Abstract
High-altitude polycythemia (HAPC) is characterized by excessive proliferation of erythrocytes, resulting from the hypobaric hypoxia condition in high altitude. The genetic variants and molecular mechanisms of HAPC remain unclear in highlanders. We recruited 141 Tibetan dwellers, including 70 HAPC patients and 71 healthy controls, to detect the possible genetic variants associated with the disease; and performed targeted sequencing on 529 genes associated with the oxygen metabolism and erythrocyte regulation, utilized unconditional logistic regression analysis and GO (gene ontology) analysis to investigate the genetic variations of HAPC. We identified 12 single nucleotide variants, harbored in 12 genes, associated with the risk of HAPC (4.7 ≤ odd ratios ≤ 13.6; 7.6E − 08 ≤ p-value ≤ 1E − 04). The pathway enrichment study of these genes indicated the three pathways, the PI3K-AKT pathway, JAK-STAT pathway, and HIF-1 pathway, are essential, which p-values as 3.70E − 08, 1.28 E − 07, and 3.98 E − 06, respectively. We are hopeful that our results will provide a reference for the etiology research of HAPC. However, additional genetic risk factors and functional investigations are necessary to confirm our results further.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12288-021-01474-1.
Keywords: High-altitude polycythemia, Highland people, SNPs
Introduction
The Qinghai-Tibet Plateau, namely the "roof of the world," is a high-altitude area with an elevation between 3000 and 5000 m. Hypoxia is one of the most critical characteristics of the high-altitude environment. High-altitude polycythemia (HAPC) is one of the chronic high-altitude diseases developed in Tibetan dwellers, characterized by an excessive number of circulating erythrocytes. The clinical diagnosis of HAPC requires a hemoglobin concentration (Hb) no less than 19 g/dL for females and 21 g/dL for males, respectively [1]. HAPC is often commonly accompanied by the symptoms, headache, dizziness, dyspnea, sleep disorders, or venous dilatation [1]. The incidence of HAPC in the Tibetan Plateau ranges from 5 to 18%, increasing with altitude, a severe public health problem in China and other Andean countries [2].
The compensation for prolonged hypobaric hypoxia is the main reason for the change of hemoglobin concentration through elevating oxygen-carrying, transportation, and exchange [3]. Elevated hemoglobin concentrations are crucial for adapting to high altitude environment. However, the pathological increased red blood cell may trigger blood viscosity, potentially slowing blood flow and inciting hypercoagulation, thrombosis, or tissue hypoxia, even further aggravate various circulatory, respiratory, digestive, and neurological diseases [4–6]. While the etiology and the pathophysiological of HAPC remain unclear, the incidence of HAPC showed significantly individual differences.
Recent studies revealed that HAPC in Tibetans or Han is associated with several gene polymorphisms, such as phosphatidylinositol-4,5-bisphosphonate 3-kinase, catalytic subunit delta gene (PIK3CD), collagen type IV α3 chain gene (COL4A3), integrin subunit alpha six gene (ITGA6), erb-b2 receptor tyrosine kinase four gene (ERBB4), EPH receptor A2 gene (EPHA2), angiotensinogen gene (AGT), and endothelial PAS domain protein one gene (EPAS1) [7–11] EPHA2 can disturb erythropoiesis by regulating EPO production, and EPAS1 contributes to genetic adaptation to high altitudes and the particular trait of the low Hb concentrations, conditions frequently observed among Tibetans [9, 12]. HAPC was found to lead to morphological changes and pathological damage to the gastric mucosa of patients. In particular, the kallikrein gene cluster (KLK1, KLK3, KLK7, KLK8, and KLK12) was upregulated > 17-fold. The elevated levels of KLK1, KLK3, KLK7, KLK8 and KLK12 may be closely associated with the hypertension, inflammation, obesity and other gastric injuries associated with polycythemia. Thus, the kallikreins are likely to potentially impact the development of HAPC [13]. Wang et al. [14] found that 19 serum proteins expressed differentially between patients and healthy controls with HAPC. Among them, C4A(The complement component 4A), C6(complement C6), CALR(Calreticulin), MASP1(Mannan Binding Lectin Serine Peptidase 1), and CNDP1(serum carnosinase) may enable researchers to use them as candidate plasma biomarkers for HAPC on account of their latent diagnostic, preventive and therapeutic values [14]. These results suggest that HAPC presents noticeable racial and significant individual differences in susceptibility.
In this study, we conducted a case–control study to investigate the association of genetic variants and the susceptibility of HAPC in the Tibetan population. We selected a group of genes considered involved in erythropoiesis and oxygen transportation by previous studies and using the target exome sequencing strategy to detect the coding variants. This work's overarching goal was to investigate associations between HAPC susceptibility and candidate genes that control oxygen metabolism in erythrocytes.
Experimental Procedures
Study Populations
HAPC was defined as males and females with Hb ≥ 21 g/dL and Hb ≥ 19 g/dL, respectively. We excluded individuals with chronic pulmonary diseases, lung cancer, or secondary polycythemia due to hypoxemia caused by certain chronic diseases. A total of 70 HAPC cases and 71 healthy controls of Tibetan inhabitants who lived in plateau areas above 2500 m were recruited from the Second People's Hospital of Tibet Autonomous Region. The cases include 35 males and 35 females. Control samples consisted of 39 males and 32 females. The age range of all samples was 20–60 years old. The Ethics Committee approved the study of the Xizang Minzu University with Ethics Number 201801.
Epidemiological and Clinical Data
Participants' information was collected through physicians or from medical chart review. All study participants provided written, informed consent for participation. Also, we obtained 5 ml of peripheral blood specimens from each participant. All persons gave their informed consent before their inclusion in the study.
Exome Sequencing and Bioinformatics of Target Genes
Targeted exome sequencing (agilent All exon kit V5 50 M) of 529 selected genes was conducted on all samples—these 529 genes with erythropoiesis and oxygen transportation by previous studies and database IPA (Ingenuity Pathway Analysis) and NCBI(National Center for Biotechnology Information), etc. [15]. Genomic DNA was extracted from the whole blood or saliva of individual subjects according to standard protocols. Double-stranded DNA was indexed and multiplexed in groups of six per lane and sequenced in 101-bp paired-end mode using the Illumina HiSeq 2000 platform. The mean on-target depth of each sample was 200X. The raw FASTQ files from the Illumina HiSeq were aligned to the human reference genome build 19 (GRCh37), and variants were called using SAMtools v0.1.18 and GATK v3.8. Low-quality variants were filtered as described previously [16]. Under the theory of "Common variants, common disease", we included the variants with minor allele frequency (MAF) > 0.05 in the Asian population HapMap database into the subsequent analysis.
Statistical Analysis
The genotype frequencies of each SNP in the control subjects were checked using the Hardy–Weinberg equilibrium (HWE). HWE values > 0.01 were further analyzed. Genes FER (ferritin), CREB5(also called CREBPA, CRE (cyclic AMP response element)-binding proteins), and TP53 with HWE values < 0.01 in the control group, failing to conform to the HWE rule, were excluded from the study.
The effects of polymorphisms on the risk of HAPC were expressed as odds ratios (ORs) with 95% confidence intervals (95% CIs), evaluated by three genetic models (dominant, recessive, and additive models) using unconditional logistic regression analysis. The Multiple comparisons were corrected by FDR and Bonferroni method. FDR and Bonferroni-corrected p < 0.05 indicated a significant between-group difference. KEGG (Kyoto Encyclopedia of Genes and Genomes)-based enrichment analysis was conducted in R software [17]. All data were analyzed by R software package GenABEL20.
Results
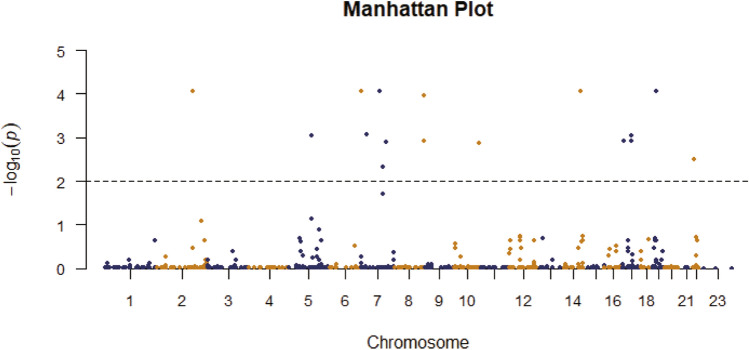
Table 1 displays the demographics of patients with HAPC and controls. Table 2 and Fig. 1 show the significantly differentiated genes, using the unconditional logistic regression analysis, false discovery rate (FDR) calculation, and Bonferroni correction. We observed significant discrepancies between the HAPC case and control groups in the dominant and additive genetic models. The results indicated the 12 genes, PDK1, RUNDC3B, EPO, MET, PTK2, RELN, TDRD1, TCL1A, STAT3, STAT5A, IL12RB1, and NF2, were associated with HAPC.
Table 1.
Demographics of healthy controls and patients with high-altitude polycythemia
| Variables | Tibetan | |
|---|---|---|
| Case (n = 70) | Control (n = 71) | |
| Male | 35 | 39 |
| Female | 35 | 32 |
Table 2.
Basic information on significant gene differences in highland people
| SNP | Gene | CHR | Model | Alleles | Case (N) | Case | HWE | Control (N) | Control | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A/B | AA | AB | BB | MAF | Case | AA | AB | BB | MAF | ||||
| rs529091195 | PDK1 | 2 | Dominant | /T | 0 | 48 | 20 | 0.3529 | 1.87E − 06 | 0 | 17 | 54 | 0.1197 |
| rs529091195 | PDK1 | 2 | Additive | /T | 0 | 48 | 20 | 0.3529 | 1.87E − 06 | 0 | 17 | 54 | 0.1197 |
| rs376837075 | FER | 5 | Dominant | /T | 0 | 66 | 2 | 0.485 | 4.13E − 17 | 0 | 33 | 35 | 0.243 |
| rs376837075 | FER | 5 | Additive | /T | 0 | 66 | 2 | 0.485 | 4.13E − 17 | 0 | 33 | 35 | 0.243 |
| rs527802276 | RUNDC3B | 7 | Dominant | /A | 0 | 61 | 7 | 0.449 | 8.42E − 13 | 0 | 29 | 41 | 0.2071 |
| rs527802276 | RUNDC3B | 7 | Additive | /A | 0 | 61 | 7 | 0.449 | 8.42E − 13 | 0 | 29 | 41 | 0.2071 |
| rs773485910 | EPO | 7 | Dominant | T/- | 0 | 46 | 22 | 0.338 | 6.99E − 06 | 0 | 22 | 49 | 0.155 |
| rs773485910 | EPO | 7 | Additive | T/- | 0 | 46 | 22 | 0.338 | 6.99E − 06 | 0 | 22 | 49 | 0.155 |
| rs397889442 | RELN | 7 | Dominant | A/- | 0 | 41 | 25 | 0.311 | 1.11E − 04 | 0 | 18 | 47 | 0.139 |
| rs397889442 | RELN | 7 | Additive | A/- | 0 | 41 | 25 | 0.311 | 1.11E − 04 | 0 | 18 | 47 | 0.139 |
| rs548208912 | CREB5 | 7 | Dominant | T/- | 0 | 61 | 3 | 0.477 | 8.11E − 15 | 0 | 33 | 34 | 0.246 |
| rs548208912 | CREB5 | 7 | Additive | T/- | 0 | 61 | 3 | 0.477 | 8.11E − 15 | 0 | 33 | 34 | 0.246 |
| rs551879100 | MET | 7 | Dominant | T/- | 0 | 58 | 10 | 0.427 | 6.20E − 11 | 0 | 33 | 38 | 0.232 |
| rs551879100 | MET | 7 | Additive | T/- | 0 | 58 | 10 | 0.427 | 6.20E − 11 | 0 | 33 | 38 | 0.232 |
| rs372806706 | PTK2 | 8 | Dominant | A/- | 0 | 61 | 6 | 0.455 | 3.86E − 13 | 0 | 31 | 39 | 0.221 |
| rs372806706 | PTK2 | 8 | Additive | A/- | 0 | 61 | 6 | 0.455 | 3.86E − 13 | 0 | 31 | 39 | 0.221 |
| rs369382658 | PTK2 | 8 | Dominant | A/- | 0 | 61 | 6 | 0.455 | 3.86E − 13 | 0 | 31 | 39 | 0.221 |
| rs369382658 | PTK2 | 8 | Additive | A/- | 0 | 61 | 6 | 0.455 | 3.86E − 13 | 0 | 31 | 39 | 0.221 |
| rs11285127 | TDRD1 | 10 | Dominant | A/- | 0 | 49 | 19 | 0.36 | 7.34E − 07 | 0 | 21 | 45 | 0.159 |
| rs11285127 | TDRD1 | 10 | Additive | A/- | 0 | 49 | 19 | 0.36 | 7.34E − 07 | 0 | 21 | 45 | 0.159 |
| rs142205645 | TCL1A | 14 | Dominant | /AG | 0 | 50 | 18 | 0.368 | 5.55E − 07 | 0 | 19 | 52 | 0.134 |
| rs142205645 | TCL1A | 14 | Additive | /AG | 0 | 50 | 18 | 0.368 | 5.55E − 07 | 0 | 19 | 52 | 0.134 |
| rs141204613 | TP53 | 17 | Dominant | /TTT | 1 | 64 | 3 | 0.485 | 1.49E − 14 | 0 | 35 | 31 | 0.265 |
| rs141204613 | TP53 | 17 | Additive | /TTT | 1 | 64 | 3 | 0.485 | 1.49E − 14 | 0 | 35 | 31 | 0.265 |
| rs558351915 | STAT3 | 17 | Dominant | A/- | 0 | 55 | 11 | 0.417 | 6.74E − 10 | 0 | 27 | 38 | 0.208 |
| rs558351915 | STAT3 | 17 | Additive | A/- | 0 | 55 | 11 | 0.417 | 6.74E − 10 | 0 | 27 | 38 | 0.208 |
| rs779456792 | STAT5A | 17 | Dominant | T/- | 0 | 52 | 16 | 0.382 | 5.58E − 08 | 0 | 26 | 45 | 0.183 |
| rs779456792 | STAT5A | 17 | Additive | T/- | 0 | 52 | 16 | 0.382 | 5.58E − 08 | 0 | 26 | 45 | 0.183 |
| rs548702753 | IL12RB1 | 19 | Dominant | /T | 2 | 47 | 16 | 0.392 | 2.66E − 05 | 0 | 18 | 48 | 0.136 |
| rs548702753 | IL12RB1 | 19 | Additive | /T | 2 | 47 | 16 | 0.392 | 2.66E − 05 | 0 | 18 | 48 | 0.136 |
| rs769771815 | NF2 | 22 | Dominant | T/- | 0 | 48 | 18 | 0.364 | 7.44E − 07 | 0 | 23 | 44 | 0.171 |
| rs769771815 | NF2 | 22 | Additive | T/- | 0 | 48 | 18 | 0.364 | 7.44E − 07 | 0 | 23 | 44 | 0.171 |
| SNP | Gene | CHR | Model | HWE | OR | 95%CI | P | P1 | P2 | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | L95 | U95 | |||||||||||
| rs529091195 | PDK1 | 2 | Dominant | 0.583 | 7.624 | 3.585 | 16.21 | 1.31E − 07 | 8.74E − 05 | 0.0004026 | |||
| rs529091195 | PDK1 | 2 | Additive | 0.583 | 7.624 | 3.585 | 16.21 | 1.31E − 07 | 8.75E − 05 | 0.0004031 | |||
| rs376837075 | FER | 5 | Dominant | 0.007 | 35 | 7.928 | 15.45 | 2.70E − 06 | 0.0009179 | 0.008261 | |||
| rs376837075 | FER | 5 | Additive | 0.007 | 35 | 7.928 | 15.45 | 2.70E − 06 | 0.000919 | 0.008272 | |||
| rs527802276 | RUNDC3B | 7 | Dominant | 0.0312 | 12.32 | 4.933 | 30.77 | 7.58E − 08 | 0.0000874 | 0.000232 | |||
| rs527802276 | RUNDC3B | 7 | Additive | 0.0312 | 12.32 | 4.933 | 30.77 | 7.58E − 08 | 0.0000875 | 0.0002323 | |||
| rs773485910 | EPO | 7 | Dominant | 0.197 | 4.657 | 2.278 | 9.519 | 2.47E − 05 | 0.004731 | 0.0757 | |||
| rs773485910 | EPO | 7 | Additive | 0.197 | 4.657 | 2.278 | 9.519 | 2.47E − 05 | 0.004737 | 0.0758 | |||
| rs397889442 | RELN | 7 | Dominant | 0.593 | 4.282 | 2.05 | 8.944 | 0.0001087 | 0.01958 | 0.3328 | |||
| rs397889442 | RELN | 7 | Additive | 0.593 | 4.282 | 2.05 | 8.944 | 0.0001087 | 0.0196 | 0.3333 | |||
| rs548208912 | CREB5 | 7 | Dominant | 0.007 | 20.95 | 5.977 | 73.42 | 1.99E − 06 | 0.0008717 | 0.006102 | |||
| rs548208912 | CREB5 | 7 | Additive | 0.007 | 20.95 | 5.977 | 73.42 | 1.99E − 06 | 0.0008729 | 0.00611 | |||
| rs551879100 | MET | 7 | Dominant | 0.015 | 6.679 | 2.95 | 15.12 | 5.26E − 06 | 0.001239 | 0.01611 | |||
| rs551879100 | MET | 7 | Additive | 0.015 | 6.679 | 2.95 | 15.12 | 5.26E − 06 | 0.001241 | 0.01613 | |||
| rs372806706 | PTK2 | 8 | Dominant | 0.016 | 12.79 | 4.887 | 33.47 | 2.08E − 07 | 0.0001061 | 0.0006368 | |||
| rs372806706 | PTK2 | 8 | Additive | 0.016 | 12.79 | 4.887 | 33.47 | 2.08E − 07 | 0.0001063 | 0.0006377 | |||
| rs369382658 | PTK2 | 8 | Dominant | 0.016 | 13.62 | 4.463 | 41.58 | 4.49E − 06 | 0.001205 | 0.01376 | |||
| rs369382658 | PTK2 | 8 | Additive | 0.016 | 13.62 | 4.463 | 41.58 | 4.49E − 06 | 0.001206 | 0.01378 | |||
| rs11285127 | TDRD1 | 10 | Dominant | 0.344 | 5.526 | 2.634 | 11.59 | 6.11E − 06 | 0.001337 | 0.01871 | |||
| rs11285127 | TDRD1 | 10 | Additive | 0.344 | 5.526 | 2.634 | 11.59 | 6.11E − 06 | 0.001338 | 0.01874 | |||
| rs142205645 | TCL1A | 14 | Dominant | 0.345 | 7.602 | 3.582 | 16.13 | 1.27E − 07 | 8.74E − 05 | 0.000389 | |||
| rs142205645 | TCL1A | 14 | Additive | 0.345 | 7.602 | 3.582 | 16.13 | 1.27E − 07 | 8.75E − 05 | 0.0003895 | |||
| rs141204613 | TP53 | 17 | Dominant | 0.003 | 19.19 | 5.475 | 67.27 | 3.90E − 06 | 0.001195 | 0.01195 | |||
| rs141204613 | TP53 | 17 | Additive | 0.003 | 19.14 | 5.477 | 66.85 | 3.75E − 06 | 0.001151 | 0.01151 | |||
| rs558351915 | STAT3 | 17 | Dominant | 0.054 | 7.037 | 3.118 | 15.88 | 2.62E − 06 | 0.0009179 | 0.008021 | |||
| rs558351915 | STAT3 | 17 | Additive | 0.054 | 7.037 | 3.118 | 15.88 | 2.62E − 06 | 0.0009191 | 0.008031 | |||
| rs779456792 | STAT5A | 17 | Dominant | 0.105 | 5.625 | 2.685 | 11.79 | 4.72E − 06 | 0.001205 | 0.01446 | |||
| rs779456792 | STAT5A | 17 | Additive | 0.105 | 5.625 | 2.685 | 11.79 | 4.72E − 06 | 0.001206 | 0.01447 | |||
| rs548702753 | IL12RB1 | 19 | Dominant | 0.592 | 8.003 | 3.688 | 17.37 | 1.43E − 07 | 8.74E − 05 | 0.0004371 | |||
| rs548702753 | IL12RB1 | 19 | Additive | 0.592 | 8.003 | 3.688 | 17.37 | 1.43E − 07 | 8.75E − 05 | 0.0004375 | |||
| rs769771815 | NF2 | 22 | Dominant | 0.189 | 5.101 | 2.434 | 10.69 | 1.59E − 05 | 0.003255 | 0.04882 | |||
| rs769771815 | NF2 | 22 | Additive | 0.189 | 5.101 | 2.434 | 10.69 | 1.59E − 05 | 0.003259 | 0.04889 | |||
SNP single-nucleotide polymorphism, MAF minor allele frequency, HWE Hardy–Weinberg equilibrium, OR odds ratio, 95% CI 95% confidence interval, P P value calculated by unconditional logistic regression analysis, P1 P value FDR-calculated, P2 P value after Bonferroni correction
Fig. 1.
Manhattan plot of the p-value of the correlation between HAPC and SNP determined by false discovery rate (FDR) calculation
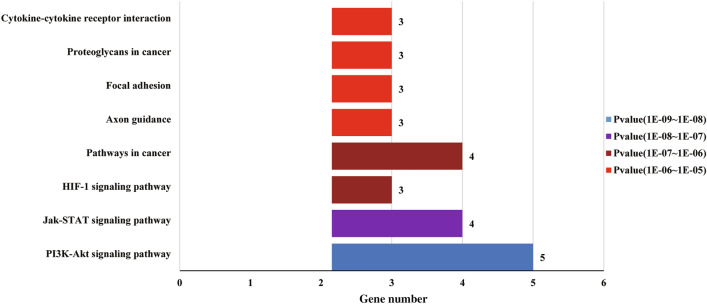
We adopted KEGG signaling pathways to determine gene-specific differences, and Table 3 and Fig. 2 display the results. Twelve genes were enriched in 16 signal pathways, including the PI3K-AKT pathway, JAK-STAT pathway, HIF-1 pathway, pathways in cancer, axon guidance, focal adhesion, proteoglycans in cancer, cytokine-cytokine receptor interaction, et al. Enrichment pathway information is shown in Figs. 3 (according to Table 2, p-value < 6.36 E − 05) and 2.
Table 3.
KEGG pathway analysis of differentially expressed genes
| Pathway | KEGG | Input | Background | p-value | Genes |
|---|---|---|---|---|---|
| ID | number | number | |||
| PI3K-Akt signaling pathway | hsa04151 | 5 | 342 | 3.70E − 08 | PTK2, MET, TCL1A |
| RELN, EPO | |||||
| Jak-STAT signaling pathway | hsa04630 | 4 | 158 | 1.28E − 07 | IL12RB1, STAT5A, |
| STAT3, EPO | |||||
| HIF-1 signaling pathway | hsa04066 | 3 | 103 | 3.98E − 06 | PDK1, STAT3, EPO |
| Pathways in cancer | hsa05200 | 4 | 397 | 4.73E − 06 | PTK2, STAT5A, |
| MET, STAT3 | |||||
| Axon guidance | hsa04360 | 3 | 176 | 1.91E − 05 | PDK1, MET, PTK2 |
| Focal adhesion | hsa04510 | 3 | 203 | 2.91E − 05 | RELN, MET, PTK2 |
| Proteoglycans in cancer | hsa05205 | 3 | 205 | 3.00E − 05 | PTK2, MET, STAT3 |
| Cytokine-cytokine receptor interaction | hsa04060 | 3 | 265 | 6.36E − 05 | IL12RB1, MET, EPO |
| Inflammatory bowel disease (IBD) | hsa05321 | 2 | 66 | 1.880E − 04 | IL12RB1, STAT3 |
| Central carbon metabolism in cancer | hsa05230 | 2 | 67 | 1.936E − 04 | STAT5A, IL12RB1 |
| Prolactin signaling pathway | hsa04917 | 2 | 72 | 2.227E − 04 | STAT5A, STAT3 |
| Bacterial invasion of epithelial cells | hsa05100 | 2 | 78 | 2.603E − 04 | MET, PTK2 |
| ErbB signaling pathway | hsa04012 | 2 | 88 | 3.293E − 04 | STAT5A, PTK2 |
| AGE-RAGE signaling pathway in diabetic complications | hsa04933 | 2 | 101 | 4.310E − 04 | STAT5A, STAT3 |
The above results were analyzed using DAVID and KOBAS online analysis tools
PI3K-Akt: Phosphatidylinositol 3 kinase(PI3K) /protein kinase B(AKT), Jak-STAT: Janus kinase (JAK)/ signal transducer and activator of transcription (STAT), HIF-1: Hypoxia inducible factor-1 (HAF-1), ErbB: Receptor tyrosine-protein kinase erbB-2, i.e. Her2, human epidermal growth factor receptor 2 (HER2), AGE-RAGE: advanced glycation end products (AGE) / receptor for dvanced glycation end products (RAGE)
Fig. 2.
KEGG pathway analysis of differentially expressed genes. The Y-axis is the name of the KEGG metabolic pathway, and the X-axis is the number of genes annotated to the pathway (according to Table 2, p-value< 6.36 E − 05)
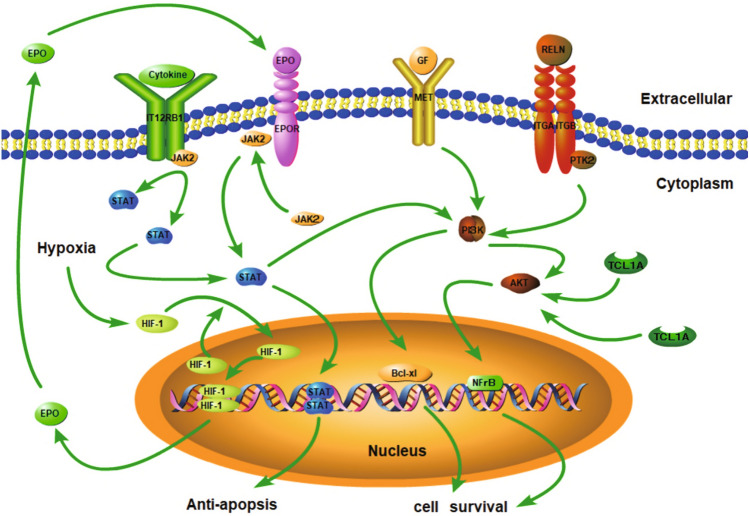
Fig. 3.
Schematic diagrams of the JAK-STAT pathway, HIF-1 pathway, and PI3K-AKT pathway. The HIF-1 signaling pathway is activated, and EPO secretion increases under hypoxic conditions. EPO then binds to the EPO receptor (EPOR). The JAK-STAT signaling pathway can also be activated and plays an anti-apoptotic role. The PI3K-Akt signaling pathway could promote the expression of the anti-apoptotic gene Bcl-xL and affect cell proliferation and differentiation. TCL1A activates the AKT pathway, which plays a role in cell survival by affecting the transcriptional activity of nuclear factor-kB (NFkB)
Discussion
Genomics and Bioinformatics Enhance the Study of HAPC
Upon completing the Human Genome Project, we will have a more comprehensive understanding of population-specific genomic variations and the interactions between genes and environmental factors. This information will help us quickly determine mutation sites related to diseases to use genetic information to establish the relationship between sequence variation and disease risk and help prevent, diagnose, and treat diseases [18, 19].
HAPC seriously intimidates the fitness of the population of the plateau. In Tibet, the increased Hb concentration enhances the efficiency of carrying oxygen to adapt to altitude hypoxia. This reaction is an imperative factor for populations, who adapt to high altitudes. We found an increased risk of HAPC associated with SNPs in EPO, STAT3, PDK1, STAT5A, IL12RB1, PTK2, MET, TCL1A, RELN, RUNDC3B, TDRD1, and NF2 in the Tibetan population. We found that these genes were mostly enriched in the PI3K-AKT, JAK-STAT, and HIF-1 pathways through KEGG pathway analysis.
Notably, the genes' distributions varied widely from the different pathways based on the KEGG pathway analysis. From the perspective of the pathways, three genes EPO, STAT3, and PDK1 were enriched in HIF-1 pathways; four genes, EPO, STAT3, STAT5A, and IL12RB1, were enriched in JAK-STAT pathways; five genes EPO, PTK2, MET, TCL1A, and RELN were enriched in PI3K-AKT pathways. Likewise, EPO gene was enriched in these three pathways; STAT3 gene was enriched in two pathways; excluding RUNDC3B, TDRD1, and NF2 genes, other genes were positioned in one pathway.
The Impact of Genes Enriched in the HIF-1 Pathway for Erythrocytes
Hypoxia-inducible factor one (HIF-1), as a transcription factor, is composed of two subunits, which include an induced expressed part of HIF-1α subunit and a control structure of HIF-1β subunit. Without hypoxia, HIF-1α subunit would never be ubiquitinated and hydrolyzed by proteasomes easily [20]. Meanwhile, complex regulation of signal transduction cascades, which are mediated by cytokines and their homologous receptors, may affect the formation of HAPC. When the body is in a low-oxygen environment, its red blood cells increase. The HIF-1 signaling pathway may activate this.
EPO is the earliest hypoxic adaptation gene, and its polymorphism is related to the formation of HAPC [21]. Our results conformed these findings as we found that the rs773485910 of the EPO gene was associated with an increased prevalence of HAPC. EPO, endogenous glycoprotein hormone, has a molecular weight of 39 KD, with 166 residues. It is a member of the hematopoietic cytokine family and is primarily involved in erythropoiesis. EPO is principally secreted by liver cells in infancy and by kidney cells in adulthood. When the body immerses in a low-oxygen environment, the acetylation level of HIF-1 alpha increase, and there is enhanced transcriptional activation of a series of target genes, stimulating increased secretion of EPO by the kidneys [22]. Via binding to the EPO receptor (EPOR) on red cell progenitors in the bone marrow, circulating EPO can arouse an increase in the amount of erythrocytes [21]. However, small amounts are also expressed in other vital organs, such as the brain, spleen, lungs, testicles, and placental tissue. At average oxygen concentrations, the EPO content of blood is low, and its primary role relates to the renewal of aging erythrocytes [23].
PDKs are serine protein kinase genes located on chromosome 2, mainly expressed in the heart, bone marrow, kidneys, and skin. There are four isomers of PDK–PDK1, PDK2, PDK3, and PDK4. PDK1, PDK2, and PDK3 are mainly involved in cellular glucose metabolism, while PDK4 is engaged primarily in cell fat metabolism. Previous studies have shown that PDK1 and PDK4 genes are associated with erythrocyte polymorphism in Chuvash decent people. Our research found that rs529091195 of the PDK gene was associated with HAPC, which might be caused by significant upregulation of HIF due to chronic hypoxia. Upregulated HIF can promote PDK1 expression, affecting the anaerobic oxidation of cells and providing energy for normal cellular life activities [24].
Moreover, by retrieving PDK1 in the KEGG pathway database, we acquire that PDK1 also brings a crucial impact in the PI3K-AKT pathway and is activated by plasma membrane intrinsic protein 3 (PIP3). Activated PDK1 fully activates adjacent protein kinase B (AKT) and regulates downstream AGC family protein kinase activity [25–27]. These actions allow PDK1 to control the physiological effects of insulin and growth factors, increase glucose uptake, promote glycogen and protein synthesis, and provide energy for cell proliferation and differentiation [28]. In this way, PDK1 influences the development of HAPC. Once activated, the PI3K-AKT pathway can also play an anti-apoptotic role, resulting in increased erythrocyte accumulation and promoting HAPC [11].
The Impact of Genes Enriched in the JAK-STAT Pathway for Erythrocytes
Important hematological factors widely involve the JAK-STAT pathway. When EPO binds to the EPO receptor, the JAK-STAT signaling pathway can also be activated [29]. Among those principal kinases involved in mediating EPO-responsive signal transduction, the Jak2 protein tyrosine kinase was identified for the first time by the researchers. JAK2 binds to EPOR at the bottom of the cytoplasm, causing JAK2 phosphorylation, which leads to tyrosine phosphorylation and coupling of STAT-5, affecting cell proliferation and differentiation [30].
STATs consist of seven separate members, STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B, and STAT6. Furthermore, they are active in many cell signaling pathways and have essential effects on innate immunity, acquired immunity, cell proliferation, differentiation, and survival [31]. We found that rs558351915 of the STAT3 gene and rs779456792 of the STAT5A gene were related to the formation of HAPC. Previous studies have shown that different STAT family members can interact in cell signaling pathways to regulate target genes' expression [32]. The JAK-STAT pathway plays an imperative role in cellular erythropoiesis, proliferation, and differentiation. STAT3 and STAT5 are essential phosphorylated kinases in the JAK-STAT cell signaling pathway. Meanwhile, EPO binds to the EPO receptor to activate and phosphorylated JAK2; and tyrosine phosphorylated and coupled with STAT5. Then, JAK1 and STAT5 upregulate the expression of membrane proteins, cytoskeleton, hematopoietic growth factor-related genes, and downstream target genes, which contain anti-apoptotic genes Bcl-xl and Bcl-2. Moreover, the PI3K-AKT pathway is a downstream effector of JAK2-STAT5 signaling. Also, STAT5 regulates and promotes hemoglobin expression, thereby induces the proliferation of red blood cells (RBC). In a word, it can increase the RBCs' count in the body's blood [33].
The Impact of Genes Enriched in the PI3K-AKT Pathway for Erythrocytes
The PI3K-Akt signaling pathway is also called the "cell survival signaling pathway," which can protect cells from inactivation and affect cell proliferation and differentiation by being activated. This signaling pathway brings a substantial impact on the process of erythropoiesis and could downregulate apoptosis by regulating apoptosis-related molecules. This process promotes the expression of the anti-apoptotic gene Bcl-xL and plays a critical anti-apoptotic role [34]. In particular, PI3K-AKT pathways affect HIF-1α transcriptional activity in hypoxia [35, 36], sequentially transactivating EPO, eventually resulting in erythrocytosis. Therefore, the PI3K-AKT signal pathway appeared to be involved in the mechanism of decreased erythroblasts apoptosis.
PTK2 genes are enriched in the PI3K-AKT signaling pathways. Upon cell-extracellular matrix (cell-ECM) contact, PTK2 can be recruited into focal plaques and is rapidly autophosphorylated to recruit other scaffolds and signal molecules to activate the downstream PI3K-AKT signaling pathways [37]. Studies also indicated that HIF-1 gene knockout could effectively inhibit the accumulation of HIF-1 protein and the expression of PTK2 mRNA. Meanwhile, this study also indicated that PTK2 activation was inhibited; the phosphorylation levels of downstream AKT were significantly reduced. These reductions indicated that PTK2 induced the phosphorylation of AKT. The transcriptional activation of PTK2, mediated by HIF-1, protects cells from inactivation and can affect cells' proliferation and differentiation by activating the cell survival signaling pathways and inhibiting the pro-apoptotic signaling pathways.
MET gene is enriched in the PI3K-AKT signaling pathways, located on chromosome 7. Its product is a sort of receptor tyrosine kinase of proteins, with about 110kB in size. MET is mainly expressed in the liver and kidneys, bone marrow cells. The mature MET protein is transmembrane, a dimer complex composed of α and β subunits. There are three structural regions, of which the intracellular domain contains the binding sites for many signal molecules MET activation of the PI3K-Akt signaling pathways can promote cell proliferation and prevent cell apoptosis [38, 39]. In addition to activating the above signaling pathways, MET can also interact with cell death receptors on cell membranes to play an anti-apoptotic role (eg. Fas, FasL). These findings illustrate that MET receptors play a direct role in preventing apoptosis.
RUNDC3B, TDRD1 and NF2 Genes Influence Erythrocytes
RUNDC3B (RUN domain containing 3B) is located on chromosome 7 and is widely expressed in the adrenal glands, brain, liver, small intestines, and other tissues. Although the biological function of RUNDC3B has not been determined, decreased expression of RUNDC3B may result in lymphoid malignancies [40]. In our study, we indicate that the rs527802276 of the RUNDC3B gene was associated with HAPC. Studies have shown that RUNDC3B and RUNDC3A (a Rap2-interacting protein) have high homology, and RUNDC3B also has a similar integration effect like RUNDC3A. The Rap protein family constitutes a subgroup of the Ras superfamily and works as a molecular "switch" regulating various cell functions, such as proliferation, differentiation, and other cell activities [41]. RUNDC3B contains a RUN domain in its N-terminal region, which is an essential component of the mitogen-activated protein kinase (MAPK) cascade. Furthermore, Rap2 interacts with MAP4K4 through its C-terminal citron homology domain. MAP4K4 is a kind of the STE20 protein kinases and regulates c-Jun N-terminal kinase (JNK). Rap2 enhances MAP4K4-induced activation of JNK. So, RUNDC3B appears to play an essential role in activating c-JUN and c-Fos transcription factors, leading to the expression of c-JUN and c-Fos in the nucleus. Its product AP1 can bind to DNA sequences and induce cell proliferation and differentiation.
The TDRD1 gene on chromosome 10 is mainly expressed in the prostate and testes, and a small amount is expressed in the kidneys. Studies have shown that TDRD1 is involved in spermatogenesis. The mutation of the TDRD1 gene is related to spermatogenesis disorder in Han males [42]. In our study, rs11285127 of the TDRD1 gene was involved in the development of HAPC.
NF2 is located on chromosome 22, and its coding product is Merlin. It has a protein with a similar structure to ERM family members. Mutation of the gene is associated with neurofibromatosis type II [43]. In our study, the rs779456792 locus of the NF2 gene was associated with HAPC. The Merlin protein may regulate the PI3K-Akt signaling pathway by interacting with PI3K and other molecules. In this way, the Merlin protein controls cell survival, proliferation, differentiation, and development of RBC, thereby affecting the formation of RBCs [44].
Conclusion
PDK1 (rs529091195), RUNDC3B (rs527802276), EPO (rs773485910), RELN (rs397889442), MET (rs551879100), TDRD1 (rs11285127), PTK2 (rs372806706, rs369382658), TCL1A (rs142205645), STAT3 (rs558351915), STAT5A (rs779456792), IL12RB1 (rs548702753) and NF2 (rs779456792) genes are associated with increased risk of HAPC. Through KEGG pathway analysis, we found that these genes were mostly enriched in the PI3K-AKT, JAK-STAT, and HIF-1 pathways.
We are hopeful that our results will provide a reference for the etiology research of HAPC. However, additional genetic risk factors and functional investigations are necessary to confirm our results further.
Supplementary Information
Below is the link to the electronic supplementary material.
Author Contribution
L.K. and S.Z. conceived, designed the study, supervised the project, and drafted the manuscript. Z.Z., L.M., X.F., K.W., participated in the design of the study, data analysis and helped to draft the manuscript. Z.Z., L.L., Y.Z., P.C., Y.L., H.Z., T.L., W.D., Z.Z., J.L., contributed to sample collection and experiments. And the manuscript has been reviewed and approved by all co-authors before submission.
Funding
This study was supported by the National Natural Science Foundation of China (No. U20A20395, 81860329, 31660307, 31460286, 31260252), the Natural Science Foundation of Xizang (Tibet) Autonomous Region (Nos. XZ201801-GB03, Z2014A09G2-3), the Science and Technology Department Project of Tibet Autonomous Region [Nos. XZ 2019 ZRG-32(Z); XZ 2019 ZRG-132; XZ2019ZRG-133; XZ2019ZRG-28(Z)], the Peak Scholars Programme of Xizang Minzu University, the Innovation Support Program for Young Teachers of Tibet Autonomous Region (Nos. QCZ2016-27; QCZ2016-29; QCZ2016-34).
Declarations
Conflict of interest
The authors declare no competing financial interests.
Ethics Approval
The study was approved by the Ethics Committee of the Xizang Minzu University. Number: 201801.
Consent to Participate
All the volunteers signed an informed consent.
Consent for Publication
All the authors approved to publish.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhiying Zhang, Lifeng Ma, and Xiaowei Fan have contributed equally to this work.
Contributor Information
Songhua Zhou, Email: fszsh1001@163.com.
Longli Kang, Email: longli_kang@163.com.
References
- 1.Leon VF, Maggiorini M, Reeves JT, et al. Consensus statement on chronic and subacute high altitude diseases. High Alt Med Biol. 2005;6:147–157. doi: 10.1089/ham.2005.6.147. [DOI] [PubMed] [Google Scholar]
- 2.Luks AM, Swenson ER, Bartsch P. Acute high-altitude sickness. Eur Respir Rev. 2017;143:160096–160099. doi: 10.1183/16000617.0096-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palmer BF. Physiology and pathophysiology with ascent to altitude. Am J Med Sci. 2010;1:69–77. doi: 10.1097/MAJ.0b013e3181d3cdbe. [DOI] [PubMed] [Google Scholar]
- 4.Guan W, Ga Q, Li R, et al. Sleep disturbances in long-term immigrants with chronic mountain sickness: a comparison with healthy immigrants at high altitude. Respir Physiol Neurobiol. 2015;206:4–10. doi: 10.1016/j.resp.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Jiang C, Liu F, Luo Y, et al. Gene expression profiling of high altitude polycythemia in Han Chinese migrating to the Qinghai-Tibetan plateau. Mol Med Rep. 2012;1:287–293. doi: 10.3892/mmr.2011.632. [DOI] [PubMed] [Google Scholar]
- 6.Reeves JT, Leon-velarde F. Chronic mountain sickness: recent studies of the relationship between hemoglobin concentration and oxygen transport. High Alt Med Biol. 2004;2:147–155. doi: 10.1089/1527029041352090. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y, Jiang C, Luo Y, et al. An EPAS1 haplotype is associated with high altitude polycythemia in male Han Chinese at the Qinghai-Tibetan plateau. Wilderness Environ Med. 2014;4:392–400. doi: 10.1016/j.wem.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 8.Fan X, Ma L, Zhang Z, et al. Associations of high-altitude polycythemia with polymorphisms in PIK3CD and COL4A3 in Tibetan populations. Hum Genomics. 2018;1:37–47. doi: 10.1186/s40246-018-0169-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu L, Zhang Y, Zhang Z, et al. Associations of high altitude polycythemia with polymorphisms in and in Chinese Han and Tibetan populations. Oncotarget. 2017;8:53234–53243. doi: 10.18632/oncotarget.18384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu J, Yang YZ, Tang F, et al. EPAS1 gene polymorphisms are associated with high altitude polycythemia in Tibetans at the Qinghai-Tibetan plateau. Wilderness Environ Med. 2015;3:288–294. doi: 10.1016/j.wem.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 11.Zhao C, Li Z, Ji L, et al. PI3K-Akt signal transduction tolecules maybe involved in downregulation of erythroblasts apoptosis and perifosine increased its apoptosis in chronic mountain sickness. Med Sci Monit. 2017;23:5637–5649. doi: 10.12659/MSM.905739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beall CM, CavalleriA GL, Deng L, et al. Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proc Natl Acad Sci USA. 2010;25:11459–11464. doi: 10.1073/pnas.1002443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li K, Gesang L, Dan Z, et al. Transcriptome reveals the over expression of a kallikrein gene cluster (KLK1/3/7/8/12) in the Tibetans with high altitude-associated polycythemia. Int J Mol Med. 2017;2:287–296. doi: 10.3892/ijmm.2016.2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, Liu F, Ye S, et al. Plasma proteome profiling of high-altitude polycythemia using TMT-based quantitative proteomics approach. J Proteomics. 2019;194:60–69. doi: 10.1016/j.jprot.2018.12.031. [DOI] [PubMed] [Google Scholar]
- 15.Tumuru L, Thein SL. Genetic control of erythropoiesis. Curr Opin Hematol. 2017;3:173–182. doi: 10.1097/MOH.0000000000000333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warden C D, Adamson A W, Neuhausen, S L, & Wu X. Detailed comparison of two popular variant calling packages for exome and targeted exon studies. PeerJ, 2014; 2, e600. [DOI] [PMC free article] [PubMed]
- 17.Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;5:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collins FS, Morgan M, Patrinos A. The Human Genome Project: lessons from large-scale biology. Science. 2003;300:286–290. doi: 10.1126/science.1084564. [DOI] [PubMed] [Google Scholar]
- 19.Watson JD. The human genome project: past, present, and future. Science. 1990;248:44–49. doi: 10.1126/science.2181665. [DOI] [PubMed] [Google Scholar]
- 20.Weidemann A, Johnson RS. Biology of HIF-1alpha. Cell Death Differ. 2008;4:621–627. doi: 10.1038/cdd.2008.12. [DOI] [PubMed] [Google Scholar]
- 21.Lappin T R, Lee F S. Update on mutations in the HIF: EPO pathway and their role in erythrocytosis. Blood Rev, 2019; 37: 100590. [DOI] [PMC free article] [PubMed]
- 22.Tanaka T, Nangaku M. Recent advances and clinical application of erythropoietin and erythropoiesis-stimulating agents. Exp Cell Res. 2012;9:1068–1073. doi: 10.1016/j.yexcr.2012.02.035. [DOI] [PubMed] [Google Scholar]
- 23.Buroker NE, Ning XH, Zhou ZN, et al. Genetic associations with mountain sickness in Han and Tibetan residents at the Qinghai-Tibetan Plateau. Clin Chim Acta. 2010;19:1466–1473. doi: 10.1016/j.cca.2010.05.043. [DOI] [PubMed] [Google Scholar]
- 24.Formenti F, Constantin TD, Emmanuel Y, et al. Regulation of human metabolism by hypoxia-inducible factor. Proc Natl Acad Sci USA. 2010;28:12722–12727. doi: 10.1073/pnas.1002339107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu Z, Coxhipkin MA, Windsor WT, et al. 3-phosphoinositide-dependent protein kinase-1 regulates proliferation and survival of cancer cells with an activated mitogen-activated protein kinase pathway. Mol Cancer Res. 2010;3:421–432. doi: 10.1158/1541-7786.MCR-09-0179. [DOI] [PubMed] [Google Scholar]
- 26.Mora A, Komander D, Van Aalten MF, et al. PDK1, the master regulator of AGC kinase signal transduction. Semin Cell Dev Biol. 2004;2:161–170. doi: 10.1016/j.semcdb.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 27.Parrales A, Lopez E, Colome AM. Thrombin activation of PI3K/PDK1/Akt signaling promotes cyclin D1 upregulation and RPE cell proliferation. Biochim Biophys Acta. 2011;10:1758–1766. doi: 10.1016/j.bbamcr.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 28.Knight ZA. For a PDK1 inhibitor, the substrate matters. Biochem J. 2011;2:e1–e2. doi: 10.1042/BJ20102038. [DOI] [PubMed] [Google Scholar]
- 29.Ma S, Chen J, Chen C, et al. Erythropoietin rescues memory impairment in a rat model of chronic cerebral hypoperfusion via the EPO-R/JAK2/STAT5/PI3K/Akt/GSK-3β pathway. Mol Neurobiol. 2018;4:3290–3299. doi: 10.1007/s12035-017-0568-5. [DOI] [PubMed] [Google Scholar]
- 30.Rane SG, Reddy EP. JAKs, STATs and Src kinases in hematopoiesis. Oncogene. 2002;21:3334–3358. doi: 10.1038/sj.onc.1205398. [DOI] [PubMed] [Google Scholar]
- 31.Ivashkiv LB, Hu X. Signaling by STATs. Arthritis Res Ther. 2004;4:159–168. doi: 10.1186/ar1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Granillo A R, Boffi J C, BaranaoL, et al (2007) STAT5 transcriptional activity is impaired by LIF in a mammary epithelial cell line. Biochem Biophys Res Commun 3: 727–732 [DOI] [PubMed]
- 33.Buitenhuis M, Coffer PJ, Koenderman L. Signal transducer and activator of transcription 5 (STAT5) Int J Biochem Cell Biol. 2004;11:2120–2124. doi: 10.1016/j.biocel.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 34.Sun G, Zhou Y, Li H, et al. Over-expression of microRNA-494 up-regulates hypoxia-inducible factor-1 alpha expression via PI3K/Akt pathway and protects against hypoxia-induced apoptosis. J Biomed Sci. 2013;20:100. doi: 10.1186/1423-0127-20-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirito K. Regulation of erythropoiesis by hypoxia inducible factors (HIFs) Rinsho Ketsueki. 2011;6:368–375. [PubMed] [Google Scholar]
- 36.Zhou J, Schmid T, Frank R, et al. PI3K/Akt is required for heat shock proteins to protect hypoxia-inducible factor 1alpha from pVHL-independent degradation. J Biol Chem. 2004;14:13506–13513. doi: 10.1074/jbc.M310164200. [DOI] [PubMed] [Google Scholar]
- 37.Bouchard V, Demers MJ, Thibodeau S, et al. Fak/Src signaling in human intestinal epithelial cell survival and anoikis: differentiation state-specific uncoupling with the PI3-K/Akt-1 and MEK/Erk pathways. J Cell Physiol. 2007;3:717–728. doi: 10.1002/jcp.21096. [DOI] [PubMed] [Google Scholar]
- 38.Chappell WH, Steelman LS, Long JM, et al. Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR inhibitors: rationale and importance to inhibiting these pathways in human health. Oncotarget. 2011;3:135–164. doi: 10.18632/oncotarget.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Xia M, Jin K, et al. Function of the c-Met receptor tyrosine kinase in carcinogenesis and associated therapeutic opportunities. Mol Cancer. 2018;1:45–49. doi: 10.1186/s12943-018-0796-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burmeister DW, Smith EH, Cristel RT, et al. The expression of RUNDC3B is associated with promoter methylation in lymphoid malignancies. Hematol Oncol. 2017;1:25–33. doi: 10.1002/hon.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rountree MR, Bachman KE, Herman JG, et al. DNA methylation, chromatin inheritance, and cancer. Oncogene. 2001;24:3156–3165. doi: 10.1038/sj.onc.1204339. [DOI] [PubMed] [Google Scholar]
- 42.Zhu XB, Lu JQ, Zhi EL, et al. Association of a TDRD1 variant with spermatogenic failure susceptibility in the Han Chinese. J Assist Reprod Genet. 2016;8:1099–1104. doi: 10.1007/s10815-016-0738-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giovannini M, Robanus ME, Vander VM, et al. Conditional biallelic Nf2 mutation in the mouse promotes manifestations of human neurofibromatosis type 2. Genes Dev. 2000;13:1617–1630. [PMC free article] [PubMed] [Google Scholar]
- 44.Petrilli AM, Fernandez VC. Role of Merlin/NF2 inactivation in tumor biology. Oncogene. 2016;5:537–548. doi: 10.1038/onc.2015.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.