Background:
Researchers routinely evaluate novel biomarkers for incorporation into clinical risk models, weighing tradeoffs between cost, availability, and ease of deployment. For risk assessment in population health initiatives, ideal inputs would be those already available for most patients. We hypothesized that common hematologic markers (eg, hematocrit), available in an outpatient complete blood count without differential, would be useful to develop risk models for cardiovascular events.
Methods:
We developed Cox proportional hazards models for predicting heart attack, ischemic stroke, heart failure hospitalization, revascularization, and all-cause mortality. For predictors, we used 10 hematologic indices (eg, hematocrit) from routine laboratory measurements, collected March 2016 to May 2017 along with demographic data and diagnostic codes. As outcomes, we used neural network-based automated event adjudication of 1 028 294 discharge summaries. We trained models on 23 238 patients from one hospital in Boston and evaluated them on 29 671 patients from a second one. We assessed calibration using Brier score and discrimination using Harrell’s concordance index. In addition, to determine the utility of high-dimensional interactions, we compared our proportional hazards models to random survival forest models.
Results:
Event rates in our cohort ranged from 0.0067 to 0.075 per person-year. Models using only hematology indices had concordance index ranging from 0.60 to 0.80 on an external validation set and showed the best discrimination when predicting heart failure (0.80 [95% CI, 0.79–0.82]) and all-cause mortality (0.78 [0.77–0.80]). Compared with models trained only on demographic data and diagnostic codes, models that also used hematology indices had better discrimination and calibration. The concordance index of the resulting models ranged from 0.75 to 0.85 and the improvement in concordance index ranged up to 0.072. Random survival forests had minimal improvement over proportional hazards models.
Conclusions:
We conclude that low-cost, ubiquitous inputs, if biologically informative, can provide population-level readouts of risk.
Keywords: cardiovascular disease, heart failure, hematology, ischemic stroke, machine learning
What is Known
Risk models often prioritize novel biomarkers for predicting adverse cardiovascular outcomes, although such biomarkers may have limited availability for model replication and deployment.
Simple biological measures, such as the complete blood count, have been shown to be associated with specific clinical events.
Artificial intelligence methods for text processing can enable rapid adjudication of events from clinical notes, thereby overcoming deficiencies in diagnostic code-based approaches.
What the Study Adds
We develop a strategy to developing time-to-event models for cardiovascular events using simple hematologic predictors from a complete blood count and artificial intelligence-adjudicated outcomes.
Models are competitive with or outperform existing models such as the pooled cohort equations.
The primary proposed clinical application of such models are population health initiatives, hospital- or payer-level financial planning, or clinical trial enrollment, where low-cost widely available predictors and ongoing event ascertainment can provide maximal utility.
Two approaches guide the development of clinical risk models.1 One strategy focuses on evaluating innovative markers, typically biomolecular, in a research cohort where banked historical samples and adjudicated outcomes are available. Recent biomarker candidates include genetic variants,2–4 protein biomarkers,5 somatic mutations,6 and serum metabolites.7 The motivation for such an approach is to improve model performance while proposing something new about disease mechanisms. One challenge for such a strategy is the lack of such markers in other cohorts, making replication a challenge. The problem of predictor availability extends to model deployment, especially when considering such models for population health initiatives. Moreover, given that contributors to disease may evolve, insights from a historical cohort may not be readily transportable to new settings.8
See Editorial by Ouyang and Cheng
An alternative strategy involves training models using existing low-cost data readily available on a high percentage of patients within a health care system. Such an approach is pragmatic and exploits model training as the first step in a systematic program to enable risk assessment for the maximal number of individuals. The output of the models could then be used to guide population health initiatives, hospital- or payer-level financial planning, or clinical trial enrollment.9 The use of widely available inputs also facilitates iteratively updating models to match changing environments.
In this work, we focus on the latter approach but use an unconventional yet widely available choice of predictors: hematologic indices available in a simple complete blood count (CBC). The CBC provides counts and distributional properties of red blood cells, platelets, and white blood cells taken in aggregate and is commonly used in screening settings. Others have shown an association of hematologic indices with all-cause mortality and cardiovascular events, including a focus on red-cell distribution width10–15 and have found similar associations with parameters based on specific leukocyte populations available in the related CBC with differential, such as the neutrophil-to-lymphocyte ratio.16
Our emphasis here was on estimating and validating time-to-event models for cardiovascular events. Unlike other risk-model efforts,17,18 which have focused on a population free of cardiovascular disease (CVD) events and not taking statins, our focus was on a broader set of patients, likely at higher risk. Our rationale was that such individuals are responsible for high resource health care utilization and could benefit from triggered therapeutic innovations to reduce risk, whether they be novel medications or population-health initiatives.
Moreover, in the spirit of enabling ease of re-training models in new settings, we use a machine-learning strategy for adjudication of acute coronary syndrome (ACS), coronary revascularization, heart failure hospitalization (HF), and ischemic stroke (IS), using discharge summaries from hospitalizations.19 We describe discrimination and calibration performance of the models across 2 institutions and compare additive models to an ensemble machine learning approach.
Methods
Data Access
Code used for data processing and analysis will be available upon request.
Overview
We used Cox proportional-hazards models to predict time to various cardiovascular outcomes. The predictors of most interest were 10 hematologic parameters produced by routine CBC. We also studied predictor sets that included demographics and disease history. Outcomes were identified and located in time from electronic health records by applying neural-network classifiers to discharge summaries. Models were developed using data from one hospital and validated using data from another hospital.
Patient Selection and Data Retrieval
Patients in the model-derivation cohort were at least 18 years old and had a CBC recorded on a Sysmex XE-5000 Automated Hematology System at the Massachusetts General Hospital during March 2016 to May 2017 (see Table S1 for a glossary of medical terms). The Massachusetts General Hospital Sysmex XE-5000 devices produce a digital file in a proprietary format for each CBC. We extracted this file from each of the 3 Massachusetts General Hospital XE-5000s during the period March 17, 2016, to May 17, 2017. We then used software supplied by Sysmex to convert the proprietary format into a collection of several nonproprietary-format files, including a result-log file. Each sample analysis on an XE-5000 produces a single result-log file, uniquely identifiable by a lab-order ID number and a timestamp. A result-log file contains various types of data, including details of the machine and operation; standard hematology parameters, such as red blood cell count and hematocrit; and more specialized hematology parameters, such as the percentage of reticulocytes or the centroid of a basophil cluster in the scattergram; and binary flags signifying pathological conditions or sample-quality problems.
Using the 595 557 result-log files generated from the XE-5000 devices during this period, we matched 452 731 unique lab-order identifiers to 136 341 unique patient identifiers. Some lab-order identifiers were associated with >1 result-log file. In 99% of cases, the files’ timestamps varied by <3 hours and mapped to a single set of hematologic parameters, so we limited analysis to the most recent file. We discarded another six files with more than one distinct set of results and another 19 files with more than one unique lab-order identifier. This process resulted in 452 706 CBC tests for 136 339 patients (Figure S1A and S1B)
For each patient, we accessed structured data from the Massachusetts General Brigham Electronic Data Warehouse (MGB-EDW), a data repository for a large multi-institutional Integrated Delivery Network in the greater Boston area. The 2 primary hospitals used in this study were Massachusetts General Hospital and Brigham and Women’s Hospital, which are 999- and 793-bed teaching hospitals, respectively, affiliated with Harvard Medica School. Together, they are the founding members of Mass General Brigham, the largest health care provider in Massachusetts. Structured data included diagnostic codes reflecting past medical history, lists of actively managed problems, the reasons for any hospital admission, and encounter diagnoses. The latter are diagnostic codes used to justify billing for a broad range of medical encounters, including laboratory draws, imaging tests, outpatient visits, and hospital admissions. In addition to structured data, we also captured medical notes focusing on discharge summaries, which represent a detailed description of the clinical course for a hospitalization encounter. Encounters, diagnostic codes, and notes could be from any of the institutions within the Massachusetts General Brigham network of hospitals.
Structured encounter information was further used to limit CBCs so that the hematologic indices more closely reflected the patient’s baseline values. Specifically, we only included CBCs drawn during an elective outpatient encounter. Furthermore, we excluded all tests that occurred within 7 days before or 30 days after an emergency department visit or hospital admission, as defined by the date of a discharge summary, as such tests may reflect some active temporary disruption in the baseline laboratory values (eg, acute infection, blood loss) which is not the intended use of our models. From the resulting 41 444 CBCs, we selected the earliest result for each patient, which also defined the patient’s time of entry into the study. This process resulted in 27 493 patient-CBC pairs.
We excluded 341 patients without demographic data and 3489 patients younger than 30 years at the time of entry into the study. Follow-up time was defined from the date of the CBC to the last encounter or note within the MGB-EDW under the following categories: encounters coded as office visit, system generated, or hospital encounter; notes coded as progress note, telephone encounter, assessment & plan, discharge summary, ED provider note, consult, MR AVS snapshot, plan of care, patient instruction, H&P. Because of the dramatic change in hospital population caused by COVID-19 in the winter of 2020, we did not consider follow-up or events after December 31, 2019. When a patient’s follow-up extended past this date or if a study outcome occurred after this date, we right-censored the patient at January 1, 2020. Finally, 437 patients with no follow-up in the MGB-EDW system were excluded, resulting in 23 226 patients.
The validation cohort consisted of adult patients who had a 10-parameter CBC (described below) collected at Brigham & Women’s Hospital between June 2015 and December 2016 and whose results were available within the MGB-EDW. As with the training data, we excluded laboratory values that occurred close in time to emergency department or hospital discharges (Figure S1C and S1D).
AI Enabled Adjudication of Cardiovascular Outcomes
We recently trained neural network-based models to classify discharge summaries as associated with one of the following 4 cardiovascular events: ACS, IS, HF, and percutaneous coronary intervention/coronary artery bypass surgery (PCI/CABG).19 Briefly, after manually labeling 1372 training notes + 592 validation notes + 1003 testing notes across 17 institutions, we trained and validated models for event adjudication, measuring area under the receiver-operating characteristic curve on the 1003 test notes as follows: ACS, 0.967; HF, 0.965; IS, 0.980; PCI/CABG, 0.998. The fraction of all discharges in our test set that received labels for CVD events was as follows: ACS, 0.054; HF, 0.122; IS, 0.059; PCI/CABG, 0.054. The 4 types of events were not mutually exclusive, and a discharge summary could receive as many as 4 positive labels. For the current work, we selected classification thresholds to maximize the F1 score (Table S2). The 4 event models were deployed on the 1 028 294 discharge summaries for the derivation and validation cohorts, and a binary event status was determined by each model for each note.
The four artificial intelligence (AI)-adjudicated clinical events, plus all-cause mortality, constituted the 5 primary outcomes for our survival analysis. In addition to these primary outcomes, we also defined 2 composite outcomes: (1) ACS or heart-failure hospitalization or IS (ACS/HF/IS) and (2) ACS/HF/IS or death (ACS/HF/IS/death). Date of death was obtained from MGB-EDW (which uses data from the Social Security Administration Death Master File) without mediation by statistical methods. We did not exclude a patient from the dataset for outcome X if they also experienced outcome Y.
Prior history of coronary artery disease (CAD), IS, or HF was established using diagnostic codes, which we previously evaluated for performance on a set of manually labeled test notes19 (Table S3). In contrast to the event models above, which map a single discharge summary to the probability of an event immediately associated with that hospital discharge, ICD10 codes were taken in aggregate across a patient’s history to generate a binary status of extant disease up to a specific date. As an alternative strategy to capture a greater number of comorbidities, we determined the Charlson comorbidity index20 for each patient using a published ICD10 code mapping21 and model weights.22
Predictors
Hematologic Predictors
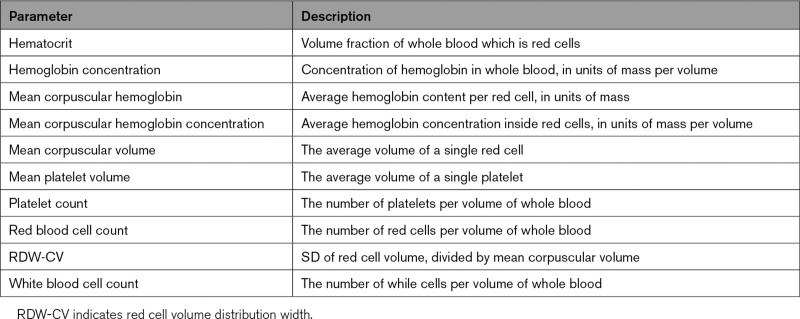
Hematologic predictors used in time-to-event models (defined in Table 1) included hematocrit, hemoglobin concentration, mean corpuscular hemoglobin, mean corpuscular hemoglobin concentration, mean corpuscular volume, mean platelet volume, platelet count, red blood cell count, and red cell volume distribution width (RDW-CV). Of the 2 red blood cell distribution width parameters calculated by the XE-5000, only RDW-CV, not RDW-SD, was available in our external data set, and thus we only included RDW-CV in our models. Entry into the study for a patient was defined by the patient’s first CBC.
Table 1.
Hematologic Parameters Used in Survival Models
Nonhematologic Predictors
Models also incorporated commonly available nonhematologic predictors:
Patient’s age at entry into the study.
Binary diagnostic codes for history of each of three conditions, at time of entry into the study: heart failure, IS, coronary artery disease, or myocardial infarction.
All predictors were centered and scaled except the 3 binary flags for disease (Table S4). Some predictor sets included first-order interactions with age. When main effects were centered and scaled, this transformation was applied before computing interaction terms (Table S5).
Derivation and Validation of the Models
For each of the 7 outcomes, we used L1-penalized Cox proportional-hazards models to estimate coefficients for each risk factor. For each outcome, we developed separate models for male and female patients. Penalty strength for each model was selected by optimizing Harrell concordance index (C-index)23 through k-fold cross validation (3≤ k ≤5). Ties were handled with Breslow method.24 Regression and penalty tuning were performed with the Python package scikit-survival v0.14.0.25
Given that except for death, one cardiovascular outcome is unlikely to preclude the development of any other, we did not look at competing risks among cardiovascular events. Death was treated as a competing risk implicitly through censoring (ie, cause-specific hazard functions for other outcomes).26
Each combination of outcome and sex was treated with 5 different models, each supported by a different set of predictors: set 1 consisted of the 10 hematologic indices (hematology-only, or HEM); set 2 added age to set 1 (age-hematology or AGE-HEM); set 3 added disease history to set 2 (age-hematology-history or AGE-HEM-HX); set 4 consisted of age and history (age-history or AGE-HX); set 5 consisted of just age (age-only or AGE). Whenever we included age in a predictor set, we also include first-order interactions between age and all main effects (Table S5).
To evaluate performance within the derivation set and provide a measure of uncertainty for the performance estimate, we repeated rounds of 5-fold cross-validation, placing the penalty-tuning loop within the model-derivation step for each fold. We then trained final models on the entire derivation data set. We externally validated each final model on the Brigham & Women’s Hospital data set, having preprocessed the validation data in the same way as the training data. We removed any patients from the validation data set who also appeared in the derivation data set.
As a sensitivity analysis, we substituted the Charlson comorbidity index for disease history, focusing on death, and ACS/HF/IS/death as outcomes. As a further evaluation of the robustness of our results, we evaluated the use of encounter diagnoses for discharge summaries as outcomes rather than AI-derived values. For the latter analysis, we focused on the AGE-HEM model, as ICD10 codes used for both past history and outcomes are likely to be correlated, and this correlation complicates interpretation of the AGE-HEM-HX models. We also evaluated model performance on an event-free population.
As a comparison to established models, for those individuals where all input parameters were available, we computed a 3-year estimate of atherosclerotic cardiovascular risk using the Pooled Cohort Equations (PCE)17 (see Methods in the Supplemental Material).
Evaluation of Model Performance
Each combination of outcome, predictors, and sex was evaluated by C-index and Brier score. Brier score was calculated using survival at 3 years post-CBC, using inverse probability of censoring weights to address censoring.27,28 Performance on the derivation data set is presented with means and CIs computed from repeated k-fold cross-validation (40 repeats of 5-fold cross-validation). Performance on the validation data set is presented with means and CIs calculated from bootstrapping the validation data set (1000 bootstrapped samples). Coefficients for final models are estimated using the entire derivation data set.
We also assessed discrimination on the external validation data set by comparing survival among quartiles of predicted risk, using an unweighted log-rank significance test as implemented in the Python package lifelines 0.26.29 For each model, we split the validation cohort into quartiles, according to the model’s predicted relative risk of the outcome for each patient. For models composed of the AGE-HEM-HX predictor set, we performed a pairwise comparison among the resulting four Kaplan-Meier curves.
Where the performance of 2 different predictor sets was compared for a single event model (eg, compare predictor sets AGE-HEM-HX and AGE-HX, for the time-to-death model on the female validation cohort), the comparison was reported as the difference between model scores for a given set of patients. For the derivation data set, this means that model scores were computed during each fold of cross-validation and compared for the patients in that fold. For the validation data set, this means that model scores were calculated on each bootstrapped sample and compared for the patients in that sample. Thus, a distribution of comparisons was compiled for each type of performance metric.
We evaluated model calibration graphically by plotting deciles of observed event probability versus predicted probability. Comparisons are made at 3 years of follow-up. Baseline hazard was estimated nonparametrically, using Breslow method,30 and represents the hazard for a patient whose predictors are all zero (ie, a patient with average hematologic indices, average age, and with no history of CAD or myocardial infarction or heart failure or stroke).
Random Survival Forest
To explore the benefit of modeling high dimensional interactions, we employed an ensemble learning method, random survival forest (RSF),31 to train models for each combination of outcome, sex, and predictor set with the help of the Python package scikit-survival v0.14.0. We performed a single round of hyperparameter tuning using k-fold cross-validation (k=5). Hyperparameters optimized included: trees per forest {50,150}; maximum depth of any tree {2,4,6,8}; number of features examined for splitting at each node {1,2,3,4}. We report the mean test-set C-index calculated over the k folds. The hyperparameter set represented by the reported C-index is the one that produced the highest mean C-index.
Other Statistical Analyses
In addition to the above analyses, we compared baseline characteristics for the derivation and validation cohorts using χ2 tests (categorical variables) or t test (continuous variables). We compared baseline clinical characteristics using t tests for cohort characteristics that are sample averages and Z tests for characteristics that are frequencies or proportions. Confidence intervals for frequency of clinical outcomes were estimated by the Clopper-Pearson method.32
Ethical Statement
This study complies with all ethical regulations and guidelines. The study protocol was approved by the Mass General Brigham institutional review board of (2019P002651).
Results
Baseline Characteristics
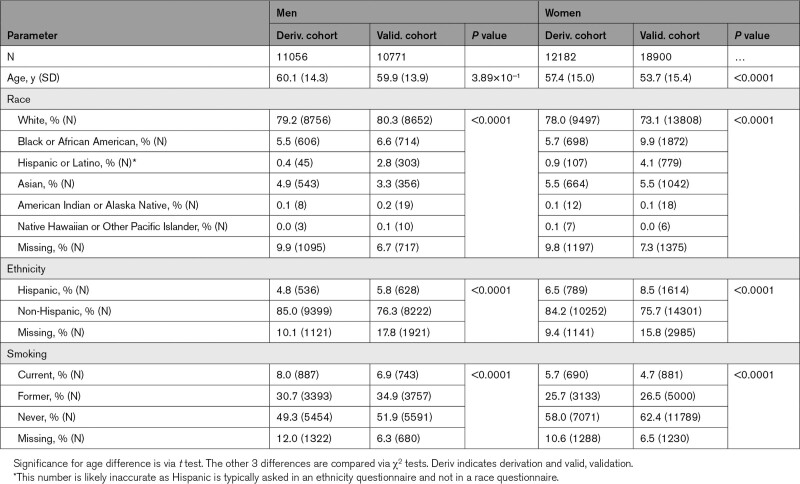
The derivation cohort included 11 056 men and 12 182 women, all of whom received at least one outpatient CBC between April 2, 2016, and May 16, 2017 (Tables 2 and 3). The external validation cohort consisted of 10 771 men and 18 900 women, all of whom received at least one outpatient CBC between June 1, 2015, and January 1, 2017 (Figure S1).17,18
Table 2.
Baseline Demographic Characteristics of Derivation and Validation Cohorts
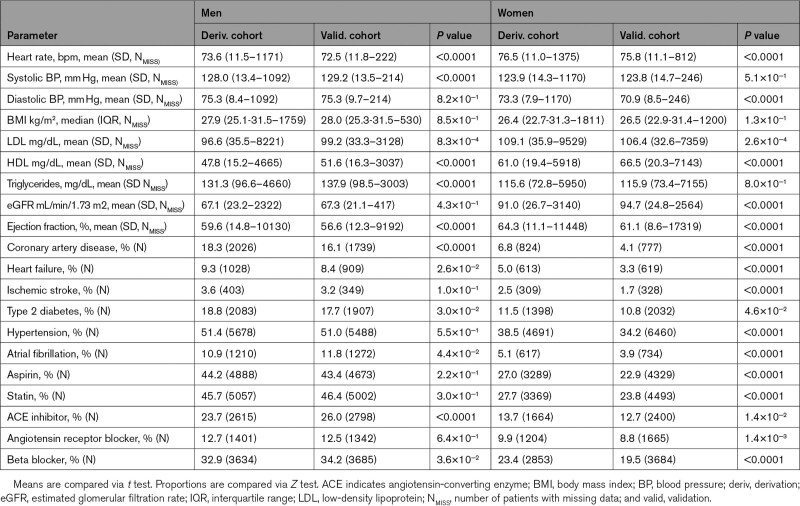
Table 3.
Baseline Clinical Characteristics of Derivation and Validation Cohorts
Median follow-up time was 3.0 years for the derivation cohort and 4.2 years for the validation cohort (Figures S2 and S3). The most notable differences between derivation and validation cohorts were a younger age of the female validation cohort than the derivation cohort (53.7 versus 57.4 years) and greater racial diversity within the validation cohort (eg, 19.6% non-White in female validation versus 12.3% in female derivation). The prevalence of CVD in the male derivation cohort was similar to that in the male validation cohort, whereas the prevalence in the female derivation cohort was higher than in the validation cohort.
Rates of CVD and Death
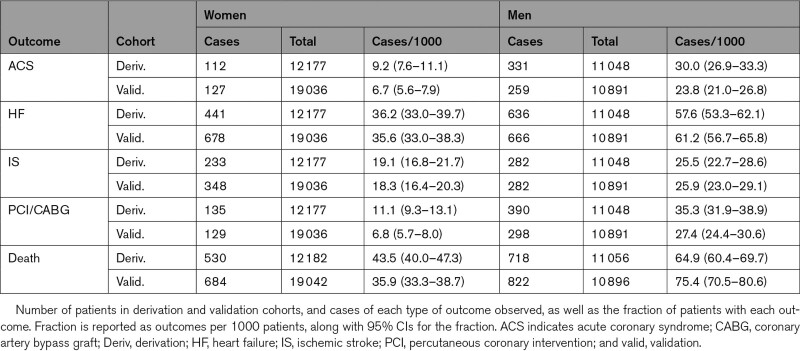
In the derivation cohort, the number of patients with one or more ACS events within the follow-up period was 443 (1.9%), with one or more HF hospitalizations was 1077 (4.6%), with one or more ISs was 515 (2.2%), with one or more coronary revascularizations was 525 (2.3%; Table 4). The number who died was 1248 (5.4%). In the validation cohort, the number of patients with one or more ACS events was 386 (1.3%), with one or more HF hospitalizations was 1344 (4.5%), with one or more ISs was 630 (2.1%), with one or more coronary revascularizations was 427 (1.4%). The number who died was 1506 (5.0%).
Table 4.
Frequency of Clinical Outcomes in Derivation and Validation Cohort
Model Performance
To evaluate the contribution of hematologic predictors, we first compare the age-only predictor set to the age-hematology predictor set. We then compare the age-hematology predictor set to the age-history predictor set. Finally, we focus on the complete model with age, medical history, and hematologic parameters.
Hematology-Only
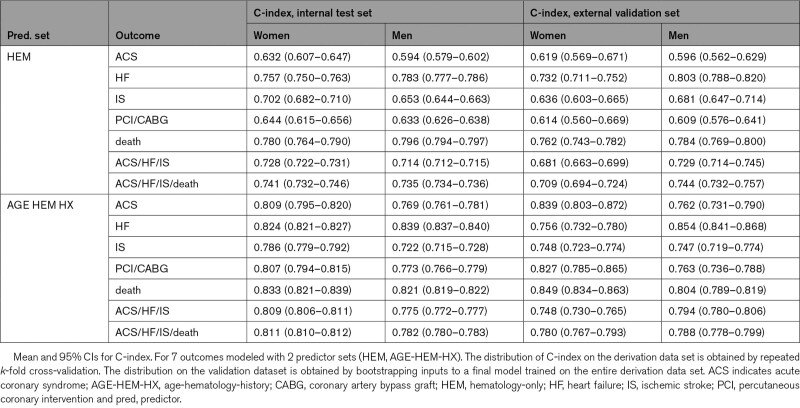
Hematologic parameters alone had utility for predicting cardiovascular outcomes (Table S6). On the internal test set, C-indices for the hematologic parameter models ranged from 0.59 to 0.80, whereas, on the validation set, C-indices ranged from 0.61 to 0.80 (Table 5). For both cohorts, the maximal discrimination was seen for HF, composite cardiovascular outcomes, and death for both sexes, whereas the poorest prediction was seen for ACS risk in men (Table 5).
Table 5.
Discrimination Performance on Internal Test Set and External Validation Set
Age-Only Compared With Age-Hematology
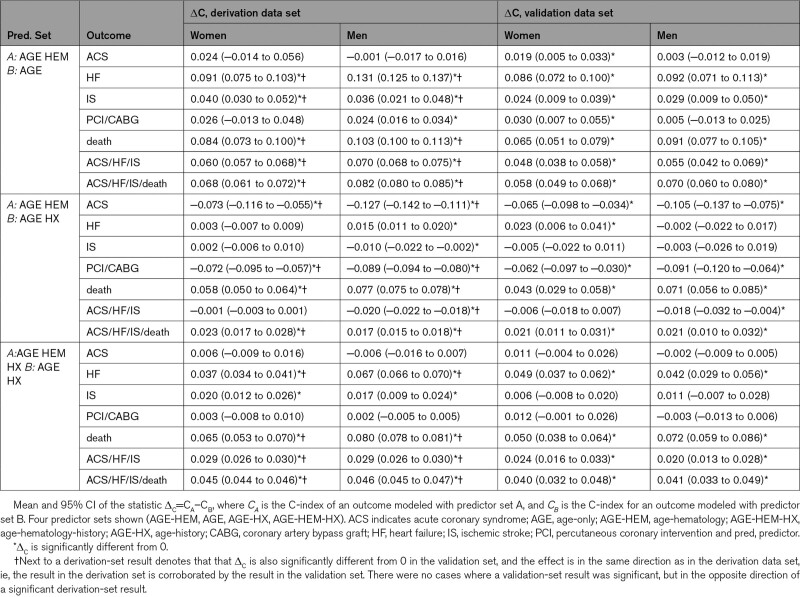
The addition of hematologic parameters to age significantly improved discrimination and calibration for nearly all outcomes (Table 6, Tables S7 through S9, AGE-HEM versus AGE), for both the internal test set and external validation set. The only models that did not benefit from adding the hematologic predictors were models for ACS and PCI/CABG, in which cases the improvement in C-index was minor or not statistically significant.
Table 6.
Comparison of Pairs of Predictor Sets According to C-Index
Age-Hematology Compared With Age-History
For several outcomes (HF, IS, composite outcomes), models with age and hematologic indices were comparable or mildly inferior to those using age and structured diagnostic codes for history of CVD and were superior in the case of death, with an increase in C-index of 0.077 (Table 6, Tables S7 and S8, AGE-HEM versus AGE-HX). In contrast, they were inferior predicting ACS and PCI/CABG, with a drop in C-index between 0.062 and 0.127, which was significant in both sexes and data sets. Calibration showed minimal difference, (Table S9, AGE-HEM versus AGE-HX), with the HF model showing the lowest magnitude change in performance (an increase of 0.003 on the external data set).
The Full Model: Age, Medical History, and Hematologic Parameters
Discrimination and Calibration
As with the age-only models, adding hematologic predictors to the age-history models yielded significantly higher C-indices for both men and women in several circumstances (Table 6, AGE-HEM-HX versus AGE-HX). For example, under the age-hematology-history predictor set, C-index for the male HF model was higher by 0.067 and C-index for the male death model was higher by 0.080, compared with the age-history predictor set. Likewise, models for death and for the 2 composite outcomes showed statistically significant improvements in both cohorts for both sexes. Improvements in Brier score followed almost the same pattern (Table S9, AGE-HEM-HX versus AGE-HX). Improvement in prediction of ACS or PCI/CABG was not statistically significant. On the validation cohort, the combined age-hematology-history models demonstrated C-indices between 0.75 and 0.85 (Table 5).
Calibration Curves
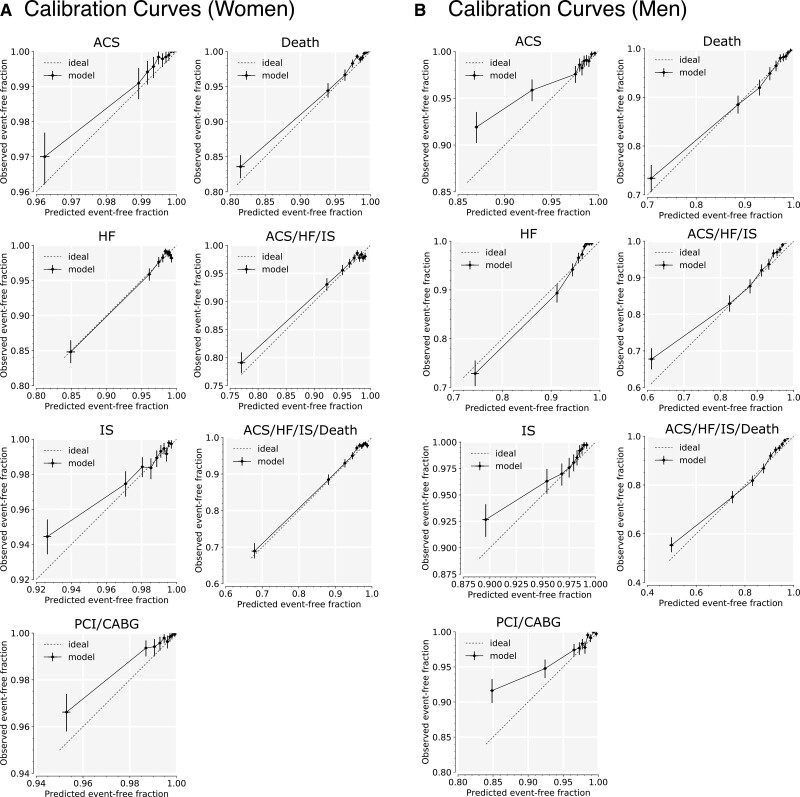
We examined calibration curves for the age-hematology-history model for all outcomes (Figure 1), as measured on the validation cohort. For women, risk was well-calibrated for all but the highest-risk decile. Over-prediction of risk for high-risk patients was more pronounced among the male models than the female models, consistent with higher Brier scores for male models.
Figure 1.
Calibration curves for age-hematology-history models in the validation cohort. Calibration curves for the 7 modeled outcomes, on women (A) and men (B) in the validation cohort. Each data point is an average over the set of 1000 bootstrapped samples. Each pair of error bars represents the middle 95% of values from the bootstrapped samples. The predictor set includes hematology and age and disease history, and interactions with age. Predicted risk is compared with observed outcomes at 3 y. ACS indicates acute coronary syndrome; CABG, coronary artery bypass graft; HF, heart failure; IS, ischemic stroke; and PCI, percutaneous coronary intervention.
Survival by Predicted-Risk Quartile
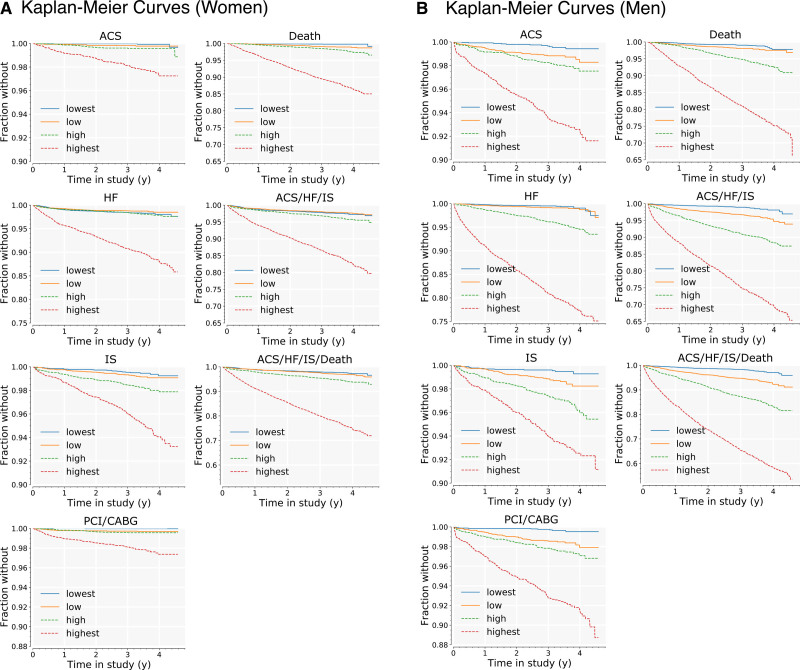
We assessed discrimination of each model by comparing survival among quartiles of the risk that it predicted. Kaplan-Meier curves are shown in Figure 2, grouped by model, one subplot per model. In each subplot, the validation cohort was split into quartiles according to the predicted relative risk of the outcome for each patient.
Figure 2.
Kaplan-Meier curves for age-hematology-history models, by quartiles of predicted risk in validation cohort. Curves are shown for all 7 outcomes in the validation cohort, women (A) and men (B). For each outcome, the cohort is split into quartiles, according to the risk model developed for that outcome on the derivation cohort. Lowest-risk quartile is shown in blue; highest-risk quartile is shown in red. ACS indicates acute coronary syndrome; CABG, coronary artery bypass graft; HF, heart failure; IS, ischemic stroke; and PCI, percutaneous coronary intervention.
For all 7 models among men, there were statistically significant differences (P<0.05) in survival between each of the 4 quartiles, with a single exception: for the HF model, survival curves for the low-risk and lowest-risk quartiles intersect at a follow-up time of ≈4 years. This could indicate poor calibration of this model for the healthiest patients, though given the low event rate after 4 years (<10/mo for the entire male cohort), it may be attributable to uncertainty in the Kaplan-Meier estimator.
Performance for the female-cohort models was similar. Survival curves for the highest risk 3 quartiles were significantly different from each other with the following exceptions: (1) the high-risk group and low-risk group were not different for the ACS model or for the PCI/CABG model; (2) the high-risk group and lowest-risk group were not different for the HF model.
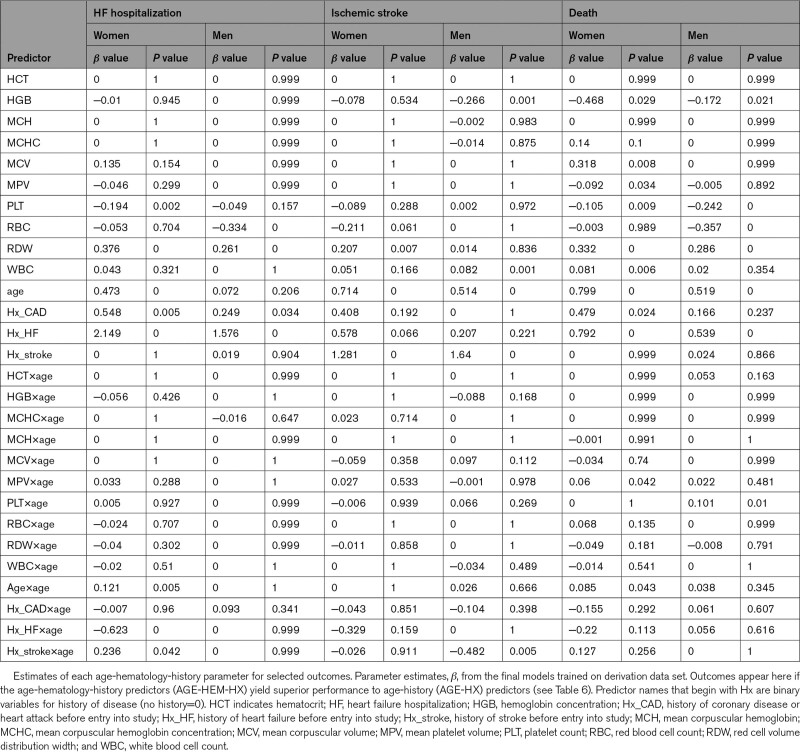
Parameter Estimates
Proportional-hazards coefficients in the age-hematology-history models were examined for the 3 outcomes (composites not included) where the predictor set had better discrimination than age-history on the internal test set (Table 7). The L1 penalization selected approximately half of the 10 hematologic indices for each of the final models. Among the highly correlated set {HCT, HGB, MCH, MCHC, MCV, red cell count}, usually more than half were eliminated, with the most retained being red cell count and HGB. Two out of the 10 hematologic indices appeared in all 6 models in Table 7: platelet count and RDW. White cell count and red cell count were retained in 5 out of 6 models. As expected, the disease-history and age terms were strong factors in most age-hematology-history models.
Table 7.
Age-History-Hematology Parameter Estimates for Selected Outcomes
Related Analyses
We explored the robustness of our results by substituting encounter diagnoses for AI-derived outcomes. Overall, C-indices were comparable and within the margin of uncertainty, though the HF and ACS/HF/IS models had higher C-indices using ICD10 codes (Tables S10 and S11). We confirmed that hematology parameters add to traditional risk factors, with C-indices and Brier scores all improving. We next looked at whether using the Charlson comorbidity index rather than the focused CVD past history codes would eliminate the additional utility of the hematology predictors, but that was not the case (Tables S12 and S13). We also looked at whether our models, which were trained using a combination of primary and secondary events as outcomes, performed as well on patients who had no CVD events before their entry into study. The incremental benefit of hematology parameters over traditional risk factors was largely intact when tested on the restricted cohort (Tables S14 and S15). For example, we saw 9 models with significant improvements in C-index when tested on the restricted cohort, instead of 12 models. We found a similar pattern with slightly more degraded performance when we tested the models on patients who had neither CVD events nor any ICD-derived history of CVD before their entry into study (Tables S16 and S17). In contrast, the number of models whose Brier score benefited from using hematology parameters on the restricted cohorts was the same as or higher than the number on the entire validation cohort.
To benchmark our hematology-based models against an established risk model, we compared the age-hematology models to a version of the PCE for ASCVD risk17 which we modified to estimate 3-year instead of 10-year risk. The age-hematology models were re-trained to predict a composite outcome, ACS/IS/death, which approximates the end point specified for the PCEs—either CHD death or the first occurrence of myocardial infarction or stroke. When tested in the derivation cohort, the age-hematology models showed superior discrimination (Table S18) and calibration (Table S19) to the PCE risk models. When tested in the validation cohort, the age-hematology model for women had superior discrimination to the PCE risk models for women. No other significant differences appeared between the 2 types of model in the validation cohort.
Random Survival Forest
Calibration of the RSF models was comparable to that of the Cox proportional hazards model (Table S20). Many of the optimized RSF models had slightly better discrimination than the Cox proportional hazards models, with advantages for RSF ranging from −0.009 to 0.050. The biggest advantage for RSF came when using the hematology predictor set, which does not include any explicit interaction terms, and which therefore, might be harder for a linear model to learn from than for a tree-based model. Improvement was less apparent for the age-hematology-history models.
Discussion
The primary result of this article is that hematologic predictors have predictive value for cardiovascular outcomes both above and beyond traditional risk factors. Our hematology-based models demonstrated success primarily for heart failure, major adverse cardiovascular events, all-cause mortality, and to a lesser extent, IS. In addition, model performance was robust to the choice of ICD10 or AI-determined outcomes.
Hematology-based models had less success predicting coronary revascularization or incident ACSs beyond what was possible using only age and prior diagnoses. Nonetheless, a model using hematologic indices alone was able to modestly predict incident ACSs (Table 5): C-index=0.62 (95% CI, 0.57–0.67) and 0.60 (95% CI, 0.56–0.63) for women and men, respectively, on the validation data set, comparable to polygenic risk scores.2 This suggests that some signal exists in the hematology data, but there is likely redundancy with age and existing CVD. Nonetheless, a strategy combining age and hematologic predictors may still be helpful in predicting coronary revascularization or incident ACSs when structured diagnoses are not as complete as those found within the MGB-EDW.
The objective of our work is fundamentally different from that of most efforts focused on training risk models: we sought to develop risk models for cardiovascular events based entirely on widely available, well-calibrated,33 and objective input features. Whereas most risk models have emphasized the utility of a diverse set of inputs, which will typically require providers to order specific laboratory tests, document an expansive set of diagnoses, and record candid responses of patients regarding such attributes as smoking status,18 our approach deliberately attempts to minimize the burden on providers. Our rationale is based partly on our own experience with EHR-based risk assessment: for example, the percentage of patients in our cohorts for which one can compute the Pooled-Cohort Equation risk score from structured data within the MGB-EDW is <30%. Even when the relevant tests may have been ordered, they may not be available as structured data in the EHR if, for example, they were collected outside of the system and were only available as scanned documents. Furthermore, most risk models focus on primary prevention—whereas our focus was on a broader patient population with anticipated high resource utilization.
We have used hematologic predictors based on the assumption that many contributors to chronic diseases such as CVD are systemic and may be reflected across multiple tissue types. This hypothesis is supported by a large amount of prior data, both experimental and observational, including the ability to estimate cardiovascular outcomes from digital retinograms,34 the association of somatic mutations in leukocytes with coronary artery disease and heart failure risk,6,35 the association of a complete-blood count-based score with mortality,10,36–39 and the modulation of CVD through leukocyte-restricted gene knockouts.40,41
The utility of RDW that we and others have observed for predicting outcomes may be related to its association with anemias of chronic disease or with clonal hematopoiesis of indeterminate potential,6 which appear to predict similar patterns of events. In both cases, the biomarkers may primarily be providing a readout for underlying systemic pathway abnormalities, with the hematopoietic lineage representing a shared upstream pathway abnormality or an index of prior exposures.
The primary limitation of our work, which is also a strength, is the deliberate use of a minimal numbers of inputs to the risk models to maximize availability. Furthermore, the use of a real-world patient population is also likely to suffer from multiple biases that may not be present in cohort studies, including unmeasured confounders. Our patient population may also exhibit some selection bias that could limit generalizability to other settings. Specifically, our model was trained on a hospital outpatient-based cohort rather than a population-based cohort. Hence, the cohort includes very few, if any, truly healthy individuals. However, the findings are in agreement with previous reports testing the association of hematologic indices with CVD.42,43 Furthermore, we would like to emphasize that our approach combining widely available inputs with automated adjudication should enable rescaling or retraining whenever it needs to be deployed on a cohort where the model does not perform well out-of-the-box. Such an approach should address if the composition of the cohort for which deployment is intended varies from that described here (eg, racial composition or comorbidity distribution). We have also not looked at how these measures of risk progress through time and in response to therapy in individual patients. Given the limited performance benefit of RSFs for predicting outcomes with these same inputs, we chose not to explore other algorithms, though some might show superior performance. The positive predictive value of our AI-adjudicated models, which in general was high, could be improved further. Nonetheless, the scale of our work, involving the analysis of >1 million notes, made a manual alternative unfeasible. Additionally, we have not provided a formal utility analysis for this model, which will depend on event rates and downstream consequences of a more widely available assessment of risk. Next, our event adjudication is likely an underestimate, as patients may have events at hospitals outside of those for which we accessed discharge summaries. Finally, for modest event rates such as those observed here (between 1% and 7.5% per person-year), measures such as C-index may not provide enough emphasis on false positives, which may be of greater interest for some applications.44,45
There is no shortage of risk models available for cardiovascular outcomes—and the challenge is motivating their use. We suspect that models such as the ones described here will primarily be attractive at an institutional or payer level, where better estimation of risk for a considerable percentage of a population has clear economic value46 or at a community level, where global identification of patients at risk is the first step in instituting preventive measures.47 We thus see our efforts aligned with the increasing emphasis on population health, whether the population is a panel within a hospital system or a group of patients within a community. Subsequent, more specialized and targeted testing can follow such an initial risk assessment rather than be a precondition for risk to be estimated in the first place. Given the deployment of hematology counters,48–51 such models could have a broad reach, though should be fine-tuned to better reflect local patterns of health outcomes.
Few efforts exist that create risk models from clinical electronic health record data or deliberately include minimal features, and none is comparable to this AI-driven approach that pushes the limits of modeling cardiovascular risk. We see our approach as complementary to other models which typically rely on individual or provider-entered data and have focused on patients not currently taking statins and free of CVD.17,18,52 Our strategy may also lend itself to the creation of continuously updating risk trajectories where the rates of change may have additional predictive utility. Our goal is to maximize the number of patients for which a system-level estimation of risk is feasible, with minimal disruption to the existing workflow of providers, a place where we see model deployment having the greatest impact.
Article Information
Sources of Funding
This work was supported by One Brave Idea.
Disclosures
Dr MacRae is a co-founder of Atman Health RCD: co-founder of Atman Health. The other authors report no conflicts.
Supplemental Materials
Methods
Figures S1–S3
Tables S1–S20
References53
Supplementary Material
Nonstandard Abbreviations and Acronyms
- ACS
- acute coronary syndrome
- AGE-HEM
- The set of predictors composed of age and hematology indices
- AGE-HEM-HX
- The set of predictors composed of age, hematology indices, and disease history
- AGE-HX
- The set of predictors composed of age and disease history
- AI
- artificial intelligence
- CABG
- coronary artery bypass graft
- CAD
- coronary artery disease
- CBC
- complete blood count
- CVD
- cardiovascular disease
- HEM
- The set of predictors composed only of hematology indices
- HEM-HX
- The set of predictors composed of hematology indices and disease history
- HF
- heart failure hospitalization
- IS
- ischemic stroke
- MGB-EDW
- Massachusetts General Brigham Electronic Data Warehouse
- PCE
- pooled cohort equation
- PCI
- percutaneous coronary intervention
- RDW-CV
- red cell volume distribution width
- RSF
- random survival forest
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/CIRCOUTCOMES.121.008007.
For Sources of Funding and Disclosures, see page 389.
Contributor Information
James G. Truslow, Email: jgtruslow@bwh.harvard.edu.
Shinichi Goto, Email: sgoto2@keio.jp.
Max Homilius, Email: mhomilius@bwh.harvard.edu.
Christopher Mow, Email: cmow1@partners.org.
John M. Higgins, Email: higgins.john@mgh.harvard.edu.
Calum A. MacRae, Email: cmacrae@bwh.harvard.edu.
References
- 1.Pencina MJ, Goldstein BA, D’Agostino RB. Prediction models - development, evaluation, and clinical application. New Engl J Med. 2020;382:1583–1586. doi: 10.1056/NEJMp2000589 [DOI] [PubMed] [Google Scholar]
- 2.Mosley JD, Gupta DK, Tan J, Yao J, Wells QS, Shaffer CM, Kundu S, Robinson-Cohen C, Psaty BM, Rich SS, et al. Predictive accuracy of a polygenic risk score compared with a clinical risk score for incident coronary heart disease. JAMA. 2020;323:627–635. doi: 10.1001/jama.2019.21782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khan SS, Cooper R, Greenland P. Do polygenic risk scores improve patient selection for prevention of coronary artery disease? JAMA. 2020;323:614–615. doi: 10.1001/jama.2019.21667 [DOI] [PubMed] [Google Scholar]
- 4.Elliott J, Bodinier B, Bond TA, Chadeau-Hyam M, Evangelou E, Moons KGM, Dehghan A, Muller DC, Elliott P, Tzoulaki I. Predictive accuracy of a polygenic risk score-enhanced prediction model vs a clinical risk score for coronary artery disease. JAMA. 2020;323:636–645. doi: 10.1001/jama.2019.22241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zethelius B, Berglund L, Sundström J, Ingelsson E, Basu S, Larsson A, Venge P, Arnlöv J. Use of multiple biomarkers to improve the prediction of death from cardiovascular causes. N Engl J Med. 2008;358:2107–2116. doi: 10.1056/NEJMoa0707064 [DOI] [PubMed] [Google Scholar]
- 6.Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, Lindsley RC, Mermel CH, Burtt N, Chavez A, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–453. doi: 10.1038/nm.2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pearl J, Bareinboim E. External validity: from do-calculus to transportability across populations. Stat Sci. 2014;29:579–595. doi: 10.1214/14-STS486 [Google Scholar]
- 9.DeSalvo KB, O’Carroll PW, Koo D, Auerbach JM, Monroe JA. Public health 3.0: time for an upgrade. Am J Public Health. 2016;106:621–622. doi: 10.2105/AJPH.2016.303063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horne BD, Anderson JL, Muhlestein JB, Ridker PM, Paynter NP. Complete blood count risk score and its components, including RDW, are associated with mortality in the JUPITER trial. Eur J Prev Cardiol. 2015;22:519–526. doi: 10.1177/2047487313519347 [DOI] [PubMed] [Google Scholar]
- 11.Laufer Perl M, Havakuk O, Finkelstein A, Halkin A, Revivo M, Elbaz M, Herz I, Keren G, Banai S, Arbel Y. High red blood cell distribution width is associated with the metabolic syndrome. Clin Hemorheol Microcirc. 2015;63:35–43. doi: 10.3233/CH-151978 [DOI] [PubMed] [Google Scholar]
- 12.Zalawadiya SK, Zmily H, Farah J, Daifallah S, Ali O, Ghali JK. Red cell distribution width and mortality in predominantly African-American population with decompensated heart failure. J Card Fail. 2011;17:292–298. doi: 10.1016/j.cardfail.2010.11.006 [DOI] [PubMed] [Google Scholar]
- 13.Felker GM, Allen LA, Pocock SJ, Shaw LK, McMurray JJ, Pfeffer MA, Swedberg K, Wang D, Yusuf S, Michelson EL, et al. ; CHARM Investigators. Red cell distribution width as a novel prognostic marker in heart failure: data from the CHARM Program and the Duke Databank. J Am Coll Cardiol. 2007;50:40–47. doi: 10.1016/j.jacc.2007.02.067 [DOI] [PubMed] [Google Scholar]
- 14.Tonelli M, Sacks F, Arnold M, Moye L, Davis B, Pfeffer M; for the Cholesterol and Recurrent Events (CARE) Trial Investigators. Relation between red blood cell distribution width and cardiovascular event rate in people with coronary disease. Circulation. 2008;117:163–168. doi: 10.1161/CIRCULATIONAHA.107.727545 [DOI] [PubMed] [Google Scholar]
- 15.Kofink D, Muller SA, Patel RS, Dorresteijn JAN, Berkelmans GFN, de Groot MCH, van Solinge WW, Haitjema S, Leiner T, Visseren FLJ, et al. ; SMART Study Group. Routinely measured hematological parameters and prediction of recurrent vascular events in patients with clinically manifest vascular disease. PLoS One. 2018;13:e0202682. doi: 10.1371/journal.pone.0202682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forget P, Khalifa C, Defour JP, Latinne D, Van Pel MC, De Kock M. What is the normal value of the neutrophil-to-lymphocyte ratio? BMC Res Notes. 2017;10:12. doi: 10.1186/s13104-016-2335-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goff DC, Jr, Lloyd-Jones DM, Bennett G, Coady S, D’Agostino RB, Sr, Gibbons R, Greenland P, Lackland DT, Levy D, O’Donnell CJ, et al. 2013 ACC/AHA guideline on the assessment of cardiovascular risk: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol. 2014;63(25 Pt B):2935–2959. doi: 10.1016/j.jacc.2013.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hippisley-Cox J, Coupland C, Brindle P. Development and validation of QRISK3 risk prediction algorithms to estimate future risk of cardiovascular disease: prospective cohort study. BMJ. 2017;357:j2099. doi: 10.1136/bmj.j2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goto S, Homilius M, John JE, Truslow JG, Werdich AA, Blood AJ, Park BH, MacRae CA, Deo RC. Artificial intelligence-enabled event adjudication: estimating delayed cardiovascular effects of respiratory viruses. medRxiv. doi: 10.1101/2020.11.12.20230706 [Google Scholar]
- 20.Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40:373–383. doi: 10.1016/0021-9681(87)90171-8 [DOI] [PubMed] [Google Scholar]
- 21.Quan H, Sundararajan V, Halfon P, Fong A, Burnand B, Luthi JC, Saunders LD, Beck CA, Feasby TE, Ghali WA. Coding algorithms for defining comorbidities in ICD-9-CM and ICD-10 administrative data. Med Care. 2005;43:1130–1139. doi: 10.1097/01.mlr.0000182534.19832.83 [DOI] [PubMed] [Google Scholar]
- 22.Charlson M, Szatrowski TP, Peterson J, Gold J. Validation of a combined comorbidity index. J Clin Epidemiol. 1994;47:1245–1251. doi: 10.1016/0895-4356(94)90129-5 [DOI] [PubMed] [Google Scholar]
- 23.Harrell FE, Jr, Califf RM, Pryor DB, Lee KL, Rosati RA. Evaluating the yield of medical tests. JAMA. 1982;247:2543–2546. [PubMed] [Google Scholar]
- 24.Breslow NE. Analysis of survival data under the proportional hazards model. Int Statistical Rev Revue Int De Statistique. 1975;43:45. doi:10.2307/1402659 [Google Scholar]
- 25.Pölsterl S. Scikit-survival: a library for time-to-event analysis built on Top of scikit-learn. J Mach Learn Res. 2020;21:1–6.34305477 [Google Scholar]
- 26.Kalbfleisch JD, Prentice RL. The Statistical Analysis of Failure Time Data. John Wiley & Sons Inc.; 2002. [Google Scholar]
- 27.Brier GW. Verification of forecasts expressed in terms of probability. Mon Weather Rev. 1950;78:1–3. [Google Scholar]
- 28.Graf E, Schmoor C, Sauerbrei W, Schumacher M. Assessment and comparison of prognostic classification schemes for survival data. Stat Med. 1999;18:2529–2545. doi: 10.1002/(sici)1097-0258(19990915/30)18:17/18<2529::aid-sim274>3.0.co;2-5 [DOI] [PubMed] [Google Scholar]
- 29.Davidson-Pilon C. Lifelines: survival analysis in Python. J Open Source Softw. 2019;4:1317. doi: 10.21105/joss.01317 [Google Scholar]
- 30.Breslow N. Discussion on Professor Cox’s Paper. J Royal Statistical Soc Ser B Methodol. 1972;34:202–220. doi: 10.1111/j.2517-6161.1950.tb00039.x [Google Scholar]
- 31.Ishwaran H, Kogalur UB, Blackstone EH, Lauer MS. Random survival forests. Ann Appl Statistics. 2008;2:841–860. doi: 10.1002/9781118445112.stat08188 [Google Scholar]
- 32.Clopper CJ, Pearson ES. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika. 1934;26:404. doi: 10.1093/biomet/26.4.404 [Google Scholar]
- 33.Miller WG. The role of proficiency testing in achieving standardization and harmonization between laboratories. Clin Biochem. 2009;42:232–235. doi: 10.1016/j.clinbiochem.2008.09.004 [DOI] [PubMed] [Google Scholar]
- 34.Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, Venugopalan S, Widner K, Madams T, Cuadros J, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402–2410. doi: 10.1001/jama.2016.17216 [DOI] [PubMed] [Google Scholar]
- 35.Dorsheimer L, Assmus B, Rasper T, Ortmann CA, Ecke A, Abou-El-Ardat K, Schmid T, Brüne B, Wagner S, Serve H, et al. Association of mutations contributing to clonal hematopoiesis with prognosis in chronic ischemic heart failure. JAMA Cardiol. 2019;4:25–33. doi: 10.1001/jamacardio.2018.3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horne BD, May HT, Muhlestein JB, Ronnow BS, Lappé DL, Renlund DG, Kfoury AG, Carlquist JF, Fisher PW, Pearson RR, et al. Exceptional mortality prediction by risk scores from common laboratory tests. Am J Med. 2009;122:550–558. doi: 10.1016/j.amjmed.2008.10.043 [DOI] [PubMed] [Google Scholar]
- 37.Horne BD, Muhlestein JB, Bennett ST, Muhlestein JB, Jensen KR, Marshall D, Bair TL, May HT, Carlquist JF, Hegewald M, et al. Extreme erythrocyte macrocytic and microcytic percentages are highly predictive of morbidity and mortality. JCI Insight. 2018;3:120183. doi: 10.1172/jci.insight.120183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anderson JL, Ronnow BS, Horne BD, Carlquist JF, May HT, Bair TL, Jensen KR, Muhlestein JB; Intermountain Heart Collaborative (IHC) Study Group. Usefulness of a complete blood count-derived risk score to predict incident mortality in patients with suspected cardiovascular disease. Am J Cardiol. 2007;99:169–174. doi: 10.1016/j.amjcard.2006.08.015 [DOI] [PubMed] [Google Scholar]
- 39.Brennan ML, Reddy A, Tang WH, Wu Y, Brennan DM, Hsu A, Mann SA, Hammer PL, Hazen SL. Comprehensive peroxidase-based hematologic profiling for the prediction of 1-year myocardial infarction and death. Circulation. 2010;122:70–79. doi: 10.1161/CIRCULATIONAHA.109.881581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Eck M, Bos IS, Kaminski WE, Orsó E, Rothe G, Twisk J, Böttcher A, Van Amersfoort ES, Christiansen-Weber TA, Fung-Leung WP, et al. Leukocyte ABCA1 controls susceptibility to atherosclerosis and macrophage recruitment into tissues. Proc Natl Acad Sci USA. 2002;99:6298–6303. doi: 10.1073/pnas.092327399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duewell P, Kono H, Rayner KJ, Sirois CM, Vladimer G, Bauernfeind FG, Abela GS, Franchi L, Nuñez G, Schnurr M, et al. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature. 2010;464:1357–1361. doi: 10.1038/nature08938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kannel WB, Anderson K, Wilson PW. White blood cell count and cardiovascular disease. Insights from the Framingham Study. JAMA. 1992;267:1253–1256. [PubMed] [Google Scholar]
- 43.Lassale C, Curtis A, Abete I, van der Schouw YT, Verschuren WMM, Lu Y, Bueno-de-Mesquita HBA. Elements of the complete blood count associated with cardiovascular disease incidence: findings from the EPIC-NL cohort study. Sci Rep. 2018;8:3290. doi: 10.1038/s41598-018-21661-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hand DJ. Evaluating diagnostic tests: the area under the ROC curve and the balance of errors. Stat Med. 2010;29:1502–1510. doi: 10.1002/sim.3859 [DOI] [PubMed] [Google Scholar]
- 45.Hand DJ. Measuring classifier performance: a coherent alternative to the area under the ROC curve. Mach Learn. 2009;77:103–123. doi: 10.1007/s10994-009-5119-5 [Google Scholar]
- 46.Trombley MJ, Fout B, Brodsky S, McWilliams JM, Nyweide DJ, Morefield B. Early effects of an accountable care organization model for underserved areas. N Engl J Med. 2019;381:543–551. doi: 10.1056/NEJMsa1816660 [DOI] [PubMed] [Google Scholar]
- 47.Kindig D, Stoddart G. What is population health? Am J Public Health. 2003;93:380–383. doi: 10.2105/ajph.93.3.380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prodjosoewojo S, Riswari SF, Djauhari H, Kosasih H, van Pelt LJ, Alisjahbana B, van der Ven AJ, de Mast Q. A novel diagnostic algorithm equipped on an automated hematology analyzer to differentiate between common causes of febrile illness in Southeast Asia. PLoS Negl Trop Dis. 2019;13:e0007183. doi: 10.1371/journal.pntd.0007183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buoro S, Seghezzi M, Vavassori M, Dominoni P, Apassiti Esposito S, Manenti B, Mecca T, Marchesi G, Castellucci E, Azzarà G, et al. Clinical significance of cell population data (CPD) on Sysmex XN-9000 in septic patients with our without liver impairment. Ann Transl Med. 2016;4:418. doi: 10.21037/atm.2016.10.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hwang DH, Dorfman DM, Hwang DG, Senna P, Pozdnyakova O. Automated nucleated RBC measurement using the sysmex XE-5000 hematology analyzer: frequency and clinical significance of the nucleated RBCs. Am J Clin Pathol. 2016;145:379–384. doi: 10.1093/ajcp/aqv084 [DOI] [PubMed] [Google Scholar]
- 51.Park SH, Park CJ, Lee BR, Nam KS, Kim MJ, Han MY, Kim YJ, Cho YU, Jang S. Sepsis affects most routine and Cell Population Data (CPD) obtained using the Sysmex XN-2000 blood cell analyzer: neutrophil-related CPD NE-SFL and NE-WY provide useful information for detecting sepsis. Int J Lab Hematol. 2015;37:190–198. doi: 10.1111/ijlh.12261 [DOI] [PubMed] [Google Scholar]
- 52.Ridker PM, Buring JE, Rifai N, Cook NR. Development and validation of improved algorithms for the assessment of global cardiovascular risk in women: the Reynolds Risk Score. JAMA. 2007;297:611–619. doi: 10.1001/jama.297.6.611 [DOI] [PubMed] [Google Scholar]
- 53.Lambert J, Chevret S. Summary measure of discrimination in survival models based on cumulative/dynamic time-dependent ROC curves. Stat Methods Med Res. 2016;25:2088–2102. doi: 10.1177/0962280213515571 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.