Abstract
A strain identified as Comamonas testosteroni I2 was isolated from activated sludge and found to be able to mineralize 3-chloroaniline (3-CA). During the mineralization, a yellow intermediate accumulated temporarily, due to the distal meta-cleavage of chlorocatechol. This strain was tested for its ability to clean wastewater containing 3-CA upon inoculation into activated sludge. To monitor its survival, the strain was chromosomally marked with the gfp gene and designated I2gfp. After inoculation into a lab-scale semicontinuous activated-sludge (SCAS) system, the inoculated strain maintained itself in the sludge for at least 45 days and was present in the sludge flocs. After an initial adaptation period of 6 days, complete degradation of 3-CA was obtained during 2 weeks, while no degradation at all occurred in the noninoculated control reactor. Upon further operation of the SCAS system, only 50% 3-CA removal was observed. Denaturing gradient gel electrophoresis (DGGE) of 16S rRNA genes revealed a dynamic change in the microbial community structure of the activated sludge. The DGGE patterns of the noninoculated and the inoculated reactors evolved after 7 days to different clusters, which suggests an effect of strain inoculation on the microbial community structure. The results indicate that bioaugmentation, even with a strain originating from that ecosystem and able to effectively grow on a selective substrate, is not permanent and will probably require regular resupplementation.
Bioaugmentation is the accelerated removal of undesired compounds from contaminated hazardous waste sites or bioreactors by using indigenous or allochthonous wild-type or genetically modified organisms (48). These inocula usually are highly efficient in the removal of the xenobiotic targets under laboratory conditions. However, under natural conditions, these laboratory strains have to compete with the established microbial community, resulting in a decrease of the amount of inoculated cells (15). This competition can be controlled by adding a carbon source that the inoculant can degrade (5) or by changing operation parameters (14). Thus far in water treatment, only a few successful cases of small-scale bioaugmentation in activated sludge by using natural or genetically modified microorganisms have been described. McClure et al. (27) obtained enhanced but incomplete degradation of 3-chlorobenzoate (3CB) in a laboratory-scale activated-sludge unit after inoculation of activated-sludge-derived bacteria (28) able to mineralize 3CB. Nüblein et al. (32) inoculated a laboratory-scale activated-sludge unit with Pseudomonas sp. strain B13 FR1(pFRC20P) and observed a drastic decrease of 3CB and 4CB 3 days after inoculation, while in the noninoculated reactor 8 and 15 days of adaptation, respectively, were needed. Selvaratnam et al. (37) inoculated a sequencing batch reactor with Pseudomonas putida PP301(pO103) to remove phenol, and ca. 85% of the phenol was degraded within 2.5 h. Little is known about the effect of bioaugmentation of activated-sludge reactors on the microbial community of 16S rRNA genes. Eichner et al. (10) used thermal gradient gel electrophoresis (TGGE) to examine the effect of a pollutant shock on the activated-sludge microbial community and observed protection by the inoculation of a genetically modified strain able to avoid formation of toxic intermediates. The addition of specialized strains to activated sludge to enhance the removal of pollutants present in the influent is not yet widely applied, because bioaugmentation is less predictable and controllable than the direct destruction of contaminants, such as incineration.
Aromatic amines, such as chloroanilines, are widely used for the production of dyes, drugs, and herbicides (22) and as a consequence of their application are released in the environment. These compounds are also introduced via the metabolism of phenylamide pesticides (17). These toxic and recalcitrant residues are considered important environmental pollutants (30). Therefore, many efforts were made to isolate bacteria capable of degrading chlorinated anilines. Moraxella sp. strain G (54) is the first isolate that was found to be able to use 4-chloroaniline as the sole source of carbon, nitrogen, and energy. Later, more chloroaniline-metabolizing strains were isolated, such as Pseudomonas sp. strain JL2 (23), Pseudomonas (now Brevundimonas) diminuta INMI KS-7 (42), Pseudomonas (now Delftia) acidovorans CA28 (24), Pseudomonas (now Delftia) acidovorans BN3.1 (8), Aquaspirillum sp. strain 2CA, and Paracoccus denitrificans 3CA (41). The first step in the degradation of chloroanilines is the deamination to chlorocatechols. These chlorinated catechols seem to be usually degraded by a modified ortho-cleavage pathway (19, 55), but recently, the use of a meta-cleavage pathway for the mineralization of 3-chlorocatechol has been determined (21, 26). In the past, the meta-cleavage intermediates of chlorinated catechols, e.g., chlorohydroxymuconic semialdehyde, were described as toxic metabolites preventing further degradation (24).
The aims of this work were to isolate and characterize the microbial component of activated sludge that is able to degrade 3-chloroaniline (3-CA) and to investigate the eventual enhanced degradation of 3-CA by the activated sludge after inoculation with the strain. In addition, the effect of inoculation on the microbial community structure of the sludge was examined.
MATERIALS AND METHODS
Media and culture conditions.
The mineral medium MMN (mineral medium without nitrogen and carbon) is derived from MMO mineral medium (39) by eliminating all nitrogen. The MMN medium contained 1,419.6 mg of Na2HPO4, 1,360.9 mg of KH2PO4, 98.5 mg MgSO4, 5.88 mg of CaCl2 · 2H2O, 1.16 mg of H3BO4, 2.78 mg of FeSO4 · 7H2O, 1.15 mg of ZnSO4 · 7H2O, 1.69 mg of MnSO4 · H2O, 0.38 mg of CuSO4 · 5H2O, 0.24 mg of CoCl2 · 6H2O, 0.10 mg of MoO3, and 3.2 mg of EDTA in 1 liter of distilled water. The liquid mineral media were supplemented with 150 to 250 mg of aniline (Sigma-Aldrich Chemie, Steinheim, Germany) or 3-CA (Fluka AG Chemische Fabrik, Buchs, Switzerland) per liter, while for the solidified media, aniline and 3-CA were added at a concentration of 500 mg/liter. Luria broth (LB) medium containing 10 g of Bacto Peptone (Difco, Detroit, Mich.), 5 g of Bacto yeast extract (Difco), and 5 g of NaCl in 1 liter of distilled water was used as a rich medium. These media were solidified with 2% agar for plate growth.
Cultures were incubated on a rotary shaker under aerobic conditions at 28°C. Growth was monitored by measuring the turbidity at 600 nm. Two hundred microliters of an overnight-grown LB culture of strain I2, washed twice in saline (0.85% NaCl), was inoculated in 200 ml of MMN medium with 3-CA (150 mg/liter) (0.1% inoculation) to monitor the transformation of 3-CA.
Isolation and characterization of Comamonas testosteroni I2.
Strain I2 was isolated from an enrichment culture obtained from activated sludge of a domestic wastewater treatment plant (Bourgoyen-Ossemeersen plant, Gent, Belgium) after repeated supplementation with 3-CA. During 6 weeks a 1-liter Erlenmeyer flask containing 200 ml of the activated sludge (4 g [dry weight] per liter) was supplemented every 7 days with 200 mg of 3-CA/liter. After 6 weeks, a 0.5-liter Erlenmeyer flask containing 200 ml of MMN with 3-CA (200 mg/liter) was inoculated with 2 ml of the activated sludge. After 6 days, 2 ml of this enrichment culture was transferred to a new 0.5-liter Erlenmeyer flask with 200 ml of MMN with 3-CA (200 mg/liter). After another 6 days, this culture was spread onto MMN–3-CA agar plates (500 mg of 3-CA/liter) and incubated at 28°C for 1 week.
Identification of the isolate.
Gas chromatographic analysis of fatty acid methyl esters (FAME) was performed as described previously (49). The FAME profiles were identified using the Microbial Identification System, version 4.0 (Microbial ID, Inc., Newark, Del.).
For sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, cells were harvested from tryptic soy agar (BBL) plates after 48 h of incubation at 37°C. Preparation of the cell extract, gel electrophoresis, and numerical comparison were performed as previously described (34) and by using the GelCompar 4.1 software package (Applied Maths, Kortrijk, Belgium).
DNA was enzymatically degraded into nucleosides as described by Mesbah et al. (29). The nucleoside mixture obtained was then separated by high-performance liquid chromatography (HPLC) using a Waters SymmetryShield C8 column thermostated at 37°C. The solvent was 0.02 M NH4H2PO4 (pH 4.0) with 1.5% acetonitrile. Nonmethylated lambda phage DNA (Sigma-Aldrich Chemie) was used as the calibration reference.
DNA-DNA hybridizations were performed with photobiotin-labeled probes in microplate wells as described by Ezaki et al. (13), using an HTS7000 BioAssay Reader (Perkin-Elmer, Norwalk, Conn.) for the fluorescence measurements. The hybridization temperature was 55°C.
Marking with gfp.
Escherichia coli strain C118 λpir(pUT-miniTn5 gfpKm) (18, 46) was obtained by transformation (9). The pUT plasmid, comprising a mini-Tn5 transposon with RP4 resolvase sites flanking the nptII gene (responsible for kanamycin resistance), was used for insertion of the gfp gene into the chromosome of strain I2. Biparental mating between the donor strain E. coli C118 λpir (18) and the recipient strain I2 with selection on LB plates with rifampin (100 mg/liter) and kanamycin (50 mg/liter) resulted in I2gfp derivatives with the nptII and gfp genes inserted in the chromosome. This was confirmed by PCR with gfp primers (see below).
SCAS reactors.
The experiments were conducted in duplicate with sludge freshly collected from a domestic wastewater treatment plant (Bourgoyen-Ossemeersen). The total bacterial count of the sludge was 4.4 × 108 bacteria/ml, determined with a Live/Dead Bacterial Viability Kit (L-13152; Molecular Probes, Eugene, Oreg.) as described by according to Boulos et al. (7). The reactors, with a volume of 1.1 liters, were operated according to the semicontinuous activated-sludge (SCAS) procedure at room temperature (ca. 21°C). The tests were conducted with synthetic influent consisting of skim milk powder (Gloria, Nestlé) dissolved in tap water. The use of skim milk powder allowed the use of a constant wastewater that was rich in nutrients, with a chemical oxygen demand (COD)/N/P ratio equal to 100:6:1. The reactors were fed every day after wastage of excess sludge and settling. The SCAS reactors were operated at a volumetric loading rate of 1 g of COD/liter · day, with a hydraulic retention time of 4 days and a sludge retention time of 11 days. All four reactors had a loading rate of 40 mg of 3-CA/liter · day, added as a daily single dose. One liter of the mixed liquor was subjected to a half-hour period of settling in an Imhoff cone to analyze the sludge volume (SV) (16). On days 1, 3, and 5 of each week, the settling was followed by decantation of 400 ml of the supernatant and addition of 500 ml of fresh influent. The wasted sludge was used for analyze as follows: at day 1 of the week, a DNA extraction of the sludge was performed, and the pH, oxygen uptake rate (OUR), and concentration of strain I2gfp were determined; daily, an HPLC sample was taken and the suspended solids (SS) and sludge volume index (SVI) were measured (16). Two duplicate reactors were inoculated with C. testosteroni I2gfp (reactors A), and two duplicate reactors were used as noninoculated controls (reactors B). No important differences could be observed between the two duplicate reactors. Hence, unless otherwise indicated, the data reported are averages for both duplicates. The reactors were operated without 3-CA for 12 days to allow the microbial community to adapt to the changed environment and growth conditions. After this period, reactors A were inoculated with C. testosteroni I2gfp to a final concentration of 3 × 106 cells/ml. The cells were pregrown overnight in LB medium containing 100 mg of 3-CA/liter, washed twice with saline, and finally resuspended in saline.
Bacterial counts.
Sludge flocs were dispersed by purging a 1-ml sample five times through a sterile syringe of 1 ml with a needle (1.2 by 40 mm). LB agar medium, supplemented with rifampin (100 mg/liter) and kanamycin (50 mg/liter) was used to count the I2gfp cells. By using green fluorescent protein fluorescence, it was possible to detect 10 CFU of strain I2gfp per ml against the background of about 103 CFU of total kanamycin- and rifampin-resistant microorganisms per ml.
UV-light microscopy.
The sludge samples were analyzed by UV-light microscopy using a Reichert-Jung Polyvar microscope equipped with an Mercury short arc lamp HBO (200 W). Samples were examined with 40× and 100× objectives, using very-low-fluorescence immersion oil.
Respirometric activity measurements.
The metabolic activity of the activated sludge in general was expressed as OUR. Therefore, activated-sludge samples (200 ml) were transferred to the vessel and saturated with oxygen by bubbling air with a pump. Once oxygen saturation (ca. 8 mg of O2/liter) was reached, the aeration was ceased and the oxygen electrode (Oxyguard Probe; Kelma, Niel, Belgium) was placed in such a way that the opening of the vessel was barely closed. Sodium acetate was added to a final concentration of 50 mg/liter. Samples were mixed with a magnetic stirrer during measurements. The method was further performed as described by Surmacz-Gorska et al. (40), calculating the activity from the constant slope of oxygen concentration over time. The activity measurements resulted in the OUR (grams of O2 per liter per day).
Analytical methods.
The supernatants of cultures were analyzed for 3-CA content by reversed-phase HPLC after centrifugation of the cells at 5,000 × g for 10 min. The HPLC system consisted of a Kontron liquid chromatograph with a DEGASYS DG-1310 system to degas the mobile phase, three Kontron 325 high-pressure pumps, a Kontron MSI 660 injector with a 20-μl loop, a Kontron DAD 495 diode array detector, and a 450 MT2/DAD software system. An Alltima C18 reversed-phase column (250 by 8 mm [inner diameter], 5-μm particle size; Alltech, Deerfield, Ill.) was used. The mobile phase consisted of CH3OH-NH4H2PO4 (0.1 M, pH 3.8)-H2O (70:25:5), with a flow rate of 0.75 ml/min. The UV detector was used at 210 nm. Quantitative data for 3-CA were obtained by comparing the peak areas of unknown concentrations with the peak areas of standards of known concentrations.
The absorption spectra were recorded on a Kontron Uvikon spectrophotometer (model 932).
DNA extraction and purification.
Total DNA was extracted from the sludge samples by a method based on the protocols described previously (11, 12). This protocol was modified as follows. Two milliliters of sludge was added to a 14-ml polypropylene round-bottom tube (Falcon). To this, 3 g of beads (0.10- to 0.11-mm-diameter) (B. Braun Biotech International, Melsungen, Germany) and 4 ml of 10 mM Tris-HCl (pH 9) were added. The mixture was beaten three times for 90 s using a bead beater (B. Braun Biotech International) at 2,000 rpm. Then, 2 ml of 4 mg of lysozyme per ml in 10 mM Tris-HCl (pH 9) was added, followed by incubation of the samples for 15 min at 28°C on a rotary shaker. Subsequently, 300 μl of 20% SDS was added, and samples were slowly mixed for 5 to 10 min. After this, 1 ml of 8 M ammonium acetate was added. The supernatant was collected after centrifugation at 7,000 × g for 15 min at 4°C. A chloroform-isoamyl alcohol (24:1) purification was done, followed by centrifugation at 7,000 × g for 15 min at 4°C. The aqueous phase was transferred to a new tube, and 0.8 volume of isopropanol was added. The precipitation was performed for 1 h at −20°C. Alternatively, 2.5 volumes of ethanol (100%) were added for overnight precipitation. The pellet (crude extract) was obtained by centrifugation at 12,000 × g for 25 min and was resuspended in 250 μl of distilled water. A 100-μl aliquot of the crude extract was further purified using Wizard PCR preps (Promega, Madison, Wis.), and the purified DNA was finally recovered in 50 μl of distilled water.
For axenic cultures, the template for PCR amplification was obtained by suspending a colony in 200 μl of sterile distilled water, boiling for 15 min, and storing at −20°C. Two microliters of the lysed cells was used in the PCR.
PCR conditions.
Two microliters of the extracted DNA was amplified by PCR with a 9600 thermal cycler (Perkin-Elmer). The PCR mixture used contained 0.5 μM each primer, 200 μM each deoxynucleoside triphosphate, 1.5 mM MgCl2, 10 μl of thermophilic DNA polymerase 10× reaction buffer (MgCl2 free), 2.5 U of Taq DNA polymerase (Promega), 400 ng of bovine serum albumin (Boehringer) per μl, and DNase- and RNase-free filter-sterilized water (Sigma-Aldrich Chemie) to a final volume of 100 μl.
Repetitive extragenic palindromic (REP)-PCR was done as described by Versalovic et al. (50) to distinguish identical isolates.
The gfp gene was amplified by PCR with a set of primers based on specific regions of the gfp sequence (GenBank accession number M62653). The set consisted of the primer gfpF (5′CCATGGCCAACACTTGTCAC3′ [forward]) and gfpR (5′CTTTCGAAAGGGCAGATTGT3′ [reverse]).
The 16S rRNA genes from sludge microbial communities were amplified by PCR as suggested by El Fantroussi et al. (12), using the forward primer P63f (5′CAGGCCTAACACATGCAAGTC3′) and the reverse primer P518r (5′ATTACCGCGGCTGCTGG3′) (31, 33). A GC clamp of 40 bp (31, 33) was added to the forward primer. The length of the expected amplified fragment with the GC clamp was 530 bp.
DGGE.
Denaturing gradient gel electrophoresis (DGGE) based on the protocol of Muyzer et al. (31) was performed using the D Gene System (Bio-Rad, Hercules, Calif.). PCR samples were loaded onto 6% (wt/vol) polyacrylamide gels in 1× TAE (20 mM Tris, 10 mM acetate, 0.5 mM EDTA, pH 7.4). The polyacrylamide gels were made with a denaturing gradient ranging from 40 to 60% (where 100% denaturant contains 7 M urea and 40% formamide). The electrophoresis was run for 14 h at 60°C and 50 V. After the electrophoresis, the gels were soaked for 5 min in fixation buffer (10% ethanol, 0.5% acetic acid) and subsequently for 10 min in SYBR GreenI nucleic acid gel stain (1:10,000 dilution; FMC BioProducts, Rockland, Maine). The stained gel was immediately photographed on a UV transillumination table with a video camera module (Vilbert Lourmat, Marne-la Vallé, France).
Analysis of DGGE patterns.
The statistical comparison of the DGGE patterns on the same gel was done with the GelCompar software 4.1 (Applied Maths). The calculation of the matrix of similarities is based on the Pearson product-moment correlation coefficient. The clustering algorithm of Ward (51) was used to calculate dendrograms.
RESULTS
Characterization of C. testosteroni I2.
The enrichment culture described above was plated on MMN medium to obtain single colonies. The purified colonies were tested in liquid MMN medium with 3-CA (200 mg/liter) and on plates with 3-CA (500 mg/liter) as the sole source of carbon, nitrogen, and energy. By further selection, five strains (named I2, I5, I6, I8, and I9) which could use 3-CA as the sole source of carbon, nitrogen, and energy were isolated. These five strains had nucleotide compositions of between 61.6 and 61.9 mol% guanosine plus cytosine and identical SDS-polyacrylamide gel electrophoresis and REP-PCR profiles (data not shown) and were identified via FAME analysis (using the commercial MIS database) as C. testosteroni. In DNA-DNA hybridization experiments, these five strains showed a DNA reassociation of between 68 and 76% with C. testosteroni LMG 1800T (25), while reassociation values within these five isolates were between 88 and 100%. The isolates were therefore identified as C. testosteroni. Since none of the techniques used could differentiate between the strains, they probably have a clonal origin.
During the growth of these 3-CA-assimilating strains in MMN medium (both liquid and solid) with 3-CA as the sole carbon, nitrogen, and energy source, a yellow coloration of the medium developed and disappeared again after prolonged incubation. On LB agar plates, the strains showed phenotypic instability, resulting in two types of colonies with different morphologies. Further purification of both types of colonies continued to yield a mixture of both types.
Degradation of 3-CA and aniline by C. testosteroni I2 in pure culture.
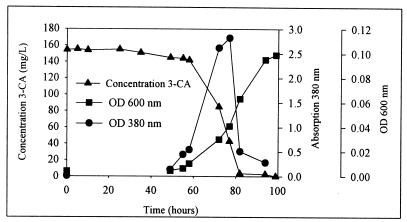
When a 0.1% inoculum is used, C. testosteroni strain I2 can metabolize aniline and 3-CA as the sole source of carbon, nitrogen, and energy in ca. 80 h (Fig. 1). In order to quantify the yellow intermediate, a wavelength scan of the medium between 200 and 400 nm was performed at different times of incubation of the growth culture. During the first 48 h, no visual changes were observed and no 3-CA was degraded. After 48 h, the culture ended the lag phase and the medium began to become yellow (Fig. 1). One day later, the cells were in the logarithmic phase and the intensity of the yellow color was high. Until 77 h, this metabolite accumulated and at the same time the concentration of 3-CA decreased equally. Once the absorption peak at 380 nm had disappeared (82 h) (Fig. 1), the total 3-CA was metabolized. An equimolar amount of chloride ions was released during the degradation of 3-CA by strain I2 (data not shown).
FIG. 1.
Growth of C. testosteroni I2 in MMN plus 3-CA during 4 days. Degradation of 3-CA in relation to the cell growth and accumulation of the intermediate (peak at 380 nm) is shown. OD, optical density.
The absorption maxima of the yellow intermediate at different pHs were examined. At pH 2, 7, and 12 the maximum absorptions of the yellow intermediate were at 323, 380, and 378 nm, respectively.
Survival and activity of C. testosteroni I2gfp in the SCAS reactors.
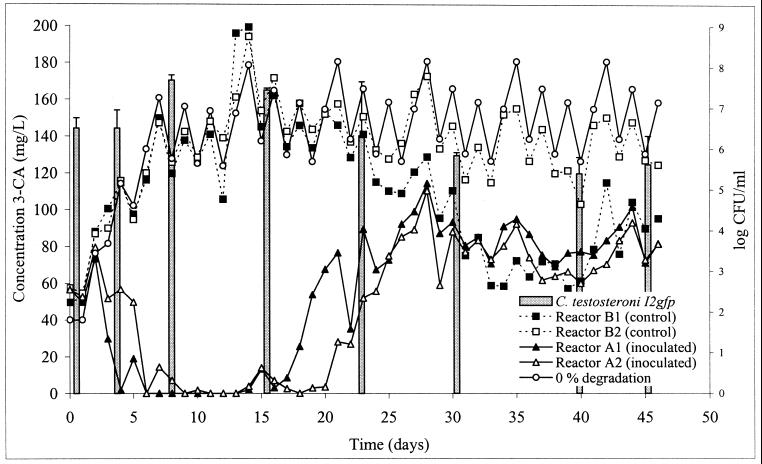
In order to monitor the survival of strain I2 in activated sludge, it was chromosomally marked with the gfp gene, which was expressed constitutively by a PpsbA-promoter (46). The insertion of the gfp gene in several transconjugants was confirmed by PCR with the gfp-specific primers gfpF and gfpR. The gfp-marked strain I2gfp showed the same degradation characteristics as the original strain in MMN medium (data not shown). Before strain I2gfp was inoculated into the sludge reactors (reactors A) and before 3-CA was added, the sludge was adapted for 12 days to the operating SCAS system. At this point (day 0 in Fig. 2), the daily supplementation with 3-CA started. During the first 3 days, no degradation was observed in any of the reactors. In the inoculated reactors A, enhanced degradation was observed from day 4 until day 7, and during the next 12 days complete degradation of 3-CA was achieved. After 3 weeks, however, the concentration of 3-CA increased and stabilized at a level corresponding with ca. 50% degradation. A mass balance calculation of 3-CA in a reactor (with 0% degradation) showed that the concentration of 3-CA fluctuated weekly, based on the daily addition and the washout. No significant removal of 3-CA occurred in the control reactors B during the first 30 days. However, from day 30, enhanced degradation was observed in reactor B1, comparable to that in the inoculated reactors. The other control duplicate, B2, did not show any enhanced degradation.
FIG. 2.
Concentration of 3-CA in the inoculated reactors A1 and A2 and the control reactors B1 and B2, together with a simulation of the 3-CA concentration if no degradation occurred (0% degradation), and survival of C. testosteroni I2gfp in reactors A (bars).
The survival and behavior of C. testosteroni I2gfp were monitored by different methods. The most sensitive way was by plating on LB medium with kanamycin and rifampin (detection limit, 10 CFU/ml). In case of background growth by indigenous bacteria, the inoculated strains could be recognized by green fluorescence protein autofluorescence under a long-wavelength UV lamp. During the first week, the concentration of I2gfp cells did not differ much from the initial value, and during weeks 2 and 3, the concentration of strain I2gfp even increased (Fig. 2). After 30 days, the inoculum size stabilized at ca. 5 × 105 CFU/ml. PCR with gfp primers was tested as an alternative technique to monitor the survival in sludge. The detection limit to obtain an amplification signal was ca. 5 × 105 CFU/ml, however, which was higher than that for the plating method. In the control reactors no amplification was observed. The distribution in the cell flocs was studied by epifluorescence microscopy. A few days after inoculation, C. testosteroni I2gfp cells were visible as green cells under UV light, although the sludge microbial community had background fluorescence. The inoculated bacteria were not randomly distributed but were observed as clusters within the sludge flocs (data not shown).
Reactor performance characteristics.
At the beginning of the 3-CA supplementation, all reactors showed the same performance parameters (OUR = 74 mg O2/liter · h; SS = 4.2 g/liter; SV = 250 ml/liter; SVI = 60 ml/liter; pH = 7.6). The OUR and pH of both reactor sets did not significantly differ during the experiment (two-tailed t test; α = 0.05). The OUR variability of different days of one week was rather high, but the values were in the same range when the same days of different weeks were compared. After a week, the concentration of SS in the control reactors B decreased about 0.6 g/liter in comparison to that in reactors A. The SV after 30 min in both reactors was significantly different after ca. 1 week (two-tailed t test; α = 0.05). During the first week, the SV of reactors A increased from 300 to 500 ml, and it remained at 500 ml for the rest of the experiment. The SV of the control reactors B was stable at 300 ml during the first 40 days, and it increased only slightly during the last week. This resulted in a significant difference between the SVI values of the reactors (two-tailed t test; α = 0.05) (data not shown).
DGGE.
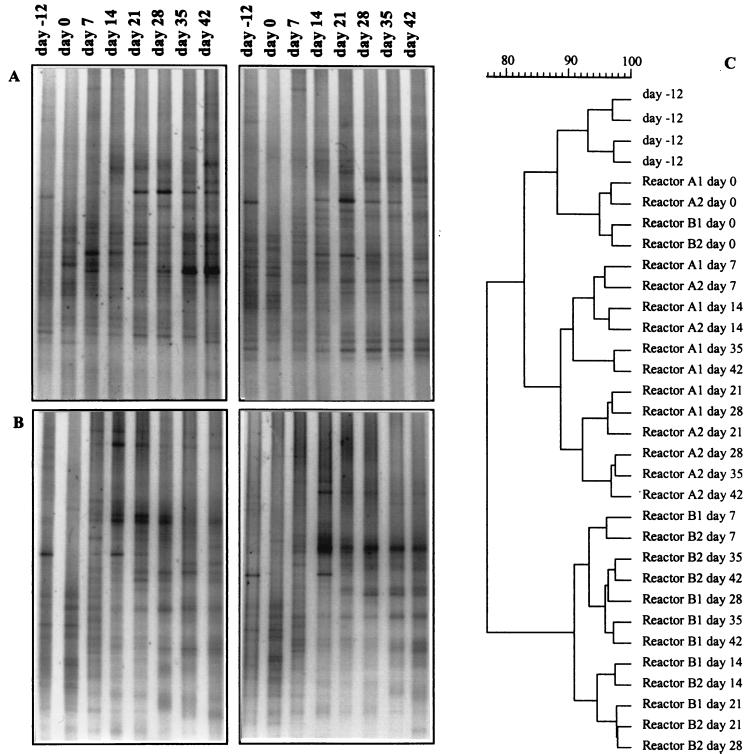
In order to monitor the changes within the microbial community of the SCAS reactors, the diversity of a 16S rRNA fragment was examined. Each week, a sample was taken from the four reactors, and after DNA extraction and purification, the PCR-amplified product was analyzed on a DGGE gel (Fig. 3A and B). The patterns of the four reactors were compared with each other after normalization. To determine the information content of the banding patterns in terms of structural diversity, they were analyzed by clustering (Fig. 3C). The cluster analysis revealed three major groups. The fingerprints showed several very strong bands, some bands of lower intensity, and an additional number of weak bands, resulting in a smear. Before the adaptation period started (day −12) and at the day of the inoculation and feeding with 3-CA (day 0), the DGGE patterns of both reactors at both times clustered together. After the first week (day 7), each reactor series began to develop a different microbial community, which was clearly separated from that of the other reactor series and from the earlier days of the experiment. On some bands, corresponding with a bacterial species, the applied treatment and the inoculation had no influence. Some bands became dominant in the reactors, while other bands vanished. The intensity of some fragments seems to be enhanced by the presence of strain I2gfp, while other species were disfavored by the inoculation.
FIG. 3.
Analysis of the DGGE profiles of the different reactors at different times; using partial 16S rRNA gene fragments. (A) DGGE gel of the inoculated reactors A1 and A2; (B) DGGE gel of the control reactors B1 and B2; (C) dendrogram of all reactors, clustered by use of the Ward method (51).
DISCUSSION
Several bacterial species capable of degrading 3-CA are known (8, 23, 24, 41, 42, 54). Strain I2, isolated and described in this study, is to our knowledge the first reported strain of C. testosteroni that is able to use 3-CA as the sole source of carbon and nitrogen. This species, which is often isolated from activated sludge, is resistant to starvation (28) and has been reported to be involved in the degradation of many aromatic products, such as (chloro)phenols (1, 2), p-toluenesulfonic acid (3), polychlorinated biphenyls (4), arylsulfonates (20), 1-(2-chlorobenzoyl)-3-(4-chlorophenyl)urea (38), and chlorinated benzenes (45).
In pure culture, complete degradation was achieved at 3-CA concentration of 200 mg/liter and was coupled with quantitative liberation of chloride. During the degradation of 3-CA, a yellow intermediate accumulated temporarily, which was further metabolized, thus indicating total metabolism of the aromatic amines. The chlorinated catechols seem to usually be degraded by a modified ortho-cleavage pathway (19, 55). Recently, the use of a meta-cleavage pathway for the mineralization of 3-chlorocatechol as central metabolite of chlorobenzene has been determined (21, 26). Riegert et al. (35) observed a yellow distal meta-cleavage product of 3-chlorocatechol with a strongly pH-dependent absorption maximum at 378 nm and found that this was a chlorohydroxymuconic semialdehyde. In our study, the λmax values at the different pHs were very similar. This comparison suggests that the degradation of 3-CA by C. testosteroni I2 also occurs by means of a distal meta-cleavage pathway for chlorocatechol. This is in contrast with the case for most other chloroaniline degraders, which have been shown to degrade 3-CA via a modified ortho-cleavage pathway (19, 55). Only one study, by Surovtseva et al. (43), mentioned a meta-cleavage of monochloroanilines by Alcaligenes faecalis; however, it is not clear if this cleavage was metabolic or cometabolic.
In this paper, the 3-CA degrading indigenous activated-sludge bacterium C. testosteroni I2 was used to accelerate the removal of 3-CA from wastewater by reinoculating the strain at high density into the same sludge system. The initial sludge microbial community was not able to effectively degrade 3-CA, although strain I2 was probably present, since it was isolated from the same activated-sludge plant. Apparently the natural level of the indigenous strain C. testosteroni I2 was too low to effect the degradation of 3-CA. To distinguish the inoculated strain from identical or similar indigenous sludge bacteria, the strain was chromosomally marked with the gfp gene. The plating method, combined with the gfp visualization with UV light, allowed sensitive and reliable monitoring of the survival of the inoculated strain and was preferred over PCR amplification of the gfp gene, which was not as reliable and sensitive. Similar difficulties with PCR-based strain detection were described by Tchelet et al. (45), who used specific primers for the 16S rRNA gene and chlorobenzene degradation (tcb) genes to monitor an inoculated Pseudomonas strain in activated sludge. Their study and ours thus show that plating can still be a reliable method when the strain has natural or inserted (gfp-Km) specific phenotypes. It is not known, however, if this culturable fraction (CFU of I2gfp) resembles the total viable count of I2gfp in the sludge.
Successful bioaugmentation depends mainly on the behavior of the inoculated strain in the environment where it is introduced. Therefore, a first criterion is good survival and retention of the strain in the system. The growth rate of the organism may be lower than washout (52) and the rate of predation, for example, by protozoa, so that the activity of grazers reduces the cell density of the inoculum species (15). In our experiment, an equilibrium seemed to be reached between washout, predation, and growth rate after 3 weeks with an inoculum concentration of ca. 5 × 105 CFU of strain I2gfp per ml. The origin and the type of inoculated strain also can play an important role in the survival of the strain. Tchelet et al. (45) used Pseudomonas sp. strain P51, originally isolated from sediments, for a bioaugmentation experiment in a soil column and sewage sludge. The survival and activity of strain P51 in the soil column were successful, but the strain was not able to maintain itself in the sludge reactors and thus no degradation was observed. McClure et al. (27) showed that a sludge isolate, AS2, was able to reach a stable level after inoculation, in contrast to other inocula tested. Strain AS2 had a characteristic flocculation, which may have been an important factor in the survival. C. testosteroni I2gfp, used in this study, also maintained a stable population in the activated-sludge system. The original strain I2, also isolated from a sludge environment, tends to form clusters within the sludge flocs. This observation, together with the unstable colony morphology on agar plates, suggests the formation of exopolysaccharide production, which was described by Bossier and Verstraete (6). Those authors described C. testosteroni A20, which expresses a phenotypic shift between mucoid-colony-forming (MCF) cells and non-MCF cells under different conditions. When the strains were cultured under unfavorable conditions, the cells shifted to the hydrophobic non-MCF-form and dense flocs were formed, providing cellular protection. Under favorable conditions, MCF cells were formed and resulted in loose associations. The possibility that strain I2gfp changes phenotype under unfavorable conditions may be an important factor in the maintenance of a stable population in the SCAS reactor.
The second criterion for successful bioaugmentation is the activity of the inoculum. In our experiment, after an initial adaptation period of 6 days, complete degradation of 3-CA was obtained during 2 weeks, while no degradation at all occurred in the noninoculated control reactor. Upon further operation of the SCAS system, 50% 3-CA removal was observed. It seems that there is a correlation between the declining population density of C. testosteroni I2gfp and the lower 3-CA removal of the inoculated reactors. Although McClure et al. (27) could establish a stable population of the introduced strain in the sludge environment, no enhanced degradation of chlorobenzoate was observed. Different authors proposed that the inability of the inoculated strains to degrade the xenobiotics may have been due to the availability of alternative substrates (5, 15, 27, 28, 36, 44). However, C. testosteroni I2gfp received daily only 40 mg of 3-CA/liter together with 1 g of COD per liter (diluted milk powder) and performed its specific activity within 2 weeks. Two preliminary SCAS experiments with the same strain I2gfp showed a very similar positive effect on 3-CA degradation during at least 2 weeks (data not shown). This observation was corroborated by the findings that when the pure culture was grown in LB medium supplemented with 100 mg of 3-CA/liter, 3-CA was not detectable after 1 day (data not shown). The degradation of 3-CA by strain I2gfp therefore is not repressed by additional nutrients. Compared with the calculated 0% degradation curve, no degradation of 3-CA was observed in either of the noninoculated control reactors B within 4 weeks. However, during the fifth week, enhanced degradation was observed in one of the reactors, probably due to the enrichment of indigenous bacteria with degradative capacities. It has been reported that in some cases indigenous bacteria become capable of removing xenobiotics after a long exposure time, either by metabolism or by cometabolism (28, 32, 47, 53). However, in our parallel SCAS reactors, the differences in degradation rates between the inoculated and control reactors were striking and stable over a prolonged time period, suggesting that the bioaugmentation was effective and not ephemeral.
The microbial community structure of the SCAS reactors was monitored by DGGE of 16S rRNA genes. The changes of the patterns over time suggest that the structure of the microbial communities was not static but rather was dynamic. After 7 days, the microbial communities in both series of reactors evolved into separate clusters. The inoculated strain could not be seen in the DGGE patterns, most probably because its proportion of the total bacterial cell count was too small and a DGGE pattern reveals only the numerically dominant populations. Eichner et al. (10) investigated the bioprotection of activated sludge from pollutant shocks by the related method TGGE. Those authors observed subtle shifts in community structure during adaptation to laboratory conditions. In their tests, the microbiota of the noninoculated control reactor collapsed after the shock load of xenobiotics, resulting in both a lower OUR and a decrease in bands in the TGGE pattern. In our studies, the diversity of bands in the pattern of the control reactor did not decrease visibly, and no drastic changes in the reactor performance were observed during the experiment. This was confirmed by the OUR measurements, where no significant differences could be observed with the inoculated reactor. In contrast to a shock load, as applied by Eichner et al. (10), the continuous supplementation of low concentrations of 3-CA in our experiment gave the sludge time to adapt. Remarkably, the DGGE technique combined with clustering analysis revealed subtle responses to the inoculation of strain I2gfp. Indeed, some species seemed to be enriched after the inoculation, while others tended to be less abundant.
This work indicates that bioaugmentation of activated-sludge systems for specific trace organics, such as 3-CA, can be achieved successfully. Moreover, this work corroborates what is often experienced in the use of activated sludge systems, i.e., that inoculation with a specific strain generally has only a transient effect. The fact that even an indigenous strain is only temporarily effective in activated-sludge communities substantiates the experience that biological supplements for such systems have to be added on a regular basis in order to assure continuous treatment efficacy. Further research will be performed to try to prolong the period of efficient degradation by the inoculum.
ACKNOWLEDGMENTS
This work was supported by project grant G.O.A. (1997–2002) from the Ministerie van de Vlaamse Gemeenschap, Bestuur Wetenschappelijk Onderzoek (Belgium), by a research grant from the Flemish Fund for Scientific Research (F.W.O. Vlaanderen), and by the EU-concerted action MAREP. E.M. Top and P. De Vos are also indebted to F.W.O. Vlaanderen for positions as Research Associate and Research Director, respectively.
We thank S. Blomme and H. Lievens for their assistance during the preliminary experiments, J. Kielemoes for microscopic analysis, and W. Dejonghe, R. Wouters, S. De Wildeman, and I. Dhaese for critically reading the manuscript.
REFERENCES
- 1.Arai H, Akahira S, Ohishi T, Maeda M, Kudo T. Adaptation of Comamonas testosteroni TA441 to utilize phenol: organization and regulation of the genes involved in phenol degradation. Microbiology. 1998;10:2895–2903. doi: 10.1099/00221287-144-10-2895. [DOI] [PubMed] [Google Scholar]
- 2.Bae H S, Lee J M, Kim Y B, Lee S T. Biodegradation of the mixtures of 4-chlorophenol and phenol by Comamonas testosteroni CPW301. Biodegradation. 1997;7:463–469. doi: 10.1007/BF00115293. [DOI] [PubMed] [Google Scholar]
- 3.Balashov S V, Balashova N V, Boronin A M. Plasmid control of p-toluenesulfonic acid degradation in Comamonas testosteroni BS1310. Microbiology. 1997;66:52–56. [Google Scholar]
- 4.Barriault D, Sylvestre M. Factors affecting PCB degradation by an implanted bacterial strain in soil microcosms. Can J Microbiol. 1993;39:594–602. doi: 10.1139/m93-086. [DOI] [PubMed] [Google Scholar]
- 5.Blumenroth P, Wagner-Döbler I. Survival of inoculants in polluted sediments: effect of strain origin and carbon source competition. Microb Ecol. 1998;35:279–288. doi: 10.1007/s002489900083. [DOI] [PubMed] [Google Scholar]
- 6.Bossier P, Verstraete W. Comamonas testosteroni colony phenotype influences exopolysaccharide production and coaggregation with yeast cells. Appl Environ Microbiol. 1996;62:2687–2691. doi: 10.1128/aem.62.8.2687-2691.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boulos L, Prevost M, Barbeau B, Coallier J, Desjardins R. LIVE/DEAD (R) BacLight (TM): application of a new rapid staining method for direct enumeration of viable and total bacteria in drinking water. J Microbiol Methods. 1999;37:77–86. doi: 10.1016/s0167-7012(99)00048-2. [DOI] [PubMed] [Google Scholar]
- 8.Brunsbach F R, Reineke W. Degradation of chloroanilines in soil slurry by specialized organisms. Appl Microbiol Biotechnol. 1993;40:2–3. doi: 10.1007/BF00902751. [DOI] [PubMed] [Google Scholar]
- 9.Chung C T, Niemela S L, Miller R H. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci USA. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eichner C A, Erb R W, Timmis K N, Wagner-Döbler I. Thermal gradient gel electrophoresis analysis of bioprotection from pollutant shocks in the activated sludge microbial community. Appl Environ Microbiol. 1999;65:102–109. doi: 10.1128/aem.65.1.102-109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El Fantroussi S, Mahillon J, Navaeu H, Agathos S N. Introduction and PCR detection of Desulfomonile tiedjei in soil slurry microcosms. Biodegradation. 1997;8:125–133. doi: 10.1023/a:1008262426800. [DOI] [PubMed] [Google Scholar]
- 12.El Fantroussi S, Verschuere L, Verstraete W, Top E M. Effect of phenylurea herbicides on soil microbial communities estimated by analysis of 16S rRNA gene fingerprints and community-level physiological profiles. Appl Environ Microbiol. 1999;65:982–988. doi: 10.1128/aem.65.3.982-988.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ezaki T, Hashimoto Y, Yabuuchi E. Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol. 1989;39:224–229. [Google Scholar]
- 14.Fujita M, Ike M, Uesugi K. Operation parameters affecting the survival of genetically engineered microorganisms in activated sludge processes. Wat Res. 1994;28:1667–1672. [Google Scholar]
- 15.Goldstein M G, Mallory L M, Alexander M. Reasons for possible failure of inoculation to enhance biodegradation. Appl Environ Microbiol. 1985;50:977–983. doi: 10.1128/aem.50.4.977-983.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenberg A E, Clesceri L S, Eaton A D, editors. Standard methods for the examination of water and wastewater. 18th ed. Washington, D.C.: American Public Health Association; 1992. [Google Scholar]
- 17.Häggblom M M. Microbial breakdown of halogenated aromatic pesticides and related compounds. FEMS Microbiol Rev. 1992;9:29–71. doi: 10.1111/j.1574-6968.1992.tb05823.x. [DOI] [PubMed] [Google Scholar]
- 18.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinteregger C, Loidl M, Streichsbier F. Characterization of isofunctional ring-leaving enzymes in aniline and 3-chloroaniline degradation by Pseudomonas acidovorans CA28. FEMS Microbiol Lett. 1992;97:261–266. doi: 10.1016/0378-1097(92)90346-p. [DOI] [PubMed] [Google Scholar]
- 20.Junker F, Cook A M. Conjugative plasmids and the degradation of arylsulfonates in Comamonas testosteroni. Appl Environ Microbiol. 1997;63:2403–2410. doi: 10.1128/aem.63.6.2403-2410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kashabek S R, Kasberg T, Müller D, Mars A E, Janssen D B, Reineke W. Degradation of chloroaromatics: purification and characterization of a novel type of chlorocatechol 2,3-dioxygenase of Pseudomonas putida GJ31. J Bacteriol. 1998;180:296–302. doi: 10.1128/jb.180.2.296-302.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kearny P C, Kaufman D D. Herbicides: chemistry, degradation and mode of action. New York, N.Y: Marcel Dekker, Inc.; 1975. [Google Scholar]
- 23.Latorre J, Reineke W, Knackmuss H J. Microbial metabolism of chloroanilines: enhanced evolution by natural genetic exchange. Arch Microbiol. 1984;140:159–165. [Google Scholar]
- 24.Loidl M, Hinteregger C, Ditzelmueller G, Ferschl A, Streichsbier F. Degradation of aniline and monochlorinated anilines by soil-borne Pseudomonas acidovorans strains. Arch Microbiol. 1990;155:56–61. [Google Scholar]
- 25.Marcus P, Talalay P. Induction and purification of alpha- and beta-hydroxysteroid dehydrogenases. J Biol Chem. 1956;218:661–674. [PubMed] [Google Scholar]
- 26.Mars A E, Kingma J, Kaschabek S R, Reineke W, Janssen D B. Conversion of 3-chlorocatechol by various catechol 2,3-dioxygenases and sequence analysis of the chlorocatechol dioxygenase region of Pseudomonas putida GJ31. J Bacteriol. 1999;181:1309–1318. doi: 10.1128/jb.181.4.1309-1318.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McClure N C, Fry J C, Weightman A J. Survival and catabolic activity of natural and genetically engineered bacteria in a laboratory-scale activated-sludge unit. Appl Environ Microbiol. 1991;57:366–373. doi: 10.1128/aem.57.2.366-373.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McClure N C, Weightman A J, Fry J C. Survival of Pseudomonas putida UWC1 containing cloned catabolic genes in a model activated-sludge unit. Appl Environ Microbiol. 1989;55:2627–2634. doi: 10.1128/aem.55.10.2627-2634.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mesbah M, Premachandran U, Whitman W B. Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol. 1989;39:159–167. [Google Scholar]
- 30.Meyer U. Biodegradation of synthetic organic colorants. London, United Kingdom: Academic Press Inc.; 1981. [Google Scholar]
- 31.Muyzer G, de Waal E C, Uitterlinden A. Profiling of complex microbial populations using denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nüßlein K, Maris D, Timmis K, Dwyer D F. Expression and transfer of engineered catabolic pathways harbored by Pseudomonas spp. introduced into activated sludge microcosms. Appl Environ Microbiol. 1992;58:3380–3386. doi: 10.1128/aem.58.10.3380-3386.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Øvreås L, Forney L, Daae F L, Torsvik V. Distribution of bacterioplankton in meromictic Lake Saelevannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol. 1997;63:3367–3373. doi: 10.1128/aem.63.9.3367-3373.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pot B, Vandamme P, Kersters K. Analysis of electrophoretic whole organism protein fingerprints. In: Goodfellow M, O'Donnel A G, editors. Modern microbial methods. Chemical methods in prokaryotic systematics. Chichester, United Kingdom: Wiley; 1994. pp. 493–521. [Google Scholar]
- 35.Riegert U, Heiss G, Fischer P, Stoltz A. Distal cleavage of 3-chlorocatechol by an extradiol dioxygenase to 3-chloro-2-hydromuconic semialdehyde. J Bacteriol. 1998;180:2849–2853. doi: 10.1128/jb.180.11.2849-2853.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt S K, Alexander M. Effects of dissolved organic carbon and second substrates on the biodegradation of organic compounds at low concentrations. Appl Environ Microbiol. 1985;49:822–827. doi: 10.1128/aem.49.4.822-827.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Selvaratnam S, Schoedel B A, McFarland B L, Kulpa C F. Application of reverse transcriptase PCR for monitoring expression of the catabolic dmpN gene in a phenol-degrading sequencing batch reactor. Appl Environ Microbiol. 1995;61:3981–3985. doi: 10.1128/aem.61.11.3981-3985.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi G H. Study on degradation and transformation of 1-(2-chlorobenzoyl)-3-(4-chlorophenyl) urea insecticide by soil microorganisms under needle and broad leaf forest. J Environ Sci. 1992;4:37–45. [Google Scholar]
- 39.Stanier R Y, Palleroni N J, Douderoff M. The aerobic Pseudomonads: a taxonomic study. J Gen Microbiol. 1966;43:159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- 40.Surmacz-Gorska J, Gernaey K, Demuynck C, Vanrolleghem P, Verstraete W. Nitrification process control in activated sludge using oxygen uptake rate measurements. Environ Technol. 1995;16:568–577. [Google Scholar]
- 41.Surovtseva E G, Ivoilov V S, Vasileva G K, Belyaev S S. Degradation of chlorinated anilines by certain representatives of the genera Aquaspirillum and Paracoccus. Microbiology. 1996;65:553–559. [Google Scholar]
- 42.Surovtseva G, Ivoilov V S, Karasevich Y N, Vacileva G K. Chlorinated anilines, a source of carbon, nitrogen and energy for Pseudomonas diminuta. Microbiologiya. 1985;54:948–952. [Google Scholar]
- 43.Surovtseva G, Vasileva G K, Vol Nova A I, Baskunov B P. Destruction of monochloroanilines by the meta-cleavage by Alcaligenes faecalis. Doklady Akademii Nauk SSSR. 1980;254:226. [PubMed] [Google Scholar]
- 44.Swindoll C M, Aelion C M, Pfaender F K. Influence of inorganic and organic nutrients on aerobic biodegradation and on the adaptation response of subsurface microbial communities. Appl Environ Microbiol. 1988;54:212–217. doi: 10.1128/aem.54.1.212-217.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tchelet R, Meckenstock R, Steinle P, van der Meer J R. Population dynamics of an introduced bacterium degrading chlorinated benzenes in a soil column and in sewage sludge. Biodegradation. 1999;10:113–125. doi: 10.1023/a:1008368006917. [DOI] [PubMed] [Google Scholar]
- 46.Tombolini R, Unge A, Davey M E, De Bruijn F J, Jansson J K. Flow cytometric and microscopic analysis of GFP-tagged Pseudomonas fluorescens bacteria. FEMS Microbiol Ecol. 1997;22:17–28. [Google Scholar]
- 47.van der Meer J R, Werlen C, Nishino S F, Spain J C. Evolution of a pathway for chlorobenzene metabolism leads to natural attenuation in contaminated groundwater. Appl Environ Microbiol. 1998;64:4185–4193. doi: 10.1128/aem.64.11.4185-4193.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Limbergen H, Top E M, Verstraete W. Bioaugmentation in activated sludge: current features and future perspectives. Appl Microbiol Biotechnol. 1998;50:16–23. [Google Scholar]
- 49.Vauterin L, Yang P, Hoste B, Vancanneyt M, Civerolo E L, Swings J, Kersters K. Differentiation of Xanthomonas campestris pv. citri strains by sodium dodecyl sulfate-polyacrylamide gel electrophoresis of proteins, fatty acid analysis, and DNA-DNA hybridization. Int J Syst Bacteriol. 1991;41:535–542. [Google Scholar]
- 50.Versalovic J, Koeuth T, Lupski J. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;24:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ward J H. Hierarchial grouping to optimize an objective function. J Am Stat Assoc. 1963;58:236–244. [Google Scholar]
- 52.Watanabe K, Yamamoto S, Hino S, Harayama S. Population dynamics of phenol-degrading bacteria in activated sludge determined by gyrB-targeted quantitative PCR. Appl Environ Microbiol. 1998;64:1203–1209. doi: 10.1128/aem.64.4.1203-1209.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weber W J J, Corseuil H X. Inoculation of contaminated subsurface soils with enriched indigenous microbes to enhance bioremediation rates. Wat Res. 1994;28:1407–1414. [Google Scholar]
- 54.Zeyer J, Kearny P C. Microbial degradation of para-chloroaniline as sole source of carbon and nitrogen. Pestic Biochem Phys. 1982;17:215–233. [Google Scholar]
- 55.Zeyer J, Wasserfallen A, Timmis K N. Microbial mineralization of ring-substituted anilines through an ortho-cleavage pathway. Appl Environ Microbiol. 1985;50:447–453. doi: 10.1128/aem.50.2.447-453.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]