Abstract
Genotyping is not routinely performed at diagnosis of von Willebrand disease (VWD). Therefore, the association between genetic variants and pathogenic mechanism or the clinical and laboratory phenotype is unknown in most patients, especially in type 1 VWD. To investigate whether genotyping adds to a better understanding of the pathogenic mechanisms and variability in phenotype, we analyzed the VWF gene in 390 well-defined VWD patients, included in the WiN study. A VWF gene variant was found in 155 patients (61.5%) with type 1, 122 patients (98.4%) with type 2, and 14 patients (100%) with type 3 VWD. Forty-eight variants were novel. For each VWF gene variant, the pathogenic mechanisms associated with reduced VWF levels was investigated using the FVIII:C/VWF:Ag and VWFpp/VWF:Ag ratios. In type 1 VWD, reduced synthesis or secretion of VWF was most frequently found in patients with nonsense variants, frameshift variants, and deletions, whereas rapid clearance of VWF was mainly found in patients with missense variants. Furthermore, type 1 VWD patients with and without a VWF gene variant were clearly distinct in their clinical features such as age of diagnosis, laboratory phenotype, and bleeding phenotype. In type 2 VWD, 81% of variants were associated with an increased clearance of VWF. To conclude, we identified the pathogenic mechanisms associated with various VWF gene variants in type 1, 2, and 3 VWD patients. Additionally, major differences in the phenotype of type 1 VWD patients with and without a variant were observed, which may be of importance for clinical management.
INTRODUCTION
von Willebrand factor (VWF) is a multimeric glycoprotein with an important role in hemostasis.1 VWF is encoded by the VWF gene, which is located on chromosome 12.1,2 The VWF gene has 52 exons and is 178 kb long.2 VWF is synthesized in endothelial cells and megakaryocytes.1,3 After synthesis, VWF undergoes posttranslational changes and dimerization in the endoplasmic reticulum.1,3 Subsequently, VWF is multimerized in the Golgi network, packed into Weibel-Palade bodies, and transferred to the cell membrane, where VWF is released into the circulation.1,3 In the circulation, VWF initiates platelet adhesion and aggregation upon vascular damage.3 VWF is also a carrier protein for factor VIII (FVIII), preventing FVIII degradation.3
Reduced VWF levels or an abnormal function of VWF is defined as von Willebrand disease (VWD).4 VWD is the most common inherited bleeding disorder and is characterized by mucocutaneous bleeding.3 VWD is categorized into three types. Type 1 VWD is characterized by a quantitative reduction of VWF and is the most common type of VWD.3 Type 2 VWD is a qualitative disorder of VWF and affects 20%–30% of VWD patients. Type 3 VWD is characterized by a complete absence of VWF and affects <5% of VWD patients.3
A large variation of mutations or deletions in the VWF gene are associated with VWD.2 The inheritance pattern of VWD is autosomal dominant in the majority of cases.3 In type 1 VWD, most patients have autosomal dominant missense mutations with an incomplete penetrance and a variable expression.3 In type 2 VWD, dominant negative missense mutations of specific VWF gene regions lead to an abnormally functioning VWF. The location of the VWF mutation determines the abnormal VWF function and, subsequently, the subtype of type 2 VWD.2 In type 3 VWD, up to 80% of patients have a null allele mutation, which is autosomal recessive and homozygous, or compound heterozygous in most patients.3 A large variation of mutations or deletions in the VWF gene are associated with VWD.2 The inheritance pattern of VWD is autosomal dominant in the majority of cases.3 In type 1 VWD, most patients have autosomal dominant missense mutations with an incomplete penetrance and a variable expression.3 In type 2 VWD, dominant negative missense mutations of specific VWF gene regions lead to an abnormally functioning VWF. The location of the VWF mutation determines the abnormal VWF function and, subsequently, the subtype of type 2 VWD.2 In type 3 VWD, up to 80% of patients have a null allele mutation, which is autosomal recessive and homozygous, or compound heterozygous in most patients.3
The inheritance pattern of VWD is in the majority of cases autosomal dominant.2,3 In some cases, recessive inheritance and incomplete penetrance with variable expression can lead to a variable disease phenotype within a family.3,5 Type 1 and type 2 VWD are mostly caused by heterozygous missense variants, whereas type 3 VWD is mostly caused by homozygous or compound heterozygous null-alleles.2
VWF gene variants can lead to reduced VWF levels in the circulation due to reduced synthesis, reduced secretion, and increased clearance of VWF.6 Although previous in vitro studies have unraveled the pathogenic mechanisms by which some VWF gene variants cause reduced VWF levels, for most variants this has not been investigated yet.2 In vitro studies are generally time consuming, only allow the investigation of a small number of VWF gene variants at the same time, and the most used techniques such as overexpression of mutant VWF in heterologous cell systems or studies on patient-derived endothelial cells cannot detect an increased clearance of VWF. It has been previously suggested that the pathophysiology of reduced VWF levels can be assessed using the FVIII activity (FVIII:C) to VWF antigen (VWF:Ag) ratio and VWF propeptide (VWFpp) to VWF:Ag ratio.6–9 A high FVIII:C/VWF:Ag ratio is a marker for reduced synthesis or secretion of VWF, whereas a high VWFpp/VWF:Ag ratio is a marker for increased clearance of VWF.6,7,9 Although these ratios have been used in several previous studies, they have not yet been used as a tool to investigate the pathophysiological defects of reduced VWF levels in individual VWF gene variants in a large cohort of VWD patients. Although, these ratios can be more easily and practically used than in vitro studies to unravel the pathogenic mechanisms associated with multiple VWF gene variants. Furthermore, although several previous studies reported that approximately 30% to 45% of type 1 VWD patients do not have a VWF gene variant, no large studies have been performed yet to investigate whether the laboratory and bleeding phenotype of type 1 VWD patients with and without a VWF gene variant are different.3,5,10–17
Therefore, we analyzed the VWF gene of a large cohort of well-defined VWD patients included in the von Willebrand in the Netherlands (WiN) study. We aimed to investigate the pathophysiological defects of reduced VWF levels associated with each VWF variant in our cohort using the FVIII:C/VWF:Ag ratio and VWFpp/VWF:Ag ratio. Second, we investigated whether the laboratory and bleeding phenotype of type 1 VWD patients with and without a VWF gene variant are different.
MATERIALS AND METHODS
We included patients from the nationwide, cross-sectional, Willebrand in the Netherlands (WiN) study.18 Patients were enrolled between 2007 and 2009. Inclusion criteria were historically lowest VWF:Ag, VWF activity or VWF collagen binding (VWF:CB) ≤0.30 IU/mL or FVIII activity (FVIII:C) ≤0.40 IU/mL (for type 2N), and a family history of VWD or personal hemorrhagic diathesis. We excluded patients with additional coagulation defects such as other bleeding disorders or acquired VWD. The study was conducted according to the declaration of Helsinki. The Medical Ethical Committees of all participating centers approved the study, and all patients signed informed consent.
Assessment methods
The assessment methods of the WiN study have been described in detail previously.7,18 Patients filled in a self-administered Tosetto bleeding score (BS), and blood was drawn to measure VWF and FVIII levels centrally and to perform genetic analyses. In children in whom we could not obtain blood samples, saliva samples were obtained for genetic analyses. VWF:Ag, VWF activity as measured by the monoclonal antibody assay (VWF:Ab), VWF:CB, and FVIII:C were measured at the Erasmus MC University Medical Center, whereas VWFpp was measured at the Leiden University Medical Center, as described before.7,18
VWF gene analysis
Genetic analysis was performed at the laboratory of hematology of the Radboud University Medical Center in Nijmegen. Ion semiconductor sequencing (Ion-Torrent) was used to analyze the VWF gene from peripheral blood obtained in EDTA tubes or from saliva samples. The 52 exons of VWF gene were analyzed including ±20 bp exon-intron boundaries. If no VWF gene variant was detected, multiplex ligation-dependent probe amplification (MLPA) analysis was performed to find large deletions or duplications. Thus, MLPA was performed in 183 patients (46.9%). All variants were confirmed with Sanger sequencing.
Single nucleotide polymorphisms, variants which have in previous studies been classified as benign, and variants classified by prediction models (such as Polyphen-2, sorting intolerant from tolerant Align Grantham Variation Grantham Deviation, and Mutation Taster) as (likely) benign (class 1 and class 2) are considered not to be pathogenic, and therefore omitted from this article.
Definitions
Type 1 VWD was defined as historically lowest VWF:Ag ≤0.30 IU/mL and VWF:Ab/VWF:Ag ratio >0.6, whereas type 2 was defined as VWF:Ab/VWF:Ag ratio ≤0.6, and type 3 as VWF:Ag and VWFpp ≤0.05 IU/mL. However, if a patient had a specific VWF gene variant, which is consistently reported in literature as a certain type of VWD, we reclassified the patient accordingly. Reduced synthesis/secretion of VWF was defined as FVIII:C/VWF:Ag ratio ≥1.9, whereas an increased clearance of VWF was defined as a VWFpp/VWF:Ag ratio of ≥2.2, as suggested before.6 Novel variants were defined as variants in the VWF gene that are not yet described in previous literature in VWD patients. To search whether variants were previously described in literature, we have consulted a recent comprehensive review by de Jong et al, two studies in which large cohorts of VWD patients were genotyped, Pubmed database (ClinVar), European Association of Haemophilia and Allied Disorders Coagulation Factor Variant Database, Human Gene Mutation Database, gnomAD, and Ensembl Database.2,10,19–21
Statistical analysis
Continuous data are described as mean ± SD or median and interquartile range (IQR), whereas categorical data are presented as number and percentage. Normality of data was assessed visually with histograms. In case of >30 patients per group, we compared continuous variables between groups using parametric tests. Categorical data were compared between groups using a Chi-square test.
Linear regression analysis was used to compare VWF and FVIII levels and BS between patients with reduced synthesis/secretion and increased clearance of VWF, and those with undetermined pathophysiology. VWF and FVIII levels and BS were also compared with linear regression between type 1 VWD patients with and without a VWF gene variant. For VWF and FVIII levels, we adjusted these analyses for age, sex, and blood group, whereas for BS we adjusted for age and sex. In type 2 VWD, the associations between VWF:Ab/VWF:Ag and VWFpp/VWF:Ag and FVIII:C/VWF:Ag ratio were also assessed with linear regression analysis. Outcomes of linear regression analysis are presented as unstandardized beta (β), 95% confidence interval, and P value. Statistical analyses were performed with SPSS Statistics version 25 (IBM Corp., Armonk, NY, USA). A P value <0.05 was considered significant.
Results
From the WiN cohort of 834 patients, we analyzed the VWF gene in 390 patients from 326 families. We analyzed all index cases (n = 326) and from families with interesting laboratory or bleeding phenotype, additional family members. Thus, 252 patients (64.6%) with type 1, 124 (32.1%) type 2, and 14 (3.3%) type 3 VWD patients were included. The patient characteristics are shown in Table 1. Most patients were female (64.1%) and had blood group O (63.0%). The mean age (±SD) at inclusion was 43 years (±19, range 2–83).
Table 1.
Patient Characteristics
| Type 1 VWD | Type 2 VWD | Type 3 VWD | Total | |
|---|---|---|---|---|
| n = 252 | n = 124 | n = 14 | n = 390 | |
| Age, mean ± SD | 44 ± 18 | 42 ± 19 | 29 ± 22 | 43 ± 19 |
| Female, n (%) | 167 (66.3) | 76 (61.3) | 7 (50.0) | 250 (64.1) |
| Blood group O, n (%) | 169 (68.4) | 67 (54.5) | 6 (42.9) | 242 (63.0) |
| Variant found, n (%) | 155 (61.5) | 122 (98.4) | 14 (100) | 291 (74.6) |
| Families, n | 226 | 94 | 13 | 326a |
| VWF:Ag | 0.38 (0.25–0.55) | 0.25 (0.16–0.37) | 0.00 (0.00–0.03) | 0.31 (0.19–0.48) |
| VWF:Ab | 0.48 (0.24–0.74) | 0.08 (0.03–0.18) | 0.00 (0.00–0.00) | 0.29 (0.11–0.60) |
| VWF:CB | 0.45 (0.24–0.69) | 0.08 (0.06–0.23) | 0.00 (0.00–0.02) | 0.29 (0.10–0.56) |
| FVIII:C | 0.68 (0.51–0.90) | 0.37 (0.25–0.50) | 0.01 (0.01–0.04) | 0.56 (0.36–0.78) |
| Bleeding score | 9 (6–15) | 11 (7–17) | 20 (13–25) | 10 (6–16) |
Data are presented as median (interquartile ranges), unless otherwise specified.
aSome relatives had different types of VWD; therefore, the sum of number of families per type is more than 326.
VWD = von Willebrand disease.
One hundred twenty-three unique VWF gene variants were found in our cohort, of which 48 (39.0%) were novel. A genetic variant was found in 155 patients (61.5%) with type 1, 122 (98.4%) patients with type 2, and 14 patients (100%) with type 3 VWD. From the type 1 and type 2 VWD patients with a VWF gene variant, 234 patients (84.4%) had a single variant, 40 (14.5%) patients had two variants, and three patients had three variants (1.1%). In type 3 VWD patients, nine patients had a single homozygous variant, one patient had a single heterozygous variant, four patients had compound heterozygous variants, from which one patient had eight different variants due to a gene conversion.
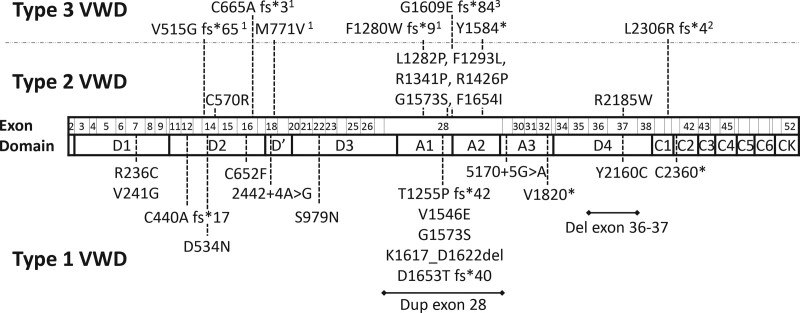
In type 1 and type 2 VWD patients with a single variant, respectively 18 and eight novel variants were identified (Figure 1). C570R, L1282P, R1426P, and G1573S were present in type 2A VWD patients with reduced high molecular weight VWF multimers. F1653I was present in a patient with type 2A, whereas R1341P was present in patients with type 2B, whom all had absence of high molecular weight VWF multimers. F1293L and R2185W were present in type 2M VWD patients with normal VWF multimers. In type 1 and type 2 VWD patients with more than one variant, respectively, 9 and 6 novel variants were identified (Suppl. Figure S1). Furthermore, seven novel variants were identified in type 3 VWD patients (Figure 1). Suppl. Table S1 provides a detailed description of each variant detected in our cohort.
Figure 1.
Novel variants in patients with a single variant found in the WiN cohort. Dotted line illustrates which exon is affected by the variant. 1Homozygous variant; 2This patient also had deletion exon 4–5; and 3This patient also had Y1570*.
Pathophysiology of reduced VWF levels in type 1 VWD
Type 1 VWD with reduced synthesis/secretion of VWF
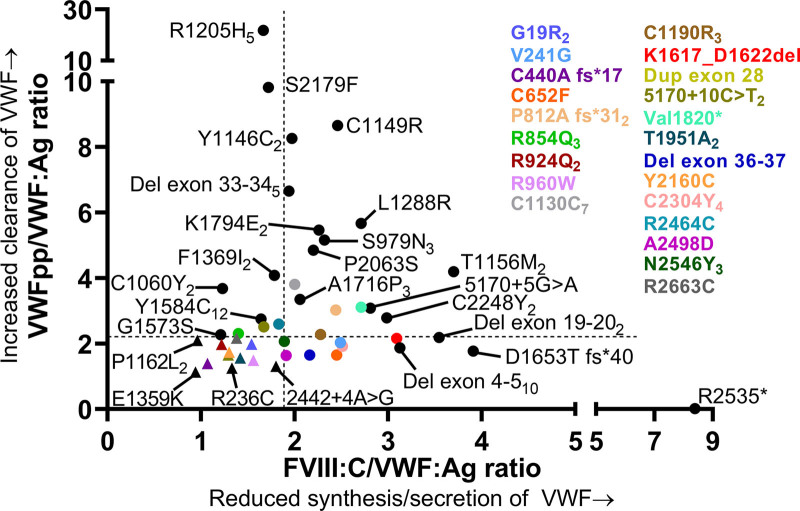
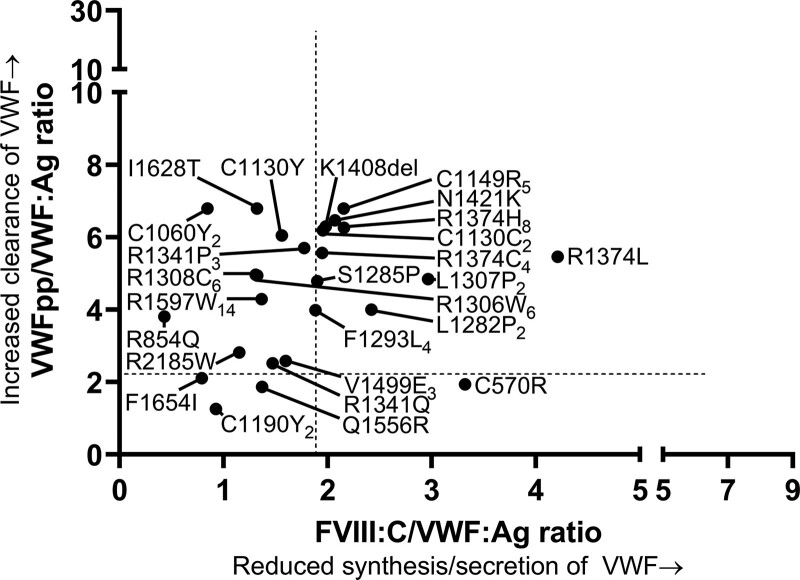
Strongly reduced synthesis/secretion of VWF with a normal clearance was found in patients with nonsense variants, frameshift variants, and deletions (Figure 2). These variants were present throughout all exons of the VWF gene (Figure 2). R2535* was present in a type 1 VWD patient with strongly reduced synthesis/secretion of VWF (FVIII:C/VWF:Ag ratio of 8.39) and centrally measured VWF:Ag of 0.06 IU/mL. Deletion of exon 4–5 was present in 14 type 1 VWD patients from 13 families in ten of whom we had data on FVIII:C/VWF:Ag and VWFpp/VWF:Ag ratios available. These patients had on average FVIII:C/VWF:Ag ratio of 3.12 and VWFpp/VWF:Ag ratio of 1.87, indicating that this variant leads to reduced synthesis/secretion of VWF. Likewise, deletion of exon 19–20, D1653T fs*40 and K1617_D1662del were associated with a very low synthesis/secretion of VWF (FVIII:C/VWF:Ag ratios above 3.0). All other type 1 VWD variants associated with reduced synthesis/secretion of VWF are illustrated in Figure 2.
Figure 2.
Pathophysiology of reduced VWF levels in patients with type 1 VWD with a single variant. Each dot represents the mean value for a variant. Triangles indicate variants with undetermined pathophysiological defects of VWF. Subscript number indicate number of patients with each variant. FVIII:C/VWF:Ag ratio ≥1.9 is defined as reduced synthesis/secretion of VWF and VWFpp/VWF:Ag ratio ≥2.2 is defined as an increased clearance of VWF. Of note, the number of patients with each variant in this figure may differ from the number of patients presented in Suppl. Table S1, because in this figure, we only present type 1 VWD patients with a single variant. VWD = von Willebrand disease.
Type 1 VWD with increased clearance of VWF
Rapid clearance of VWF with normal synthesis and secretion was mostly found in patients with missense variants in VWF exons 22 to 38 (VWF domains D3-A1-A2-A3-D4; Figure 2). As expected, R1205H (Vicenza) was the variant with the fastest clearance of VWF, followed by S2179F (Figure 2). C1149R and Y1146C were associated with type 1 VWD with a rapid clearance of VWF and VWFpp/VWF:Ag ratios of respectively 8.66 and 8.26. Of note, C1149R was also associated with reduced synthesis/secretion of VWF based on an increased FVIII:C/VWF:Ag ratio of 2.46. Except for S2179F which affected the D4 domain of VWF, all variants associated with a rapid clearance of VWF with VWFpp/VWF:Ag ratios above 6, affected the D3 domain of VWF (R1205H, C1149R, and Y1146C).
Furthermore, in-frame deletion exon 33–34, K1794E, P2063S, and S979N were also associated with a fast clearance of VWF (VWFpp/VWF:Ag ratio >4.8). Y1584C was present in 17 type 1 VWD patients from different families in 12 of whom we had data available on FVIII:C/VWF:Ag and VWFpp/VWF:Ag ratios. In patients with Y1584C, the average VWFpp/VWF:Ag ratio was 2.75 (range 1.6–5.6), suggesting that this variant is also associated with an increased clearance of VWF. Finally, A1716P was present in three patients from one family, all with an increased clearance of VWF with VWFpp/VWF:Ag ratios ranging from 2.6 to 3.9.
Type 1 VWD with reduced synthesis/secretion and increased clearance of VWF
Both reduced synthesis/secretion and increased clearance of VWF was mostly found in patients with missense mutations of the D3, A3, and D4 domains of VWF (Figure 2). C1190R, which was present in five unrelated patients and T1156M present in four patients from two families, had FVIII:C/VWF:Ag ratio from 3.1 to 4.5 and VWFpp/VWF:Ag ratio ranging from 2.8 to 5.1, suggesting that both variants are associated with reduced synthesis/secretion and increased clearance of VWF (Figure 2). Furthermore, we found two patients from one family with L1288R with mean FVIII:C/VWF:Ag ratio of 2.7 and mean VWFpp/VWF:Ag ratio of 5.7, indicating that this variant also leads to reduced synthesis/secretion and increased clearance of VWF (Figure 2).
Type 1 VWD variants with undetermined pathophysiological defects of VWF
In 20 patients with historically lowest VWF levels ≤0.30 IU/mL (and a single VWF variant), we identified 12 different variants with undetermined synthesis, secretion, and clearance defects of VWF based on FVIII:C/VWF:Ag ratio <1.9 and VWFpp/VWF:Ag ratio <2.2, respectively (Figure 2; Suppl. Table S2). These variants were found throughout all exons of the VWF gene. Mean VWFpp was normal in these patients: 0.94 ± 0.22 (range 0.56–1.29). The pathophysiologic mechanism leading to reduced VWF levels remains unclear in these variants.
Pathophysiological mechanism and phenotype in type 1 VWD
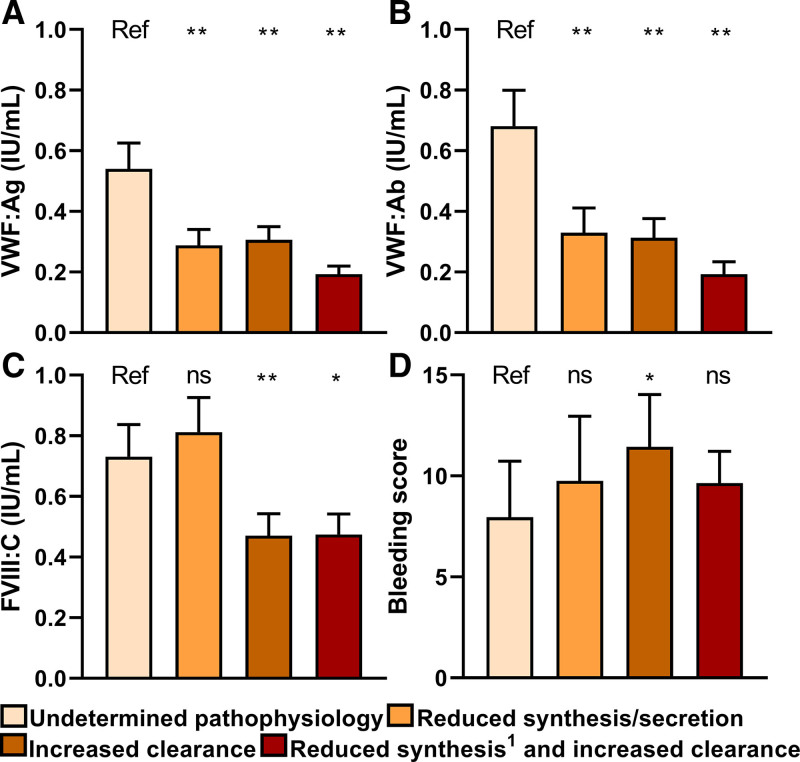
In type 1 VWD patients with a VWF gene variant, patients with reduced synthesis/secretion or increased clearance of VWF had lower VWF:Ag, VWF:Ab, and FVIII:C compared with patients with undetermined pathophysiological defects of VWF (Figure 3A–C). Also, patients with a variant leading to an increased clearance of VWF had 2.1 points higher BS compared to patients with undetermined pathophysiological defects of VWF (95% CI: 0.7; 4.0, P = 0.043) adjusted for age and sex (Figure 3D).
Figure 3.
Pathophysiology of reduced VWF levels are associated with VWF and FVIII levels and BS in type 1 VWD patients with a variant. (A, B) Patients with reduced synthesis/secretion or increased clearance have lower VWF:Ag and VWF:Ab than patients with undetermined pathophysiological defects of VWF. (C) Patients with increased clearance and both reduced synthesis/secretion and increased clearance of VWF also have lower FVIII:C compared to patients with undetermined pathophysiology. (D) BS is higher in patients with increased clearance of VWF compared with those with undetermined pathophysiological defects of VWF. Data are presented as mean and 95% CI and 1reduced secretion of VWF. *P < 0.05, **P < 0.001. ns compared with patient with undetermined pathophysiological defects of VWF. Linear regression is used to compare each group with patients with undetermined pathophysiological defects of VWF. Regression analysis are adjusted for age, sex and blood group. Analysis with BS as outcome are not adjusted for blood group. BS = bleeding score; CI = confidence intervals; ns = not significant; Ref = reference; VWD = von Willebrand disease.
Difference in type 1 VWD patients with and without a VWF gene variant
Difference in clinical phenotype
Type 1 VWD patients with a VWF gene variant were diagnosed at a younger age (26 ± 18 versus 32 ± 14, P = 0.003) and were more often referred because of a positive family history compared with patients without a VWF gene variant, who were more often referred because of a personal bleeding diathesis (P < 0.001; Table 2). In type 1 VWD patients with blood group O, a VWF gene variant was found in 95 of 169 (56.2%) patients, whereas in patients with blood group non-O a variant was found in 55 of 78 (70.5%) of patients (P = 0.032). Centrally measured VWF levels were lower in patients with a VWF gene variant compared to those without a VWF gene variant (0.27 IU/mL versus 0.53 IU/mL, P < 0.001; Table 2).
Table 2.
Differences in Type 1 VWD Patients With and Without a VWF Gene Variant
| With Variant | Without Variant | P | |
|---|---|---|---|
| n = 155 | n = 97 | ||
| Age diagnosis, mean ± SD | 26 ± 18 | 32 ± 14 | 0.003 |
| Age inclusion, mean ± SD | 41 ± 19 | 48 ± 15 | <0.001 |
| Female, n (%) | 100 (64.5%) | 67 (69.1%) | 0.457 |
| Blood group O, n (%) | 95 (63.3%) | 74 (76.3%) | 0.032 |
| Positive family history | 114 (91.9%) | 37 (56.9%) | <0.001 |
| Reason for referral | |||
| Bleeding | 71 (49.3%) | 69 (75.8%) | <0.001 |
| Family history | 73 (50.7%) | 22 (24.2%) | |
| VWF:Ag historically lowesta | 0.28 (0.20–0.38) | 0.40 (0.31–0.46) | 0.023 |
| VWF:Ab historically lowesta | 0.20 (0.10–0.28) | 0.24 (0.20–0.28) | <0.001 |
| VWF:CB historically lowesta | 0.21 (0.10–0.30) | 0.27 (0.20–0.37) | 0.006 |
| FVIII:C historically lowest | 0.46 (0.34–0.66) | 0.51 (0.40–0.66) | 0.288 |
| VWF:Agb | 0.27 (0.19–0.39) | 0.53 (0.41–0.66) | <0.001 |
| VWF:Abb | 0.26 (0.17–0.48) | 0.67 (0.53–0.93) | <0.001 |
| VWF:CBb | 0.27 (0.17–0.46) | 0.62 (0.52–0.86) | <0.001 |
| FVIII:Cb | 0.56 (0.39–0.77) | 0.86 (0.67–1.05) | <0.001 |
| FVIII:C/VWF:Ag ratioc | 1.94 (1.56–2.48) | 1.63 (1.43–1.78) | <0.001 |
| VWFpp/VWF:Ag ratiod | 2.74 (1.77–4.43) | 1.97 (1.60–2.23) | <0.001 |
| Bleeding score | 9 (5–14) | 10 (6–18) | 0.007 |
Data are presented as median (interquartile range), unless otherwise specified.
aIn all patients, either historically lowest VWF:Ag, VWF:Ab, or VWF:CB was below 0.30 IU/mL.
bCentrally measured in the WiN study.
cNormal ratio <1.9.
dNormal ratio <2.2. Independent t-test for continuous variables and chi-square for categorical variables.
VWD = von Willebrand disease.
Despite higher VWF and FVIII levels, patients without a VWF gene variant had a higher BS compared to patients with a VWF gene variant, respectively 10 (6–18) versus 9 (5–14) (P = 0.007). After adjustment for age and sex, this difference remained significant: β = 1.9 (0.1; 3.6), P = 0.039.
Difference in pathophysiology
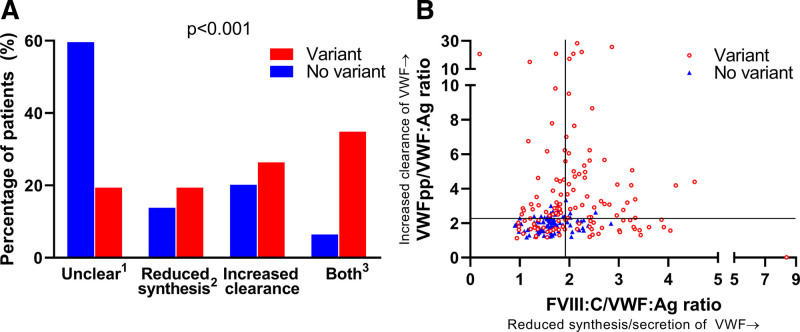
Overall, type 1 VWD patients with a VWF gene variant had a higher FVIII:C/VWF:Ag ratio and VWFpp/VWF:Ag ratio compared with patients without a VWF gene variant (P < 0.001; Table 2). In 55 of 92 patients (59.8%) without a VWF gene variant, the pathophysiological defect of VWF was unclear, whereas this was the case for 26 of 131 patients (19.8%) with a VWF gene variant (P < 0.001; Figure 4A). Also, only 6 of 92 patients (6.5%) without a VWF gene variant had both reduced synthesis/secretion and increased clearance of VWF, whereas this was the case in 45 of 131 patients (34.4%) with a VWF gene variant (P < 0.001; Figure 4A). Finally, very high FVIII:C/VWF:Ag ratios and VWFpp/VWF:Ag ratios, indicative for very low synthesis/secretion and rapid clearance of VWF, were only present in type 1 VWD patients with a VWF gene variant (Figure 4B).
Figure 4.
There is a clear distinction in pathophysiology of type 1 VWD patients with and without a VWF gene variant. (A) Most patients without a VWF gene variant had an undermined pathophysiologic defect of VWF based on the FVIII:C/VWF:Ag ratio and VWFpp/VWF:Ag ratio, whereas in patients with a variant most had a synthesis/secretion and clearance defect. P value outcomes of ANOVA test. 1Undetermined pathophysiological defect of VWF, 2or reduced secretion of VWF, 3Combination of reduced synthesis/secretion and increased clearance of VWF. (B) Extreme values of FVIII:C/VWF:Ag and VWFpp/VWF:Ag were only seen in patients with a variant. Each dot represents a patient. VWF = Von Willebrand factor.
Pathophysiology of reduced VWF levels in type 2 and 3 VWD
In type 2 VWD patients with a single variant, 13 variants were associated with an increased clearance of VWF with normal synthesis and secretion (Figure 5). These variants affected a wide range of VWF domains (D′ to D4). Nine variants were associated with both reduced synthesis/secretion and increased clearance of VWF (Figure 5). Interestingly, all these variants affected the VWF D3 domain or A1 domain. C570R was the only variant which resulted in reduced synthesis/secretion of VWF with normal clearance (Figure 5). In C1190Y, Q1556R and F1654I the pathophysiological defects of reduced VWF levels could not be determined with the FVIII:C/VWF:Ag and VWFpp/VWF:Ag ratios (Figure 5). Overall, lower VWF:Ab/VWF:Ag ratio was associated with higher VWFpp/VWF:Ag ratio (β = 2.0 [0.9–3.2]; P = 0.001) and FVIII:C/VWF:Ag ratio (β = 1.1 [0.8–1.4]; P < 0.001), both adjusted for age, sex, and blood group.
Figure 5.
Pathophysiology of reduced VWF levels in patients with type 2 VWD with a single variant. Each dot represents the mean value for a variant. Subscript number indicate number of patients with each variant. FVIII:C/VWF:Ag ratio ≥1.9 is defined as reduced synthesis/secretion of VWF and VWFpp/VWF:Ag ratio ≥2.2 is defined as increased clearance of VWF. Of note, the number of patients with each variant in this figure may differ from the number of patients presented in Suppl. Table S1, because in this figure we only present type 2 VWD patients with a single variant. VWD = von Willebrand disease; VWF = Von Willebrand factor.
Since type 3 VWD was defined as VWFpp ≤0.05, all patients had undetectable VWFpp levels, which is indicative for no synthesis of VWF. Interestingly, one type 3 VWD patient with a novel heterozygous Y1584* variant, in whom no deletions were found with MLPA, had VWF:Ag of 0.03 IU/mL, undetectable VWF:Ab, FVIII:C of 0.45 IU/mL, FVIII:C/VWF:Ag ratio of 15, was diagnosed at an older age (35) compared with other type 3 patients (median 1 year [IQR 0–3.8]), and had a lower BS (9) compared with other type 3 VWD patients (median of 20 [IQR 15–25]).
DISCUSSION
In this study, we analyzed the VWF gene in a large cohort of well-defined VWD patients. A genetic variant in the VWF gene was found in 60.7% of type 1, 98.4% of type 2, and 100% of type 3 patients. Forty-seven variants were novel. We have presented for all VWF gene variants in our cohort whether they are associated with reduced VWF levels due to reduced synthesis/secretion or increased clearance of VWF. Furthermore, in type 1 VWD, the pathophysiology of reduced VWF levels is associated with the laboratory and bleeding phenotype of patients. Finally, type 1 VWD patients with and without a VWF gene variant were clearly distinct in their pathophysiology of reduced VWF levels and their clinical features such as reason for referral, age of diagnosis, centrally measured VWF levels, and BS.
Reduced VWF:Ag may be caused by reduced synthesis/secretion or increased clearance of VWF. Identification of the pathophysiological mechanism of each variant may contribute to a better understanding of the laboratory and bleeding phenotype of patients. In line with previous studies, we have found that R1205H and S2179F were the variants with highest clearance of VWF, confirming the validity of our approach using FVIII:C/VWF:Ag and VWFpp/VWF:Ag ratios.8,19 It has previously been suggested to identify type 1 VWD variants with a rapid clearance of VWF, such as S2179F, as a distinct group of type 1 VWD.8,19 Based on the pathophysiological mechanisms observed in our current study, one may also include C1149R, Y1146C, and deletion exon 33–34 as such variants, as they also lead to a rapid clearance of VWF. Furthermore, we have shown that the pathophysiological mechanism of reduced VWF levels are strongly associated with VWF, FVIII, and BS. This illustrates that similar to type 1 Vicenza, the pathophysiology of other VWF gene variants may also explain the heterogeneity of the laboratory and bleeding phenotype of VWD patients.8,22
The findings of this study may serve as a starting point for in vitro studies investigating the pathophysiology of reduced VWF levels in VWD patients. For instance, in agreement with our study it has recently been shown in HEK293 cells that in-frame deletions of VWF lead to reduced VWF levels due to reduced synthesis or secretion of VWF.23 Although it was found that deletion of exons 33–34 resulted in a mild synthesis defect of VWF, clearance defects of VWF could not be investigated in previous studies.23 In this study, we have observed that patients with exon 33–34 have a markedly increased clearance of VWF based on the VWFpp/VWF:Ag ratio, illustrating the additional value of our approach.23 Other in vitro studies have found similar results as our current study for the pathophysiologic mechanisms of several variants, including R924Q, C1149R, and S1285P.2,24 Therefore, this study may lay the foundation for future in vitro studies on VWF gene variants and their association with reduced synthesis/secretion or increased clearance of VWF.
Furthermore, in 39.3% of patients with type 1 VWD, defined as historically lowest VWF levels ≤0.30 IU/mL, no VWF gene variant was detected. We found a slightly lower prevalence of VWF gene variants compared to some previous studies, probably because we have excluded synonymous variants and benign variants.5,10–17 In line with our study, it was previously described in one study that in VWD patients with normal multimers, patients without a VWF gene variant had higher VWF:RCo compared with patients with a VWF gene variant.5 However, no significant difference in the BS of both groups were found, probably due to the small number of included patients.5 In the current study, we have compared various aspects of type 1 VWD patients with and without a VWF gene variant, and found important differences in the pathophysiology, laboratory, and clinical phenotype of patients. This raises the question whether we should perform genetic analysis in type 1 VWD patients to discriminate between patients with and without a VWF gene variant. Importantly, there is no evidence yet which indicates that the management of type 1 VWD patients with and without a VWF gene variant should be different. Therefore, studies are needed to investigate whether the presence or absence of a VWF gene variant influences treatment response in type 1 VWD patients.
Furthermore, we have identified 47 novel VWF gene variants, of which eight were present in type 2 VWD patients with a single variant and seven in type 3 VWD patients. Interestingly, we identified a novel heterozygous Y1584* variant leading to a laboratory phenotype of type 3 VWD, however, with FVIII:C of 0.45 IU/mL and a milder bleeding phenotype compared to other type 3 patients. Since type 2 and 3 VWD are almost always caused by a defect in the VWF gene, it is likely that these variants are pathogenic. Novel variants in type 1 VWD may either be benign or pathogenic.5,11,12,17 Therefore, functional studies are needed to assess the pathogenicity of these novel variants.
The main of strength of this study is that we performed genetic analysis of the complete VWF gene in a large cohort of well-defined VWD patients. Therefore, we were able to study the association between various VWF gene variants and their pathogenic mechanisms of reduced synthesis/secretion or increased clearance of VWF. This is the first study in which the FVIII:C/VWF:Ag ratio and VWFpp/VWF:Ag ratio are used as a tool to investigate the pathophysiological defects of reduced VWF levels in each VWF gene variant found in a large cohort of VWD patients. It is also the first study to demonstrate important differences between the laboratory and bleeding phenotype of type 1 VWD patients with and without a VWF gene variant. The most important limitation of this study is that we did not confirm our findings in in vitro studies. However, our results are comparable to previous in vitro studies, as discussed above. Moreover, it was not feasible to investigate the pathogenic mechanisms of all detected variants in in vitro studies. Another limitation of this study is that FVIII:C/VWF:Ag ratio cannot distinguish between a synthesis or secretion defect of VWF. Based on previous studies, we hypothesize that an increased FVIII:C/VWF:Ag ratio with normal VWFpp/VWF:Ag ratio in patients with null-alleles reflect reduced synthesis, whereas both an increased FVIII:C/VWF:Ag ratio and VWFpp/VWF:Ag ratio in patients with missense variants reflect reduced secretion of VWF.6,23 Future in vitro studies are needed to investigate this hypothesis. Finally, patients with type 2N VWD may have a lower FVIII:C/VWF:Ag ratio because of a decreased binding of VWF to FVIII. Therefore, the FVIII:C/VWF:Ag ratio may not be indicative for synthesis/secretion defects of VWF in patients with type 2N VWD. Similarly, VWFpp/VWF:Ag ratio may not reflect VWF clearance if there are variants identified in VWFpp.
To conclude, in this study, we present for various VWF gene variants their pathogenic mechanisms leading to reduced VWF levels. Therefore, this study forms a basis for future studies on VWF gene variants and their pathogenic mechanisms of reduced VWF levels. In addition, we have demonstrated that type 1 VWD patients with and without a VWF gene variant are two clearly distinct groups with regard to their pathophysiology, laboratory phenotype, and bleeding phenotype. The increasing knowledge about phenotype-genotype correlation in VWD may be important for clinical management and may provide reasons to routinely perform genetic analysis in VWD patients.
ACKNOWLEDGMENTS
List of WiN study members: C. J. Fijnvandraat, M. Coppens, J. de Meris, L. Nieuwenhuizen, K. Meijer, R. Y. J. Tamminga, P. F. Ypma, H. C. J. Eikenboom, J. G. van der Bom, F. J. W. Smiers, K. P. M. van Galen, F. C. J. I. Heubel-Moenen, P. Brons, B. A. P. Laros-van Gorkom, F. W. G. Leebeek (principal investigator), M. H. Cnossen, J. Boender, and F. Atiq.
AUTHORSHIP CONTRIBUTIONS
FA designed the study, performed statistical analysis, interpreted data, and wrote the article. JB, WH, JT, SS, and SK designed the study, collected data, interpreted data, and critically revised the article. MC, BL, JM, KF, JB, KM, KG, and JE designed the study, interpreted data, and critically revised the article. FL conceived of and designed the study, interpreted data, and critically revised the article. All authors gave their consent to the final version of the article.
DISCLOSURES
FA received the CSL Behring-professor Heimburger Award 2018 and a travel grant from Sobi. JB started working at Sobi after finishing this research project. WLvH reports speaker and consultant and travel fees from Takeda, Bayer, CSL Behring, and Sobi. He is also cofounder and CSO of Enzyre. MHC has received investigator-initiated research grants over the years from the Netherlands Organisation for Scientific Research (NWO), the Netherlands Organization for Health Research and Development (ZonMw), the Dutch “Innovatiefonds Zorgverzekeraars,” Baxter/Baxalta/Shire, Pfizer, Bayer Schering Pharma, CSL Behring, Sobi Biogen, Novo Nordisk, Novartis, and Nordic Pharma, and has served as a steering board member for Roche and Bayer. All grants, awards, and fees go to the Erasmus MC as institution. The institution of KF has received unrestricted research grants from CSL Behring, Sobi, and NovoNordisk, and her institution received consultancy fees from Grifols, Takeda, Novo Nordisk, and Roche. KM received research support from Bayer, Sanquin, and Pfizer; speaker fees from Bayer, Sanquin, Boehringer Ingelheim, BMS, and Aspen; consulting fees from uniQure, of which all fees go to the institution. BAPL-vG has received unrestricted educational grants from Baxter and CSL Behring. JE received research support from CSL Behring, and he has been a teacher on educational activities of Roche. KPMvG received unrestricted research support from CSL Behring and Bayer and speakers fee from Takeda. JGvdB has been a teacher on educational activities of Bayer and received consultancy fees from Novo Nordisk, paid to the Leiden University Medical Center. FWGL received research support from CSL Behring and Shire/Takeda for performing the Willebrand in the Netherlands (WiN) study and uniQure for a study not related to this article, and is consultant for uniQure, Sobi, Biomarin, and Shire/Takeda, of which the fees go to the institution, and has received a travel grant from Sobi. He is also a DSMB member for a study by Roche. All the other authors have no conflicts of interest to disclose.
SOURCES OF FUNDING
The WiN study was supported (in part) by research funding from the Dutch Hemophilia Foundation (Stichting Haemophilia), Shire (Takeda) and CSL Behring (unrestricted grant).
Supplementary Material
Footnotes
The members of WiN study group are listed in Acknowledgments.
Supplemental digital content is available for this article.
REFERENCES
- 1.Lenting PJ, Christophe OD, Denis CV. von Willebrand factor biosynthesis, secretion, and clearance: connecting the far ends. Blood. 2015;125:2019–2028. [DOI] [PubMed] [Google Scholar]
- 2.de Jong A, Eikenboom J. Von Willebrand disease mutation spectrum and associated mutation mechanisms. Thromb Res. 2017;159:65–75. [DOI] [PubMed] [Google Scholar]
- 3.Leebeek FW, Eikenboom JC. Von Willebrand’s disease. N Engl J Med. 2016;375:2067–2080. [DOI] [PubMed] [Google Scholar]
- 4.James PD, Connell NT, Ameer B, et al. ASH ISTH NHF WFH 2021 guidelines on the diagnosis of von Willebrand disease. Blood Adv. 2021;5:280–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodeve A, Eikenboom J, Castaman G, et al. Phenotype and genotype of a cohort of families historically diagnosed with type 1 von Willebrand disease in the European study, Molecular and Clinical Markers for the Diagnosis and Management of Type 1 von Willebrand Disease (MCMDM-1VWD). Blood. 2007;109:112–121. [DOI] [PubMed] [Google Scholar]
- 6.Eikenboom J, Federici AB, Dirven RJ, et al. VWF propeptide and ratios between VWF, VWF propeptide, and FVIII in the characterization of type 1 von Willebrand disease. Blood. 2013;121:2336–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanders YV, Groeneveld D, Meijer K, et al. von Willebrand factor propeptide and the phenotypic classification of von Willebrand disease. Blood. 2015;125:3006–3013. [DOI] [PubMed] [Google Scholar]
- 8.Haberichter SL, Balistreri M, Christopherson P, et al. Assay of the von Willebrand factor (VWF) propeptide to identify patients with type 1 von Willebrand disease with decreased VWF survival. Blood. 2006;108:3344–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haberichter SL, Castaman G, Budde U, et al. Identification of type 1 von Willebrand disease patients with reduced von Willebrand factor survival by assay of the VWF propeptide in the European study: molecular and clinical markers for the diagnosis and management of type 1 VWD (MCMDM-1VWD). Blood. 2008;111:4979–4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veyradier A, Boisseau P, Fressinaud E, et al. A laboratory phenotype/genotype correlation of 1167 french patients from 670 families with von Willebrand disease: a new epidemiologic picture. Medicine (Baltimore). 2016;95:e3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cumming A, Grundy P, Keeney S, et al. An investigation of the von Willebrand factor genotype in UK patients diagnosed to have type 1 von Willebrand disease. Thromb Haemost. 2006;96:630–641. [PubMed] [Google Scholar]
- 12.James PD, Notley C, Hegadorn C, et al. The mutational spectrum of type 1 von Willebrand disease: results from a Canadian cohort study. Blood. 2007;109:145–154. [DOI] [PubMed] [Google Scholar]
- 13.Johansson AM, Halldén C, Säll T, et al. Variation in the VWF gene in Swedish patients with type 1 von Willebrand disease. Ann Hum Genet. 2011;75:447–455. [DOI] [PubMed] [Google Scholar]
- 14.Robertson JD, Yenson PR, Rand ML, et al. Expanded phenotype-genotype correlations in a pediatric population with type 1 von Willebrand disease. J Thromb Haemost. 2011;9:1752–1760. [DOI] [PubMed] [Google Scholar]
- 15.Budde U, Schneppenheim R, Eikenboom J, et al. Detailed von Willebrand factor multimer analysis in patients with von Willebrand disease in the European study, molecular and clinical markers for the diagnosis and management of type 1 von Willebrand disease (MCMDM-1VWD). J Thromb Haemost. 2008;6:762–771. [DOI] [PubMed] [Google Scholar]
- 16.Federici AB, Bucciarelli P, Castaman G, et al. Management of inherited von Willebrand disease in Italy: results from the retrospective study on 1234 patients. Semin Thromb Hemost. 2011;37:511–521. [DOI] [PubMed] [Google Scholar]
- 17.Batlle J, Pérez-Rodríguez A, Corrales I, et al. Molecular and clinical profile of von Willebrand disease in Spain (PCM-EVW-ES): proposal for a new diagnostic paradigm. Thromb Haemost. 2016;115:40–50. [DOI] [PubMed] [Google Scholar]
- 18.de Wee EM, Mauser-Bunschoten EP, Van Der Bom JG, et al. Health-related quality of life among adult patients with moderate and severe von Willebrand disease. J Thromb Haemost. 2010;8:1492–1499. [DOI] [PubMed] [Google Scholar]
- 19.Borràs N, Batlle J, Pérez-Rodríguez A, et al. Molecular and clinical profile of von Willebrand disease in Spain (PCM-EVW-ES): comprehensive genetic analysis by next-generation sequencing of 480 patients. Haematologica. 2017;102:2005–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stenson PD, Mort M, Ball EV, et al. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet. 2014;133:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yates AD, Achuthan P, Akanni W, et al. Ensembl 2020. Nucleic Acids Res. 2020;48:D682–D688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castaman G, Tosetto A, Rodeghiero F. Reduced von Willebrand factor survival in von Willebrand disease: pathophysiologic and clinical relevance. J Thromb Haemost. 2009;7(Suppl 1):71–74. [DOI] [PubMed] [Google Scholar]
- 23.Cartwright A, Webster SJ, de Jong A, et al. Characterization of large in-frame von Willebrand factor deletions highlights differing pathogenic mechanisms. Blood Adv. 2020;4:2979–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berber E, James PD, Hough C, et al. An assessment of the pathogenic significance of the R924Q von Willebrand factor substitution. J Thromb Haemost. 2009;7:1672–1679. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.