Abstract
As the only ribosomally encoded N-substituted amino acid, proline promotes distinct secondary protein structures. The high proline content in collagen, the most abundant protein in the human body, is crucial to forming its hallmark structure: the triple-helix. For over five decades, proline has been considered compulsory for synthetic designs aimed at recapitulate collagen’s structures and properties. Here we describe that N-substituted glycines (N-glys), also known as peptoid residues, exhibit a general triple-helical propensity similar to or greater than proline, enabling synthesis of stable triple-helical collagen mimetic peptides (CMPs) with unprecedented sidechain diversity. Supported by atomic-resolution crystal structures, as well as circular dichroism and computational characterizations spanning over 30 N-gly-containing CMPs, we discovered that N-glys stabilize the triple-helix primarily by sterically preorganizing individual chains into the polyproline-II helix. We demonstrated that N-glys with exotic sidechains including a ‘click’-able alkyne and a photo-sensitive sidechain enable CMPs for functional applications including spatio-temporal control of cell adhesion and migration. The structural principles uncovered in this study open up opportunities for a new generation of collagen mimetic therapeutics and materials.
Graphical Abstract
INTRODUCTION
Among the twenty canonical amino acids, proline (Pro) stands out as the single residue featuring an N-substitution. Pro’s unique cyclic sidechain mandates restricted backbone dihedral angles; when incorporated into a protein chain, its tertiary amide group lacks the N-hydrogen atom to donate a hydrogen bond. As a result, Pro disrupts secondary conformations favored by most amino acids, such as α-helices and β-sheets, and promotes distinct folding patterns, such as β-turns and polyproline helices. From a chemical perspective, there are numerous N-substituted α-amino acids as possible protein building blocks,1 of which evolution has sampled only one candidate for ribosomal protein expression: Pro. The rules governing the folding of N-substituted amino acids and their relationship to Pro’s unique conformation are intriguing topics for both fundamental science and practical applications of de novo protein design.2
Collagen, the most abundant protein in vertebrates, best exemplifies a natural protein structure dictated by Pro’s folding. A collagen chain consists mostly of a repetitive sequence of Gly-X-Y triplets, where the X and Y positions are often occupied by Pro and its post-translational hydroxylation product, 4(R)-hydroxyproline (Hyp).3 As many as 22% of all residues in human collagen are either Pro or Hyp4 and, therefore, the defining structural motif of collagen is the intertwining of three polyproline II-type helices.5-6 This unique triple-helical motif is responsible for collagen’s remarkable properties, including higher-order assembly,7 mechanical strength,8 resistance to proteases, and binding with numerous cell receptors.9 The triple-helix has also inspired many synthetic designs aimed at recapitulating nature’s supramolecular chemistry.10-11
Because collagens are large, insoluble proteins that are difficult to study holistically, many research groups have turned to synthetic collagen mimetic peptides (CMPs).3,6,12 CMPs featuring GlyProPro or GlyProHyp triplets have been synthesized as models of the collagen triple-helix since the late 1960s.13 Studies of CMPs revealed that all natural amino acids destabilize the triple-helix when replacing Pro in the GlyProHyp triplet,14 while hydroxylation of Pro at the Y position stabilizes the structure.3 Throughout the 1990s12 and until more recently.15-16 a substantial portion of CMP studies have focused on uncovering and exploiting the stabilizing effects of the post-translational modification of Pro by leveraging synthetic Pro derivatives.3,15-19
Despite more than five decades of vigorous research, there have been few attempts to produce a stable collagen triple-helix without Pro, and the structural forces imposed on the triple-helical conformation by unnatural N-substituted amino acids remain almost entirely unexplored. As probably the only precedent, Goodman and coworkers showed that replacing Pro with N-isobutylglycine (Nleu) within CMPs results in a stable triple-helices.20-24 They suggested that the stabilization comes from interchain interactions between the hydrophobic sidechains of Nleu and the adjacent Pro.21-23 Meanwhile, peptoids, which are synthetic oligomers of N-substituted glycines (e.g., Nleu), have been studied for decades as a promising class of synthetic peptidomimetics in biomedicine and material science.25-26 Repetitive peptoid sequences have been shown to form robust conformations resembling polyproline helices,27-31 as well as a multitude of secondary and higher order structures including helical bundles,32 superhelices,33 and two-dimensional sheets.34-35 Yet so far, the structural influences of peptoid residues upon collagen triple-helicity remain unclear.
Here we present a systematic study on the triple-helical stability of N-substituted glycines (N-glys, Figure 1a) and demonstrate their potential for making stable collagen triple-helices with extraordinarily diverse sidechain structures. Using host-guest peptide systems for both collagen-mimetic and polyproline peptides, we examined peptides featuring more than 30 different N-glys for their triple-helical and polyproline-II-helix (PPII) propensities. We acquired atomic-resolution crystal structures of N-gly containing CMPs and performed molecular dynamics (MD) simulations, providing insights into the structure of peptoid residues within collagen triple-helices and their mechanism of stabilization. To demonstrate utility, we presented stable triple-helices featuring exotic N-gly sidechains including light sensitive ones, which enabled spatiotemporal control of cell adhesion and migration on a collagen substrate. So far, Pro and its derivatives have been indispensable for constructing synthetic collagen mimetics. The design principles uncovered in this study drastically expand the library of residues with high triple-helical propensity, and have immense implications for a new generation of collagen-mimetic therapeutics and materials.
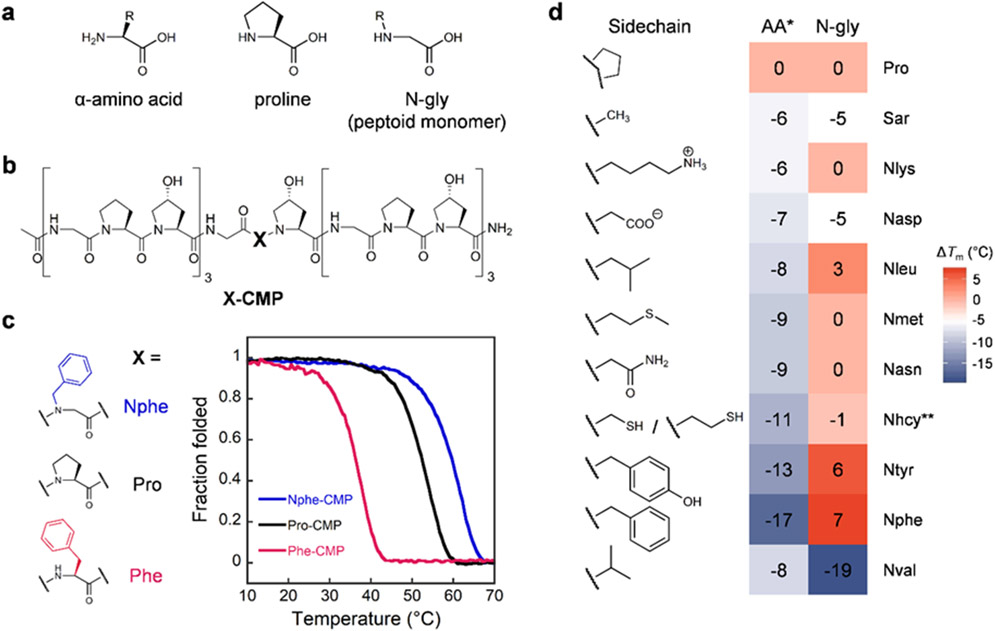
Figure 1. A single N-substituted glycine (N-gly), also known as a peptoid residue, in the central Pro position within (GlyProHyp)7 shows high triple-helical propensity.
a, General structures of an α-amino acid, Pro, and an N-gly. b, Chemical structure of X-CMP, a host-guest collagen mimetic peptide with the sequence of a (GlyProHyp)7, where the central X-position Pro is substituted, for example, with an N-gly residue. c, An X-CMP triple-helix dissociates into single strands under gradual heating monitored under CD, where the middle point of this two-state transition is defined as the X-CMP’s melting temperature (Tm). The Nphe sidechain at the X position resulted in a CMP triple-helix considerably more stable than Pro (ΔTm: +7 °C). In comparison, Phe with the same sidechain was highly destabilizing (ΔTm: −17 °C). d, Triple-helical stabilities of X-CMPs featuring N-glys with canonical sidechains, quantitated in ΔTm (in comparison to the host peptide: Pro-CMP). Generally, peptoid residues (right column) have higher stabilities than their amino acid counterparts (left column), and many have stabilities similar to or better than Pro. *ΔTm for amino acid residues taken from reference21. **Nhcy (homocysteine) is a similar but not exact equivalent to cysteine.
RESULTS AND DISCUSSION
Triple-helical stability of CMPs with N-glys.
To investigate the triple-helical propensity of peptoid residues, we inserted a series of N-gly guests into the central X position of a conventional CMP host peptide with the sequence: Ac-(GlyProHyp)3-Gly-X-Hyp-(GlyProHyp)3-NH2 (designated as X-CMP, Figure 1b; see Supporting Information for methods).36 We measured the X-CMPs’ triple-helical stability via thermal unfolding experiments monitored by circular dichroism (CD, Figure 1c, Table S1 and Section 4). We first investigated a group of N-glys with sidechains selected from canonical amino acids (Figure 1d). It is known that Pro is the most stabilizing amino acid at position X, and substitution from Pro to another canonical amino acid clearly reduces Tm by 4-17 °C (Figure 1d).14 Surprisingly, we found that almost all N-gly residues with canonical sidechains were more stable than their amino acid counterparts with the biggest Tm difference seen between Nphe- and Phe-CMP (Figure 1c), and many were as stable as Pro (Figure 1d, exception: Nval). These results demonstrate that, in the X position, shifting the sidechain from the Cα carbon to the nitrogen (i.e., transforming a canonical amino acid to its peptoid analogue) may improve triple-helical stability.
Crystal structures of N-gly-CMPs.
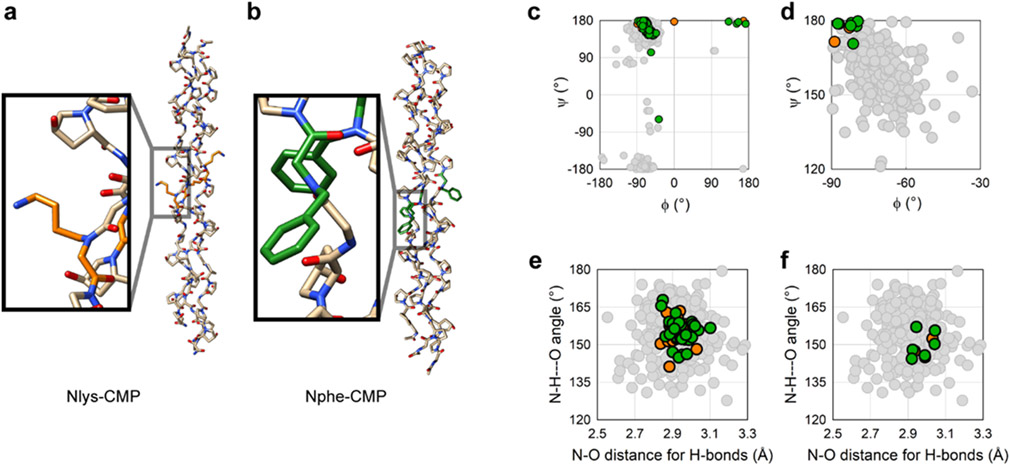
To decipher the precise molecular structure of N-glys within the CMP, we determined the X-ray crystal structures of Nlys-CMP and Nphe-CMP. We chose these X-CMPs for crystallographic analysis because of their high stabilities and differing sidechain properties. We solved their structures to 0.95 Å (Nlys-CMP) and 1.10 Å (Nphe-CMP) resolution (Figure 2a,b; Table S2, Figures S1 and S2). The overall conformation in both crystal structures was consistent with those reported for conventional collagen peptides in the Protein Data Bank (PDB, Table S3), with the average (φ, ψ) angles and standard deviations of all non-terminal Pro, Hyp, and Gly residues being (−70 ± 6°, 162 ± 6°), (−58 ± 5°, 147 ± 7°), and (−69 ± 3°, 174 ± 5°), respectively (Figure 2c, Figure S3). The dihedral angles of the two N-glys [Nlys: (−86 ± 3°, 176 ± 3°), Nphe: (−82 ± 3°, 177 ± 3°)] were also within the range populated by the X-position residues of collagen peptides in PDB (Figure 2d), though shifted from the average angles (Δφ = −13°, Δψ = +21°). This deviation did not alter the backbone structures and there was neither kinking nor bending of the triple-helix near the N-gly in either crystal structure. Furthermore, the inter-strand hydrogen bonding pattern formed along the length of the triple-helix was consistent with those seen in collagen peptides in the PDB (Figure 2e,f). This suggests that the triple-helical stability of N-gly-CMPs may not originate from improved hydrogen bonding37 of N-glys.
Figure 2. CMPs containing peptoid residues adopt a triple-helical conformation consistent with native collagens.
a,b Crystal structures of Nlys-CMP and Nphe-CMP, each showing a characteristic collagen triple-helix (Nlys in orange, Nphe in green). c, A Ramachandran plot showing the (φ, ψ) angles of all amino acid residues in the crystal structure of Nlys-CMP (orange) and Nphe-CMP (green), overlaid on angles from reported CMP crystal structures in PDB (grey, Table S3). d, A Ramachandran plot showing (φ, ψ) angles from the N-gly residues (Nlys in orange, Nphe in green), overlaid with angles (in grey) from the X position of Gly-X-Y repeats in reported CMP crystal structures. The peptoid residues adopt a conformation common to the X position in reported Gly-X-Y collagen sequences, but with shifted average angles (Δφ: −13°, Δψ: +21°). e, A plot of the N-H-O angle versus the N-O distance of all interstrand amide-amide H-bonds in the crystal structures of Nlys-CMP (orange) and Nphe-CMP (green) overlaid on H-bonds from reported CMP structures (grey). f, A plot, same as e but showing only H-bonds associated with N-gly residues (Nlys in orange, Nphe in green), whose parameters are consistent with reported crystal structures of collagen peptides.
Probing the structural factors for N-glys’ triple-helical stability.
Next, we introduced a series of guest N-glys into X-CMPs to understand how sidechain structures affect triple-helical stability (Figure 3). We first focused on hydrophobic sidechains, which previous reports from Goodman and coworkers21-22 suggested as the main stabilizing factor in N-gly-CMPs; recent studies also indicated that hydrophobic lipid moieties attached to Pro can stabilize the CMP triple-helix.38 We found that the aliphatic Nchx and the aromatic Nphe produced triple-helices with stability notably higher than Pro (Figure 3a). In addition, Nphe derivatives with electron-rich (Ntyr) or poor (Nnbz) aromatic rings generated triple-helices with similar stability, suggesting that neither the aromaticity nor its electron density is crucial to triple-helix stabilization. Meanwhile, although structurally similar, the hydrophilic Ndxn and the deprotonated Ntyr (pH 12) were both more stabilizing than Pro and only slightly less stabilizing than their hydrophobic analogs in the group (Figure 3a). These results suggest that the hydrophobicity of the peptoid residue might not be the main driving force for triple-helix stabilization. Furthermore, we synthesized a pair of CMPs featuring central guest triplets of GlyNpheHyp and GlyNpheIle to examine the hydrophobic interactions between adjacent residues (Table S4). Introduction of Ile did not provide any additional stabilizing effect, further de-emphasizing the hydrophobicity factor.
Figure 3. The structure of the peptoid sidechain affects triple-helical propensity.
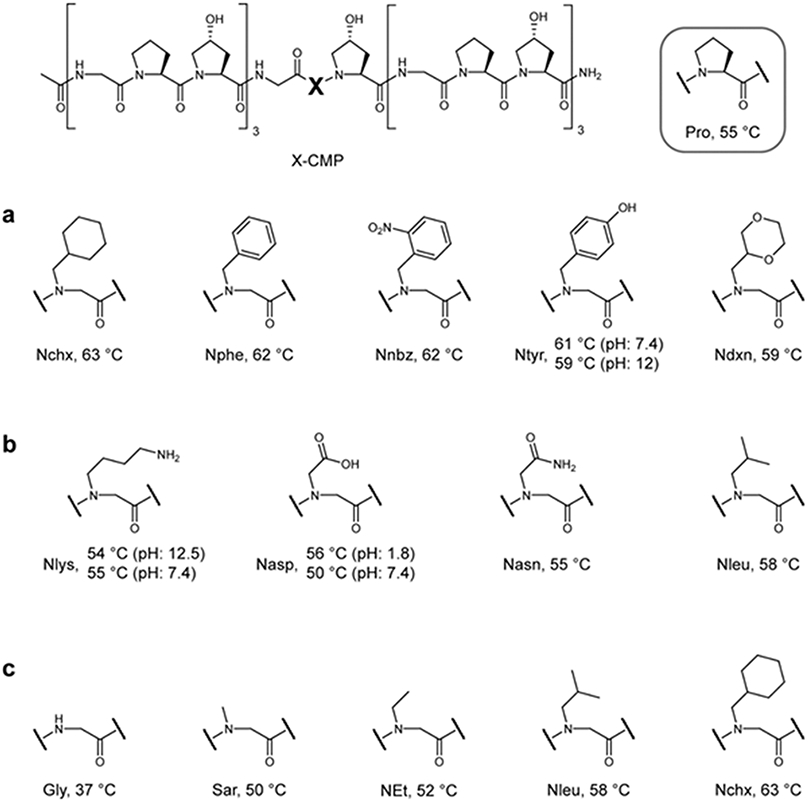
a, Tm values for a series of X-CMPs with N-Cα-substituted 6-membered rings. There is little difference in triple-helical stability among the aliphatic, aromatic, and hydrophilic rings, and every peptide had a Tm higher than the host: Pro-CMP. b, Although charged groups destabilized the triple-helix at physiological pH, when the sidechains were uncharged, both Nlys and Nasp, as well as the hydrophilic Nasn, were as stabilizing as Pro. c, A series of X-CMPs with aliphatic sidechains of increasing size showing concomitantly increasing Tm values, implying an robust steric effect. Although the hydrophobicity in this series increases as the aliphatic sidechains increase in size, according to series a and b, hydrophobicity has only a minor effect on stability. The results suggest that the tertiary amide structure N-gly (which hinders the α helix and β sheet conformations) and the size of the N-substituted sidechain play the most critical roles in stabilization of N-gly-CMP triple-helices.
Next, we investigated the charged and hydrophilic peptoid sidechains (Figure 3b). Although charged Nasp and Nlys modestly destabilized the triple-helices through electrostatic repulsions, when we altered pH so their sidechains were uncharged, both X-CMPs were as stable as Pro-CMP. Also, peptoid residues with similar sidechain configurations conferred strikingly similar Tm values (Figure 3b: Nasp, Nasn, and Nleu; Figure 3a: Nchx, Nphe, Nnbz, Ntyr, Ndxn), and within each group, Tm values of the hydrophobic residues were only higher by 2-4 °C.
Lastly, we found that progressively increasing the size of the aliphatic sidechains produced a concomitant increase in Tm for the corresponding X-CMPs (Figure 3c). Knowing that the hydrophobicity has only a modest stabilizing effect for triple-helices, we reasoned that the large increase in stability from Gly-CMP (Tm, 37 °C) to Nchx-CMP (Tm, 63 °C) cannot be explained by hydrophobicity alone, and were prompted to look into other mechanisms by which peptoid residues stabilize the collagen triple-helix.
Polyproline-II-helix (PPII) propensity of N-glys.
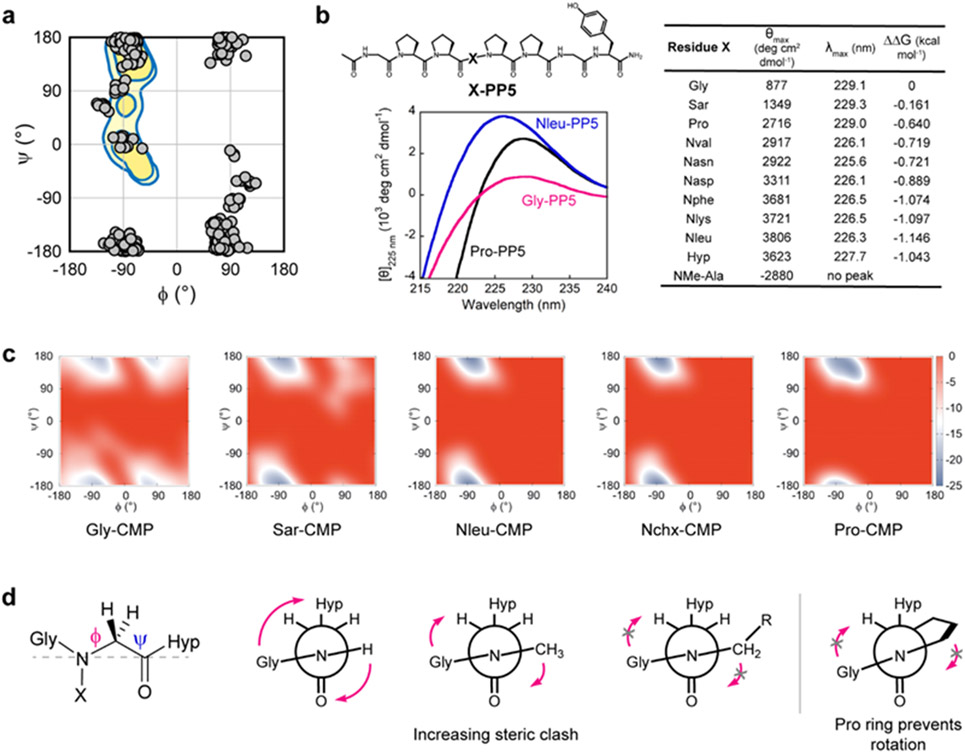
It is generally accepted that Pro and Hyp stabilize the collagen triple-helix by pre-organizing individual collagen strands into the PPII conformation.39-40 Therefore, we hypothesized that N-glys, with their Pro-like tertiary amide structure, could induce similar pre-organizational effects in N-gly-CMPs. To test this hypothesis, we first surveyed a large set of small molecule crystal structures containing N-glys from the Cambridge Structural Database (Tables S5 and S6). We found that the dihedral angles of these peptoid residues largely fall within the most probable range for Pro, with the highest density present in the area corresponding to polyproline helices (Figure 4a). This suggests that similar to Pro, N-glys may have a high natural propensity for the PPII conformation.
Figure 4. N-glys have a strong polyproline-II helix (PPII) propensity which stabilizes the X-CMP triple-helix.
a, A survey of dihedral angles from N-glys in small molecule crystal structures (from Cambridge Structure Database), showing their natural propensity for a left- or right-handed polyproline helix (watermark: regions populated by Pro). b, Structure of the PPII host-guest peptide X-PP5 and comparative CD spectra of Gly-PP5, Pro-PP5, and Nleu-PP5 showing PPII-characteristic signals near 228 nm. All N-glys, except Sar, had a θmax higher than Pro indicating peptoids’ strong PPII propensity. No peak was observed near 228 nm for NMe-Ala (Supporting Information Page S58). c, Metadynamics calculations of the free energy (kcal/mol) landscape of the guest residues in Gly-, Sar-, Nleu-, Nchx-, and Pro-CMPs. Each model yielded a native state consistent with the triple-helical and PPII conformations (−70°, 170°). However, peptides with small guest residues (Gly, Sar) showed a secondary non-PPII energy well. d, Newman-like projections of N-glys (grey dashed line: the line of sight for the projection). N-glys with larger sidechains may experience major steric clashes with their carbonyl group and have reduced conformational flexibility. This can help preorganize individual chains into a PPII conformation, thereby stabilizing the triple-helical assembly.
To systematically study the N-glys’ propensity for the PPII conformation, we employed another host-guest peptide: Ac-GlyProPro-X-ProProGlyTyr-NH2 designated as X-PP5, where X represents the guest residue replacing the central Pro within the (Pro)5 sequence. This short, proline-rich host peptide was previously utilized to measure the PPII propensity of the twenty canonical amino acids based on their CD intensities near 228 nm.41-42 Except for Sar-PP5, all N-gly-PP5s showed PPII CD intensity higher than Pro-PP5 (Figure 4b) and their amino acid counterparts.41 Notably, the PPII propensities of Nphe, Nlys, and Nleu were even higher than Hyp, which is known for having the highest PPII propensity among all natural amino acids.43 These results indicate that N-glys can promote the PPII conformation of individual peptide strands, thereby improving triple-helical stability. The free energy bonus arising from PPII preorganization by just a few bulky N-glys (Figure 4b, ΔΔG) can add up to roughly an extra amide-amide hydrogen bond (−2.0 kcal/mol)44 in the triple-helix.
Previous studies have shown that peptoids with specific N-substitutions can form stable polyproline helices.27-31,45 So far, the frontier of peptoid research has largely focused on control over the cis-trans isomerization of peptoid amide bonds (i.e., adoptions of PPI or PPII conformation) through specific steric or stereoelectronic effects encoded by N-gly sidechains.27-31,46 In this work, we quantitated the general PPII propensity of peptoid residues. We learned that although Pro has the highest PPII propensity of all canonical amino acids, it is on the low side among N-substituted α-amino acids (Figure 4b).
Simulations and steric effects of the N-substitution.
We conducted MD simulations for triple-helical X-CMPs incorporating guest residues Gly, Sar, Nleu, Nchx, or Pro (Figure 4c). We based our simulations on the crystal structures of Nphe- and Nlys-CMPs (Figure 2) and chose N-glys with simple aliphatic sidechains that lack potential for electrostatic and hydrogen bonding interactions. The simulation results demonstrated that Nleu and Nchx displayed energy landscapes very similar to Pro, with a low-energy valley confined to the regions (φ, ψ angles) for PPII and the triple-helix. In comparison, Sar and Gly, bearing a smaller or no sidechain, showed more conformational flexibility (Figure 4c). These simulation results are in agreement with our CD experiments which showed that the size of an N-gly’s sidechain correlates with its PPII propensity (i.e., Gly < Sar < Nleu) and the triple-helical stability of its corresponding X-CMP (Figures 3c and 4c). As a plausible explanation for this effect, a bulky sidechain can impose a steric restraint to the N-gly’s backbone carbonyl, hindering the rotation around its nitrogen-carbon bond and constraining the φ angle to values appropriate for triple-helical folding (Figure 4d). Additionally, we believe that an N-gly’s ψ angle is less sterically influenced by the N-substitution because it is located further from the sidechain; it is expected to assume a value close to ± 180° because the α-carbon has no sidechain.
The pyrrolidine ring of Pro has been assumed to be a structural requisite for collagen mimetics for decades.3,47 In an attempt to replace Pro in CMPs, it was found that converting Pro to N-methyl-L-alanine (NMe-Ala), removing only the γ carbon and eliminating the ring structure, substantially destabilizes the triple-helix48 (Figure S4). It was concluded from this observation that the conformational restrictions imposed by the Pro ring are more important for triple-helical stability than the presence of an N-substitution.48 In contrast, our work supported that N-glys with bulky sidechains can form well-folded collagen triple-helices without Pro’s cyclic sidechain. We believe that the low triple-helicity of NMe-Ala48 is due to its poor PPII propensity (Figure 4b). Without the ring, steric repulsion between the two adjacent methyl groups of NMe-Ala precludes formation of the dihedral angles necessary to form the polyproline-II helix, thereby nullifying any stabilizing effect from the N-methyl group (Figure S4).
In addition to the φ, ψ dihedral angles, the ω angle (i.e., the cis-trans isomerization of the Pro amide bond) is a key parameter determining the backbone folding of a CMP: the higher the trans:cis ratio of the Pro amide bonds in a single strand, the more stable the resulting triple-helix.3 This rule is well accepted for CMPs based on Pro, Hyp, and modified prolines (e.g. fluoroproline, azidoproline),17,49 whose φ, ψ angles are largely restrained by the pyrrolidine ring. However, for peptoid residues, how the trans:cis ratio of the N-gly’s amides affects the triple-helix stability now becomes a topic for further investigation. In the current study, although residues Sar, NEt, and Nphe all seem to favor the trans-amide less than Pro (Table S8), they generally stabilize the triple-helix, suggesting that the effect of the cis-trans isomerization may be more complex for residues that do not contain a proline ring.
N-Cα chiral branching.
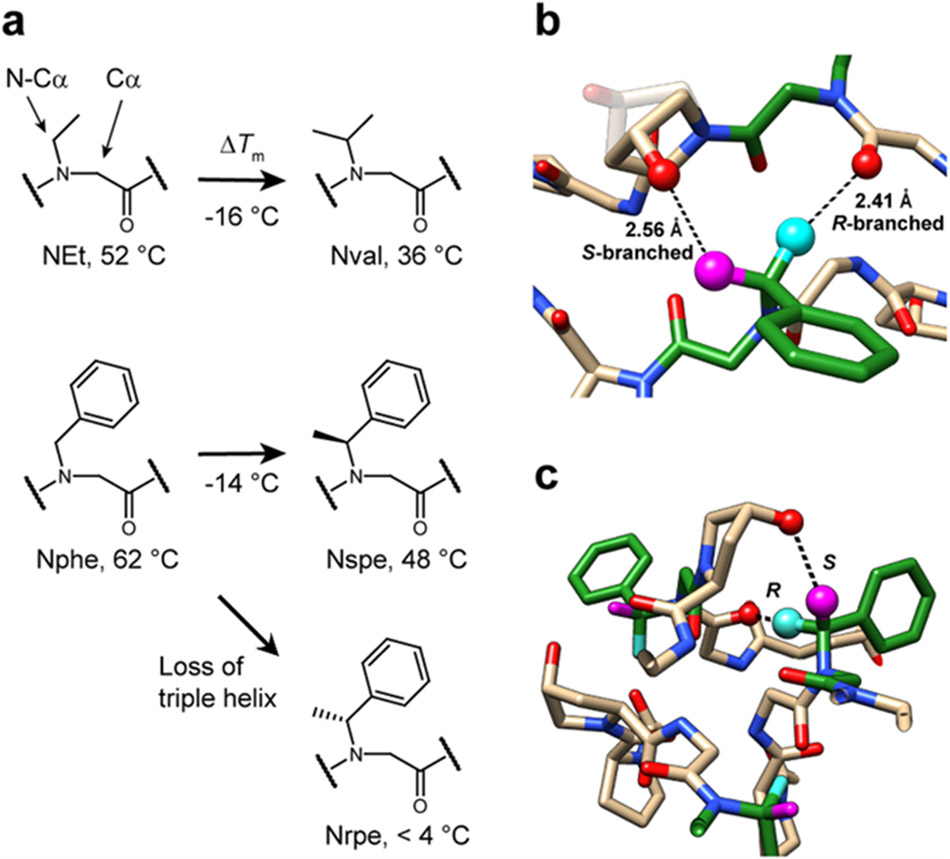
To understand why Nval, the only peptoid residue with an N-Cα-branched sidechain in our library, has unexpectedly low triple-helical stability (Tm: 36 °C, Figure 1d), we created a group of X-CMPs featuring guest residues NEt, Nval, Nphe, Nspe and Nrpe (Figure 5). Their Tm values (Figure 5a) indicated that adding an N-Cα methyl group to an N-gly destabilizes the resulting triple-helix with all branched residues having lower stabilities than their unbranched analogs. Additionally, triple-helical folding was greatly affected by the chirality of the N-Cα-branching (Figure 5a, Nspe versus Nrpe). Structural 3D modeling revealed chirality-specific locations of steric clashes between the branching methyl group and an adjacent peptide strand (Figure 5b,c). Since an N-Cα branch abolishes triple helix formation with an R-chiral branch, we expect Nval to adopt a geometry similar to the S-chiral Nspe in the triple helix. However, its branched structure and potential steric clash provides one explanation for Nval-CMP’s low Tm despite Nval having a PPII propensity higher than Pro (Figure 4b).
Figure 5. N-Cα branching of peptoid residues affects triple-helical folding.
a, Nval- and Nspe-CMPs, each containing an N-Cα branch, had Tm values 14-16 °C lower than their unbranched analogs: NEt and Nphe. Moreover, while Nspe conferred a triple-helix with a Tm of 48 °C, its enantiomer, Nrpe completely abolished the triple-helical folding. b,c, Molecular modeling based on the crystal structure of Nphe-CMP showing potential steric clashes within the triple-helix introduced by the N-Cα methyl branch. The R-branch of Nrpe (cyan), pointing directly toward the inner core of the triple-helix, may clash with the backbone carbonyl of the cross-chain Gly and interfere with proper backbone assembly. In contrast, the S-branch (magenta) in Nspe may clash with the hydroxyl group of a cross-chain Hyp, which may be less destabilizing since the steric repulsion may be alleviated by changing the Hyp’s ring pucker (shown with half-transparency in b).
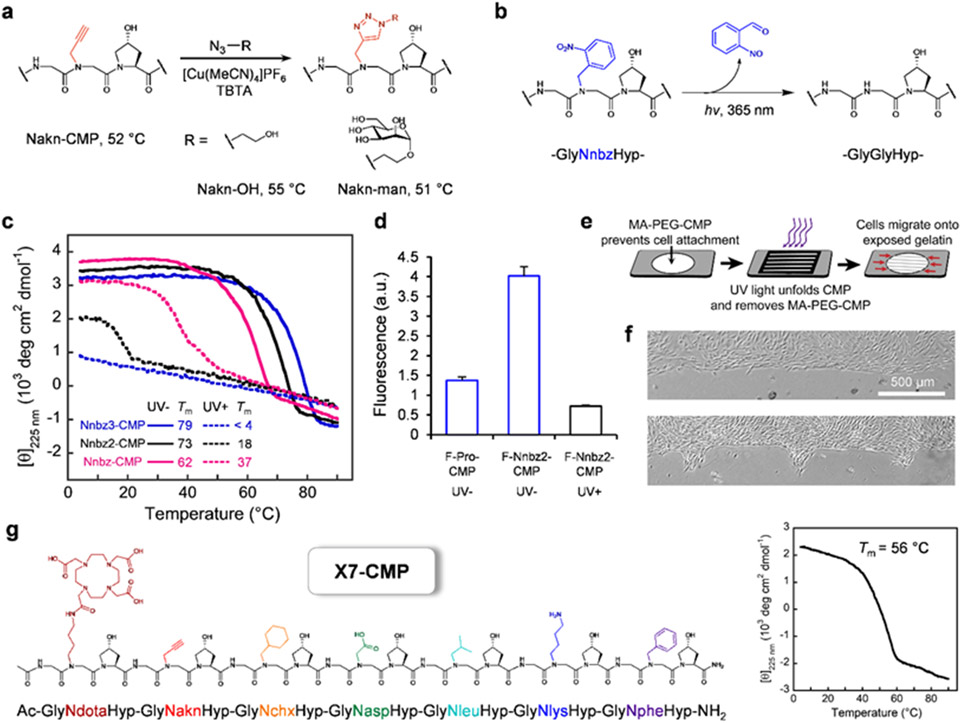
Sidechain functionalization.
After a series of mechanistic investigations into triple-helical folding, we turned to leveraging the diverse pool of peptoid residues to produce functionalized collagen mimetics for potential applications. As one example, we demonstrated facile sidechain functionalization through ‘click’ chemistry, utilizing an alkyne-bearing peptoid residue (Nakn, Figure 6a, Supplementary Discussion and Methods in Supporting Information). In another example, we synthesized Nnbz-CMP, an X-CMP featuring a UV-cleavable N-o-nitrobenzyl (nbz) sidechain.50 We hypothesized that Nnbz-CMP would form a robust triple-helix, but UV irradiation would convert the stabilizing Nnbz residue into Gly, thereby dramatically weakening the triple-helix (Figure 6b). CD measurements confirmed this hypothesis (Figure 6c). Moreover, characterization of CMPs bearing more than one Nnbz residue (e.g., Nnbz2,3, Table S7) indicated that (i) the stabilizing effect of N-glys may be additive, and (ii) the increased number of Pro → Nnbz substitutions resulted in even wider Tm differences before and after UV irradiation (Figure 6c).
Figure 6. X-CMPs unlock unprecedented exotic functional designs.
a, Facile modification of the alkyne-sidechain through ‘click’ chemistry, forming spatially-demanding triazole units without destabilizing the triple-helix (Supplementary Discussion). b, Conversion of the triple-helix-stabilizing Nnbz to the destabilizing Gly via photo-cleavage of the N-o-nitrobenzyl group. c, CD melting curves showing widening of Tm difference before and after UV irradiation as more Nnbz residues replace Pro in the triple-helix (Table S7). d, Fluorescence of gelatin films treated with fluorescein-labeled X-CMPs, demonstrating F-Nnbz2-CMP’s higher affinity to gelatin, and its ability to unfold and unbind upon UV irradiation. e, Schematic of UV-patterning of a gelatin substrate enabled by Nnbz2-CMP conjugated to MA-PEG. f, Light micrographs of MDA-MB-231 cells grown on a gelatin film migrating from a defined boundary (top) into selected areas where the gelatin-bound MA-PEG-CMP had been removed by UV light through a photomask (1 d after UV, bottom). g, Pro-free X7-CMP hosting seven peptoid residues with diverse sidechain moieties showcasing a hyperstable triple-helical collagen peptide with the greatest sidechain diversity to date.
Next, we examined the Nnbz-substituted CMP’s capacity to hybridize with denatured collagen and their release by UV irradiation. Previously, we reported that single-strand CMPs [e.g., (GlyProHyp)9] can form hybrid triple-helices with collagens that are denatured by heat, proteases, or mechanical damage.8,50 Using Nnbz2-CMP labeled with fluorescein (F-Nnbz2-CMP), we found that Nnbz2-CMP displayed a drastically higher level of binding to gelatin (heat-denatured collagen) than the host peptide Pro-CMP, consistent with its stronger triple-helical propensity (Figure 6d). Furthermore, upon brief UV irradiation, over 80% of the bound F-Nnbz2-CMP was released from the gelatin matrix (Figure 6d).
Spatio-temporal modification of cell culture substrates.
To further demonstrate X-CMP’s applications in biomaterials, we conjugated Nnbz2-CMP to a multi-arm poly(ethylene glycol) (MA-PEG) polymer (Figure S5a,b). The resulting conjugate MA-PEG-CMP readily formed a hydrogel that was physically crosslinked via its Nnbz2-CMP triple-helices, and showed the ability to dissolve and release its bound contents on-demand as the CMP crosslinks unfold upon UV irradiation (Figure S5c). Next, taking advantage of PEG’s ability to repel cell adhesion combined with Nnbz2-CMP’s UV-triggered unbinding from gelatin, we demonstrated that a gelatin film can be coated with MA-PEG-CMP to accurately photo-pattern cell attachment in selected areas (Figure S6), or to trigger cell migration into defined regions at a designated time (Figure 6e,f). These proof-of-concept experiments showcase the practical use of X-CMPs for creating complex cell culture substrates with spatio-temporal controls.
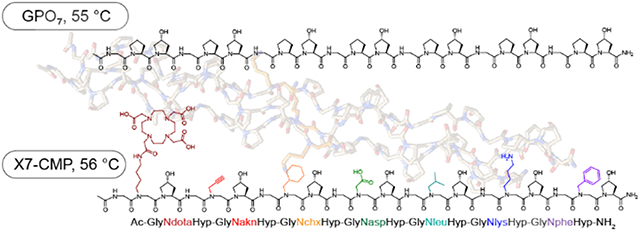
Billions of stable, Pro-free triple-helices.
Finally, we synthesized X7-CMP, a (Gly-X-Hyp)7 sequence where each X position was populated with a different peptoid residue (Figure 6g). With a Tm 1 °C higher than (GlyProHyp)7, X7-CMP represents a hyperstable, synthetic collagen triple-helix with the greatest sidechain-diversity to date. As hundreds of amines are commercially available for efficient solid-phase peptoid synthesis, this work expands the amino-acid library for stable CMPs by more than an order of magnitude. Even with a modest library of 20 N-glys incorporated into a short (Gly-X-Hyp)7 sequence, over 1.2 billion (207) triple-helical peptides can be generated. The design principles discovered in this study open the door for not only advancing our understanding of the fundamental molecular interactions (e.g., amide cis-trans isomerization, n→π*)51-52 in polyproline and triple-helical folding, but also developing a new class of collagen-mimetic therapeutics and biomaterials with remarkable functionalities. With the unprecedented structural diversity offered by N-gly-containing CMPs, we anticipate the long-awaited new era of de novo design of functional collagen peptidomimetics.
Supplementary Material
Acknowledgements
The authors thank Rodrigo Galindo-Murillo and Xiaolei Zhu for consultation on the simulations of X-CMP structures and Hendra Wahyudi for assistance in the synthesis of Fmoc-Nnbz-OH. This research was funded by grants from the National Institutes of Health (R01AR071358, R21EY029430, and R21OD026618) awarded to S.M.Y.
Footnotes
Supporting Information: Supplementary discussion, additional experimental information, HPLC, MALDI, and CD for all peptides, as well as chemical structures of host-guest X-CMPs, X-ray crystallography data, visualization and Ramachandran plots of the solved crystal structures, schematic of MA-PEG-CMP conjugation, and pre-patterned MA-PEG-CMP showing spatial control of cell adhesion. This material is provided free of charge via the Internet at http://pubs.acs.org. In addition, complete crystallographic information for both solved crystal structures can be downloaded free of charge from the Protein Data Bank at http://www.rcsb.org (PDB, IDs: 7JX4 and 7JX5).
Competing interests
The authors declare no competing interests.
REFERENCES
- 1.Horne WS; Grossmann TN, Proteomimetics as protein-inspired scaffolds with defined tertiary folding patterns. Nat. Chem 2020, 12 (4), 331–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang P; Boyken SE; Baker D, The coming of age of de novo protein design. Nature 2016, 537 (7620), 320–327. [DOI] [PubMed] [Google Scholar]
- 3.Shoulders MD; Raines RT, Collagen structure and stability. Annu. Rev. Biochem 2009, 78 (1), 929–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramshaw JAM; Shah NK; Brodsky B, Gly-X-Y tripeptide frequencies in collagen: A context for host–guest triple-helical peptides. J. Struct. Biol 1998, 122 (1), 86–91. [DOI] [PubMed] [Google Scholar]
- 5.Ramachandran GN; Kartha G, Structure of collagen. Nature 1955, 176 (4482), 593–595. [DOI] [PubMed] [Google Scholar]
- 6.Bella J; Eaton M; Brodsky B; Berman HM, Crystal and molecular structure of a collagen-like peptide at 1.9 A resolution. Science 1994, 266 (5182), 75–81. [DOI] [PubMed] [Google Scholar]
- 7.Orgel JPRO; Irving TC; Miller A; Wess TJ, Microfibrillar structure of type I collagen in situ. Proc. Natl. Acad. Sci. U. S. A 2006, 103 (24), 9001–9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zitnay JL; Li Y; Qin Z; San BH; Depalle B; Reese SP; Buehler MJ; Yu SM; Weiss JA, Molecular level detection and localization of mechanical damage in collagen enabled by collagen hybridizing peptides. Nat. Commun 2017, 8, 14913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leitinger B, Transmembrane collagen receptors. Annu. Rev. Cell. Dev. Biol 2011, 27 (1), 265–290. [DOI] [PubMed] [Google Scholar]
- 10.O'Leary LER; Fallas JA; Bakota EL; Kang MK; Hartgerink JD, Multi-hierarchical self-assembly of a collagen mimetic peptide from triple helix to nanofibre and hydrogel. Nat. Chem 2011, 3, 821–828. [DOI] [PubMed] [Google Scholar]
- 11.Tanrikulu IC; Forticaux A; Jin S; Raines RT, Peptide tessellation yields micrometre-scale collagen triple helices. Nat. Chem 2016, 8, 1008–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmgren SK; Taylor KM; Bretscher LE; Raines RT, Code for collagen's stability deciphered. Nature 1998, 392 (6677), 666–667. [DOI] [PubMed] [Google Scholar]
- 13.Sakakibara S; Kishida Y; Kikuchi Y; Sakai R; Kakiuchi K, Synthesis of poly-(L-prolyl-L-prolylglycyl) of defined molecular weights. Bull. Chem. Soc. Jpn 1968, 41 (5), 1273–1273. [Google Scholar]
- 14.Persikov AV; Ramshaw JAM; Kirkpatrick A; Brodsky B, Amino acid propensities for the collagen triple-helix. Biochemistry 2000, 39 (48), 14960–14967. [DOI] [PubMed] [Google Scholar]
- 15.Maaßen A; Gebauer JM; Theres Abraham E; Grimm I; Neudörfl J-M; Kühne R; Neundorf I; Baumann U; Schmalz H-G, Triple-helix-stabilizing effects in collagen model peptides containing PPII-helix-preorganized diproline modules. Angew. Chem. Int. Ed 2020, 59 (14), 5747–5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aronoff MR; Egli J; Schmitt A; Wennemers H, Alkylation of γ-azaproline creates conformationally adaptable proline derivatives for pH-responsive collagen triple helices. Chem. Eur. J 2020, 26 (22), 5070–5074. [DOI] [PubMed] [Google Scholar]
- 17.Erdmann RS; Wennemers H, Functionalizable collagen model peptides. J. Am. Chem. Soc 2010, 132 (40), 13957–13959. [DOI] [PubMed] [Google Scholar]
- 18.Siebler C; Erdmann RS; Wennemers H, Switchable proline derivatives: Tuning the conformational stability of the collagen triple helix by pH changes. Angew. Chem. Int. Ed 2014, 53 (39), 10340–10344. [DOI] [PubMed] [Google Scholar]
- 19.Hentzen NB; Smeenk LEJ; Witek J; Riniker S; Wennemers H, Cross-linked collagen triple helices by oxime ligation. J. Am. Chem. Soc 2017, 139 (36), 12815–12820. [DOI] [PubMed] [Google Scholar]
- 20.Goodman M; Melacini G; Feng Y, Collagen-like triple helices incorporating peptoid residues. J. Am. Chem. Soc 1996, 118 (44), 10928–10929. [Google Scholar]
- 21.Feng Y; Melacini G; Goodman M, Collagen-based structures containing the peptoid residue N-Isobutylglycine (Nleu): synthesis and biophysical studies of Gly-Nleu-Pro sequences by circular dichroism and optical rotation. Biochemistry 1997, 36 (29), 8716–8724. [DOI] [PubMed] [Google Scholar]
- 22.Melacini G; Feng Y; Goodman M, Collagen-based structures containing the peptoid residue N-Isobutylglycine (Nleu): conformational analysis of gly-nleu-pro sequences by 1H-NMR and molecular modeling. Biochemistry 1997, 36 (29), 8725–8732. [DOI] [PubMed] [Google Scholar]
- 23.Goodman M; Bhumralkar M; Jefferson EA; Kwak J; Locardi E, Collagen mimetics. Biopolymers 1998, 47 (2), 127–142. [DOI] [PubMed] [Google Scholar]
- 24.Johnson G; Jenkins M; McLean KM; Griesser HJ; Kwak J; Goodman M; Steele JG, Peptoid-containing collagen mimetics with cell binding activity. J. Biomed. Mater. Res 2000, 51 (4), 612–624. [DOI] [PubMed] [Google Scholar]
- 25.Simon RJ; Kania RS; Zuckermann RN; Huebner VD; Jewell DA; Banville S; Ng S; Wang L; Rosenberg S; Marlowe CK, Peptoids: a modular approach to drug discovery. Proc. Natl. Acad. Sci. U. S. A 1992, 89 (20), 9367–9371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robertson EJ; Battigelli A; Proulx C; Mannige RV; Haxton TK; Yun L; Whitelam S; Zuckermann RN, Design, synthesis, assembly, and engineering of peptoid nanosheets. Acc. Chem. Res 2016, 49 (3), 379–389. [DOI] [PubMed] [Google Scholar]
- 27.Wu CW; Sanborn TJ; Huang K; Zuckermann RN; Barron AE, Peptoid oligomers with α-chiral, aromatic side chains: sequence requirements for the formation of stable peptoid helices. J. Am. Chem. Soc 2001, 123 (28), 6778–6784. [DOI] [PubMed] [Google Scholar]
- 28.Wu CW; Kirshenbaum K; Sanborn TJ; Patch JA; Huang K; Dill KA; Zuckermann RN; Barron AE, Structural and spectroscopic studies of peptoid oligomers with α-chiral aliphatic side chains. J. Am. Chem. Soc 2003, 125 (44), 13525–13530. [DOI] [PubMed] [Google Scholar]
- 29.Stringer JR; Crapster JA; Guzei IA; Blackwell HE, Extraordinarily robust polyproline type I peptoid helices generated via the incorporation of α-chiral aromatic N-1-Naphthylethyl side chains. J. Am. Chem. Soc 2011, 133 (39), 15559–15567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roy O; Dumonteil G; Faure S; Jouffret L; Kriznik A; Taillefumier C, Homogeneous and robust polyproline type I helices from peptoids with nonaromatic α-chiral side chains. J. Am. Chem. Soc 2017, 139 (38), 13533–13540. [DOI] [PubMed] [Google Scholar]
- 31.Shah NH; Butterfoss GL; Nguyen K; Yoo B; Bonneau R; Rabenstein DL; Kirshenbaum K, Oligo(N-aryl glycines): a new twist on structured peptoids. J. Am. Chem. Soc 2008, 130 (49), 16622–16632. [DOI] [PubMed] [Google Scholar]
- 32.Lee B-C; Zuckermann RN; Dill KA, Folding a nonbiological polymer into a compact multihelical structure. J. Am. Chem. Soc 2005, 127 (31), 10999–11009. [DOI] [PubMed] [Google Scholar]
- 33.Murnen HK; Rosales AM; Jaworski JN; Segalman RA; Zuckermann RN, Hierarchical self-assembly of a biomimetic diblock copolypeptoid into homochiral superhelices. J. Am. Chem. Soc 2010, 132 (45), 16112–16119. [DOI] [PubMed] [Google Scholar]
- 34.Mannige RV; Haxton TK; Proulx C; Robertson EJ; Battigelli A; Butterfoss GL; Zuckermann RN; Whitelam S, Peptoid nanosheets exhibit a new secondary-structure motif. Nature 2015, 526 (7573), 415–420. [DOI] [PubMed] [Google Scholar]
- 35.Nam KT; Shelby SA; Choi PH; Marciel AB; Chen R; Tan L; Chu TK; Mesch RA; Lee B-C; Connolly MD; Kisielowski C; Zuckermann RN, Free-floating ultrathin two-dimensional crystals from sequence-specific peptoid polymers. Nat. Mater 2010, 9 (5), 454–460. [DOI] [PubMed] [Google Scholar]
- 36.Zuckermann RN; Kerr JM; Kent SBH; Moos WH, Efficient method for the preparation of peptoids [oligo(N-substituted glycines)] by submonomer solid-phase synthesis. J. Am. Chem. Soc 1992, 114 (26), 10646–10647. [Google Scholar]
- 37.Zhang Y; Malamakal RM; Chenoweth DM, Aza-glycine induces collagen hyperstability. J. Am. Chem. Soc 2015, 137 (39), 12422–12425. [DOI] [PubMed] [Google Scholar]
- 38.Egli J; Esposito C; Müri M; Riniker S; Wennemers H, Influence of lipidation on the folding and stability of collagen triple helices—an experimental and theoretical study. J. Am. Chem. Soc 2021, 143 (15), 5937–5942. [DOI] [PubMed] [Google Scholar]
- 39.Adzhubei AA; Sternberg MJE; Makarov AA, Polyproline-II helix in proteins: Structure and function. J. Mol. Biol 2013, 425 (12), 2100–2132. [DOI] [PubMed] [Google Scholar]
- 40.Cram DJ, The design of molecular hosts, guests, and their complexes. Science 1988, 240 (4853), 760–767. [PubMed] [Google Scholar]
- 41.Brown AM; Zondlo NJ, A propensity scale for type II polyproline helices (PPII): aromatic amino acids in proline-rich sequences strongly disfavor PPII due to proline–aromatic interactions. Biochemistry 2012, 51 (25), 5041–5051. [DOI] [PubMed] [Google Scholar]
- 42.Pandey AK; Thomas KM; Forbes CR; Zondlo NJ, Tunable control of polyproline helix (PPII) structure via aromatic electronic effects: an electronic switch of polyproline helix. Biochemistry 2014, 53 (32), 5307–5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horng J-C; Raines RT, Stereoelectronic effects on polyproline conformation. Protein Sci. 2006, 15 (1), 74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jenkins CL; Vasbinder MM; Miller SJ; Raines RT, Peptide bond isosteres: Ester or (E)-alkene in the backbone of the collagen triple helix. Org. Lett 2005, 7 (13), 2619–2622. [DOI] [PubMed] [Google Scholar]
- 45.Butterfoss GL; Renfrew PD; Kuhlman B; Kirshenbaum K; Bonneau R, A preliminary survey of the peptoid folding landscape. J. Am. Chem. Soc 2009, 131 (46), 16798–16807. [DOI] [PubMed] [Google Scholar]
- 46.Crapster JA; Guzei IA; Blackwell HE, A peptoid ribbon secondary structure. Angew. Chem. Int. Ed 2013, 52 (19), 5079–5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Egli J; Schnitzer T; Dietschreit JCB; Ochsenfeld C; Wennemers H, Why proline? Influence of ring-size on the collagen triple helix. Org. Lett 2020, 22 (2), 348–351. [DOI] [PubMed] [Google Scholar]
- 48.Kersteen EA; Raines RT, Contribution of tertiary amides to the conformational stability of collagen triple helices. Biopolymers 2001, 59 (1), 24–28. [DOI] [PubMed] [Google Scholar]
- 49.Bretscher LE; Jenkins CL; Taylor KM; DeRider ML; Raines RT, Conformational stability of collagen relies on a stereoelectronic effect. J. Am. Chem. Soc 2001, 123 (4), 777–778. [DOI] [PubMed] [Google Scholar]
- 50.Li Y; Foss CA; Summerfield DD; Doyle JJ; Torok CM; Dietz HC; Pomper MG; Yu SM, Targeting collagen strands by photo-triggered triple-helix hybridization. Proc. Natl. Acad. Sci. U. S. A 2012, 109 (37), 14767–14772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Newberry RW; Raines RT, The n→π* Interaction. Acc. Chem. Res 2017, 50 (8), 1838–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gorske BC; Stringer JR; Bastian BL; Fowler SA; Blackwell HE, New strategies for the design of folded peptoids revealed by a survey of noncovalent interactions in model systems. J. Am. Chem. Soc 2009, 131 (45), 16555–16567. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.