Abstract
We analyzed the impact of surfactant addition on hydrocarbon mineralization kinetics and the associated population shifts of hydrocarbon-degrading microorganisms in soil. A mixture of radiolabeled hexadecane and phenanthrene was added to batch soil vessels. Witconol SN70 (a nonionic, alcohol ethoxylate) was added in concentrations that bracketed the critical micelle concentration (CMC) in soil (CMC′) (determined to be 13 mg g−1). Addition of the surfactant at a concentration below the CMC′ (2 mg g−1) did not affect the mineralization rates of either hydrocarbon. However, when surfactant was added at a concentration approaching the CMC′ (10 mg g−1), hexadecane mineralization was delayed and phenanthrene mineralization was completely inhibited. Addition of surfactant at concentrations above the CMC′ (40 mg g−1) completely inhibited mineralization of both phenanthrene and hexadecane. Denaturing gradient gel electrophoresis of 16S rRNA gene segments showed that hydrocarbon amendment stimulated Rhodococcus and Nocardia populations that were displaced by Pseudomonas and Alcaligenes populations at elevated surfactant levels. Parallel cultivation studies revealed that the Rhodococcus population can utilize hexadecane and that the Pseudomonas and Alcaligenes populations can utilize both Witconol SN70 and hexadecane for growth. The results suggest that surfactant applications necessary to achieve the CMC alter the microbial populations responsible for hydrocarbon mineralization.
Surfactants have been successfully used to enhance the apparent solubility of nonpolar organic contaminants (NOC) as well as their subsequent removal from soil. There is, however, a great deal of conflicting information regarding the ability of surfactants to enhance the bioavailability and biodegradation of NOC (10, 21, 28). For most surfactants, an increase in NOC solubility is achieved only at surfactant concentrations greater than the critical micelle concentration (CMC), where the majority of surfactant molecules are aggregated into micelles. Surfactant micelles contain a hydrophobic core with a high affinity for NOC, resulting in increases in the apparent solubility of NOC. However, surfactant application at concentrations approaching and exceeding the CMC often result in significant decreases in rates of microbial NOC degradation. Potential mechanisms of inhibition include reduction in NOC bioavailability when bound in surfactant micelles (9, 13, 27), inhibition of microbial attachment at mineral and organic surfaces (4, 7, 23), and surfactant toxicity (3, 25, 26, 33). With a few exceptions, the majority of studies investigating the influence of surfactants on NOC biodegradation involve pure cultures. Furthermore, those using mixed cultures primarily address the impact of surfactant dose on the kinetics of NOC degradation. To our knowledge, there has been no attempt to evaluate changes in microbial community structure across a range of surfactant applications. Consequently, the goal of this study was to examine shifts of NOC-degrading populations as a function of surfactant application at concentrations below and above the CMC.
In a previous study, Macur and Inskeep (15) showed that degradation of phenanthrene and hexadecane by indigenous soil microorganisms was markedly inhibited at concentrations of Witconol SN70 (a nonionic, alcohol ethoxylate) above the CMC. Inhibition of phenanthrene and hexadecane degradation occurred despite a significant increase in NOC solubility at concentrations above the CMC. Although CO2 evolution data suggested that this was not caused by gross toxicity of the surfactant, it was suggested that specific inhibition of hydrocarbon-degrading populations could have occurred. Based on these prior results, we utilized a similar experimental design to reevaluate our observations on a microbial population level.
We hypothesized that surfactant applications spanning sub- and supra-CMC conditions would result in observable changes in the soil microbial community that might correlate with changes in NOC degradation rates. Experiments were conducted to evaluate microbial degradation rates of [14C]phenanthrene and [14C]hexadecane as a function of surfactant application in batch vessels containing soil. Over time, soil subsamples were subjected to DNA extraction, PCR amplification of a portion of the 16S rRNA gene, and separation of the PCR products using denaturing gradient gel electrophoresis (DGGE) (16). DGGE is a cultivation-independent technique that has been used previously to demonstrate changes in microbial communities in systems undergoing bioremediation (8, 20, 30). Since the phylogenetic information obtained from 16S rRNA genes gives little insight on the physiology of the microorganisms present, enrichment cultures were established to obtain isolates of hydrocarbon- and surfactant-degrading organisms. The 16S rRNA genes from the isolates were also analyzed using DGGE and compared to the molecular fingerprints of the soil treatments to assess whether we successfully isolated microorganisms that appeared to be relevant in the soil community and that were possibly responsible for in situ degradation of hydrocarbons and surfactant.
MATERIALS AND METHODS
Soil.
Mineralization experiments were conducted with the surface horizon of a Wheeling loam soil collected near Dover, Ohio. After air drying, the soil was passed through a 2-mm sieve and stored at 4°C. The soil contained 1.46% organic C (17), 0.2% total Kjeldahl N, 36.2 mg of extractable P kg−1 (18), 52% sand, 32% silt, and 16% clay. The 1:1 pH and gravimetric water content at 33 kPa were determined to be 8.3 and 20.9%, respectively (19). Plate counts on nutrient-rich agar (35) yielded 8.5 × 106 CFU g−1.
Chemicals.
Phenanthrene (aqueous solubility = 1.29 mg liter−1 [14]) and n-hexadecane (solubility = 3.6 μg liter−1) and their radiolabeled forms ([9-14C]phenanthrene and [1-14C]hexadecane) were obtained from Sigma Chemical (St. Louis, Mo.). The purity of both radiolabeled compounds was >98%, and the specific activities were 8.3 mCi mmol−1 for phenanthrene and 2.6 mCi mmol−1 for hexadecane. Witconol SN70, a linear alcohol ethoxylate {[CH3(CH2)x(OCH2CH2)yOH], where x is from 10 to 14 (primarily 10) and y is 5 (average molecular weight = 392 g mol−1; specific gravity = 0.98 g cm−3}, was obtained from Witco (Houston, Tex.). The CMC for Witconol SN70 was determined to be 75 mg liter−1 in deionized water and 237 mg liter−1 in soil solution equivalent (SSE) medium (2). For the purposes of this study the CMC in soil (CMC′) is defined as the surfactant concentration necessary to achieve an aqueous surfactant concentration equal to the CMC and accounts for surfactant sorption by the soil (the CMC′ was determined to be 13 mg g−1 in Wheeling loam soil) (15).
Hydrocarbon mineralization.
Experiments designed to measure degradation of the NOC were conducted in triplicate under unsaturated soil conditions in 30-ml glass bottles at 25 ± 2°C. Phenanthrene was dissolved in hexadecane and added to 5 g of air-dried Wheeling soil to yield a final concentration of 0.25 mg of phenanthrene g−1 and 15 mg of hexadecane g−1, with 10,000 dpm g−1 for [14C]phenanthrene or [14C]hexadecane. The solutions were added dropwise from a 20-μl syringe onto soil, with simultaneous mixing on a vortex shaker. Appropriate volumes of deionized water and concentrated solutions of Witconol SN70 were added to soil to achieve a gravimetric water content of 20% and final surfactant concentrations of 0, 2, 10, and 40 mg g of soil−1. Humidified CO2-free air was pumped through each vessel at a rate of approximately 2 ml min−1 and passed through 10 ml of 0.5 N NaOH to trap 14CO2. Base traps were exchanged every 1 to 4 days, depending on mineralization activity. One-milliliter aliquots of the trap solutions were added to 4 ml of ScintiSafe Plus 50% (Fisher Scientific, Fair Lawn, N.J.) and analyzed for 14CO2 with a Tricarb CA2200 scintillation analyzer (Packard Instruments, Meriden, Conn.). After the experiments were completed, 1-g soil subsamples were combusted in a biological oxidizer (model OX-300; R.J. Harvey Instrument, Hillsdale, N.J.) and the residual 14C was measured using scintillation analysis. Mass balance determinations based on total evolved 14CO2 and residual 14C resulted in average recoveries of radiolabeled phenanthrene and hexadecane of 89.3 and 85.2%, respectively.
Enrichment and isolation of hexadecane and surfactant degraders.
Wheeling loam soil (0.5 g) was added to a series of 250-ml Erlenmeyer flasks containing hexadecane and/or surfactant in 50 ml of SSE liquid medium. Four flasks contained 237 mg of hexadecane liter−1 plus 0, 23.7, 237, or 2,370 mg of surfactant liter−1 to bracket the CMC of Witconol SN70 (CMC in SSE medium = 237 mg liter−1). A fifth flask contained only 2,370 mg of surfactant liter−1. This enrichment series was duplicated, using soil taken from the high-surfactant-concentration treatments described above after a 60-day incubation. This resulted in a total of 10 enrichment flasks. Flasks containing hexadecane also received [14C]hexadecane (approximately 100,000 dpm) as a tracer. Evolution of 14CO2 in the enrichments containing hexadecane was monitored by frequent sampling of 0.5 N NaOH from wells suspended from rubber stoppers and scintillation analysis as described above. After 6 weeks, 10-fold serial dilutions were performed for each of the 10 flasks and 100-μl aliquots were spread onto two sets of SSE plates solidified with 1.5% Noble agar (Difco, Detroit, Mich.). One set of plates was supplemented with 2 g of Witconol SN70 liter−1, and the other set was sprayed with a hexadecane solution (2% in acetone) following inoculation. This resulted in the hexadecane being present as an opaque layer after acetone evaporation (11). Colonies that grew on either hexadecane or Witconol SN70 medium were picked and restreaked for isolation on 25% YEPG (0.05 g of yeast extract liter−1, 0.25 g of peptone liter−1, 0.5 g of glucose liter−1, and 0.05 g of NH4NO3 liter−1) plates. Surfactant-degrading isolates were also tested for their ability to grow on hexadecane spray plates. All isolates were regrown in 25% YEPG liquid medium and frozen at −80°C in a 50% glycerol solution until further use.
DNA extraction.
At various intervals, 0.5-g (dry wt) soil samples were removed from replicate batch mineralization treatments based on the 14CO2 evolution patterns. DNA was extracted with the FastDNA SPIN kit for soil (Bio 101, Vista, Calif.) per the manufacturer's instructions. Briefly, the soil samples were added to lysing reagents in a Multimix 2 tissue matrix tube that contained silica and ceramic beads of various sizes. The tubes were shaken at a speed of 6.5 m s−1 for 45 s in a Savant FastPrep instrument (Savant, Farmingdale, N.Y.) and then centrifuged. The supernatant was transferred to a tube containing a protein-precipitating solution and centrifuged. The supernatant was removed, combined with a DNA-binding matrix, and centrifuged through a spin filter. The filter was then washed with a salt-ethanol solution and centrifuged. After drying, the DNA on the spin filters was eluted in 100 μl of sterile water (Sigma), electrophoresed in a 1% SeaKem GTG agarose gel (FMC BioProducts, Rockland, Maine), and stained with ethidium bromide for quantification. DNA was extracted from hexadecane- and phenanthrene-degrading isolates by the same protocol, with the exception that four to five colonies were picked from a plate and subjected to the extraction procedure.
PCR amplification.
DNA extracts from soil samples and isolates were amplified using primer 1070f (Macromolecular Resources, Ft. Collins, Colo.), which targets the domain Bacteria (Escherichia coli positions 1055 to 1070), and primer 1392r-GC, which targets a universally conserved region (E. coli positions 1392 to 1406) and contains a 40-base GC clamp (5, 8). Reactions were carried out in 50-μl volumes containing 5 μl of template DNA (0.2 to 5 ng/μl), 50 mM KCl, 10 mM Tris HCl (pH 8.3), 1.5 mM MgCl2, 200 μM concentrations of deoxynucleoside triphosphates, a 0.5 μM concentration of each primer, and 1.25 U of Taq polymerase (FisherBiotech, Fair Lawn, N.J.). The polymerase was added after an initial 2-min denaturation step at 94°C, followed by 30 cycles of 94°C, 55°C, and 72°C for 45 s each, with a 7-min extension at 72°C in the final step. PCR products and a mass ladder (Gibco BRL, Grand Island, N.Y.) were visualized with a 3% NuSieve 3:1 agarose gel (FMC BioProducts) following ethidium bromide staining.
DGGE analysis.
The technique of Muyzer et al. (16) was used to separate and visualize PCR products from soil treatments and isolates. The specific methods used were as described in detail by Ferris et al. (5), with the following exceptions. Each lane of the gradient gel received approximately 120 ng of PCR product. Gels contained 8% acrylamide and were poured with a urea-formamide (UF) gradient of 40 to 60% or 35 to 65% and run at a constant voltage of 80 mV for 12 h. Gels were stained with SYBR Green II (Molecular Probes, Eugene, Oreg.) and photographed. Bands that appeared unique to a given treatment and bands that comigrated with those of isolates were stabbed with a sterile pipette tip that was placed in 20 μl of sterile water and rinsed repeatedly. This served as a template for further cycles of PCR-DGGE in order to obtain single bands of sufficient purity for sequencing. When two or more bands comigrated to a similar position on a gel, a 45 to 55% UF gradient was used to facilitate better separation.
DNA sequencing.
To obtain phylogenetic information on isolates and predominant bands, 16S ribosomal DNA (rDNA) PCR products were sequenced using an ABI Prism 310 (PE Applied Biosystems, Foster City, Calif.) and a BigDye Terminator Cycle Sequencing Ready Reaction kit. Bidirectional sequencing reactions were carried out as described by the manufacturer using primers 1114f (6) and 1392r (without the GC clamp). The sequences were aligned using the Sequencher 3.1.1 software program (Gene Codes Corporation, Ann Arbor, Mich.) and compared to sequences deposited in GenBank by performing a BLAST search (1).
RESULTS
Hydrocarbon mineralization.
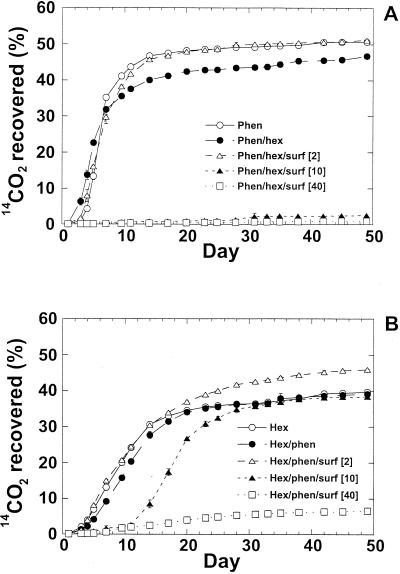
In the absence of Witconol SN70, mineralization of phenanthrene and hexadecane occurred rapidly in this soil, with the maximum rate of mineralization occurring around day 7 for most treatments (Fig. 1). Addition of 2 mg of surfactant g of soil−1 had little or no effect on the mineralization rate of either hydrocarbon. Increasing the surfactant concentration to 10 mg g of soil−1 (a concentration similar to the CMC′) completely inhibited the mineralization of phenanthrene. In contrast, although there was an extended lag period (11 days) prior to hexadecane mineralization, there were no differences in the extent or rate of hexadecane mineralization in the presence of 10 mg of surfactant g of soil−1. Surfactant applications above the CMC′ (40 mg g−1) completely inhibited the mineralization of both hydrocarbons.
FIG. 1.
Mineralization of radiolabeled phenanthrene (A) and hexadecane (B) in the presence and absence of surfactant. Each point is the mean of three replicate experiments; the error bars represent the standard errors. Abbreviations: Phen or phen, phenanthrene; Hex or hex, hexadecane; and surf, surfactant. The concentration of surfactant added (in milligrams per gram of soil) is indicated in brackets.
Molecular analysis of soil treatments.
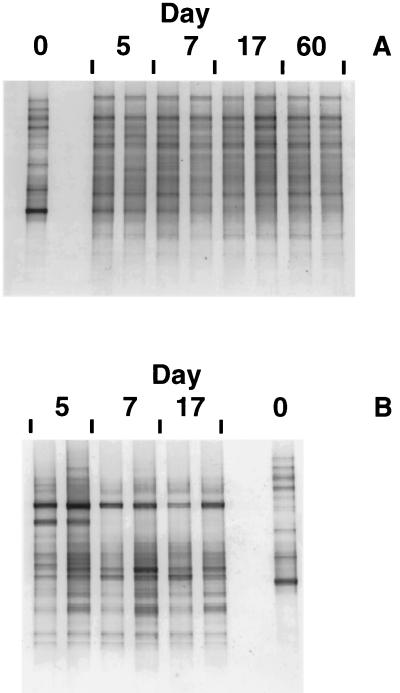
All DGGE profiles discussed in this study were obtained using replicate samples to demonstrate the reproducibility of the molecular techniques. For example, gels from uncontaminated control treatments displayed relatively stable and reproducible banding patterns throughout the 60-day experiment that differed somewhat from the patterns generated with stored soil (day 0) (Fig. 2A), probably reflecting effects of wetting and/or warming. Replicate gels from soil amended with 40 mg of Witconol SN70 g−1 also revealed reproducible differences in the DGGE community profile, including a few bands that disappeared between days 5 and 7 (Fig. 2B). Differences in prominent DGGE bands between control and surfactant treatments (40 mg g−1) suggested significant changes in microbial populations as a function of surfactant application.
FIG. 2.
Denaturing gradient gels (40 to 60% UF) of PCR-amplified 16S rDNA fragments from uncontaminated control soil (A) and soil amended with surfactant (40 mg/g) (B). The results of two independent replicate experiments are shown for each sampling to demonstrate the reproducibility of DGGE. Day 0 is the stored soil used in all experiments and is shown on both gels for reference.
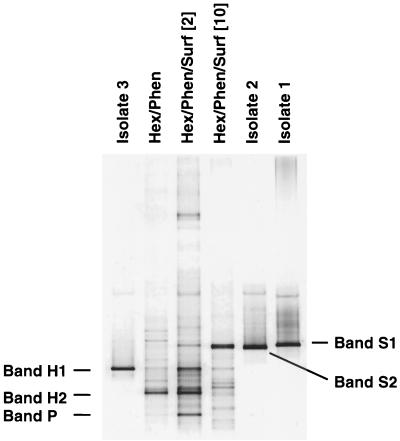
DGGE analysis of all surfactant, phenanthrene, and hexadecane treatments at day 7 revealed distinct banding patterns and trends associated with some specific treatments (Fig. 3). We did not observe the emergence of any prominent bands in response to the addition of phenanthrene alone, a result in contrast to those obtained with treatments containing hexadecane or surfactant. The treatment containing hexadecane alone resulted in increased intensity of one prominent band, H2. Two additional bands, H1 and P, emerged when hexadecane and phenanthrene were added together. Addition of surfactant at a concentration below the CMC′ (2 mg g−1) in the presence of both hydrocarbons resulted in further intensification of bands H1 and P, as well as the emergence of new bands S1 and S2. While S1 and S2 appear to constitute a single band, they are, in fact, two closely migrating bands that represent two very different sequences (see below). Surfactant additions at a concentration approaching the CMC′ (10 mg g−1) resulted in diminished intensity of bands P, H1, and H2 and an increased intensity of bands S1 and S2 prior to (day 7) and during (day 17) hexadecane mineralization. A further increase in surfactant concentration (40 mg g−1) enriched primarily bands S1 and S2, both of which were also pronounced in treatments that received only surfactant.
FIG. 3.
Denaturing gradient gel (40 to 60% UF) of PCR-amplified 16S rDNA fragments from day 7 of all soil treatments and from day 17 of the sample containing hexadecane, phenanthrene, and 10 mg of surfactant per g. Prominent bands associated with some amendments are labeled and described in the text. The abbreviations for the soil treatments are described in the legend for Fig. 1.
We sequenced the 16S rRNA gene fragments from the prominent bands revealed by DGGE analysis and conducted a BLAST search of the GenBank database to determine their phylogenetic type. The partial sequences obtained from surfactant-associated bands (S1 and S2) were found to be identical to 16S rRNA sequences of the gram-negative organisms Pseudomonas pavonaceae and Alcaligenes xylosoxidans (Table 1). In contrast, bands associated with hydrocarbon mineralization (H1, H2, and P) were found to be contributed by gram-positive organisms. The 16S rDNA sequences of bands H1 and H2 were identical to those found in Rhodococcus sp. strain Q15 and Nocardia uniformis, respectively. The band P sequence was 99.7% similar to the 16S rDNA sequence of another Rhodococcus sp. in the region analyzed.
TABLE 1.
Similarities between hexadecane- and surfactant-degrading isolates and their (respective) closest GenBank relatives
| Band | Isolatea | Closest GenBank relative | Percent similarityb | GenBank accession no. |
|---|---|---|---|---|
| S1 | 1 | Pseudomonas pavonaceae | 100 | D84019 |
| S2 | 2 | Alcaligenes xylosoxidans | 100 | D87096 |
| H1 | 3 | Rhodococcus sp. strain Q15 | 100 | AF046885 |
| H2 | None | Nocardia uniformis | 100 | Z46752 |
| P | None | Rhodococcus sp. | 99.7 | AJ007001 |
| 4 | Enterobacter nimipressuralis | 100 | Z96077 | |
| 5 | Bartonella vinsonii | 99 | Z31352 | |
| 6 | Methylobacterium sp. strain F73 | 100 | D32237 | |
| 7 | Xanthomonas campestris | 100 | X95917 | |
| 8 | Pseudomonas sp. strain BKME-9 | 100 | AJ011504 | |
| 9 | Pseudomonas sp. strain Dha51 | 100 | AJ011507 | |
| 10 | Ralstonia sp. strain CT14 | 99.7 | D88001 | |
| 11 | Pseudomonas putida strain MTB5 | 100 | AF131104 |
Isolates listed with a corresponding band are identical to that band in the region sequenced. Bands were prominent in DGGE analysis performed following various soil treatments.
Percent similarity between band and/or isolate and the closest GenBank relative.
Hexadecane- and surfactant-degrading isolates.
Following a 6-week enrichment in flasks containing varying hexadecane-to-surfactant ratios, approximately 60 isolates that could grow on SSE agar with hexadecane or surfactant as the sole C source were obtained. We focused our enrichments on isolating hexadecane- and surfactant-degrading microorganisms, since the treatments containing only hexadecane or surfactant resulted in stronger evidence of population selection (i.e., more obvious DGGE profile differences) than did the treatment with only phenanthrene (Fig. 3).
When the 16S rDNA fragments from the isolates were run on denaturing gradient gels, a number of the fragments comigrated with prominent bands present in the treatments (Fig. 4), whereas others did not appear to match any bands. Based on DNA sequence analysis of the 322-base region used for DGGE, the 60 isolates contained 11 unique sequences (Table 1). Comparison of these sequences revealed that three isolates had 100% 16S rDNA sequence identity with comigrating bands that were pronounced in several of the different soil amendments. A. xylosoxidans-like isolates corresponding to band S2 were recovered on plates containing either surfactant or hexadecane. P. pavonaceae-like isolates corresponding to band S1 were recovered only on plates with hexadecane as the C source but did have the ability to grow on plates containing Witconol SN70 as the sole C source. Rhodococcus sp. strain Q15-like isolates corresponding to band H1 were only obtained on plates with hexadecane as the sole C source. None of the other eight unique 16S rDNA sequences of isolates matched prominent bands present in the soil treatments.
FIG. 4.
Denaturing gradient gel (35 to 65% UF) of PCR-amplified 16S rDNA fragments from isolates 1, 2, and 3, which comigrate with prominent bands in selected soil treatments. The abbreviations for the soil treatments are explained in the legend for Fig. 1, and the isolates are described in Table 1.
DISCUSSION
Amending soil with hexadecane or Witconol SN70 resulted in obvious shifts in DGGE profiles, whereas the influence of phenanthrene was less pronounced (Fig. 3). Presumably, the differential effects of the amendments were at least partially due to the lower amount of C present as phenanthrene (0.24 mg g of soil−1) relative to hexadecane or surfactant C (from 1.29 to 25.74 mg g of soil−1). Particularly noteworthy was the prominence of a few bands that corresponded to hexadecane and surfactant amendments. DGGE bands H2 and, to a lesser extent, H1 were conspicuous in treatments exhibiting hexadecane mineralization in the absence of Witconol SN70 and when the surfactant was present at a low concentration (2 mg g of soil−1). This low-surfactant-concentration treatment is unique in that bands H1, H2, and P are intense, and bands S1 and S2 appear to be emerging. This potentially represents an environment where surfactant levels are sufficient to enrich for surfactant degraders but not high enough to inhibit hydrocarbon-degrading populations. At surfactant applications similar to or higher than CMC′ band intensity shifted from those bands associated with hydrocarbon mineralization to bands corresponding to surfactant addition, S1 and S2. This shift coincided with the inhibition of phenanthrene and hexadecane mineralization (Fig. 1).
Supporting our assertion that molecular analysis reveals important shifts in microbial populations is the fact that we obtained isolates capable of utilizing hexadecane and/or surfactant that corresponded to prominent bands in the DGGE profiles. For example, one hexadecane-utilizing Rhodococcus-like isolate (isolate 3) corresponded to band H1 (Fig. 4). It is notable that the closest relative to our isolate is a psychrotrophic n-alkane degrader cultivated from Lake Ontario, Rhodococcus sp. strain Q15 (31, 32). We also successfully obtained surfactant degraders (P. pavonaceae-like and A. xylosoxidans-like isolates) that corresponded to bands S1 and S2. In fact, it was not until we examined these isolates using DGGE that we realized S1 and S2 were two distinct bands; the subtle difference in band mobility is evident in Fig. 4, lanes 5 and 6, and was verified by using a 45 to 55% UF gradient as well. Although bands S1 and S2 were observed primarily in response to surfactant addition, corresponding isolates have the capacity to utilize both hexadecane and Witconol SN70 for growth. This potentially important finding would not have been revealed by kinetic data or molecular analysis alone. The fact that populations matching hexadecane or surfactant degraders whose DGGE bands were pronounced in these environments suggests that we are likely to have isolated relevant microorganisms that are responsible for hexadecane and surfactant degradation in situ.
We also obtained isolates that did not correspond to prominent bands in amended soil (Table 1). Three of the isolates (isolates 8, 9, and 11) obtained on hexadecane- and/or surfactant-amended plates but not observed in DGGE fingerprints of soil treatments only differed from the P. pavonaceae-like S1 band by one or two bases in the region analyzed. While this slight difference may seem negligible, ecologically distinct populations that differ by a single base, using the same 16S rDNA region analyzed as this study, have been detected in hot spring communities (6, 29) and phenanthrene enrichments (8). Based on the lack of corresponding bands in amended soils, the majority of the isolates, 8 out of 11, do not appear to be relevant hexadecane or surfactant degraders in situ and their relevance might be overestimated in the absence of any cultivation-independent analysis. These isolates were most likely obtained because our enrichment techniques favored their selection. Interestingly, all eight isolates are affiliated with the proteobacterial line of descent, many of whose members are noted for their ability to grow rapidly on diverse substrates.
We also observed some prominent bands for which we obtained no corresponding isolates. For example, the fact that we did not isolate a hexadecane-degrading microorganism that comigrated with the N. uniformis-like band (H2) tempers our initial inference, based solely on DGGE patterns, that band H2 represents a population with a significant role in hexadecane mineralization. However, it is possible that our enrichment strategy did not favor the selection of this population. While we did attempt to duplicate the soil amendment conditions by adding hexadecane alone and with Witconol SN70 at concentrations bracketing the CMC, our enrichments surely did not fully represent the niche diversity of the soil environment. Recent work by Friedrich et al. (8) showed that phenanthrene-degrading mycobacterial populations were enriched from two different soils when phenanthrene was presented on a sorbing phase that reduced its bioavailability. However, phenanthrene presented in the absence of a sorbing phase selected for Burkholderia-like populations. If the N. uniformis-like population is similarly adapted to low bioavailability environments like the mycobacterial populations, then the lack of a sorbing phase may have excluded its enrichment and isolation.
While we did observe surfactant-driven population shifts in this soil, the mechanism of inhibition of hexadecane and phenanthrene mineralization is still obscure. One commonly cited reason for inhibition is surfactant toxicity (3, 25, 26, 33). Witconol SN70, a nonionic, alcohol ethoxylate, was chosen both for its prior success in enhancing pyrene mineralization (24) and its biodegradability (12, 34). Macur and Inskeep (15) previously suggested that total CO2 evolution data did not indicate gross toxicity of Witconol SN70 to all organisms in this soil. However, there were clearly dramatic effects on the DGGE profiles when soils were amended with high concentrations of Witconol SN70 (Fig. 2 and 3). The disappearance of bands H1, H2, and P could be interpreted as evidence that this surfactant is toxic to a subset of hydrocarbon degraders in this soil; however, the absence of bands does not provide conclusive evidence of toxicity. The amendment obviously changed the environment to the advantage of surfactant-degrading populations S1 and S2, and this could have influenced PCR results. It is conceivable that the dominance of populations S1 and S2 might have biased PCR amplification, reducing the intensity of bands from other populations below detection limits. However, DGGE profiles, in combination with cultivation results, provide clear evidence that surfactant-degrading bacteria (S1 and S2 populations) were enriched upon Witconol SN70 amendment.
The DGGE profiles make us aware of the complex microbial population dynamics that might underlie the mineralization results we have observed. For instance, inhibition of hexadecane and phenanthrene metabolism by surfactant addition is consistent with selective toxicity, but inhibition could also involve resource competition between hydrocarbon- and surfactant-degrading populations (22). The kinetics of hexadecane mineralization in the 10-mg g−1 Witconol SN70 treatment (Fig. 1B) are interesting in this regard. While we observed an 11-day delay in hexadecane mineralization, DGGE results did not indicate that any new and prominent populations arose with the onset of hexadecane mineralization (compare profiles for days 7 and 17 in Fig. 3). Since surfactant-degrading isolates corresponding to bands S1 and S2 also degrade hexadecane, the delay in hydrocarbon mineralization could be a diauxic effect where Witconol SN70 is a preferred C source.
The mechanism of inhibition of hydrocarbon-degrading bacteria may also include physical changes in the soil microenvironment when Witconol SN70 was applied at concentrations similar to or higher than the CMC′. It has been argued that NOC solubilized into surfactant micelles may be unavailable to microorganisms (9, 13, 27). If hydrocarbon bioavailability was limiting in our system, the availability of micelle-bound phenanthrene should increase as the surfactant is degraded. However, phenanthrene mineralization was not observed in the presence of 10 or 40 mg of surfactant g−1 at any time throughout the incubation. One further explanation proposed by others is that surfactants inhibit bacterial attachment which may be important for the degradation of NOC (4, 7, 23). Macur and Inskeep (15) found that a surfactant application rate of 13 mg g−1 was necessary to achieve the effective CMC in this soil (CMC′), and that approximately 90% (11 mg g−1) of the surfactant is sorbed at this dosage. This surface coverage likely represents a significant perturbation of interfacial properties of colloidal surfaces, which may disrupt contact between cells and NOC. Because we used a complex soil system that mimics nature, it is possible that a number of these potential mechanisms contribute to the inhibition of NOC mineralization in the presence of Witconol SN70.
Molecular analysis revealed that surfactant addition indeed caused dramatic shifts in the microbial populations present in a hydrocarbon-amended soil. These shifts coincided with the inhibition of hydrocarbon mineralization, the apparent disappearance of select hydrocarbon-mineralizing populations and the emergence of populations capable of degrading both the surfactant and hexadecane. Although the mechanism of surfactant-induced inhibition of hydrocarbon mineralization is unclear, results presented here demonstrate the importance of linking microbial population dynamics with mineralization rate data in complex soil-contaminant systems. Specifically, interpretations regarding effects of surfactants on degradation of NOC in soils should not be based on the assumption that microbial populations remain constant over a range of surfactant concentrations.
ACKNOWLEDGMENTS
This work was supported by NSF as part of a Joint Program on Bioremediation (DEB-9729857), by a grant from the Great Plains Hazardous Substance Research Center at Kansas State University (94-9), and by the Montana Agricultural Experiment Station. We also acknowledge the Montana State University Thermal Biology Institute and NSF-EPSCoR for contributions to sequencing.
We thank Mary Bateson and Jacob Wheeler for technical assistance and three anonymous reviewers for their constructive comments during the review of the manuscript.
Footnotes
Journal Series number 2000-33 of the Montana Agricultural Experiment Station.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angle J S, McGrath S P, Chaney R L. New culture medium containing ionic concentrations of nutrients similar to concentrations found in the soil solution. Appl Environ Microbiol. 1991;57:3674–3676. doi: 10.1128/aem.57.12.3674-3676.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aronstein B N, Calvillo Y M, Alexander M. Effect of surfactants at low concentrations on the desorption and biodegradation of sorbed aromatic compounds in soil. Environ Sci Technol. 1991;25:1728–1731. [Google Scholar]
- 4.Efroymson R A, Alexander M. Biodegradation by an Arthrobacter species of hydrocarbons partitioned into an organic solvent. Appl Environ Microbiol. 1991;57:1441–1447. doi: 10.1128/aem.57.5.1441-1447.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferris M J, Muyzer G, Ward D M. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl Environ Microbiol. 1996;62:340–346. doi: 10.1128/aem.62.2.340-346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferris M, Ward D M. Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat community examined by denaturing gradient gel electrophoresis. Appl Environ Microbiol. 1997;63:1375–1381. doi: 10.1128/aem.63.4.1375-1381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foght J M, Gutnick D L, Westlake D W S. Effect of emulsan on biodegradation of crude oil by pure and mixed bacterial cultures. Appl Environ Microbiol. 1989;55:36–42. doi: 10.1128/aem.55.1.36-42.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedrich M, Grosser R G, Kern E A, Inskeep W P, Ward D M. Effect of model sorptive phases on phenanthrene biodegradation: molecular analysis of enrichments and isolates suggests selection based on bioavailability. Appl Environ Microbiol. 2000;66:2703–2710. doi: 10.1128/aem.66.7.2703-2710.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guha S, Jaffe P R. Biodegradation kinetics of phenanthrene partitioned into the micellar phase of nonionic surfactants. Environ Sci Technol. 1996;30:605–611. [Google Scholar]
- 10.Jordan R N, Cunningham A B. Surfactant-enhanced bioremediation: a review of the effects of surfactants on the bioavailability of hydrophobic organic chemicals in soils. In: Block J C, Baveye P, Goncharuk V V, editors. Bioavailability of xenobiotics in the environment and practical consequences for bioremediation. Dodrecht, The Netherlands: Kluwer; 1999. pp. 463–496. [Google Scholar]
- 11.Kiyohara H, Nagao K, Yana K. Rapid screen for bacteria degrading water-insoluble, solid hydrocarbons on agar plates. Appl Environ Microbiol. 1982;43:454–457. doi: 10.1128/aem.43.2.454-457.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kravetz L. Biodegradation of nonionic ethoxylates. J Am Oil Chem Soc. 1981;58:58A–65A. [Google Scholar]
- 13.Laha S, Luthy R G. Inhibition of phenanthrene mineralization by nonionic surfactants in soil-water systems. Environ Sci Technol. 1991;25:1920–1930. [Google Scholar]
- 14.Mackay D, Shiu W Y. Aqueous solubility of polynuclear aromatic hydrocarbons. J Chem Eng Data. 1977;22:399–402. [Google Scholar]
- 15.Macur R E, Inskeep W P. Effects of a nonionic surfactant on biodegradation of phenanthrene and hexadecane in soil. Environ Toxicol Chem. 1999;18:1927–1931. [Google Scholar]
- 16.Muyzer G, de Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes encoding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson D W, Sommers L E. Total carbon, organic carbon and organic matter. In: Page A L, editor. Methods of soil analysis. Part 2. Chemical and microbiological properties. Madison, Wis: American Society of Agronomy; 1982. pp. 539–579. [Google Scholar]
- 18.Olsen S R, Sommers L E. Phosphorus. In: Page A L, editor. Methods of soil analysis. Part 2. Chemical and microbiological properties. Madison, Wis: American Society of Agronomy; 1982. pp. 403–430. [Google Scholar]
- 19.Richards L A. Physical condition of water in soil. In: Black C A, editor. Methods of soil analysis. Part 1. Physical and mineralogical properties. Madison, Wis: American Society of Agronomy; 1965. pp. 128–152. [Google Scholar]
- 20.Rooney-Varga J N, Anderson R T, Fraga J L, Ringelberg D, Lovley D R. Microbial communities associated with anaerobic benzene degradation in a petroleum-contaminated aquifer. Appl Environ Microbiol. 1999;65:3056–3063. doi: 10.1128/aem.65.7.3056-3063.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rouse J D, Sabatini D A, Suflita J M, Harwell J H. Influence of surfactants on microbial degradation of organic compounds. Crit Rev Environ Sci Technol. 1994;24:325–370. [Google Scholar]
- 22.Steffensen W S, Alexander M. Role of competition for inorganic nutrients in the biodegradation of mixtures of substrates. Appl Environ Microbiol. 1995;61:2859–2862. doi: 10.1128/aem.61.8.2859-2862.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stelmack P L, Gray M R, Pickard M A. Bacterial adhesion to soil contaminants in the presence of surfactants. Appl Environ Microbiol. 1999;65:163–168. doi: 10.1128/aem.65.1.163-168.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thibault S L, Anderson M, Frankenberger W T., Jr Influence of surfactants on pyrene desorption and degradation in soils. Appl Environ Microbiol. 1996;62:283–287. doi: 10.1128/aem.62.1.283-287.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tiehm A. Degradation of polycyclic aromatic hydrocarbons in the presence of synthetic surfactants. Appl Environ Microbiol. 1994;60:258–263. doi: 10.1128/aem.60.1.258-263.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsomides H J, Hughes J B, Thomas J M, Ward C H. Effect of surfactant addition on phenanthrene biodegradation in sediments. Environ Toxicol Chem. 1995;14:953–959. [Google Scholar]
- 27.Volkering F, Breure A M, van Andel J G, Rulkens W H. Influence of nonionic surfactants on bioavailability and biodegradation of polycyclic aromatic hydrocarbons. Appl Environ Microbiol. 1995;61:1699–1705. doi: 10.1128/aem.61.5.1699-1705.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Volkering F, Breure A M, Rulkens W H. Microbiological aspects of surfactant use for biological soil remediation. Biodegradation. 1998;8:401–417. doi: 10.1023/a:1008291130109. [DOI] [PubMed] [Google Scholar]
- 29.Ward D M. A natural species concept for prokaryotes. Curr Opin Microbiol. 1998;1:271–277. doi: 10.1016/s1369-5274(98)80029-5. [DOI] [PubMed] [Google Scholar]
- 30.Watanabe, K., M. Teramoto, H. Futamata, and S. Harayama. Molecular detection, isolation, and physiological characterization of functionally dominant phenol-degrading bacteria in activated sludge. Appl. Environ. Microbiol. 64:4396–4402. [DOI] [PMC free article] [PubMed]
- 31.Whyte L G, Greer C W, Iniss W E. Assessment of the biodegradation potential of psychrotrophic microorganisms. Can J Microbiol. 1996;42:99–106. doi: 10.1139/m96-016. [DOI] [PubMed] [Google Scholar]
- 32.Whyte L G, Hawari J, Zhou E, Bourbonniere L, Iniss W E, Greer C W. Biodegradation of variable-chain-length alkanes at low temperatures by a psychrotrophic Rhodococcus sp. Appl Environ Microbiol. 1998;64:2578–2584. doi: 10.1128/aem.64.7.2578-2584.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willumsen P A, Karlson U. Effect of calcium on the surfactant tolerance of a fluoranthene degrading bacterium. Biodegradation. 1998;9:369–379. doi: 10.1023/a:1008357904624. [DOI] [PubMed] [Google Scholar]
- 34.Zhang C, Valsaraj K T, Constant W D, Roy D. Aerobic biodegradation kinetics of four anionic and nonionic surfactants at sub- and supra-critical micelle concentrations (CMCs) Wat Res. 1999;33:115–124. [Google Scholar]
- 35.Zuberer D A. Recovery and enumeration of viable bacteria. In: Weaver R W, editor. Methods of soil analysis. Part 2. Chemical and microbiological properties. Madison, Wis: Soil Science Society of America; 1994. pp. 119–144. [Google Scholar]