are stabilized via AtUBP12/13-mediated deubiquitination. As a Solanaceous orthologue of AtUBP12, Nicotiana tabacum UBP12 (NtUBP12) has also been found to influence Cf-9-triggered hypersensitive responses negatively.13 Recently, we isolated CaUBP12, which has high sequence homology with AtUBP12/13 and NtUBP12 from Capsicum annuum belonging to the same Solanaceae family as tobacco.19 CaUBP12 plays a positive role in the drought resistance of pepper plants, in which it appears to enhance the protein stability of CaSnRK2.6, an orthologue of AtSnRK2.6/OST1 that functions as a positive regulator of ABA signaling.20 These observations indicate that UBP12 functions in the responses of plants to biotic and abiotic stresses via its deubiquitination activity.
In this study, we isolated the UBP12 gene from another Solanaceae plant, N. benthamiana. This NbUBP12 gene consists of a 3,351-bp open reading frame that encodes a 1,116-amino acid protein of molecular weight 130.75 kDa and an isoelectric point of 5.58. Multiple alignment and phylogenetic tree analyses showed that NbUBP12 was clustered with AtUBP12/13, CaUBP12, and NtUBP12 proteins (Figure 1a). We searched conserved motifs and domains in NbUBP12 using the webtools, Motif Scan (https://myhits.sib.swiss/cgi-bin/motif_scan) and InterProSan (https://www.ebi.ac.uk/interpro). Like other UBP12 proteins, NbUBP12 also contains highly conserved sequence regions, including a unique meprin and TRAF homology domain and specific catalytic Cys- and His-boxes essential for DUB activity (Figure 1b).13,14 Since the function of protein is associated with its subcellular localization in cells, we analyzed subcellular location of NbUBP12 protein. When continuously expressed in the leaf epidermal cells of N. benthamiana plants, green fluorescent protein (GFP)-tagged NbUBP12 predominantly targeted to the nucleus and cytoplasm (Figure 1c). Next, we investigated expression patterns of NbUBP12 gene. Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analysis revealed that the NbUBP12 gene was expressed in all examined tissues of tobacco plants (Figure 1d left). The expression level was relatively lower in petioles and flowers and significantly higher in seeds. Furthermore, in the leaves of tobacco plants, the NbUBP12 gene was found to be highly induced at 24 h after subjecting plants to drought stress, but not in response to abscisic acid (ABA) treatment (Figure 1d right). This drought stress-induced expression was consistent with that observed for CaUBP12,19 suggesting that NbUBP12 may also be associated with drought resistance.
Figure 1.
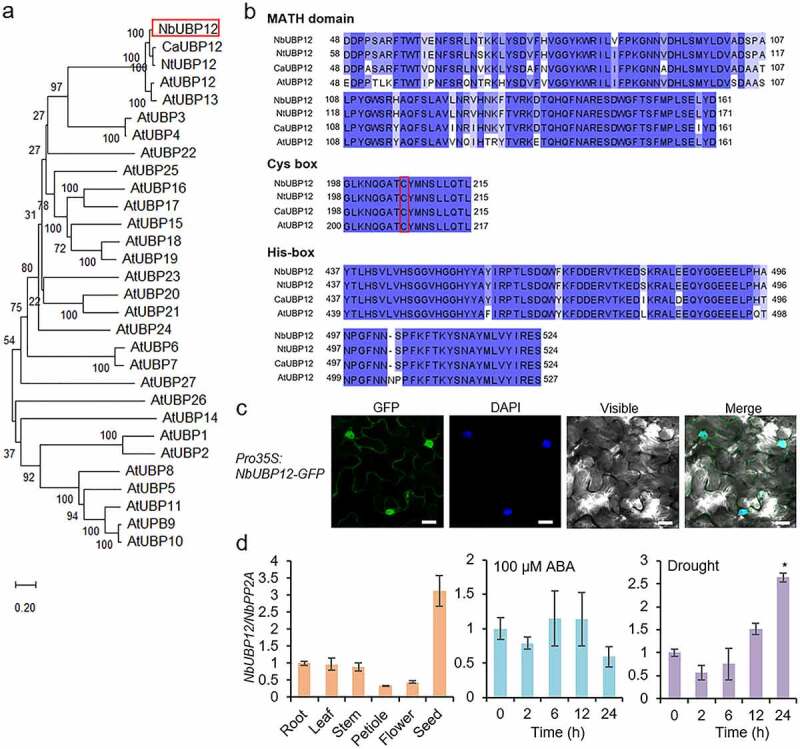
(a) Phylogenetic tree analysis of NbUBP12 with UBP proteins. The phylogenetic tree was drawn using MEGA software (version 10.1) using the deduced amino acid sequences of UBP12 from Arabidopsis, pepper, and tobacco plants. (b) Multiple alignment of UBP12 enzymatic domain within Nicotiana benthamiana UBP12 (NbUBP12) with Nicotiana tabacum (NtUBP12), Capsicum annuum (CaUBP12), and Arabidopsis thaliana (AtUBP12) using MEGA system and the web tool Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo). Red box indicates the Cys residue that is required for enzymatic activity. (c) Subcellular localization of NbUBP12 in the leaf epidermal cells of Nicotiana benthamiana plants. The scale bar represents 20 μm. (d) Quantitative RT-PCR analysis of NbUBP12 expression in the various tissues (left), ABA treatment (middle), and under the drought stress by withholding water (right). Total RNAs from the stressed samples were extracted from the third leaf of each plant. All data represent the mean ± standard deviation of three independent experiments. Asterisks indicate significant differences than 0 h (Student’s t-test; *P < .05).
NbUBP12 shows high sequence homology with CaUBP12 (89.6% identity and 94.5% similarity), and given the known function of CaUBP12, we speculated as to whether NbUBP12 might function similarly under drought stress. To verify this assumption, we initially performed a TRV-based virus-induced gene silencing (VIGS) assay to generate NbUBP12-knockdown plants. We designed a gene-specific target region (1000–1300 bp) for silencing NbUBP12 using the VIGS tool available from the Solanaceae Genomics Network (http://solgenomics.net; Nicotiana benthamiana ver.1.0 database) (Figure 2a). Two weeks after agroinfiltration, there were no phenotypic differences between NbUBP12-silenced (TRV2:NbUBP12) and control (TRV:00) plants (Figure 2b). To ascertain the efficacy of the TRV-based VIGS system in N. benthamiana plants, we used the phytoene desaturase (PDS) gene as a positive control, as the silencing of PDS typically leads to photobleaching.21 In line with the expectations, we observed that TRV2:PDS plants exhibited a leaf-bleached phenotype (Figure 2b). The VIGS system accordingly proved successful in inducing the silencing of the NbUBP12 gene, and qRT-PCR analysis revealed that the levels of NbUBP12 expression were approximately 70% lower in the leaves of TRV2:NbUBP12 plants than in those of TRV2:00 plants (Figure 2c).
Figure 2.
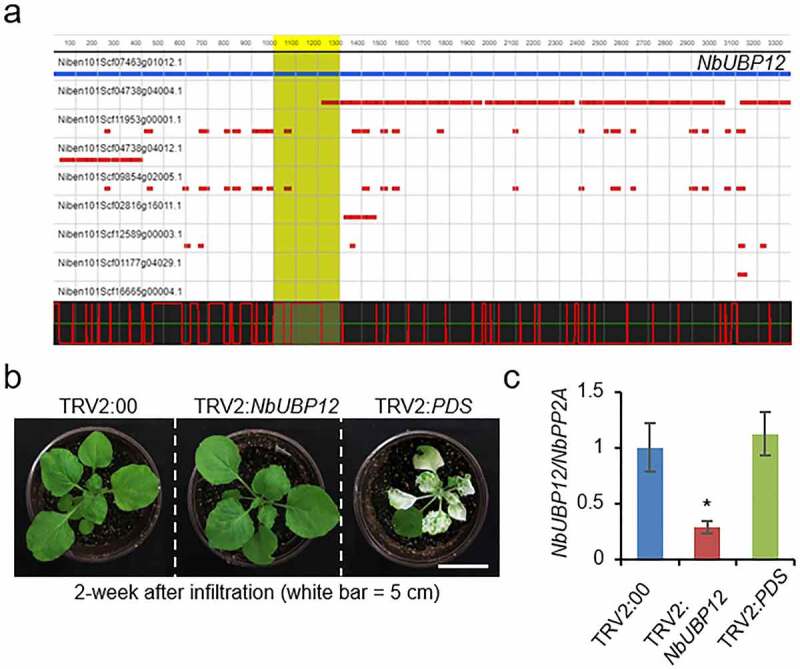
(a) Graphical representation of the virus-induced gene silencing (VIGS) target region of NbUBP12. Analyses were performed using the SGN VIGS tool (http://vigs.solgenomics.net/) with red bars indicating the siRNA of off-target genes. (b and c) NbUBP12 VIGS system analysis using empty vector control tobacco plants (TRV:00) and tobacco plants containing a widely used VIGS system positive control gene (TRV2:PDS). (b) Photographs showing the phenotypes of control and knockdown plants at 2 weeks after agroinfiltration. (c) Confirmation of the knockdown of NbUBP12 transcripts based on RT-PCR analysis. The total RNA used for analysis was extracted from the third leaf of each plant line at 2 weeks after agroinfiltration. Asterisks indicate significant differences between TRV2:NbUBP12 and TRV2:00 plants (Student’s t-test; *P < .05).
Using NbUBP12-silenced tobacco plants, we examined whether NbUBP12 plays a role in drought stress response (Figure 3). At two weeks after agroinfiltration, we subjected TRV2:NbUBP12 and TRV:00 plants to drought stress by withholding watering for 13 days. Under well-watered conditions, we detected no differences in the phenotypes of the two tobacco lines (Figure 3a, left panel). However, compared with the control plants, TRV2:NbUBP12 plants showed significantly wilted phenotypes in response to drought treatment and rewatering (Figure 3a, middle and right panels). Consistently, 38.75% of the TRV2:NbUBP12 plants resumed growth at 3 days after rehydration, whereas TRV2:00 plants showed a 72.5% survival rate (Figure 3a and b). Since the phenotypic difference between TRV2:NbUBP12 and TRV:00 plants was easily observed 8 days after drought stress, we harvested leaf samples at this time, and measured the level of malondialdehyde (MDA), which is widely used as a membrane lipid peroxidation marker. As shown in Figure 3c, the MDA content was higher in TRV2:NbUBP12 than in TRV:00 only after drought stress treatment. We also measured delayed fluorescence (DF) of TRV2:NbUBP12 and TRV:00 leaves. DF, emitted from photosystem II, can be used as a sensitive indicator for chlorophyll content as well as the physiological state of the plants exposed to environmental stresses, such as drought and salinity, and generally DF is reduced by drought stress.22–25 At 8 days after drought stress, DF intensity was much lower in TRV2:NbUBP12 leaves than in TRV:00 leaves, indicating that drought stress severely reduced the photosynthetic rate of TRV2:NbUBP12, compared to the control (Figure 3d and e). These results suggested that NbUBP12 plays a positive role in the regulation of drought stress tolerance mechanisms.
Figure 3.
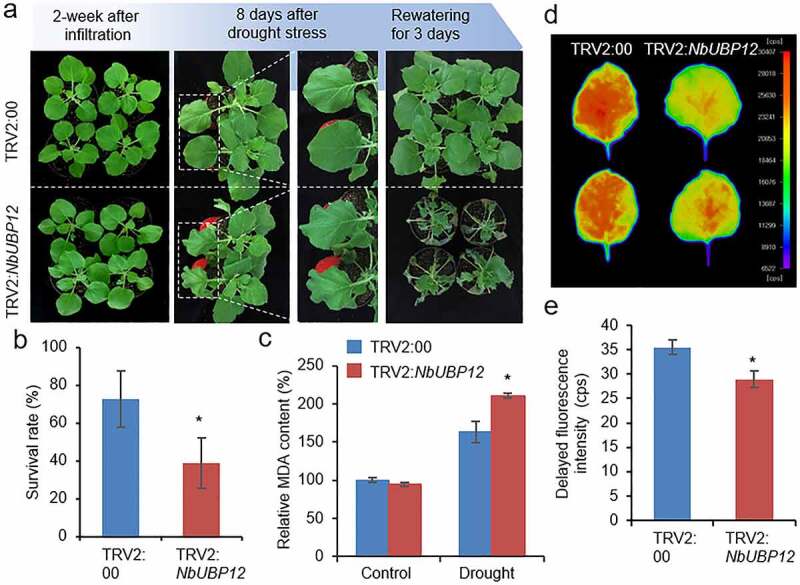
(a and b) Drought sensitivity of NbUBP12-silenced (TRV2:NbUBP12) and control (TRV2:00) plants. Two weeks after agroinfiltration, healthy plants of each line (n = 32) were subjected to drought stress by withholding watering for 13 days and rewatering for 3 days. Representative images of plants were obtained at 0 and 8 days after drought stress, and 3 days after rewatering (a), and the percentages of surviving plants were calculated (b). All data represent the means ± standard deviation of three independent experiments. (c) Measurement of malondialdehyde (MDA) in the leaves of TRV2:NbUBP12 and TRV2:00 plants treated with drought stress for 8 days. (d and e) Delayed fluorescence (DF) of the leaves of TRV2:NbUBP12 and TRV2:00 plants treated with drought stress for 8 days. Using NightShade LB 985 In vivo Plant Imaging System, representative images were obtained (d) and DF intensity was calculated (e). The closer the color is to red (high intensity), the higher the chlorophyll content. The data represent the means ± standard error of three independent experiments. Asterisks indicate significant differences between TRV2:NbUBP12 and TRV2:00 plants (Student’s t-test; *P < .05).
We further speculated as to whether this enhanced drought sensitivity of TRV2:NbUBP12 is associated with ABA-mediated stomatal responses. In this regard, it has previously been demonstrated that stomatal closure is associated with an increase in the surface temperature of leaves, as a consequence of reduced evaporative cooling.26,27 Accordingly, to verify our conjecture, we measured the leave surface temperature of TRV2:NbUBP12 and TRV:00 plants subsequent to spraying with 100 μM ABA. Prior to ABA treatment, we detected no apparent difference in the leaf temperatures of the two lines (Figure 4a and b). However, at 6 h after ABA treatment, we found that leaf temperatures were significantly higher in TRV:00 plants than in TRV2:NbUBP12 plants. We subsequently measured stomatal pore sizes on the leaves of TRV2:NbUBP12 and TRV:00 plants in the absence and presence of ABA. Leaf peels were harvested from these plant lines 2 weeks after infiltration and then incubated in a stomatal opening solution containing 0, 10, or 20 μM ABA. While no significant differences between TRV2:00 and TRV2:NbUBP12 plants were detected in the absence of ABA (Figure 4 c and d), ABA treatment was found to promote stomatal closure in both plant lines. However, consistent with the leaf temperature data, we observed that stomatal pore sizes in TRV2:NbUBP12 plants were notably larger than those in TRV2:00 plants. Moreover, we analyzed expression levels of ABA-responsive genes, such as RAB18, RD29B, DREB, and LEA homologs from tobacco, in both plant lines. After treatment with 100 μM ABA, TRV2:NbUBP12 leaves showed lower induction of all the genes than those of TRV2:00, but not before treatment (Figure 4e). These observations thus indicate that NbUBP12 contributes to drought resistance by modulating ABA-mediated stomatal closure and ABA-resposive gene expression.
Figure 4.
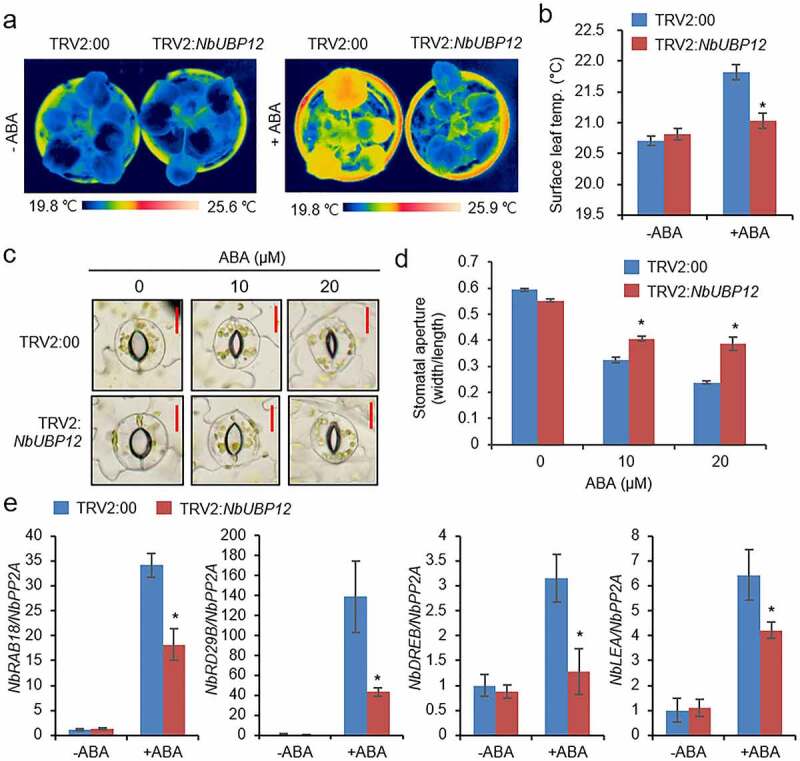
(a and b) Leaf temperatures of TRV2:NbUBP12 and TRV2:00 plants after exposure to abscisic acid (ABA). Representative thermographic images were obtained at 6 h after treatment with 100 μM ABA (a) and mean leaf temperatures were determined from the three largest leaves of plants from each line (n = 12) (b). (c and d) ABA-induced stomatal closure in TRV2:NbUBP12 and TRV2:00 plants. Stomatal apertures were measured 3 h after treatment with 0, 10, or 20 μM ABA. Representative images of the stomata were obtained for the leaves of each line (c) and the apertures of 100 randomly selected stomata were measured (d). The scale bar represents 10 μm. (e) Quantitative RT-PCR analysis of ABA-responsive genes in TRV2:NbUBP12 and TRV2:00 plants after treatment with ABA. Total RNAs from the stressed samples were extracted from the third leaf of each plant. All data represent the mean ± standard deviation of three independent experiments. Asterisks indicate significant differences between TRV2:NbUBP12 and TRV2:00 plants (Student’s t-test; *P < .05).
In conclusion, we demonstrated that NbUBP12 functions as a positive regulator of drought resistance and that this regulatory effect is associated with ABA-mediated stomatal regulation, which is consistent with the previously established function of CaUBP12. Our data would accordingly tend to imply the functional conservation of UBP12 proteins across the members of the Solanaceae family, at least with respect to the drought stress response. Moreover, it is conceivable that these UBP12 proteins target common substrates, including SnRK2.6/OST1, to modulate drought resistance. We thus speculate that gaining an understanding of the detailed mechanisms through which UBP12-mediated deubiquitination stabilizes substrate proteins will contribute to developing a novel approach for the modulation of drought resistance. Furthermore, given the negative role played by NbUBP12 in plant immunity, the functional differences of NbUBP12 with respect to biotic and abiotic stress responses could be resolved by isolating stress-specific substrates.
Funding Statement
This work was supported by a grant from the Agriculture & Technology Development (Project No. PJ01589201), Rural Development Administration, Republic of Korea, and by the National Research Foundation of Korea (NRF) grant funded by the Korea Government (MSIT) (No. 2021R1A2C2006338), Republic of Korea.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Shu K, Yang W.. E3 ubiquitin ligases: ubiquitous actors in plant development and abiotic stress responses. Plant Cell Physiol. 2017;58:1–7. doi: 10.1093/pcp/pcx071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verma V, Ravindran P, Kumar PP.. Plant hormone-mediated regulation of stress responses. Bmc Plant Biol. 2016;16:86. doi: 10.1186/s12870-016-0771-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 4.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 5.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta -Mol Cell Res. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards RA. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Hu M, Li PW, Li MY, Li WY, Yao TT, Wu JW, Gu W, Cohen RE, Shi YG. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111:1041–1054. doi: 10.1016/S0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- 8.Johnston SC, Larsen CN, Cook WJ, Wilkinson KD, Hill CP. Crystal structure of a deubiquitinating enzyme (human UCH-L3) at 1.8 A resolution. Embo J. 1997;16:3787–3796. doi: 10.1093/emboj/16.13.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balakirev MY, Tcherniuk SO, Jaquinod M, Chroboczek J. Otubains: a new family of cysteine proteases in the ubiquitin pathway. EMBO reports. 2003;4:517–522. doi: 10.1038/sj.embor.embor824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan N, Doelling JH, Falbel TG, Durski AM, Vierstra RD. The ubiquitin-specific protease family from arabidopsis. AtUBP1 and 2 are required for the resistance to the amino acid analog canavanine. Plant Physiol. 2000;124:1828–1843. doi: 10.1104/pp.124.4.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Wang F, Zhang H, He H, Ma L, Deng XW. Functional characterization of the Arabidopsis ubiquitin-specific protease gene family reveals specific role and redundancy of individual members in development. Plant J. 2008;55:844–856. doi: 10.1111/j.1365-313X.2008.03557.x. [DOI] [PubMed] [Google Scholar]
- 12.Zhou H, Zhao J, Cai J, Patil SB. UBIQUITIN-SPECIFIC PROTEASES function in plant development and stress responses. Plant Mol Biol. 2017;94:565–576. doi: 10.1007/s11103-017-0633-5. [DOI] [PubMed] [Google Scholar]
- 13.Ewan R, Pangestuti R, Thornber S, Craig A, Carr C, O’Donnell L, Zhang C, Sadanandom A. Deubiquitinating enzymes AtUBP12 and AtUBP13 and their tobacco homologue NtUBP12 are negative regulators of plant immunity. New Phytol. 2011;191:92–106. doi: 10.1111/j.1469-8137.2011.03672.x. [DOI] [PubMed] [Google Scholar]
- 14.Cui X, Lu F, Li Y, Xue Y, Kang Y, Zhang S, Qiu Q, Cui X, Zheng S, Liu B, et al. Ubiquitin-specific proteases UBP12 and UBP13 act in circadian clock and photoperiodic flowering regulation in Arabidopsis. Plant Physiol. 2013;162:897–906. doi: 10.1104/pp.112.213009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeong JS, Jung C, Seo JS, Kim JK, Chua NH. The deubiquitinating enzymes UBP12 and UBP13 positively regulate MYC2 levels in jasmonate responses. Plant Cell. 2017;29:1406–1424. doi: 10.1105/tpc.17.00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.An Z, Liu Y, Ou Y, Li J, Zhang B, Sun D, Tang W. Regulation of the stability of RGF1 receptor by the ubiquitin-specific proteases UBP12/UBP13 is critical for root meristem maintenance. Proc Natl Acad Sci USA. 2018;115:1123–1128. doi: 10.1073/pnas.1714177115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee CM, Li MW, Feke A, Liu W, Saffer AM, Gendron JM. GIGANTEA recruits the UBP12 and UBP13 deubiquitylases to regulate accumulation of the ZTL photoreceptor complex. Nat Commun. 2019;10:3750. doi: 10.1038/s41467-019-11769-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park SH, Jeong JS, Seo JS, Park BS, Chua NH. Arabidopsis ubiquitin-specific proteases UBP12 and UBP13 shape ORE1 levels during leaf senescence induced by nitrogen deficiency. New Phytol. 2019;223:1447–1460. doi: 10.1111/nph.15879. [DOI] [PubMed] [Google Scholar]
- 19.Lim CW, Baek W, Lim J, Hong E, Lee SC. Pepper ubiquitin-specific protease, CaUBP12, positively modulates dehydration resistance by enhancing CaSnRK2.6 stability. Plant J. 2021;107:1148–1165. In press. doi: 10.1111/tpj.15374. [DOI] [PubMed] [Google Scholar]
- 20.Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell. 2002;14:3089–3099. doi: 10.1105/tpc.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu YL, Schiff M, Marathe R, Dinesh-Kumar SP. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 2002;30:415–429. doi: 10.1046/j.1365-313X.2002.01297.x. [DOI] [PubMed] [Google Scholar]
- 22.Guo Y, Tan JL. A biophotonic sensing method for plant drought stress. Sens Actuators B Chem. 2013;188:519–524. doi: 10.1016/j.snb.2013.07.020. [DOI] [Google Scholar]
- 23.Wang JS, Xing D, Zhang LR, Jia L. A new principle photosynthesis capacity biosensor based on quantitative measurement of delayed fluorescence in vivo. Biosens Bioelectron. 2007;22:2861–2868. doi: 10.1016/j.bios.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 24.Zhang LR, Xing D. Rapid determination of the damage to photosynthesis caused by salt and osmotic stresses using delayed fluorescence of chloroplasts. Photoch Photobio Sci. 2008;7:352–360. doi: 10.1039/b714209a. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Fu Y, Fan J, Li Q, Francis F, Chen JL. Comparative transcriptome and histological analyses of wheat in response to phytotoxic aphid Schizaphis graminum and non-phytotoxic aphid Sitobion avenae feeding. Bmc Plant Biol. 2019;19:547. doi: 10.1186/s12870-019-2148-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim CW, Baek W, Lee SC. The pepper RING-type E3 ligase CaAIRF1 regulates ABA and drought signaling via CaADIP1 protein phosphatase degradation. Plant Physiol. 2017;173:2323–2339. doi: 10.1104/pp.16.01817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park SY, Peterson FC, Mosquna A, Yao J, Volkman BF, Cutler SR. Agrochemical control of plant water use using engineered abscisic acid receptors. Nature. 2015;520:545–548. doi: 10.1038/nature14123. [DOI] [PubMed] [Google Scholar]


