Abstract
The ability of terminal restriction fragment (T-RFLP or TRF) profiles of 16S rRNA genes to provide useful information about the relative diversity of complex microbial communities was investigated by comparison with other methods. Four soil communities representing two pinyon rhizosphere and two between-tree (interspace) soil environments were compared by analysis of 16S rRNA gene clone libraries and culture collections (Dunbar et al., Appl. Environ. Microbiol. 65:1662–1669, 1998) and by analysis of 16S rDNA TRF profiles of community DNA. The TRF method was able to differentiate the four communities in a manner consistent with previous comparisons of the communities by analysis of 16S rDNA clone libraries. TRF profiles were not useful for calculating and comparing traditional community richness or evenness values among the four soil environments. Statistics calculated from RsaI, HhaI, HaeIII, and MspI profiles of each community were inconsistent, and the combined data were not significantly different between samples. The detection sensitivity of the method was tested. In standard PCRs, a seeded population comprising 0.1 to 1% of the total community could be detected. The combined results demonstrate that TRF analysis is an excellent method for rapidly comparing the relationships between bacterial communities in environmental samples. However, for highly complex communities, the method appears unable to provide classical measures of relative community diversity.
Rapid analysis of diversity in complex microbial communities has remained an elusive but important goal in microbial ecology. Community diversity can be examined at several levels. The most simple analyses use DNA profiles (generated by PCR and sometimes followed by restriction digestion of amplification mixtures) to identify differences in the composition of communities. More refined approaches describe differences not only in community composition but also in community organization by measuring the number (richness) and relative abundance (structure or evenness) of species or phylotypes. The richness and evenness of biological communities reflect selective pressures that shape diversity within communities. Measuring these parameters is most useful when assessing treatment effects (e.g., physical disturbance, pollution, nutrient addition, predation, climate change, etc.) on community diversity. Diversity statistics can also indicate the ability of a community to recover from disturbance and utilize resources efficiently (4). An ideal method for analysis of diversity in complex microbial communities would enable the simultaneous measurement of composition, phylotype richness, and community structure. The method would be rapid and reproducible and would permit flexible sampling of the entire microbial community.
Direct amplification of bacterial 16S rRNA genes from extracted soil DNA provides the most comprehensive and flexible means of sampling bacterial communities. Analysis of clone libraries of 16S rRNA genes amplified from different environments can provide relative measures of diversity that are, in general, consistent with qualitative relationships determined from traditional culture collections (9). However, analysis of individual 16S rRNA gene clones in multiple libraries is an expensive and extremely inefficient approach for comparison of numerous bacterial communities in replicated field experiments. Other methods, such as thermal or denaturing gradient gel electrophoresis (DGGE) (12, 14, 16, 21, 23, 28), heteroduplex analysis (8, 11), or terminal restriction fragment (T-RFLP or TRF) analysis (3, 6, 18, 20), assess the diversity of 16S rRNA gene mixtures more crudely than cloning and sequencing but are far more rapid and therefore more amenable to field-scale experiments in which replication is important. DGGE and the TRF method were recently shown to identify similar relationships among marine communities (20). DGGE has also been shown to provide estimates of cyanobacterial richness consistent with estimates based on direct observation of cell morphological types in cyanobacterial mat communities (22). Although cyanobacteria comprise a small phylogenetic group, these findings support the idea that rapid fingerprinting techniques might be capable of assessing the richness and evenness of microbial communities in general.
We calibrated the TRF method by comparing the composition, relative species richness, and evenness of four soil microbial communities that had been analyzed previously by cultivation and by 16S rDNA cloning (9). The four soils were from pinyon rhizosphere and between-tree (interspace) environments at two sites 19 km apart in northern Arizona (17). Both sites are pinyon-juniper woodlands but differ dramatically in soil type (7, 17). Here we show that the TRF method successfully demonstrated relationships between the four samples consistent with previous comparisons of 801 16S rRNA gene clones from the samples. However, calculations from TRF profiles provided variable values for comparison of phylotype richness and community evenness and depended on the restriction enzyme used to derive the profile. The data demonstrate both the strengths and limitations of the TRF method for analysis of natural communities that are highly complex.
MATERIALS AND METHODS
Field sites and soil samples.
Soil samples were collected from a site in the Coconino National Forest near the town of Cosnino and another site 19 km due north at Sunset Crater National Monument (17). The sites differ dramatically in soil type, but have similar dominant plant communities (pinyon-juniper woodlands), elevation, and general weather patterns (7, 13, 17). At Cosnino, the soil is a light sandy loam (13), and the areas between widely spaced trees (interspaces) are sparsely covered with grass and forb species. In contrast, Sunset Crater soil consists largely of black, coarse-textured, well-drained cinders, and the interspace regions between trees are typically barren (7). Composite soil samples were collected from interspace areas unassociated with plant roots (C0 and S0 samples) and the rhizospheres of pinyon trees (Pinus edulis Englm.) that were matched for age (C1 and S1 samples) at Cosnino and Sunset Crater, respectively, as previously described (17).
16S rDNA clone libraries.
A 200-member 16S rDNA clone library was constructed for each of the four Arizona soil samples as previously described (17). Briefly, DNA was extracted using a four-step procedure including three cycles of freezing-thawing, a 70°C heat incubation with sodium dodecyl sulfate, bead mill homogenization, and ethanol precipitation. The concentration of PCR-inhibiting materials was found to be too high in the precipitated DNA. Therefore, the DNA was cleaned further by phenol-chloroform extraction, passage through Sephadex G-200 spin columns, and then precipitated again with ethanol. The resulting high-molecular-weight DNA was stored at −20°C and was used as a template in PCR with 16S rRNA gene primers 8-27f (pA; 5′-AGAGTTTGATCCTGGCTCAG) (10) and 1507-1492r (5′-TACCTTGTTACGACTT) (29). For each soil DNA, 16S rDNA amplicons from 10 independent PCRs were pooled, ligated into the pGEM-T plasmid vector (Promega, Madison, Wis.), and transformed into Escherichia coli DH10β Electromax cells (Gibco BRL, Gaithersburg, Md.). For each soil, at least 200 clones containing inserts of the correct size (approximately 1,500 bp) were stored in 20% glycerol at −70°C.
TRF analysis of C0 and S0 16S rDNA clones.
Cloned 16S rDNA sequences from the C0 and S0 clone libraries (representing the interspace areas at Cosnino and Sunset Crater, respectively) were amplified using the primers M13-20 (5′-GTAAAACGACGGCCAGT) and M13-24 (5′-AACAGCTATGACCATG). Each 25-μl reaction mixture contained 30 mM Tris (pH 8.4), 50 mM KCl, 1.5 mM MgCl2 (24), 50 μM concentrations of each deoxynucleoside triphosphate, 25 pmol of each primer, and 0.75 U of Taq polymerase (AmpliTaq; Perkin-Elmer, Foster City, Calif.). Frozen cells (1 μl) from 20% glycerol stocks were added as template in each PCR. PCRs were performed in a Perkin-Elmer 9600 thermal cycler with the following cycling conditions: 2 min of denaturation at 94°C, 25 cycles of 30 s at 50°C, 1 min at 72°C, 10 s at 94°C, and a final cycle of annealing at 55°C for 1 min and extension at 72°C for 5 min. One microliter of each reaction mixture was used as template in a second PCR containing forward primer 8-27f fluorescently labeled with TET (4,7,2′,7′-tetrachloro-6-carboxyfluorescein; ABI, Perkin-Elmer) and reverse primer 1507-1492r. Reaction conditions were the same as those described above except that only 10 cycles of PCR were used instead of 25. Fluorescent amplification products were ethanol precipitated and resuspended in 25 μl of sterile, distilled water, and 8 μl was digested with 5 U of RsaI (New England Biolabs, Beverly, Mass.) in 12-μl reaction mixtures. Following restriction digestion, 1 μl of each digest was dried, suspended in 1.75 μl of loading buffer containing 0.25 μl of Genescan 2500 TAMRA size standard (ABI), a 5:1 mixture of deionized formamide-blue dextran, and 25 mM EDTA, and then denatured at 94°C for 2 min. Fragments were separated by electrophoresis in denaturing 4% polyacrylamide gels with an ABI 377 DNA sequencer. Reagents for polyacrylamide gel electrophoresis were purchased from Bio-Rad (Hercules, Calif.). TRF sizes were determined using Genescan version 2.02 analytical software (ABI).
TRF profiles for C0, C1, S0, and S1 soil DNA samples.
The four soil DNA templates used for TRF analysis were the same DNA preparations from which 16S rDNA was amplified in 1994 for construction of clone libraries (17). These DNA preparations were stored frozen between construction of the clone libraries and the TRF analyses (approximately 4 years). 16S rDNA for TRF analysis was amplified with primer 8-27f fluorescently labeled with TET and with primer 1507-1492r. Each 50-μl reaction mixture contained 30 mM Tris (pH 8.4), 50 mM KCl, 1.5 mM MgCl2 (24), 50 μM concentrations of each deoxynucleoside triphosphate, 50 pmol of each primer, and 0.75 U of LD Taq polymerase (AmpliTaq; Perkin-Elmer). Cycling conditions were as follows: 2 min of denaturation at 94°C, 35 cycles of 30 s at 50°C, 1 min at 72°C, 10 s at 94°C, and a final cycle of annealing at 55°C for 1 min and extension at 72°C for 5 min. Three independent PCRs were performed for each sample and combined. PCR products were separated by electrophoresis in 1% Nusieve agarose (FMC, Rockland, Maine), and the DNA band approximately 1,500 bp in size was excised and purified as had been done previously for the construction of clone libraries (17).
16S rDNA from the four soil DNA samples was also amplified with a FAM (5-carboxyfluorescein; ABI)-labeled forward primer (YOGA31F, 5′-GATCCTGGCTCAGAATC, E. coli positions 15 to 31) that is specific for members of the Acidobacterium division (2) and with reverse primer 1507-1492r. Amplification conditions were the same as above except that a 42°C annealing temperature was used. Three independent PCRs were performed for each sample, and PCR products were combined and purified with a Qiaquick PCR cleanup kit (Qiagen, Inc., Chatsworth, Calif.). Purified amplicons were digested with the enzymes HaeIII, HhaI, RsaI, and MspI, and fragments were separated by electrophoresis in polyacrylamide gels as described above. For each sample, two or three aliquots of each digest were applied to separate gels to obtain replicate profiles.
Analysis of TRF profiles.
TRF profiles were analyzed as follows, using S-PLUS version 3.2 (MathSoft, Inc., Seattle, Wash.). First, replicate profiles of each sample were compared to identify the subset of reproducible fragment sizes (i.e., peaks that appeared in every replicate profile of a sample). The average height of each reproducible peak of a sample was calculated, and the set of reproducible peaks with newly calculated average heights was assigned as the average profile of the sample for use in all subsequent analyses. Second, the DNA quantity analyzed for each of the four samples was compared and standardized to the lowest quantity. To standardize the DNA quantities, the sum of peak heights in each average profile of a sample was calculated as a representation of the total DNA quantity. The sum-of-peak-height values were standardized between samples by proportionally decreasing the height of each peak in the average profiles until the sum of peak heights (total fluorescence) for each profile equaled the lowest value represented among the samples. This procedure was performed to allow comparisons between samples of equal size (equal amounts of DNA). Adjustment of larger sample sizes usually resulted in the elimination of one or more peaks from a profile as some adjusted peak heights dropped below the noise threshold (height, 25).
Community composition comparison.
For comparison of community similarity between the four different soil environments, peak heights from TRF profiles were converted to binary data (presence or absence of a peak). Profiles generated from different enzymes were combined in a tandem array, and a Jaccard similarity matrix was calculated for the set of samples.
Calculation of traditional diversity indices.
To evaluate richness and evenness, diversity statistics were calculated from each standardized, average enzyme profile of a sample by using the number and height of peaks in each average profile as representations of the number and relative abundance of different phylotypes in a sample. It is understood that any given TRF fragment may represent sequences from multiple phylogenetic groups and may therefore not represent a true phylotype in the traditional sense. We use the term phylotype to indicate groups for richness calculations and also for the sake of consistency during comparisons of the TRF method with restriction fragment length polymorphism (RFLP) analysis of 16S rRNA gene clones. Phylotype richness (S) was calculated as the total number of distinct TRF sizes (between 94 and 827 bp) in a profile. The Shannon-Weiner diversity index (19) was calculated as follows: H = −Σ(pi)(log2 pi), where p is the proportion of an individual peak height relative to the sum of all peak heights. Simpson's index of diversity (27) was calculated as follows: D = 1 − Σ(pi)2. The scale for D ranges from 1 to Dmax, where Dmax = 1 − 1/(S). Evenness (19) was calculated from the Shannon-Weiner diversity function: E = H/Hmax, where Hmax = log2(S).
Detection sensitivity.
Detection assays were performed by amplification and TRF analysis of 16S rRNA genes from a soil DNA sample that had been mixed with different concentrations of a cloned 16S rRNA gene prior to PCR amplification. The soil DNA was extracted from a Cosnino interspace soil sample collected in 1998. The cloned 16S gene was selected from the S0 16S rRNA gene clone library (17) based on its unique RsaI TRF size which was not observed in previous TRF profiles of Cosnino interspace soil DNA samples. Plasmid DNA containing the cloned 16S rRNA gene was extracted by alkaline lysis (25). PCRs were performed with primers 8-27f (labeled with FAM) and 1507-1492r as described above, with 1 ng of Cosnino interspace soil DNA template and plasmid DNA dilutions ranging from 1 to 0.001 pg. Amplification products were purified using a Qiaquick PCR Purification Kit (Qiagen), gel quantified, digested with RsaI, and subjected to TRF analysis as described above.
RESULTS AND DISCUSSION
TRF analysis is increasingly popular as a fingerprinting method for analysis of microbial communities, although the abilities of the method are still being explored. We examined the capabilities of the TRF method by calibrating the method with data from a previous RFLP analysis of 801 16S rDNA clones from four soil communities.
Community composition.
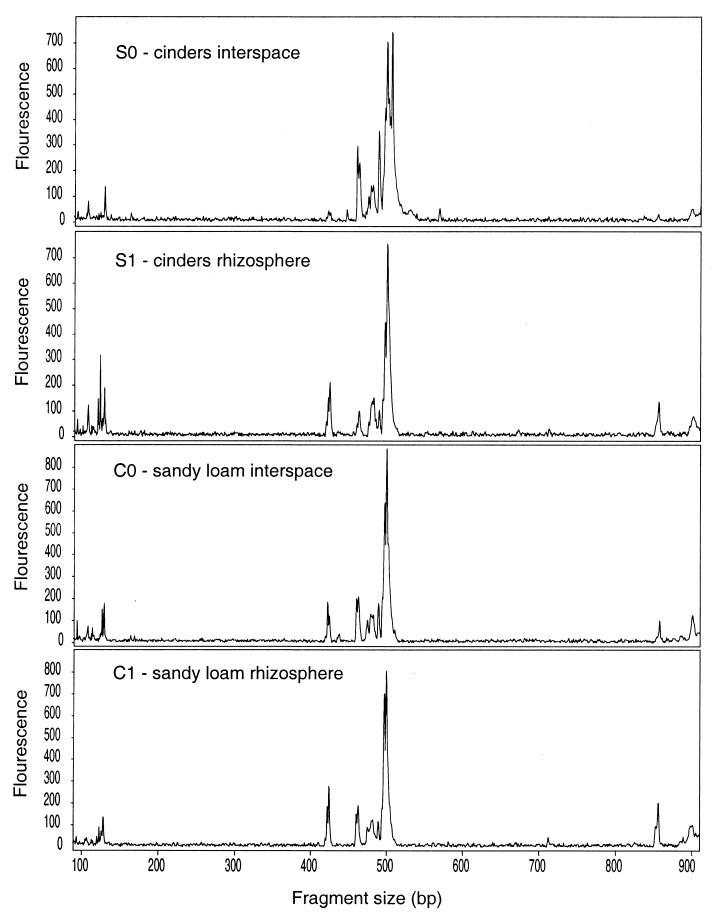
Figure 1 illustrates the typical extent of differences we observed in TRF profiles of the C0, C1, S0, and S1 DNA. Profiles from the C0, C1, and S1 samples were visually quite similar regardless of the enzyme used for restriction digestion of amplified 16S rDNA. In contrast, the profiles from the S0 environment were noticeably different. The striking differences (or similarities) in the height of peaks in profiles from different environments have not yet been incorporated into our comparison of the composition of samples. Although we are developing analytical procedures that will include peak height information, at present only the presence or absence of TRFs in community profiles is used to compare community composition.
FIG. 1.
RsaI TRF profiles of 16S rDNA amplified directly from S0, S1, C0, and C1 soil DNA samples.
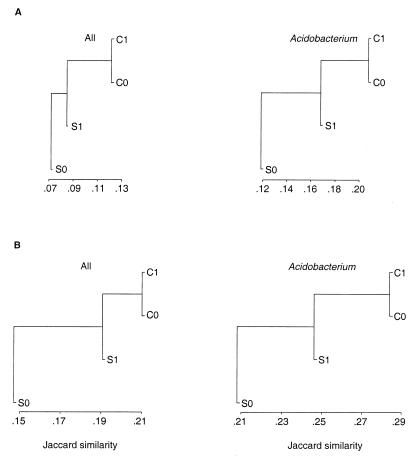
The similarity of the four environments (C0, C1, S0, and S1), based on a distance matrix analysis of community TRF profiles, is illustrated in Fig. 2. The analysis indicated that the sandy loam rhizosphere (C1) and interspace (C0) soil communities at Cosnino were the most similar in composition, whereas the cinders interspace (S0) community was the most different. These results were consistent with expectations based on the physical conditions of the four environments. The soil conditions (texture, moisture, and nutrient capacity) in the two sandy loam environments at Cosnino are very similar but differ substantially from the conditions in the cinders soil environments at Sunset Crater (7). The cinders interspace (S0) environment is primarily gravel in texture and is very hot, dry, and nutrient-poor (7), and the bacterial community in the extreme S0 environment was expected to be the most divergent. On the cinder field, nutrient levels are higher in the pinyon rhizospheres than in the interspaces. The cinders rhizosphere (S1) environment exhibits conditions intermediate between the Cosnino sandy loam soil and the cinders interspace (S0) environment. The pinyon pine roots in the cinders probably modulate but do not entirely eliminate the effects of the cinders soil type.
FIG. 2.
Dendrograms based on Jaccard similarity comparisons of the C0, C1, S0, and S1 soil communities. (A) Dendrograms based on all RsaI plus BstUI RFLP data from 16S rDNA clone libraries or only RsaI plus BstUI RFLP patterns representing members of the Acidobacterium division. (B) Dendrograms based on TRF profiles of 16S rDNAs amplified from soil DNA with conserved bacterial primers 8-27f and 1507-1492r or with specific Acidobacterium division primers YOGA31f and 1507-1492r.
The TRF dendrograms in Fig. 2 illustrate the same relationship between the four bacterial communities that had previously been identified by comparison of the four communities by using RsaI plus BstUI RFLP patterns of approximately 200 individual 16S rDNA clones from each environment (9). RFLP analysis (or sequence analysis) of clones from 16S rDNA clone libraries provides the most detailed, reliable information about the composition of microbial communities but is the most time-consuming and expensive method of community analysis. The finding that rapid comparison of microbial community similarity by TRF analysis provides results consistent with the much slower procedure of comparing clone libraries validates use of the TRF method. Moeseneder et al. (20) demonstrated that the TRF method and DGGE provided consistent results in differentiating marine communities. The combined data demonstrate that TRF profiles can be used to effectively investigate natural communities in field-scale experiments and that the TRF method is interchangeable with other molecular techniques.
A second TRF analysis of the four soil environments was performed to compare the composition of the Acidobacterium division in each environment. For the TRF analysis of total bacteria (using primer set 8-27f and 1507-1492r), the soil DNA templates, PCR conditions, and amplicon purification procedures were the same as those used to create the 16S rDNA clone libraries. TRF analysis performed using a primer set specific for members of the Acidobacterium division (2) was conducted using somewhat different PCR amplification and amplicon purification procedures. Nonetheless, the Acidobacterium TRF results demonstrated the same relationships between the four environments as the TRF analysis for total bacteria, indicating that the C0 and C1 environments were the most similar and the S0 environment was the most distinct (Fig. 2). The topology of the Acidobacterium TRF dendrogram was also consistent with the topology of a dendrogram based on comparison of RFLP patterns of Acidobacterium division clones from the four 16S rDNA clone libraries (division-level affiliation of the clones was determined by sequence analysis) (9, 17; J. Dunbar, S. M. Barns, J. Davis, G. Fisher, and C. R. Kuske, unpublished data). The agreement between results from RFLP analysis of 16S rDNA clone libraries and two different TRF analyses of the original soil DNA templates indicates that the TRF method is robust in identifying relationships (based on composition) among natural communities. This illustrates the capacity of the TRF method to explore differences in community composition between environments without the need to separately examine individual members of the community (by 16S rDNA gene cloning).
Method resolution.
Although the overall relationships between the four soil environments were consistent between the TRF analysis and the combined results from RFLP analysis of individual 16S rDNA clones, the resolution (i.e., the extent of discrimination between the four communities) differed between the two methods. The degree of similarity among the four soil environments (C0, C1, S0, and S1) was higher according to TRF profiles than according to RFLP data from the 16S rDNA clone libraries. The similarity values from TRF analysis ranged from 15 to 21% (Fig. 2). In contrast, the communities appeared to be only 7 to 12% similar based on RsaI plus BstUI RFLP data from clone libraries (9).
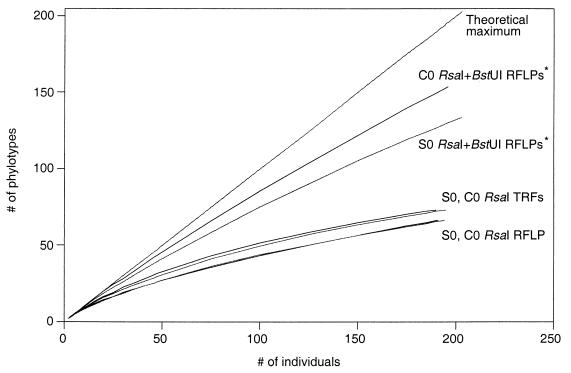
To better evaluate the resolution provided by the TRF method, we determined the ability of the method to measure diversity in the C0 and S0 16S rDNA clone libraries (Table 1; Fig. 3). The 16S rDNA clone libraries are a subset of the total bacterial diversity in the C0 and S0 soil environments. The clone libraries have a known number of members and can be considered two defined communities since the diversity of these two communities was characterized previously by RFLP analysis of each individual clone (9). A total of 154 and 134 RsaI plus BstUI RFLP patterns (i.e., patterns from RsaI digests that were further differentiated by comparison of patterns from separate BstUI digests) were previously identified in the C0 and S0 clone libraries, respectively (9). Only a fraction of this diversity was detected by obtaining an RsaI TRF profile for each of the clones in the C0 or S0 libraries. A total of 73 and 75 distinct RsaI TRF sizes were observed, respectively, from 190 C0 clones and 182 S0 clones (Table 1; Fig. 3). The lower resolution of the TRF method does not appear to result from examining only the length variation of one restriction fragment (the 5′ TRF) instead of all the restriction fragments that are produced by digestion of the 16S rDNA (as in a standard RFLP). As shown in Fig. 3, for example, the number of RsaI TRFs and RsaI RFLP patterns observed among the clones was similarly low. Only 66 distinct RsaI RFLP patterns were identified for each set of clones. The number of RsaI TRFs was slightly larger (73 and 75) than the number of RsaI RFLP patterns (66 and 66) due to the higher resolution (± 0.5 bp) of the TRF method in fragment size determination compared to the conventional agarose gel RFLP method. Based on these data, the lower resolution of the TRF method when compared to sequentially digested (RsaI plus BstUI) individual clones appears to result from measuring the diversity of 16S rRNA gene sequences with only a single enzyme.
TABLE 1.
Comparison of phylotype richness, diversity and evenness values for the C0 (sandy loam interspace) and S0 (cinder interspace) bacterial communities, derived from three different methods
| Parameter | 16S rDNA clone libraries
|
Community TRF
|
||||
|---|---|---|---|---|---|---|
| C0 clone, RsaI+BstUI RFLPs | S0 clone, RsaI+BstUI RFLPs | C0 clone, RsaI TRF | S0 clone, RsaI TRF | C0 soil DNA, RsaI TRF | S0 soil DNA, RsaI TRF | |
| Sa | 154 | 134 | 73 | 75 | 20 (15)b | 20 (14) |
| Hc | 7.067 | 6.612 | 5.586 | 5.436 | 3.709 | 3.765 |
| H/Hmaxd | 0.972 | 0.936 | 0.903 | 0.873 | 0.858 | 0.871 |
Phylotype richness, S, was calculated as the total number of distinct TRF sizes (peaks between 94 and 827 bp) in a profile.
Number in parentheses indicates the number of TRF sizes in the profile from soil DNA that were also observed among 16S rDNA clones obtained from the same soil DNA 4 years previously. The discrepancy between the number of RsaI TRFs observed among the clones (73 and 75 TRFs) and the number observed in the profiles from soil DNA (20 TRFs in each) may have been the result of degradation of the soil DNA template during storage for 4 years.
Shannon-Weiner diversity index (19) was calculated as follows: H = −Σ(pi)(log2 pi), where p is the proportion of an individual peak height relative to the sum of all peak heights.
Evenness (19) was calculated from the Shannon-Weiner diversity function as follows: E = H/Hmax, where Hmax = log2(S).
FIG. 3.
Sampling curves showing diversity of the C0 and S0 clone libraries assessed by RsaI plus BstUI RFLP patterns, RsaI RFLP patterns, and RsaI TRFs. Curves were constructed by rarefaction (15, 26). Curves marked with asterisks were reprinted from reference 9 with permission from the publisher.
Use of multiple enzymes for either RFLP or TRF analysis of individual clones in a 16S rDNA library increases the resolution by increasing the number of different fragments observed and simultaneously decreasing the number of recorded similarities. In contrast, use of multiple enzymes will typically not provide substantial increases in resolution for community TRF analysis of a mixture of 16S rDNAs amplified from a soil DNA template. This is not a limitation of the TRF method, per se, but is instead a limitation of analyzing mixed 16S rDNA sequences. For mixed 16S rDNAs amplified from two environmental samples, data from an additional enzyme digest may increase the differences observed between the two samples, but the total number of similarities will also increase. Thus, the resolution measured as the percent difference that can be detected between the two samples may not change substantially by use of multiple enzyme digests. The value of using multiple enzymes for TRF analysis is to increase confidence that the similarity relationships identified between samples are not the result of biases in the way that a single enzyme samples diversity.
Phylotype richness.
The phylogenetic resolution of different TRFs observed in TRF profiles of microbial communities is expected to vary. Whereas one TRF size in a profile may be derived uniquely from a small, phylogenetically coherent group of bacteria (i.e., a true phylotype; for example, see reference 5), another TRF size may represent a broader, more distantly related set of organisms. The lack of phylogenetic resolution in the latter category of TRFs will contribute noise when TRF profiles are used to measure the relative phylotype richness of different communities. We sought to determine whether useful measures of relative phylotype richness could be obtained from TRF profiles despite the noise contributed by TRFs of low phylogenetic resolution. The ability of TRF profiles to document relative phylotype richness in the four soil bacterial communities was evaluated in two ways.
First, phylotype richness values from RFLP analysis of clones in the C0 and S0 16S rDNA libraries were compared with the number of distinct RsaI TRFs observed among the same clones or observed in profiles from the C0 and S0 soil DNA samples (Table 1). RsaI plus BstUI RFLP analysis of clones more accurately distinguishes phylotypes. Therefore, we expected richness values calculated from TRF profiles to be lower for the S0 community when compared to the C0 community since previous RFLP analysis of 16S rDNA clone libraries had indicated this relationship. However, this relationship was not apparent from the number of distinct RsaI TRFs obtained from individual C0 and S0 16S rDNA clones or from the number of TRFs in RsaI profiles generated directly from C0 and S0 soil DNA (Table 1). In both cases, an identical or nearly identical number of RsaI TRFs were obtained for the C0 and S0 communities.
Second, trends among the richness values derived from TRF profiles of all four soil DNA samples (Table 2) were evaluated. Once again, we expected the S0 environment to have the lowest richness value based on previous comparisons of the four communities (9) and the community similarity analysis (present study; Fig. 1). However, richness values from the TRF profiles of community DNA failed to reveal substantial or consistent differences in richness between the S0 community and the other communities (Table 2). In fact, the estimates of community richness we obtained from TRF profiles of the four soil communities lacked any consistent trends. For example, among the MspI profiles, the S1 sample had the highest richness value (S = 34) while the S0 sample had the lowest value (S = 24). In contrast, the S1 sample had the lowest value (S = 14) among HaeIII profiles while the S0 sample had a higher richness value (S = 18). Although the average S values demonstrated a pattern consistent with data from clone library RsaI plus BstUI RFLP data, they were not statistically different from one another due to the large variance that arose from averaging results of multiple enzymes that measure sequence diversity to different extents.
TABLE 2.
Diversity statistics calculated from TRF profiles of 16S rDNAs amplified from C0, C1, S0, and S1 soil DNA
| Enzyme | Index | Diversity statisticsa
|
|||
|---|---|---|---|---|---|
| C0 | C1 | S0 | S1 | ||
| HaeIII | Sb | 21 | 19 | 18 | 14d |
| HhaI | S | 14 | 13 | 12 | 11 |
| MspI | S | 33 | 27 | 24 | 34 |
| RsaI | S | 20 | 22 | 20 | 20 |
| Avg S | 22 | 20 | 19 | 20 | |
| HaeIII | Hc | 4.13 | 3.92 | 3.74 | 3.49 |
| HhaI | H | 3.68 | 3.52 | 3.51 | 3.32 |
| MspI | H | 4.78 | 4.48 | 4.19 | 4.72 |
| RsaI | H | 3.71 | 3.74 | 3.77 | 3.72 |
| Avg H | 4.07 | 3.91 | 3.80 | 3.81 | |
C0, Cosnino interspace; C1, Cosnino rhizosphere; S0, Sunset Crater interspace; S1, Sunset Crater rhizosphere.
Phylotype richness, S, was calculated as the total number of distinct TRF sizes (between 94 and 827 bp) in a profile.
Shannon-Weiner diversity index (19) was calculated as follows: H = −Σ(pi)(log2 pi), where p is the proportion of an individual peak height relative to the sum of all peak heights.
Boldface type indicates the lowest value in each series.
Use of TRF profiles to provide relative measures of phylotype richness for comparison of bacterial communities was originally projected as a capability of the method (18). The TRF method has previously been shown to be capable of assessing phylotype richness in simple, artificial communities containing only four or six members (1, 18, 20). It is possible that use of TRF profiles to measure relative phylotype richness is only possible for simple communities. The same variability and inconsistency that we observed in richness values for our soil samples are apparent in richness values reported for 20 marine samples (20). Based on these findings, the TRF method appears ineffective in comparing the relative richness of extremely complex communities.
Community complexity can be artificially reduced by use of group-specific PCR primers instead of universal primers. For example, Nübel et al. (22) used PCR primers specific for members of the cyanobacteria division to amplify 16S rDNA sequences from eight cyanobacterial mat communities. DGGE analysis of cyanobacterial and plastid 16S rDNA sequences successfully provided estimates of phylotype richness congruent with estimates based on the diversity of cell morphologies observed in each sample. In the same manner, we decreased the complexity of the C0, C1, S0, and S1 environments by using specific primers to amplify 16S rRNA genes from members of the Acidobacterium division only. However, diversity indices calculated from Acidobacterium TRF profiles were as variable as the values from bacterial TRF profiles created with the primers 8-27f and 1507-1492r (data not shown). It is possible that the Acidobacterium division is still too complex a subcommunity. Unlike the cyanobacteria division, this division is phylogenetically very broad (17), and Acidobacterium division sequences accounted for approximately 40% of the RsaI plus BstUI RFLP patterns identified in the C0, C1, S0, and S1 clone libraries. For broad divisions, primer sets that are specific for smaller subgroups may be required to effectively detect differences in the richness and structure of complex communities. Alternatively, use of the method for comparing richness in communities should be confined to the most simple natural communities or to experimental communities for which the initial species composition is known.
Community evenness.
In parallel with the above comparisons of richness, we evaluated community evenness values derived from TRF profiles of C0 and S0 16S rDNA clones and from profiles of all four soil DNA samples (Table 1). The Shannon-Weiner diversity index (H; Table 1) and Simpson's diversity index (D; data not shown), both of which emphasize phylotype richness but also measure structure, indicated that the S0 clone library was less diverse than the C0 clone library when calculated from TRF data from 372 individual clones. The evenness index (H/Hmax) of TRFs from the C0 and S0 clone libraries indicated that the frequency distribution in the S0 library was more skewed than that in the C0 library (0.873 versus 0.903). Although the numerical differences were small, they were consistent with our previous comparison of evenness calculated from RsaI plus BstUI RFLP data from the two clone libraries (9). In contrast, evenness statistics calculated from the TRF profiles of C0 and S0 total community DNA indicated the opposite trend, suggesting that the S0 environment was more diverse and less skewed than the C0 environment. Additional contradictions and inconsistencies were apparent among evenness values from TRF profiles of all four (C0, C1, S0, and S1) community DNA samples. The combined results suggest that diversity indices calculated from community TRF profiles of highly complex communities may not be adequate to accurately measure relative community structure.
The inability of TRF profiles to provide reliable measures of phylotype richness and community structure is not completely surprising. The lower resolution of the TRF method (i.e., the substantial probability of multiple phylotypes being represented by a single fragment size in a TRF profile) would tend to obscure differences in phylotype richness and evenness that might be detected by other methods with higher phylogenetic resolution. Use of TRF profiles to measure richness and community structure is also hampered by inherent variation in the extent to which different restriction enzymes reveal sequence variation. Each enzyme used to create a TRF profile represents a fundamentally different sampling technique. Thus, TRF profiles of a single sample can vary both in richness (total number of TRFs in a profile) and in evenness (frequency distribution of TRFs) depending on the enzyme used. Combining data from different enzyme profiles in valid ways that yield statistically informative results is therefore difficult.
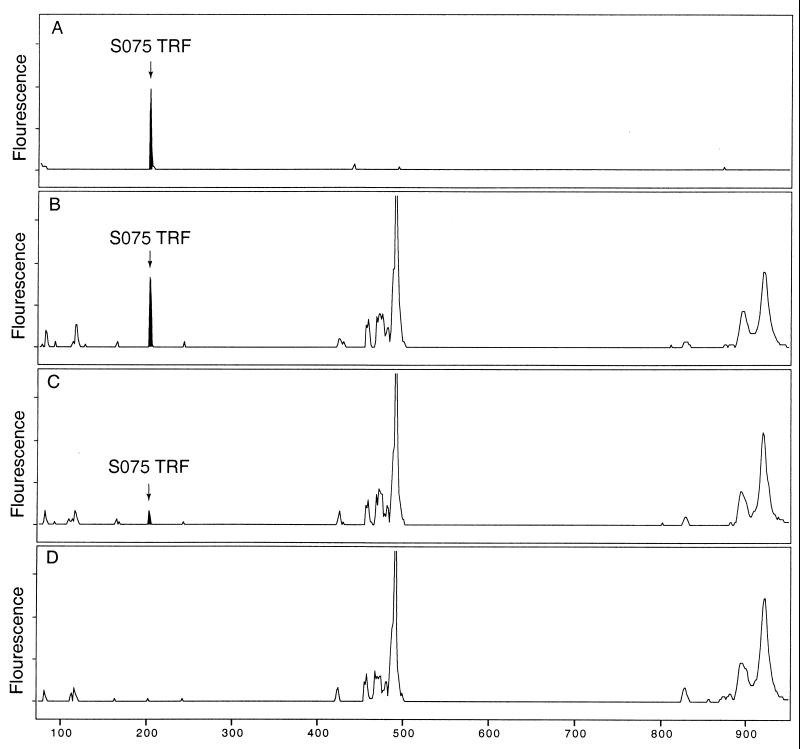
Detection sensitivity.
The detection sensitivity of the TRF method was tested using soil DNA templates spiked with dilutions of a 5-kb plasmid purified from a clone (labeled S075) from the S0 16S rRNA gene clone library (Fig. 4). In PCR extinction dilution experiments, the detection threshold for 16S rDNA (that is, the lowest dilution of DNA from which a visible 16S rDNA amplicon could be obtained) with soil DNA was 1,000-fold higher than with the cloned plasmid DNA (data not shown). Therefore, we converted DNA quantities to genome equivalents, assuming an average bacterial genome size of 5 Mb and an average of one copy of the rRNA gene per genome. The conversion of DNA quantities better illustrates the ratio of target to nontarget DNA in the PCRs. As shown in Fig. 4, 0.01 pg of the 16S rRNA gene clone spiked in 1 ng of soil DNA was detected in an RsaI TRF profile. This represented a ratio of approximately 2,000 genomes to 200,000 genomes (1% of the total). In the absence of soil DNA, the detection limit of the pure 16S rRNA gene clone remained at approximately 2,000 genome equivalents per PCR. Similar results were obtained when 0.1 pg of the S0 clone was mixed with 10 ng of soil DNA in PCRs (data not shown). The S0 clone was also detected at concentrations of 0.001 pg (200 genome equivalents) in the presence of 1 ng of soil DNA per PCR (Fig. 4C). However, at this concentration, detection of the target was variable in contrast to the reproducible detection of 0.01 pg of the S0 clone. The data suggest that the detection sensitivity in these assays was determined by the target concentration alone and was not substantially affected by the background concentration of soil DNA. The data also suggest that populations comprising between 0.1 and 1% of a bacterial community could be detected in TRF profiles. The detection sensitivity of the TRF method is comparable to other DNA-based community analysis techniques. Using DGGE, Muyzer et al. (21) reported the detection of a population comprising 1% of a mixture of DNA from five organisms. Although their DNA mixture was far less complex than the soil DNA used in our assays, the detection sensitivities of DGGE and the TRF method appear to be consistent.
FIG. 4.
Detection of a 16S rDNA clone (5-kb plasmid from clone S075) in a background of 1 ng of C0 soil DNA. (A) RsaI TRF profile of 16S rDNA amplified from 0.01 pg of S075 plasmid DNA. (B) RsaI TRf profile of 16S rDNA amplified from a mixture of 0.01 pg of plasmid S075 and 1 ng of C0 soil DNA. (C) RsaI TRf profile of 16S rDNA amplified from a mixture of 0.001 pg of plasmid S075 and 1 ng of C0 soil DNA. (D) RsaI TRF profile of 16S rDNA amplified from 1 ng of C0 soil DNA.
Summary.
The calibration we performed of TRF analysis of four soil microbial communities and RFLP data from 801 clones from the same environments demonstrated strengths and limitations of the TRF method. For the complex soil communities compared in this study, TRF profiles were unable to provide reliable information describing relative phylotype richness and evenness. However, the method was very effective in elucidating similarity relationships between communities and has good detection sensitivity. The TRF method should be especially useful for rapid analysis of replicate samples in field-scale studies. Eventual incorporation of peak height data into analyses of community similarity will further enhance the method and its power to reveal differences between communities. While data from TRF profiles must be cautiously interpreted in some contexts, the method should in general prove to be a useful new tool for microbial ecology research.
ACKNOWLEDGMENTS
We thank Tom Whitham and Catherine Gehring for their collaboration at the Sunset Crater study site and Joseph Busch for help generating the 16S rDNA clone libraries.
This work was supported in part by the Department of Energy Program for Ecosystem Research.
REFERENCES
- 1.Avaniss-Aghajani E, Jones K, Chapman D, Brunk C. A molecular technique for identification of bacteria using small subunit ribosomal RNA sequences. BioTechniques. 1994;17:144–149. [PubMed] [Google Scholar]
- 2.Barns S M, Takala S L, Kuske C R. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl Environ Microbiol. 1999;65:1731–1737. doi: 10.1128/aem.65.4.1731-1737.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunk C F, Avaniss-Aghajani E, Brunk C A. A computer analysis of primer and probe hybridization potential with bacterial small-subunit rRNA sequences. Appl Environ Microbiol. 1996;62:872–879. doi: 10.1128/aem.62.3.872-879.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapin F S, Walker B H, Hobbs R J, Hooper D U, Lawton J H, Sala O E, Tilman D. Biotic control over the functioning of ecosystems. Science. 1997;277:500–504. [Google Scholar]
- 5.Chin K J, Lukow T, Stubner S, Conrad R. Structure and function of the methanogenic archaeal community in stable cellulose-degrading enrichment cultures at two different temperatures (15° and 30°C) FEMS Microbiol Ecol. 1999;30:313–326. doi: 10.1111/j.1574-6941.1999.tb00659.x. [DOI] [PubMed] [Google Scholar]
- 6.Clement B G, Kehl L E, DeBord K L, Kitts C L. Terminal Restriction Fragment Patterns (TRFPs), a rapid, PCR-based method for the comparison of complex bacterial communities. J Microbiol Methods. 1998;31:135–142. [Google Scholar]
- 7.Cobb N S, Mopper S, Gehring C A, Caouette M, Christensen K M, Whitham T G. Increased moth herbivory associated with environmental stress of pinyon pine at local and regional levels. Oecologia. 1997;109:389–397. doi: 10.1007/s004420050098. [DOI] [PubMed] [Google Scholar]
- 8.Delwart E L, Shpaer E G, Louwagie J, McCutcham F E, Grez M, Rubsamen-Waigmann H, Mullins J I. Genetic relationships determined by a DNA heteroduplex mobility assay analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 9.Dunbar J, Takala S, Barns S M, Davis J A, Kuske C R. Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl Environ Microbiol. 1999;65:1662–1669. doi: 10.1128/aem.65.4.1662-1669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards U, Rogall T, Blocker H, Emde M, Bottger E C. Isolation and direct complete determination of entire genes. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Espejo R T, Feijoo C G, Romero J, Vasquez M. PAGE analysis of the heteroduplexes formed between PCR-amplified 16S rRNA genes: estimation of sequence similarity and rDNA complexity. Microbiology. 1998;144:1611–1617. doi: 10.1099/00221287-144-6-1611. [DOI] [PubMed] [Google Scholar]
- 12.Ferris M J, Muyzer G, Ward D M. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl Environ Microbiol. 1996;62:340–346. doi: 10.1128/aem.62.2.340-346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hendricks D M. Arizona soils. Tucson: University of Arizona Press; 1985. [Google Scholar]
- 14.Heuer H, Krsek M, Baker P, Smalla K, Wellington E M H. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl Environ Microbiol. 1997;63:3233–3241. doi: 10.1128/aem.63.8.3233-3241.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hurlbert S H. The nonconcept of species diversity: a critique and alternative parameters. Ecology. 1971;52:577–586. doi: 10.2307/1934145. [DOI] [PubMed] [Google Scholar]
- 16.Kowalchuk G A, Stephen J R, Boer W D, Prosser J I, Embley T M, Woldendorp J W. Analysis of ammonia-oxidizing bacteria of the beta subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl Environ Microbiol. 1997;63:1489–1497. doi: 10.1128/aem.63.4.1489-1497.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuske C R, Barns S M, Busch J D. Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl Environ Microbiol. 1997;63:3614–3621. doi: 10.1128/aem.63.9.3614-3621.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu W, Marsh T L, Cheng H, Forney L J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Margalef R. Information theory in ecology. General Systems. 1958;3:36–71. [Google Scholar]
- 20.Moeseneder M M, Arrieta J M, Muyzer G, Winter C, Herndl G J. Optimization of terminal-restriction fragment length polymorphism analysis for complex marine bacterioplankton communities and comparison with denaturing gradient gel electrophoresis. Appl Environ Microbiol. 1999;65:3518–3525. doi: 10.1128/aem.65.8.3518-3525.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muyzer G, Waal E C D, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nübel U, Gracia-Pichel F, Kühl M, Muyzer G. Quantifying microbial diversity: morphotypes, 16S rRNA genes, and carotenoids of oxygenic phototrophs in microbial mats. Appl Environ Microbiol. 1999;65:422–430. doi: 10.1128/aem.65.2.422-430.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ovreas L, Forney L, Daae F L, Torsvik V. Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol. 1997;63:3367–3373. doi: 10.1128/aem.63.9.3367-3373.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ponce M R, Michol J L. PCR amplification of long DNA fragments. Nucleic Acids Res. 1992;20:623. doi: 10.1093/nar/20.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Simberloff D. Use of rarefaction and related methods in ecology. In: Dickson K L, Cairns J J, Livingston R J, editors. Biological data in water pollution assessment: quantitative and statistical analyses. West Conshohocken, Pa: American Society for Testing and Materials; 1978. pp. 150–165. [Google Scholar]
- 27.Simpson E H. Measurement of diversity. Nature. 1949;163:688. [Google Scholar]
- 28.Ward D M, Santegoeds C M, Nold S C, Ramsing N B, Ferris M J, Bateson M M. Biodiversity within hot spring microbial mat communities: molecular monitoring of enrichment cultures. Antonie Leeuwenhoek. 1997;71:143–150. doi: 10.1023/a:1000131426164. [DOI] [PubMed] [Google Scholar]
- 29.Wilson K H, Blitchington R B, Green R C. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J Clin Microbiol. 1990;28:1942–1946. doi: 10.1128/jcm.28.9.1942-1946.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]