Abstract
Bacteriophage lysins (Ply), or endolysins, are phage-encoded cell wall lytic enzymes which are synthesized late during virus multiplication and mediate the release of progeny virions. Bacteriophages of the pathogen Listeria monocytogenes encode endolysin enzymes which specifically hydrolyze the cross-linking peptide bridges in Listeria peptidoglycan. Ply118 is a 30.8-kDa l-alanoyl-d-glutamate peptidase and Ply511 (36.5 kDa) acts as N-acetylmuramoyl-l-alanine amidase. In order to establish dairy starter cultures with biopreservation properties against L. monocytogenes contaminations, we have introduced ply118 and ply511 into Lactococcus lactis MG1363 by using a pTRKH2 backbone. The genes were expressed under control of the lactococcal promoter P32, which proved superior to other promoters (P21 and P59) tested in this study. High levels of active enzymes were produced and accumulated in the cytoplasmic cell fractions but were not released from the cells at significant levels. Therefore, ply511 was genetically fused with the SPslpA nucleotide sequence encoding the Lactobacillus brevis S-layer protein signal peptide. Expression of SPslpA–ply511 from pSL-PL511 resulted in secretion of functional Ply511 enzyme from L. lactis cells. One clone expressed an unusually strong lytic activity, which was found to be due to a 115-bp deletion that occurred within the 3′-end coding sequence of SPslpA–ply511, which caused a frameshift mutation and generated a stop codon. Surprisingly, the resulting carboxy-terminal deletion of 80 amino acids in the truncated Ply511Δ(S262–K341) mutant polypeptide strongly increased its lytic activity. Proteolytic processing of the secretion competent SPSlpA-Ply511 propeptide following membrane translocation had no influence on enzyme activity. Immunoblotting experiments using both cytoplasmic and supernatant fractions indicated that the enzyme was quantitatively exported from the cells and secreted into the surrounding medium, where it caused rapid lysis of L. monocytogenes cells. Moreover, transformation of pSL-PL511ΔC into L. lactis Bu2-129, a lactose-utilizing strain that can be employed for fermentation of milk, also resulted in secretion of functional enzyme and showed that the vector is compatible with the native lactococcal plasmids.
Listeria monocytogenes is widely distributed in the environment and, during the last decade, was recognized as an important food-borne pathogen. Various foods, such as meat, milk and other dairy products, and vegetables contaminated with L. monocytogenes have been linked with human listeriosis (6, 33). Listeriosis occurs primarily in certain high-risk groups, including pregnant women, neonates, and immunocompromised adults. Unlike other common food-borne diseases, listeriosis is associated with a mortality rate of 20% or higher (38). These properties in conjunction with the involvement of industrially processed foods have resulted in renewed attention to the importance of L. monocytogenes as a food-borne human pathogen.
Lactic acid bacteria play an important role in the manufacturing of fermented foods, especially dairy products. These bacteria are responsible not only for the development of flavor and texture but also for the preservation of many products (see reference 12). In recent years, much research was performed to genetically modify lactic starter strains in order to improve their characteristics and allow new applications (see reference 10). The availability of heterologous gene expression systems for lactic acid bacteria is of increasing interest because these organisms are generally recognized as safe. Genetic optimization of starter cultures, leading to a protective effect against food-borne pathogens, is an attractive approach for increased protection against hazardous contaminations. Several reports have described the production of bacteriocins and other antimicrobial metabolites by lactic acid bacteria, which are active against such organisms as Listeria, Clostridium, and Bacillus species (1, 11). The antimicrobial effects of bacteriocins in foods, such as nisin and pediocin, were the subject of several investigations (2, 7). However, many bacteriocins not only act against the target organism (e.g., L. monocytogenes) but may also affect a wide range of other sensitive bacteria. Thus, the broad-range inhibitors might negatively influence the “normal” micro-ecosystem by inhibiting the organisms responsible for the ripening process.
Endolysins are cell-wall-hydrolyzing enzymes synthesized during late gene expression in the lytic cycle of phage multiplication and enable the release of progeny virions from infected cells through degradation of the bacterial peptidoglycan. We have previously isolated and characterized the Ply endolysins from L. monocytogenes bacteriophages (22). Ply118 represents a 30.8-kDa enzyme from bacteriophage A118 which cleaves between the l-alanine and d-glutamate residues of the Listeria peptidoglycan, whereas the ply511 gene product encodes an N-acetylmuramoyl-l-alanine amidase with a molecular mass of 36.5 kDa. Both enzymes have a high substrate specificity and, with very few exceptions, exclusively lyse Listeria cells. Cloning and expression of ply118 has enabled several biotechnological applications, such as rapid lysis of Listeria cells from without (21) and programmed self-destruction of intracellular attenuated Listeria cells within the cytosol of macrophages (4).
The aim of this study was to introduce the endolysin-encoding genes from Listeria bacteriophages into lactococcal starter organisms in order to obtain organisms with biopreservation properties against L. monocytogenes. We report here the cloning and expression of the endolysin genes ply118 and ply511 in Lactococcus lactis under control of lactococcal promoters and describe the use of a Lactobacillus brevis signal peptide to obtain secretion of functional Ply511 from L. lactis cells.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli was grown in Luria-Bertani broth or in brain heart infusion broth at 37°C with shaking. L. lactis strains were grown in M17 medium (Merck, Darmstadt, Germany) supplemented with 0.5% glucose (GM17) or 0.5% lactose (LM17) at 30°C without shaking. L. monocytogenes was grown in tryptose broth at 30°C without shaking. The ability to ferment lactose was tested on bromocresol purple-lactose indicator agar (BAG, Lich, Germany). The following antibiotics were added as selective agents when appropriate: erythromycin (5 μg ml−1 [Lactococcus spp.], 150 μg ml−1 [E. coli]) or ampicillin (100 μg ml−1 [E. coli]).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strain | ||
| E. coli | ||
| XL1-Blue MRF′ | Cloning host: Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac [F′ proAB lacZqΔM15, Tn10(Tetr)] | Stratagene |
| DH5α MCR | Cloning host: F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 supE44 λ− thi-1 gyrA96 relA1 | Life Technologies |
| L. lactis | ||
| MG1363 | Plasmid-free derivative of SH4109, Lac− Prt− | 9 |
| Wg2 | Industrial starter strain | 28 |
| Bu2-129 | Lac+ Cit− | 26 |
| L. brevis DSM/20556 | Same as ATCC 8287 | DSM |
| L. monocytogenes WSLC 1001 | Same as ATCC 19112, serovar 1/2c | Weihenstephan Listeria Collection |
| Plasmid | ||
| pBluescript II SK(−) | Cloning and expression vector; AmprlacZ | Stratagene |
| pTRKH2 | Shuttle cloning vector; Emr, lac, gram-negative ori-p15A, gram-positive ori-pAMβ1, high copy number in Lactococcus spp. | 27 |
| pBPL118 | ply118 inserted into the EcoRI/SalI site of pBluescript II SK(−) | This study |
| pBPL118-P21 | Promoter P21 inserted into the XbaI/BamHI site of pBPL118 | This study |
| pBPL118-P32 | Promoter P32 inserted into the XbaI/BamHI site of pBPL118 | This study |
| pBPL118-P59 | Promoter P59 inserted into the XbaI/BamHI site of pBPL118 | This study |
| pLC-PL118-P21 | P21–ply118 cassette from pBPL118-P21 inserted into the XbaI/SalI site of pTRKH2 | This study |
| pLC-PL118-P32 | P32–ply118 cassette from pBPL118-P32 inserted into the XbaI/SalI site of pTRKH2 | This study |
| pLC-PL118-P59 | P59–ply118 cassette from pBPL118-P59 inserted into the XbaI/SalI site of pTRKH2 | This study |
| pLC-PL511 | ply511 inserted into the PstI/SalI site of pLC-PL118-P32, replacing ply118 | This study |
| pSL-ΔSPNuc | SPslpA-ΔSPnuc fusion inserted into PstI/SalI digested pLC-PL118-P32, replacing ply118 | This study |
| pSL-PL118 | SPslpA–ply118 fusion inserted into PstI/SalI digested pLC-PL118-P32, replacing ply118 | This study |
| pSL-PL511 | SPslpA–ply511 fusion inserted into PstI/SalI digested pLC-PL118-P32, replacing ply118 | This study |
| pSL-PL511ΔC | pSL-PL511 encoding C terminally truncated SPSlpA-Ply511Δ(S294–K373) | This study |
DNA manipulation.
Plasmid DNA was purified from E. coli using anion-exchange columns (Qiagen, Hilden, Germany). Plasmids of L. lactis were isolated in a similar way, except that degradation of the cell wall was carried out by prior addition of 30 mg of lysozyme per ml and incubation for 30 min at 37°C. Chromosomal DNA was isolated as described elsewhere (46). Restriction enzymes and other DNA-modifying enzymes from various sources were used according to the suppliers' recommendations. All relevant DNA sequences were verified by nucleotide sequencing on automated ABI 373A DNA sequencers (Perkin-Elmer Biosystems). The program DNAsis for Windows, version 2.10 (Hitachi), was used for analysis of nucleotide and amino acid sequences. E. coli and L. lactis strains were transformed by electroporation (5, 45) using a Gene-Pulser apparatus (Bio-Rad), cuvettes with an electrode gap of 2.0 mm, a single pulse of 12.5 kV cm−1, a capacity setting of 25 μF, and a 200-Ω resistance. Plasmid-containing clones were selected by the addition of antibiotics to growth media.
Cloning and expression of ply118 and ply511 genes in L. lactis.
We began by amplifying ply118 from purified DNA of L. monocytogenes phage A118 (22) by using PCR and the primers ply118-5′-ex and ply118-3′-ex (listed in Table 2). An artificial ribosome binding site (boldface) and spacer sequence (5′-GGAGGATTTAAAATG-3′) was added upstream of the ATG start codon (underlined) via the 5′ primer. The product was then digested with EcoRI and SalI and cloned into the EcoRI/SalI site of the pBluescript II SK(−) backbone (pBPL118) using E. coli XL1-Blue MRF′ as the host. In the next step, three different lactococcal promoters (41) were obtained by PCR amplification from purified chromosomal DNA of L. lactis Wg2 (28); the primers are shown in Table 2. The products were digested with XbaI and BamHI and cloned into the corresponding site of pBPL118, resulting in pBPL118-P21, pBPL118-P32, and pBPL118-P59.
TABLE 2.
Oligonucleotide primers used in this study
| Oligonucleotide | Sequence (5′→3′)a | Application |
|---|---|---|
| ply118-5′-ex | ATATGAATTCTGGAGGATTTAAATGACAAGTTATTATTATAGTAGAA | Amplification of ply118 for pLC-PL118 |
| ply118-3′-ex | AAGTGTCGACCTAAATCTTTTTAACAAACTTCGTGT | |
| ply511-5′-ex | ATATCTGCAGTGGAGGATTTAAAATGGTAAAATATACCGTAGAGAAC | Amplification of ply511 for pLC-PL511 |
| ply511-3′-ex | AAGTGTCGACTTATTTTTTGTAACTGCTCCTGTAC | |
| P21-5′ | ATATTCTAGACAAATCCCACACTCTATCGTCAAACAA | Amplification of promoter P21 |
| P21-3′ | AAGTGGATCCCTTAATCCTGCGCGTCTGCCAATTCCG | |
| P32-5′ | ATATTCTAGAATAGTTTTAGCTATTAATCTTTTTTTA | Amplification of promoter P32 |
| P32-3′ | AAGTGGATCCCCGAATATTTTTTTACCTACCTAGTAT | |
| P59-5′ | ATATTCTAGAAGAAAATACACTAACCAAAGAAGCGCG | Amplification of promoter P59 |
| P59-3′ | AAGTGGATCCCTTTATTCAGTTTTCAATGGTCTAGCT | |
| SPslpA-5′ | ATATCTGCAGTGGAGGATTTAAAATGCAATCAAGTTTAAAGAAATCTCTTTACTT | Amplification of the slpA signal sequence |
| SPslpA-3′ | AAGTGACGTCTTAGCTGAAGCAGTCGTTGAAAC | |
| ΔSPnuc-5′ | ATATGACGTCTGATCCAACAGTATATAGTGCAACTTCAACTA | Amplification of ΔSPnuc for pSLΔSP-Nuc |
| ΔSPnuc-3′ | AAGTGTCGACTTATTGACCTGAATCAGCGTTGTCTTC | |
| ply118-5′-sec | ATATGACGTCTTATTATTATAGTAGAAGTTTAGCGAATGTA | Amplification of ply118 for pSL-PL118 |
| ply511-5′-sec | ATATGACGTCTGTAAAATATACCGTAGAGAACAAAATTATTGCAGGA | Amplification of ply511 for pSL-PL511 |
Restriction sites are underlined; translation start and stop codons are printed in boldface.
To construct the endolysin expression vectors, the three individual promoter ply118 fragments were removed from pBPL118-P21, pBPL118-P32, and pBPL118-P59 by XbaI/SalI digestion and inserted into the E. coli-Lactococcus shuttle vector pTRKH2 (27) digested with XbaI/SalI. The resulting plasmids pLC-PL118-P21, pLC-PL118-P32, and pLC-PL118-P59 were initially propagated in E. coli, checked for the correct sequence, and transformed into L. lactis MG1363 (9). Vector pLC-PL511 was constructed by replacement of the PstI/SalI ply118 sequence in pLC-PL118-P32 with the corresponding ply511 gene amplified from phage A511 DNA (22), using the primers listed in Table 2.
Construction of the staphylococcal nuclease secretion probe vector.
The secretion probe vector is based on the signal peptide sequence of the L. brevis surface layer protein SlpA (43) and the nuc gene for Staphylococcus aureus nuclease (37), devoid of its export signal (ΔSPnuc). The SPslpA signal sequence (with PstI and AatII restriction sites added at the 5′ and 3′ ends) was amplified using the primers shown in Table 2, using chromosomal DNA of L. brevis as template. Vector pFUN (30) was used as a template for amplification of ΔSPnuc with primers ΔSPnuc-5′ and ΔSPnuc-3′ (Table 2), to which AatII and SalI restriction sites were added at the 5′ and 3′ ends, respectively. The two PCR fragments were digested with AatII and subsequently ligated with T4 DNA ligase (Roche), resulting in a 513-bp fragment with PstI and SalI recognition sites at the 5′ and 3′ ends, respectively. This fragment was then reamplified using primer pairs SPslpA-5′ and ΔSPnuc-3′. Finally, the complete SPslpA-ΔSPnuc cassette was digested with PstI and SalI and cloned into pLC-PL118-P32 digested with PstI and SalI, resulting in the secretion probe vector pSL-ΔSPNuc. E. coli DH5α was used as an intermediate recipient, and recombinant E. coli transformants were screened for nuclease activity on agar plates containing single-stranded DNA as a substrate and toluidine blue as an indicator dye, as described previously (15). A plasmid from a nuclease-positive E. coli clone was recovered and checked for correct sequence and then electroporated into L. lactis MG1363. The L. lactis(pSL-ΔSPNuc) transformants also secreted Nuc activity, and the identity of the recovered plasmid was checked again. E. coli DH5α(pFUN) and L. lactis MG1363(pFUN) were used as negative controls in this assay.
Construction of endolysin secretion vectors.
Plasmid pLC-PL118-P32 was used as the backbone for secretion vectors pSL-PL118 and pSL-PL511. In both vectors, the signal sequence of SPslpA was fused to ply118 and ply511. In order to obtain an in-frame fusion between the 3′ end of the SPslpA and the AatII site at the 5′ end of ply genes, primers ply118-5′-sec and ply118-3′-ex and primers ply511-5′-sec and ply511-3′-ex, respectively, were used for amplification (Table 2). The two endolysin genes and the SPslpA fragments were digested with AatII and, after ligation, yielded products of 967 bp (SPslpA–ply118) and 1,147 bp (SPslpA–ply511), respectively. These were then reamplified with the two primer pairs SPslpA-5′/ply118-3′-ex (generating SPslpA–ply118) and SPslpA-5′/ply511-3′-ex (SPslpA–ply511). The resulting in-frame genetic fusions were digested with PstI and SalI and inserted into PstI/SalI-digested pLC-PL118-P32, resulting in pSL-PL118 and pSL-PL511. These constructs were electroporated into L. lactis MG1363 and into the lactose-utilizing strain L. lactis Bu2-129 (26), which is suitable for fermentation of milk and production of cheese.
Photometric assay for endolysin activity.
Aliquots (50 ml) of overnight cultures of L. lactis MG1363 carrying either pLC-PL118-P32 or pLC-PL511 were harvested by centrifugation at 4°C and washed once with 10 ml of SM buffer (50 mM Tris HCl, 100 mM NaCl, 10 mM MgSO4; pH 7.5) (34). Cells were resuspended in 2 ml of SM buffer and disrupted by double passage through a French press at a 100-MPa pressure (SLM Aminco). Cellular debris was removed by centrifugation (15,000 × g, 4°C). The clear supernatants were sterile filtered (0.2-μm [pore-size] PES filter; Pall-Gelman Sciences), and the protein concentration was determined with a colorimetric protein assay (Nanoquant; Carl Roth, Karlsruhe, Germany), using bovine serum albumin as a standard. The cell extracts were stored at −20°C. For preparation of substrate cells, L. monocytogenes WSLC 1001 was grown overnight in tryptose broth in a volume of 500 ml and then harvested by centrifugation. Cells were washed once in SM buffer, resuspended in a 1/50 volume of buffer, and stored frozen in 1-ml portions. For quantitative determination of lysin activity, 900 μl of Listeria cells (diluted with SM to an optical density at 600 nm [OD600] of approximately 1.5) were mixed in a standard 1-cm cuvette with 100 μl of endolysin preparation, i.e., the cytoplasmic extract of recombinant lactococci. The decrease in OD was monitored over the following 20 min at room temperature (22 to 25°C). One unit of activity has been defined as the amount of endolysin necessary to decrease the OD600 by 0.01 per minute (22).
Endolysin activity plate test.
Screening for Ply-secreting L. lactis clones was carried out by plating transformants on GM17 agar containing sufficient L. monocytogenes cells to obtain a clearly visible turbidity of the medium. After incubation for 15 to 20 h at 30°C, clones secreting active endolysin were detected by the formation of a clear zone (halo) around the lactococcal colonies.
Immunological detection of Ply118 and Ply511.
In order to assay the production as well as the secretion of Ply118 and Ply511 from L. lactis cells, rabbit polyclonal antibodies were raised against purified Ply118 (21) according to a standard 70-day protocol. The rabbit serum contained a high titer of reactive antibodies and could directly be used for immunological detection of both endolysins, since the antibodies showed strong cross-reaction with Ply511. Cell extracts and supernatants of the different L. lactis recombinant clones were examined by Western blotting. For supernatant fractionation, 15-ml cultures were grown for 12 h, and cells were pelleted by centrifugation. The supernatants were carefully removed and sterile filtered, and proteins were concentrated by ultrafiltration (Fugisep-Maxi; cutoff, 10 kDa; Sevatec, Witten, Germany). Cell extracts and supernatant fractions were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and subsequently electroblotted onto polyvinylidene difluoride membranes (14). After blocking of the membranes in Tris-buffered saline (50 mM TrisCl, 150 mM NaCl; pH 7.5) containing 1% purified casein blocking reagent (chemiluminescent western-blotting kit; Roche), immunological detection was carried out according to the manufacturer's recommendations using anti-Ply118 (1:5,000 dilution), a secondary antibody (anti-rabbit immunoglobulin G conjugated to horseradish peroxidase), and luminol as a chemiluminescent peroxidase substrate.
RESULTS
Cloning and expression of functional phage endolysins in L. lactis.
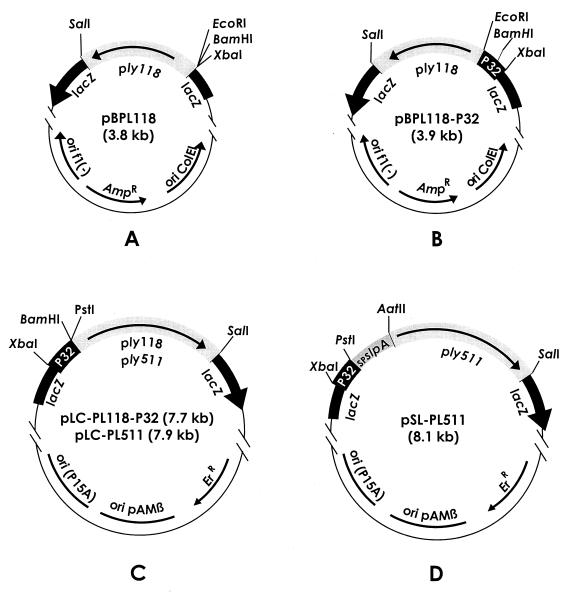
Our initial goal was the construction of an endolysin expression vector for L. lactis. We started by cloning an 871-bp ply118 fragment, equipped with a suitable ribosome-binding site, into pBluescript, yielding pBPL118 (Fig. 1A). For strong gene expression, different L. lactis promoters (P21, P32, and P59) were initially tested. They were introduced upstream of ply118, resulting in pBPL118-P21, pBPL118-P32 (Fig. 1B), and pBPL118-P59, respectively. The individual expression cassettes were then inserted into pTRKH2, a high-copy-number E. coli-Lactococcus shuttle vector. The resulting plasmids pLC-PL118-P21, pLC-PL118-P32 (Fig. 1C), and pLC-PL118-P59 were first established in E. coli before transformation into L. lactis. Expression of ply118 under control of the three individual promoters could then be tested and compared using lactococcal cell extracts for lysis of L. monocytogenes cell suspensions in a photometric assay. L. lactis MG1363(pLC-PL118-P32) cell extract contained the highest level of endolysin activity, whereas expression from P21 and P59 yielded significantly lower activity (data not shown). This indicated that P32 was the best-suited promoter for expression of ply in the lactococcal background and so it was used for all further plasmid constructs described here. Plasmid pLC-PL511 (Fig. 1C) was constructed by replacing ply118 in pLC-PL118-P32 with ply511.
FIG. 1.
Schematic illustration of the vectors used for construction (A and B), intracellular production (C), and secretion (D) of endolysin enzymes. Only the relevant coordinates and some important properties are shown; details are described in the text. Abbreviations: Ampr and Err, genes specifying resistance to ampicillin and erythromycin, respectively; P32, lactococcal promoter; SPslpA, signal sequence of L. brevis S-layer protein A; ply511 and ply118, endolysin genes from Listeria bacteriophages A511 and A118, respectively (22).
Production of Ply118 and Ply511 enzymes in L. lactis.
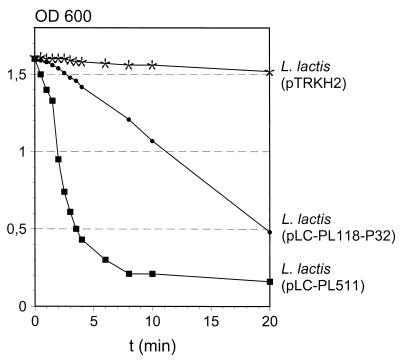
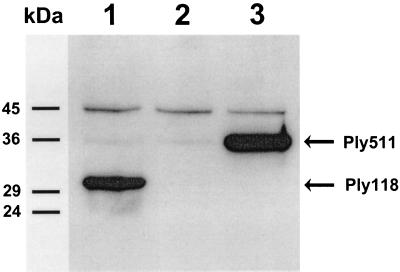
Expression of ply511 and ply118 in Lactococcus sp. and production of the corresponding gene products was analyzed by activity assay and by immunoblotting. The total protein contents of the extracts from recombinant cells were standardized to equal concentrations of 0.5 mg/ml. These preparations were then used in the photometric activity assays (Fig. 2). After a few minutes, the Listeria cell suspensions appeared almost clear. The extract from MG1363(pLC-PL511) showed a significantly stronger activity than did Ply118. Calculation of the enzyme activity revealed values of 60 U/ml for Ply118 and 180 U/ml for Ply511 in the standardized extracts. No lysis was seen with the control strain L. lactis (pTRKH2). It should also be noted that supernatants from these cultures contained no lytic activity (data not shown). For further analysis, the individual cell extracts were subjected to Western blotting. Because Ply118 and Ply511 show distinctive regions of amino acid sequence identity within the central to C-terminal polypeptide domains (22), anti-Ply118 showed strong cross-reaction with Ply511 and could therefore be used for the detection of both endolysins. Figure 3 shows that in the cytoplasmic extract of L. lactis pLC-PL118-P32 a single protein band of 30 kDa reacted with the antibody. A band of approximately 36 kDa was detected in the corresponding fraction of L. lactis (pLC-PL511). These results agree well with the predicted mass of Ply118 (30.8 kDa) and Ply511 (36.5 kDa). These findings showed that the Ply enzymes (i) are synthesized in L. lactis as active, full-length products, (ii) are not proteolytically degraded or otherwise inactivated in the lactococcal intracellular environment, and (iii) are not released or liberated from the cells under the culture conditions used here.
FIG. 2.
Decrease of the OD of a suspension of L. monocytogenes WSLC 1001 cells following the addition of cell extracts of L. lactis MG1363 carrying either pLC-PL118-P32, pLC-PL511, or the control vector pTRKH2 (see Materials and Methods).
FIG. 3.
Detection of recombinant Ply118 (30.8 kDa) and Ply511 (36.5 kDa), respectively, in the cytoplasmic fractions of overnight cultures of recombinant L. lactis MG1363 (indicated by arrows). Proteins from the cell extracts were separated by SDS-PAGE and detected by Western blotting with anti-Ply antibodies. Lane 1, MG1363(pLC-PL118-P32); lane 2, negative control MG1363(pTRKH2); lane 3, MG1363(pLC-PL511). The positions of molecular mass markers (in kilodaltons) are indicated on the left.
Staphylococcal nuclease as a reporter for SPSlpA-mediated secretion.
The S. aureus nuclease (SNase, Nuc) is a useful reporter for the protein secretion ability of gram-positive bacterial cells (30, 36). We have used a truncated Nuc protein (lacking its own signal peptide) as a reporter for SPSlpA-mediated secretion from L. lactis cells. For this purpose, the export signal peptide coding sequence of L. brevis S-layer protein A (SlpA) was genetically fused to the truncated Nuc protein. The pSL-ΔSPNuc vector was constructed by replacing ply118 in pLC-PL118-P32 with SPslpA-ΔSPnuc and transformed into L. lactis MG1363. Colonies that developed on erythromycin-containing media showed nuclease activity in the agar plate diffusion test (15) (results not shown). This confirmed that SPSlpA can be used for secretion of heterologous proteins in Lactococcus sp.
SPSlpA enables membrane translocation of active Ply511.
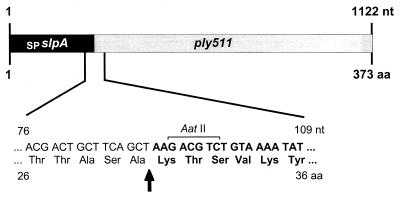
The ply118 and ply511 coding sequences, devoid of their own start codons, were fused in frame with the SPslpA sequence. The resulting SPslpA–ply118 and SPslpA–ply511 cassettes were cloned into the pTRKH2 backbone equipped with promoter P32, replacing ply118 in pLC-PL118-P32 (Fig. 1D). Figure 4 shows the genetic fusion of the signal sequence SPslpA and the endolysin gene ply511 and the corresponding amino acid sequence, including the protease cleavage site. After processing by a lactococcal signal peptide protease proximal to Lys-31, the amino-terminal (native) methionine of Ply is replaced by the addition of three residues (NH2-Lys-Thr-Ser-…). The two vectors were designated pSL-PL118 and pSL-PL511 (Table 1). However, all of the plasmids recovered from E. coli clones revealed more or less severe mutations within the ply gene cassettes and did not produce lytic activity (data not shown). Therefore, ligation reactions were directly transformed into MG1363 cells. Transformants were plated on GM17 erythromycin agar plates to which heat-inactivated L. monocytogenes cells were added at high density in order to assay for production and secretion of functional Ply118 and Ply511 enzymes from the developing colonies. Despite multiple attempts, however, we were unable to obtain transformants exporting active Ply118. Subsequent analysis of plasmids from several individual clones again revealed deletions and nucleotide substitutions in the SPslpA–ply118 sequence. In contrast, colonies of cells carrying pSL-PL511 formed clear zones on the turbid agar, indicating the production and secretion of functional, active Ply511 from the cells into the surrounding medium (Fig. 5). One specific clone exhibited unusually large halos around the colonies, i.e., the released lytic enzymes resulted in large, very distinctive clearing zones on the indicator medium. The corresponding plasmid pSL-PL511ΔC revealed a 115-bp deletion that occurred within the 3′-end coding sequence of SPslpA–ply511, which shifted the reading frame and generated a stop codon 12 bp downstream of the deletion site. Surprisingly, the resulting deletion of 80 amino acids from the Ply511 C terminus strongly increased the lytic activity. Prior to processing and secretion, the polypeptide represents an SPSlpA-Ply511Δ(S294–K373) mutant and, in the processed form, a Ply511Δ(S262–K341) mutant. This truncated endolysin was designated Ply511ΔC.
FIG. 4.
Schematic representation of the genetic fusion of the SPslpA signal sequence and ply511. The corresponding nucleotide sequence and amino acid sequence of the region joining both fragments in SPslpA–ply511 is shown enlarged. The arrow indicates the proposed signal peptide cleavage site of SlpA (43). The ply gene region is shown in boldface, and the restriction site used for genetic fusion (AatII) is also indicated.
FIG. 5.
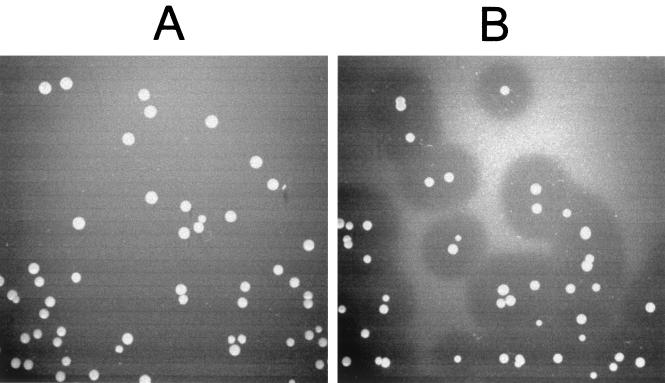
Colonies of recombinant L. lactis grown on GM17 agar medium containing suspended L. monocytogenes cells. The control strain MG1363(pLC-PL511) shows no lytic effect (A), whereas strain L. lactis MG1363(pSL-PL511ΔC) secreting the C terminally truncated Ply511 enzyme shows clear zones of lysis around the individual colonies (B).
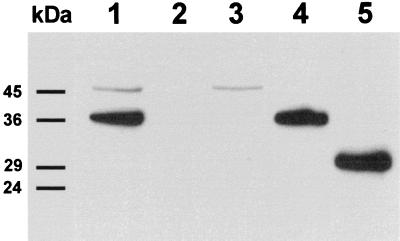
Probing the cell-free supernatants of L. lactis strains carrying pSL-PL511 or pSL-PL511ΔC with anti-Ply antibodies illustrated the quantitative secretion of the corresponding proteins from the cells (Fig. 6). The supernatant of a liquid culture of L. lactis carrying pSL-PL511 revealed one distinct protein band of the expected size (36.5 kDa) that was indistinguishable from the one observed in cell extract from L. lactis carrying pLC-PL511. This indicated that the SlpA signal peptide (30 amino acids, 2.9 kDa) must have been proteolytically removed during processing and secretion of the enzyme. The supernatant of L. lactis carrying pLC-PL511 revealed no signal, indicating that no unspecific release of the intracellular enzyme occurred. A smaller protein band (ca. 29 kDa) was detected in the supernatant of L. lactis carrying pSL-PL511ΔC, which corresponds well to the 28.5 kDa predicted for the truncated Ply511ΔC enzyme.
FIG. 6.
Detection of Ply in cell extracts and supernatants of recombinant L. lactis cultured for 12 h by immunoblotting with Ply-specific antibodies. Lanes 1 and 2, cell extract and supernatant fraction (respectively) of L. lactis MG1363(pLC-PL511); lanes 3 and 4, cell extract and supernatant fraction (respectively) of MG1363(pSL-PL511); lane 5, supernatant of MG1363(pSL-PL511ΔC). The positions of molecular mass markers (in kilodaltons) are indicated on the left.
Lactose utilization and endolysin secretion.
The ability to ferment lactose and produce lactic acid is the most prominent function of lactic acid bacteria used in the dairy industry. Because of this, we transformed pSL-PL511ΔC into L. lactis Bu2-129, a lactose-utilizing strain that can be employed for the fermentation of milk. As with the plasmid-free laboratory strain MG1363, colonies of the transformants gave rise to clearing zones on the turbid agar containing Listeria cells, thus indicating the secretion of functional Ply511. Also, lactose utilization did not seem to be negatively influenced by the endolysin expression plasmid; colonies of recombinant cells also showed yellow (acid) halos on bromocresol purple-lactose indicator agar which were indistinguishable from those observed with Bu2-129.
DISCUSSION
In this study we have demonstrated that the lytic enzyme- encoding genes ply118 and ply511 from Listeria bacteriophages can be cloned and expressed in L. lactis. Comparison of three different lactococcal promoters, P21, P32, and P59, indicated that the highest endolysin activity levels were obtained under the transcriptional control of P32, which normally drives expression of the gene for fructose 1,6-biphosphate aldolase in this organism (42). This observation is in contrast to the study of van der Vossen et al. (41), in which P59 showed the strongest expression levels. The latter promoter has also been employed for the production of other heterologous proteins in L. lactis: a B. subtilis protease (39), hen egg white lysozyme (40), and colicin V (24). In contrast to several other lactococcal promoters (13, 35, 44, 45), P32 is a constitutive promoter, with no need for specific induction of expression. Because of these advantageous properties, we have used P32 for all constructs.
Previous experiments employing the recombinant L. lactis BU2-129(pLC-PL118) as a protective measure against Listeria contamination and growth during the ripening process of artificially contaminated Camembert cheese (16) showed that the plasmid had no detrimental effect on growth of the cultures, viable cell counts, and acid production (i.e., the final pH of the cheese surface). However, it was also found that the slow, “natural” lysis of the lactococcal cells during stationary phase (see references 10 and 35) is insufficient to mediate efficient release of the “intracellular” endolysin onto the cheese surface (16). This finding is supported by our results (Fig. 6), in which no endolysin could be detected in the supernatant of cells expressing the native Ply511 protein. Thus, it was necessary to ensure more effective release of the lytic enzyme. Efficient membrane translocation could be achieved by construction of a secretion-competent fusion protein using the L. brevis slpA signal peptide. It should be noted that, in most cases, membrane passage of phage endolysins is dependent on the accumulation of holin proteins, which are thought to form pores in the bacterial cytoplasmic membrane and thereby allow release of the enzymes (22, 47). We have shown here that endolysins may also be exported with the aid of a signal peptide. However, with some putative exceptions (20), this situation has not yet been shown to naturally occur in phages, presumably due to the paramount importance of the independently expressed holins for lysis timing.
Although the layouts of the three gene fusions SPslpA-ΔSPnuc, SPslpA–ply118, and SPslpA–ply511 were identical, cells carrying pSL-PL118 were unable to secrete active endolysin. All of the SPslpA–ply118 transformants exhibited severe mutations within the signal sequence and/or the ply118 coding sequence. However, we have shown that cytoplasmic production of Ply118 without secretion is fully compatible with L. lactis, i.e., it did not result in growth impairment or plasmid modifications. These findings suggest that the deleterious event takes place during secretion of the enzyme, which involves membrane translocation and proteolytic processing to yield the active Ply118 enzyme. Although this l-alanoyl-d-glutamate peptidase does not visibly lyse L. lactis cells (22), there is a possibility that direct contact of Ply118 with the lactococcal murein during export and processing affects some function which is vital for growth and cell division. This hypothesis agrees well with our results that L. lactis carrying pLC-PL511 produces higher levels of lytic activity compared to cells carrying pLC-PL118-P32. This is in contrast to the production of these enzymes in E. coli, where Ply118 is synthesized at much higher levels (21, 22). The effect may be explained by the different enzymatic activities of Ply118 and Ply511 and supports our hypothesis that Ply118, when expressed from a constitutive promoter, impairs lactococcal viability. This problem may be circumvented by the use of an alternative promoter which can be specifically induced and allows lower expression levels (13, 35).
Introduction of pSL-PL511 into L. lactis resulted in strong production and quantitative secretion of Ply511 from the recombinant cells. The mutations that occurred in the SPslpA–ply511 cassette were mostly silent and did not result in amino acid changes or decreased activity. However, the truncated polypeptide specified by pSL-PL511ΔC exhibited strongly increased lytic activity. The 80-amino-acid deletion in Ply511ΔC [Ply511Δ(S262–K341)] corresponds to the C-terminal 24% of the native protein. The observation that C-terminal deletions can improve endolysin activity corresponds well to other results from our laboratory: In two endolysins from S. aureus phages, C-terminal deletions of up to 75% also strongly increased the lytic activities (18, 19). We have recently determined that the Ply118 and Ply511 enzymes show a modular design, in which the catalytic activity is located in the N-terminal region, whereas the C-terminal part harbors the cell wall binding domain (unpublished data). Although it is still unclear why the lytic activity is increased in the truncated proteins lacking part or most of their cell wall binding domains, our results suggest that it may be possible to further optimize the desired enzymatic properties through protein engineering.
Cloning of pSL-PL511ΔC into the lactose-metabolizing strain L. lactis Bu2-129 showed that (i) the cloning vector is compatible with the native plasmids of this organism and that (ii) nonlaboratory, wild-type strains can also produce and secrete the functional endolysins. For application in foods, however, genetically modified organisms should be devoid of markers such as antibiotic resistance. For this purpose, a number of “food-grade cloning” systems were developed, based on various selective markers such as nisin resistance (8), thymidylate synthetase (32), the lacF gene (23, 29), or nonsense suppressors of mutations in the lactococcal purine biosynthetic pathway (3). In order to prevent segregational instability of the plasmid, chromosomal integration of the modified ply gene or the entire vectors may be considered (17, 25, 31). Therefore, further research is planned in order to establish food-grade cloning of ply and to determine the inhibitory effect of the recombinant starter cultures on L. monocytogenes during the ripening process of contaminated soft cheese.
ACKNOWLEDGMENTS
We are grateful to Todd Klaenhammer for supplying vector pTRKH2, to Isabelle Poquet for providing vector pFUN, and to Nataša Vukov for valuable discussions.
REFERENCES
- 1.Cardinal M J, Meghrous J, Lacroix C, Simard R E. Isolation of Lactococcus lactis strains producing inhibitory activity against Listeria. Food Biotechnol. 1997;11:129–146. [Google Scholar]
- 2.Cintas L M, Rodríguez J M, Fernández M F, Sletten K, Nes I F, Hernández P E, Holo H. Isolation and characterization of pediocin L50, a new bacteriocin from Pediococcus acidilactici with a broad inhibitory spectrum. Appl Environ Microbiol. 1995;61:2643–2648. doi: 10.1128/aem.61.7.2643-2648.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dickely F, Nilsson D, Bech-Hansen E, Johansen E. Isolation of Lactococcus lactis nonsense suppressors and construction of a food-grade cloning vector. Mol Microbiol. 1995;15:839–847. doi: 10.1111/j.1365-2958.1995.tb02354.x. [DOI] [PubMed] [Google Scholar]
- 4.Dietrich A, Bubert A, Gentschev I, Sokolovic Z, Simm A, Catic A, Kaufmann S H E, Hess J, Szalay A A, Goebel W. Delivery of antigen-encoding plasmid DNA into the cytosol of macrophages by attenuated suicide Listeria monocytogenes. Nat Biotechnol. 1998;16:181–185. doi: 10.1038/nbt0298-181. [DOI] [PubMed] [Google Scholar]
- 5.Dower W J, Miller J F, Ragsdale C W. High-efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farber J M, Peterkin P I. Listeria monocytogenes, a food-borne pathogen. Microbiol Rev. 1991;55:476–511. doi: 10.1128/mr.55.3.476-511.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foegeding P M, Thomas A B, Pilkington D H, Klaenhammer T R. Enhanced control of Listeria monocytogenes by in situ-produced pediocin during dry fermented sausage production. Appl Environ Microbiol. 1992;58:884–890. doi: 10.1128/aem.58.3.884-890.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Froseth B R, McKay L L. Development and application of pFM011 as a possible food-grade cloning vector. J Dairy Sci. 1991;74:1445–1453. [Google Scholar]
- 9.Gasson M. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gasson M J, de Vos W M, editors. Genetics and biotechnology of lactic acid bacteria. London, United Kingdom: Chapman and Hall, Ltd.; 1994. [Google Scholar]
- 11.Holzapfel W H, Geisen R, Schilling U. Biological preservation of foods with references to protective cultures, bacteriocins and food-grade enzymes. Int J Food Microbiol. 1995;24:343–362. doi: 10.1016/0168-1605(94)00036-6. [DOI] [PubMed] [Google Scholar]
- 12.Johnson M E, Steele J L. Fermented dairy products. In: Doyle M P, Beuchat L R, Montville T J, editors. Food microbiology: fundamentals and frontiers. Washington, D.C.: ASM Press; 1997. pp. 581–594. [Google Scholar]
- 13.Kleerebezem M, Beerthuyzen M M, Vaughan E E, de Vos W M, Kuipers O P. Controlled gene expression systems for lactic acid bacteria: transferable nisin-inducible expression cassettes for Lactococcus, Leuconostoc, and Lactobacillus spp. Appl Environ Microbiol. 1997;63:4581–4584. doi: 10.1128/aem.63.11.4581-4584.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kyhse-Andersen J. Electroblotting of multiple gels: a simple apparatus without tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J Biochem Biophys Methods. 1984;10:203–209. doi: 10.1016/0165-022x(84)90040-x. [DOI] [PubMed] [Google Scholar]
- 15.Lachica R V F, Genigeorgis C, Hoeprich P D. Metachromatic agar-diffusion methods for detecting Staphylococcus nuclease activity. Appl Microbiol. 1971;21:585–587. doi: 10.1128/am.21.4.585-587.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lang-Halter E. Einfluss von Phagenlysin auf das Wachstum von Listeria durch rekombinante Starter- und Reifungskulturen auf der Oberfläche von Weichkäsen. Diploma thesis. Freising-Weihenstephan, Germany: Technical University of Munich; 1998. [Google Scholar]
- 17.Lillehaug D, Nes I F, Birkeland N-K. A highly efficient and stable system for site-specific integration of genes and plasmids into the phage φLC3 attachment site (attB) of the Lactococcus lactis chromosome. Gene. 1997;188:129–136. doi: 10.1016/s0378-1119(96)00798-6. [DOI] [PubMed] [Google Scholar]
- 18.Loessner M J, Gaeng S, Scherer S. Evidence for a holin-like protein gene fully embedded out-of-frame in the endolysin gene of Staphylococcus aureus bacteriophage 187. J Bacteriol. 1999;181:4452–4460. doi: 10.1128/jb.181.15.4452-4460.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loessner M J, Gaeng S, Wendlinger G, Maier S K, Scherer S. The two-component lysis system of Staphylococcus aureus phage Twort: a large TTG-start holin and an associated amidase endolysin. FEMS Microbiol Lett. 1998;162:265–274. doi: 10.1111/j.1574-6968.1998.tb13008.x. [DOI] [PubMed] [Google Scholar]
- 20.Loessner M J, Maier S K, Daubek-Puza H, Wendlinger G, Scherer S. Three Bacillus cereus bacteriophage endolysins are unrelated but reveal high homology to cell wall hydrolases from different bacilli. J Bacteriol. 1997;179:2845–2851. doi: 10.1128/jb.179.9.2845-2851.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loessner M J, Schneider A, Scherer S. Modified Listeria bacteriophage lysin genes (ply) allow efficient overexpression and one-step purification of biochemically active fusion proteins. Appl Environ Microbiol. 1996;62:3057–3060. doi: 10.1128/aem.62.8.3057-3060.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loessner M J, Wendlinger G, Scherer S. Heterogeneous endolysins in Listeria monocytogenes bacteriophages: a new class of enzymes and evidence for conserved holin genes within the siphoviral lysis cassettes. Mol Microbiol. 1995;16:1231–1241. doi: 10.1111/j.1365-2958.1995.tb02345.x. [DOI] [PubMed] [Google Scholar]
- 23.MacCormick C A, Griffin H G, Gasson M J. Construction of a food-grade host/vector system for Lactococcus lactis based on the lactose operon. FEMS Microbiol Lett. 1995;127:105–109. doi: 10.1111/j.1574-6968.1995.tb07457.x. [DOI] [PubMed] [Google Scholar]
- 24.McCormick J K, Klaenhammer T R, Stiles M E. Colicin V can be produced by lactic acid bacteria. Lett Appl Microbiol. 1999;29:37–41. doi: 10.1046/j.1365-2672.1999.00571.x. [DOI] [PubMed] [Google Scholar]
- 25.McIntyre L L, Harlander S K. Construction of first-generation lactococcal integrative cloning vectors. Appl Microbiol Biotechnol. 1993;40:348–355. doi: 10.1007/BF00170391. [DOI] [PubMed] [Google Scholar]
- 26.Neve H, Geis A, Teuber M. Conjugation, a common plasmid transfer mechanism in lactic acid bacteria streptococci of dairy starter cultures. Syst Appl Microbiol. 1987;9:151–157. [Google Scholar]
- 27.O'Sullivan D, Klaenhammer T R. High- and low-copy-number Lactococcus shuttle cloning vectors with features for clone screening. Gene. 1993;137:227–231. doi: 10.1016/0378-1119(93)90011-q. [DOI] [PubMed] [Google Scholar]
- 28.Otto R, de Vos W M, Gavrieli J. Plasmid DNA in Streptococcus cremoris Wg2: influence of pH on selection in chemostats of a varient lacking a protease plasmid. Appl Environ Microbiol. 1982;43:1272–1277. doi: 10.1128/aem.43.6.1272-1277.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Platteeuw C, van Alen-Boerrigter I, van Schalkwijk S, de Vos W M. Food-grade cloning and expression system for Lactococcus lactis. Appl Environ Microbiol. 1996;62:1008–1013. doi: 10.1128/aem.62.3.1008-1013.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poquet I, Ehrlich D, Gruss A. An export-specific reporter designed for gram-positive bacteria: application to Lactococcus lactis. J Bacteriol. 1998;180:1904–1912. doi: 10.1128/jb.180.7.1904-1912.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romero D A, Klaenhammer T R. IS946-mediated integration of heterologous DNA into the genome of Lactococcus lactis subsp. lactis. Appl Environ Microbiol. 1992;58:699–702. doi: 10.1128/aem.58.2.699-702.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross P, O'Gara F, Condon S. Thymilate synthase gene from Lactococcus lactis as a genetic marker: an alternative to antibiotic resistance genes. Appl Environ Microbiol. 1990;56:2164–2169. doi: 10.1128/aem.56.7.2164-2169.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryser E T, Marth E H, editors. Listeria, listeriosis, and food safety. New York, N.Y: Marcel Dekker; 1999. [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Sanders J W, Venema G, Kok J. A chloride-inducible gene expression cassette and its use in induced lysis of Lactococcus lactis. Appl Environ Microbiol. 1997;63:4877–4882. doi: 10.1128/aem.63.12.4877-4882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Savijoki K, Kahala M, Palva A. High level heterologous protein production in Lactococcus and Lactobacillus using a new secretion system based on the Lactobacillus brevis S-layer signals. Gene. 1997;186:255–262. doi: 10.1016/s0378-1119(96)00717-2. [DOI] [PubMed] [Google Scholar]
- 37.Shortle D. A genetic system for analysis of staphylococcal nuclease. Gene. 1983;22:181–189. doi: 10.1016/0378-1119(83)90102-6. [DOI] [PubMed] [Google Scholar]
- 38.Slutsker L, Schuchat A. Listeriosis in humans. In: Ryser E T, Marth E H, editors. Listeria, listeriosis, and food safety. New York, N.Y: Marcel Dekker; 1999. pp. 75–95. [Google Scholar]
- 39.van de Guchte M, Kodde J, van der Vossen J M B, Kok J, Venema G. Heterologous gene expression in Lactococcus lactis subsp. lactis: synthesis, secretion, and processing of the Bacillus subtilis neutral protease. Appl Environ Microbiol. 1990;56:2606–2611. doi: 10.1128/aem.56.9.2606-2611.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van de Guchte M, van der Vossen J M B, Kok J, Venema G. Construction of a lactococcal expression vector: expression of hen egg white lysozyme in Lactococcus lactis subsp. lactis. Appl Environ Microbiol. 1989;55:224–228. doi: 10.1128/aem.55.1.224-228.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Vossen J M B, van der Lelie D, Venema G. Isolation and characterization of Streptococcus cremoris Wg2-specific promoters. Appl Environ Microbiol. 1987;53:2452–2457. doi: 10.1128/aem.53.10.2452-2457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Venema G. Molecular biology and genetic modification of lactococci. J Dairy Sci. 1993;76:2133–2144. [Google Scholar]
- 43.Vidgrén G, Palva I, Pakkanen R, Lounatmaa K, Palva A. S-layer protein gene of Lactobacillus brevis: cloning by polymerase chain reaction and determination of the nucleotide sequence. J Bacteriol. 1992;174:7419–7427. doi: 10.1128/jb.174.22.7419-7427.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wegmann U, Klein J R, Drumm I, Kuipers O P, Henrich B. Introduction of peptidase genes from Lactobacillus delbrueckii subsp. lactis into Lactococcus lactis and controlled expression. Appl Environ Microbiol. 1999;65:4729–4733. doi: 10.1128/aem.65.11.4729-4733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wells J M, Wilson P W, Le Page R W F. Improved cloning vectors and transformation procedure for Lactococcus lactis. J Appl Bacteriol. 1993;74:629–636. doi: 10.1111/j.1365-2672.1993.tb05195.x. [DOI] [PubMed] [Google Scholar]
- 46.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Stuhl K, editors. Current protocols in molecular biology. I. New York, N.Y: John Wiley & Sons; 1997. pp. 2.4.1–2.4.5. [DOI] [PubMed] [Google Scholar]
- 47.Young R, Bläsi U. Holins: form and function in bacteriophage lysis. FEMS Microbiol Rev. 1995;17:191–205. doi: 10.1111/j.1574-6976.1995.tb00202.x. [DOI] [PubMed] [Google Scholar]