Abstract
Antimicrobial resistance (AMR) continues to spread at an alarming rate worldwide. Novel approaches are needed to mitigate its deleterious impact on antibiotic efficacy. Antibiotic stewardship aims to promote the appropriate use of antibiotics through evidence-based interventions. One paradigm is precision medicine, a medical model in which decisions, practices, interventions, and therapies are adapted to the individual patient based on their predicted response or risk of disease. Precision medicine approaches hold promise as a way to improve outcomes for patients with myriad illnesses, including infections such as bacteraemia and pneumonia. This review describes the latest advances in precision medicine as they pertain to antibiotic stewardship, with an emphasis on hospital-based antibiotic stewardship programmes. The impact of the COVID-19 pandemic on AMR and antibiotic stewardship, gaps in the scientific evidence, and areas for further research are also discussed.
Introduction
The overuse of antibiotics is one of the main contributors to the ongoing spread of antimicrobial resistance (AMR). The CDC estimates that about 30% of all antibiotics prescribed in US acute care hospitals are either unnecessary or suboptimal.1 The WHO estimates that only 50% of antibiotics are used correctly globally and that drug-resistant infections cause at least 700 000 deaths worldwide per year, a number that could rise to 10 million deaths by 2050 if no corrective measures are taken.2 Antibiotic stewardship focuses on reducing inappropriate prescribing of antibiotics while promoting their prudent use. Compelling evidence has emerged for the effectiveness of antibiotic stewardship programmes in reducing inappropriate antimicrobial use, hospital length of stay, healthcare costs, rates of resistance, and hospital-acquired infections.3
Despite these successes, ongoing challenges exist for hospital-based antibiotic stewardship programmes. Some of these include the conundrum of differentiating bacterial from viral infections, delays in pathogen diagnosis and antibiotic susceptibility due to technological limitations, and discerning infectious from non-infectious conditions. The COVID-19 pandemic has caused difficulties for appropriate antimicrobial use and stewardship, such as over-prescribing antibiotics to prevent bacterial coinfections.4 Indeed, approximately 75% of patients diagnosed with COVID-19 received antibiotics during the early stages of the pandemic.5 COVID-19 has led to increased hospitalizations, including mechanical ventilation and extended duration of stay, both of which increase the risk for acquiring AMR infections (Table 1).
Table 1.
Risk factors for AMR infections
| Risk factor |
|---|
| Recent or ongoing antibiotic use |
| Colonization with drug-resistant organism(s) |
| Increased hospital length of stay |
| Surgical procedures |
| Mechanical ventilation |
| Medical devices (urinary catheters, IV catheters, surgical drains) |
| High-risk sexual activity |
| Influenza |
| COVID-19 |
| Immunodeficiency (HIV/AIDS, cystic fibrosis) |
| Chronic kidney disease |
| Recent travel to or receipt of healthcare in a country with high endemicity of AMR |
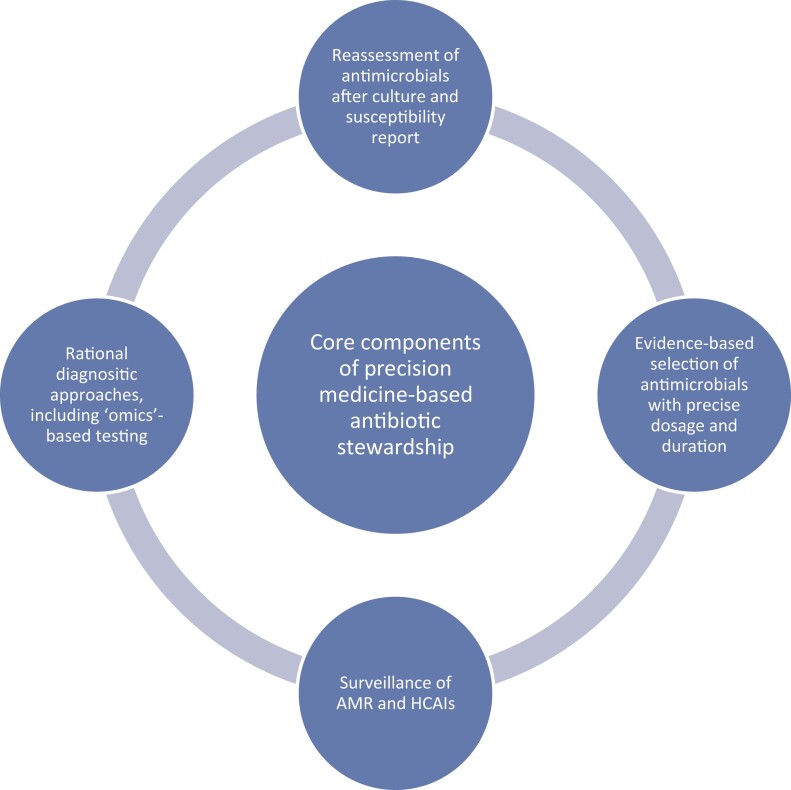
Precision medicine is one paradigm that might address some of the challenges currently facing antibiotic stewardship programmes. It is defined as ‘medical care designed to optimize efficiency or therapeutic benefit for particular groups of patients using genetic or molecular profiling’.6 More practically, precision medicine involves rapidly identifying altered biology within a patient and using the findings to guide therapy. Precision medicine was first developed in the field of oncology and has since moved into myriad areas in medicine.7 Many precision medicine modalities employ ‘omics’-based biomarkers such as proteomics, metabolomics and lipidomics to estimate disease prognosis and predict treatment response, like WGS and whole exome sequencing (WES). Furthermore, ‘omics’-based testing is predicted to evolve in complexity and scope so that clinicians will be able use a single ‘omics’ test to simultaneously inform the treatment pathway, therapy choice and disease prognosis.8 While the use of precision medicine in infectious diseases is at an early stage, it is also increasingly showing potential for improving clinical outcomes, such as in sepsis4 and viral pneumonia.9 For example, precision medicine approaches can help distinguish viral from bacterial pathogens in pneumonia, thus limiting the unnecessary use of antibacterial agents, a key goal in antibiotic stewardship.10 Figure 1 presents an overview of the key components of antibiotic stewardship using a precision medicine approach. The technology with precision medicine is more rapid than traditional laboratory methods such as cultures, which may reduce the time to diagnosis and lead to more appropriate antibiotic therapy.
Figure 1.
Core components of antimicrobial stewardship using a precision medicine approach. HCAIs, healthcare-associated infections.
This review presents a comprehensive update on the use of precision medicine in antibiotic stewardship. Studies in English on precision medicine in adult patients that pertain to antibiotic stewardship were identified using PubMed, with a focus on those published within the last 5 years. Potential barriers and challenges for implementing precision medicine approaches in antibiotic stewardship programmes are discussed, along with areas where further investigation is warranted.
Rapid diagnostic testing
Over half of the antibiotics given to hospitalized patients are for three conditions: lower respiratory tract infection [e.g. community-acquired pneumonia (CAP) or ventilator-associated pneumonia (VAP)], urinary tract infection (UTI) and soft tissue infection.11 Thus, improving the speed and accuracy of testing methods for these infections would be a considerable improvement leading to a multitude of beneficial downstream effects. Current methods for diagnosing pneumonia (e.g. routine microbiological cultures and multiplexed syndromic panels) have several limitations including difficulty distinguishing colonization from infection and identifying whether the causative pathogen is of viral or bacterial aetiology. One promising approach involves measuring peripheral blood host gene expression. Using a host gene expression-based assay that quantified the expression of 45 host messenger RNAs in patients with pneumonia, Ko et al.12 found the test had a sensitivity of 89.8% (95% CI, 77.8%–96.2%), a specificity of 82.1% (95% CI, 77.4%–86.6%) and a negative predictive value (NPV) of 97.9% (95% CI, 95.3%–99.1%) for bacterial infection. It also had an NPV of 98.9% (95% CI, 96.1%–100%) for bacterial infection when a viral aetiology was strongly suspected. The test correctly identified 30 of 33 participants (90.9%) with acute COVID-19 as having a viral infection. Notably, the results were available in less than an hour. These findings support a previous study that found host gene expression can distinguish between bacterial, viral and non-infectious illnesses in immunocompromised patients.13 However, one important caveat when evaluating tests that discriminate bacterial from viral infections is the absence of a diagnostic criterion standard, such as microbiological confirmation.
Biomarkers are key components of precision medicine. A biomarker is defined as ‘a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacological responses to a therapeutic intervention’.14 The most widely used biomarker to distinguish bacterial from viral pneumonia is procalcitonin (PCT). A peptide produced by the parafollicular cells of the thyroid, PCT is also released from adipocytes and neuroendocrine cells in the lungs and intestine as a result of the inflammatory cascade triggered during bacterial infection.9 Viral infections, in contrast, cause an increase in interferon γ, which inhibits production of PCT. Clinical studies on the use of PCT to distinguish viral from bacterial infection in pneumonia and to guide the use of antibiotic therapy have been mixed, with some showing it to be beneficial15,16 while others did not.17,18 A recent study found that PCT had a sensitivity for bacterial pneumonia of only 28.6% (95% CI, 16.2%–40.9%), while the specificity was 87.0% (95% CI, 82.7%–90.7%).12 The American Thoracic Society and IDSA CAP guidelines recommend that empirical antibiotic therapy should be initiated in adults with clinically suspected and radiographically confirmed CAP, regardless of the initial serum PCT level.19 The COVID-19 pandemic, which has resulted in a large increase in unnecessary antibiotic use, has led to a re-examination of PCT. A retrospective single-centre study found that most patients admitted with COVID-19 had low PCT (<0.25 ng/mL).20 Among the patients with low PCT, 57% had their antibiotics discontinued early (within 24 h) and their cultures remained negative for bacterial infection throughout their hospitalization. Furthermore, overall hospital length of stay was shorter in the low PCT group compared with the high PCT group (6 days versus 9 days, respectively). Another study on non-critically ill patients also showed a reduction in overall antibiotic use (mean duration 4.4 versus 5.4 days) when a PCT algorithm was followed.21 Reports on using PCT for intra-abdominal infections have also been mixed, although it appears to be most useful in diagnosing postoperative infections and necrotizing pancreatitis.22
The COVID-19 pandemic has brought renewed focus on the need for biomarkers besides PCT to curtail unnecessary antibiotic use, especially for pneumonia. VAPrapid2 was a multicentre, randomized, controlled trial (RCT) on ICU patients with suspected VAP that measured levels of IL-1β and IL-8 in bronchoalveolar lavage (BAL) fluid, with the aim of improving antibiotic stewardship.23 No significant difference in the primary outcome, distribution of antibiotic-free days, in the 7 days following BAL was found between the intervention and control groups. However, BAL is not a gold standard diagnostic test for VAP and clinician skepticism about the technique may have impacted the results. Another RCT compared daily monitoring of C-reactive protein (CRP) to best clinical practices in ICU patients for determining the need for antibiotics.24 The median duration of antibiotic treatment was 6.0 days for the CRP group and 7.0 days in the control group (P = 0.011). However, clinicians must be cautious when using CRP to evaluate for bacterial coinfection with COVID-19 after dexamethasone and tocilizumab treatment. This is due to the fact that CRP is suppressed by immunomodulatory agents.25 In a recent prospective trial from China, Xiao et al.26 measured levels of presepsin, the soluble form of CD14, in patients with sepsis to determine when it was safe to discontinue antibiotic therapy. Compared with standard of care, patients in the presepsin group had significantly more days without antibiotics [14.54 days (SD 9.01) compared with 11.01 days (SD 7.73), respectively; P < 0.001]. Mortality in the presepsin group showed no difference versus the control group at Days 28 (17.7% versus 18.2%; P = 0.868) and 90 (19.9% versus 19.5%; P = 0.891). Furthermore, patients in the presepsin group had a significantly shorter hospital length of stay and lower healthcare costs compared with the control group.
Besides biomarkers, another promising precision medicine approach to improve antibiotic stewardship is the use of phages to detect bacterial pathogens. A recombinant luciferase reporter bacteriophage has been designed that detects MRSA nasal colonization.27 The assay positively identified 97.7% of 390 clinical MRSA isolates at low bacterial concentrations, with a turnaround-time of 6 hours. Reporter phage-based detection assays have been developed for a number of other human pathogens including Escherichia coli, Listeria spp, Bacillus anthracis, and Mycobacterium spp.28 The major limitation of phage assays is the restricted host ranges of many phages that are intrinsically linked to phage resistance. While no reporter phage-based assay has yet been cleared for clinical use by the FDA, they may prove to be an inexpensive and rapid alternative to culture-based and molecular diagnostics. Further improvements in these assays are likely to be made through genetic engineering.
Differentiating UTIs from asymptomatic bacteriuria is challenging but has important implications for antibiotic stewardship. Indeed, the very definition of UTI is problematic and based on the antiquated notion that bacteria are not normal inhabitants of the urinary tract.29 In clinical practice, the ambiguity associated with UTIs means many patients with a range of non-specific symptoms (e.g. change in mental status, fever, weakness) that may or may not be accompanied by a change in the colour or appearance of their urine are prescribed antibiotics unnecessarily. The term ‘urinary tract dysbiosis’ has been proposed as an alternative to UTI.30 It better describes otherwise well patients with urinary tract symptoms and reflects current understanding of the urinary microbiome. This paradigm follows the principles of precision medicine more than the standard one (i.e. treating most urinary symptoms as UTI) by acknowledging that individuals have their own unique urinary microbiome and returning it to its normal state of equilibrium should be paramount. Thus, antibiotic stewardship has a crucial role to play in reducing overtreatment of bacteriuria. Saatchi et al.31 investigated the rates of recurrent UTI cases and prescriptions in the presence of antimicrobial stewardship efforts in British Colombia between 2008 and 2018. During the course of the study, UTI guidelines and community-based antimicrobial stewardship programmes were incorporated into evidence-based practice to help physicians prescribe antibiotics appropriately. Data for recurrent UTIs in adult women were collected from a provincial database, a physician billing system and a consolidation file to combine antibiotic prescribing, diagnoses and patient demographic information. Overall the prevalence of recurrent UTI cases declined by 59%, while recurrent UTI-associated antibiotic prescribing decreased by 73%. The greatest decrease was seen in quinolones (87%). Misclassification bias due to the inclusion of an ambiguous ICD-9 code, absence of laboratory data and lack of patient outcomes are important limitations to the study. Nonetheless, it seems reasonable to conclude that the reduction in recurrent UTIs and overall antibiotic prescribing was primarily due to antibiotic stewardship efforts. Another study found that an intervention as simple as an order set in the electronic medical record (EMR) that requires practitioners to choose an indication for the type of urine study can decrease the number of urine cultures performed and days of antibiotic therapy.32
Artificial intelligence and machine learning algorithms hold great promise for integrating precision medicine approaches with antibiotic stewardship. There are three main areas in which investigators are focused: (i) predicting AMR using genomic data; (ii) gaining insights into the cellular functions disrupted by antibiotics to understand mechanisms of action and develop novel antibiotics; and (iii) informing antimicrobial stewardship decisions using data extracted from the EMR.33 Although important advances have been made in using genomic data to predict AMR and the fact that genotypic methods are faster than phenotypic ones, significant limitations still exist. For example, Pseudomonas aeruginosa has phenotypic plasticity and complex AMR patterns that can obfuscate genotypic predictions.34 Shortcomings in the accuracy of culture-based antibiotic susceptibility testing can also derail genome-based predictions.35 The EMR is a rich source of data for machine learning models that aim to benefit antibiotic stewardship. Feretzakis et al.36 designed five machine learning algorithms to make antibiotic susceptibility predictions using patient demographic data, culture results and antibiotic susceptibility tests. The best model was capable of distinguishing between two classes of antibiotic susceptibility with an accuracy of 75.8% based on the type of the examined sample, the Gram stain classification of the pathogen and prior antibiotic susceptibility results. Another study sought to improve comparisons between hospitals for accurately identifying inpatient antimicrobial use.37 Two machine learning models were designed to analyse data from the EMR. They found the addition of more complex variables increased the accuracy of identifying inpatient antimicrobial use, with the largest improvements occurring as a result of adding diagnosis information. Machine learning algorithms have been applied to outpatient UTI prescribing. In a cohort of 3629 patients presenting between 2014 and 2016, the algorithm achieved a 67% reduction in the use of second-line antibiotics and reduced inappropriate antibiotic therapy by 18% relative to clinicians.38 Further randomized trials are necessary to evaluate the utility of machine learning algorithms in antibiotic stewardship, followed by implementation science to disseminate the research findings into clinical practice.
Antibiotic treatment using a precision medicine approach
Clinicians must take a number of factors into account when deciding on an appropriate antibiotic regimen. These include the source of infection, the most likely pathogens, patient comorbidities, allergy history, recent antibiotic use, concurrent medications for possible interactions, organ function (e.g. renal and hepatic), and local antimicrobial resistance patterns.6 The major classes of antibiotics with some commonly used examples in clinical practice are listed in Table 2. Unfortunately, AMR has been reported for every one of these agents. Using antibiotics appropriately is crucial for preserving their long-term effectiveness and preventing a new post-antibiotic era. Socioeconomic status and patient demographics can also impact antibiotic prescribing and further investigation into these factors is warranted.39
Table 2.
Major classes of antibiotics used in clinical practice
| Antibiotic class | Commonly used examplesa | Major resistance mechanisms |
|---|---|---|
| Penicillins | Amoxicillin, ampicillin, nafcillin | β-Lactamases, reduced affinity to PBPs |
| Monobactams | Aztreonam | β-Lactamases |
| Cephalosporins | Cefdinir, ceftriaxone, cefepime, ceftaroline, cefiderocol | Reduced affinity to PBPs |
| Tetracyclines | Doxycycline, omadacycline, minocycline | Ribosomal mutation, efflux pumps, enzymatic inactivation |
| Quinolones | Ciprofloxacin, levofloxacin, delafloxacin | Efflux pumps, mutations in DNA gyrase or topoisomerase IV, plasmid-mediated resistance genes |
| Lincosamides | Clindamycin | Methyltransferases, target mutation, efflux pumps |
| Glycopeptides | Vancomycin, dalbavancin | Alterations in terminal amino acid residues |
| Aminoglycosides | Amikacin, gentamicin, tobramycin, streptomycin | Efflux pumps, ribosomal mutations |
| Carbapenems | Imipenem, meropenem, ertapenem | Carbapenemases, efflux pumps, altered PBPs |
| Sulphonamides | Sulfamethoxazole, dapsone | Plasmid-associated resistance enzymes |
| Macrolides | Azithromycin, clarithromycin, fidaxomicin | Efflux pumps, ribosomal mutations |
| Oxazolidinones | Linezolid, tedizolid | Ribosomal mutations |
| Cyclic lipopeptides | Daptomycin | Decreased cell membrane permeability |
| Pleuromutilins | Lefamulin | Ribosomal mutations |
| Rifamycins | Rifampicin, rifabutin | Mutations in RNA polymerase |
| Polymyxins | Colistin, polymyxin B | Modifications of LPS that inhibit drug binding |
PBPs, penicillin-binding proteins; LPS, lipopolysaccharide.
Not a comprehensive listing of all agents in the class.
Optimizing antibiotic dosing for individual patients can not only lead to less toxicity but may also reduce the spread of AMR.40 Therapeutic drug monitoring (TDM) offers a way to use antibiotics more precisely with the goal of achieving pharmacokinetic/pharmacodynamic targets that are designed to suppress the emergence of resistance.41 Some of these include preventing the upregulation of efflux pumps and reducing the frequency of secondary mutational events. The application of pharmacokinetic/pharmacodynamic target-guided dosing with the support of dosing software has been shown to reduce nephrotoxicity, shorten the length of therapy and help improve TDM in patients dosed using either routine TDM or AUC-targeted approaches.42
Although broad spectrum antibiotics often have an important role early in the course of infection when the offending pathogen is not yet known, they also have detrimental effects on the host microbiome and create selection pressure for the development of more resistant bacteria. De-escalation of broad spectrum antibiotics to a narrower spectrum agent based on antibiotic susceptibility testing is a priority of antibiotic stewardship. An alternative to broad spectrum antibiotics is precision antimicrobials, which specifically inhibit a step in pathogenesis that disrupts maintenance and/or persistence of the pathogen in the host, or selectively kills the pathogenic organism with minimal off-target effects.43 There are at least 17 precision or narrow spectrum antibiotics already approved for use or in various stages of clinical development.44
Another exciting area for precision medicine is the possibility of using the clustered regularly interspersed short palindromic repeats-CRISPR associated protein (CRISPR-Cas) system to target specific bacterial pathogens. CRISPR-Cas is a bacterial adaptive immune system that uses DNA-encoded, RNA-mediated or DNA-targeting processes to counter the invasion of bacteria by employing foreign genetic material and mobile genetic elements, such as plasmids and phages.45 CRISPR-Cas systems can differentiate pathogenic from commensal bacteria and are potentially capable of selectively removing AMR genes from bacterial populations and bacterial virulence factors.46 They can also sensitize bacteria to an antibiotic by eliminating plasmids harbouring antibiotic resistance genes.47
Monitoring therapeutic response and conversion to oral therapy
The efficacy of serial PCT monitoring for antibiotic stewardship has been investigated in a number of studies.48–51 The main conclusions of these are that PCT monitoring decreases antimicrobial consumption, reduces length of stay, lowers hospitalization costs, causes no differences in infection relapse, re-hospitalization rate or mortality, and reduces the incidence of MDR organisms and invasive fungal infections. Optimizing the duration of therapy can be especially important because many studies show infections are often treated for longer than guidelines recommend and that each additional day of antibiotics increases the risk of patient harm.52 Converting antibiotics from an IV route of administration to oral has several advantages including lower costs, reduced hospital length of stay, fewer complications, and less inconvenience for patients.53 The following criteria can be used to decide if conversion to oral therapy is indicated: (i) susceptibility to an appropriate oral agent is demonstrated; (ii) the patient is hemodynamically stable; (iii) reasonable source control measures have occurred; and (iv) concerns about insufficient intestinal absorption are not present.54
Conclusions and future directions
Antibiotic stewardship is a crucial component in the overall effort for combating the spread of AMR. Precision medicine approaches appear to be effective at optimizing antibiotic stewardship practices. Some methods such as decision-making tools and algorithms are readily available for implementation, whereas others like ‘omics’ still require further development or validation. The majority of available data focuses on ICU patients in high-income countries, yet most antibiotic prescribing occurs in outpatient settings where data are sparse. The mismatch between data and clinical need is even more evident for disadvantaged populations and in resource-limited settings such as sub-Saharan Africa and Asia, where disparities in outcomes for infections and the burden of AMR are highest.55 However, improving outcomes in a narrow field is a reductive approach that ignores the complexity of how health conditions interact and co-occur. Future studies on the intersection of health equity, antibiotic stewardship and precision medicine are necessary so that data collection systems can be designed that will allow for accurate generalizations in diverse populations.
The impact of the COVID-19 pandemic on AMR and the emergence of new MDR organisms is complex and not yet fully elucidated. Bacterial coinfections and secondary infections following severe COVID-19 are important factors causing novel shifts in AMR patterns. Hopefully as the pandemic recedes, precision medicine approaches will increasingly demonstrate their value in improving antibiotic stewardship and reducing the global burden of AMR.
Transparency declarations
None to declare.
References
- 1. CDC . Core Elements of Hospital Antibiotic Stewardship Programs. US Department of Health and Human Services, 2019. https://www.cdc.gov/antibiotic-use/core-elements/hospital.html.
- 2. WHO . Tackling Antimicrobial Resistance. 2019. https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance.
- 3. Van Dort BA, Penm J, Ritchie A et al. The impact of digital interventions on antimicrobial stewardship in hospitals: a qualitative synthesis of systematic reviews. J Antimicrob Chemother 2022. 10.1093/jac/dkac112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Patel A. Tackling antimicrobial resistance in the shadow of COVID-19. mBio 2021; 12: e0047321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Langford BJ, So M, Raybardhan S et al. Antibiotic prescribing in patients with COVID-19: rapid review and meta-analysis. Clin Microbiol Infect 2021; 27: 520–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Watkins RR, Bonomo RA, Rello J. Managing sepsis in the era of precision medicine: challenges and opportunities. Expert Rev Anti Infect Ther 2022; 20: 871–880. [DOI] [PubMed] [Google Scholar]
- 7. Chen W, Anothaisintawee T, Butani D et al. Assessing the cost-effectiveness of precision medicine: protocol for a systematic review and meta-analysis. BMJ Open 2022; 12: e057537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yan S-K, Liu R-H, Jin H-Z et al. “Omics” in pharmaceutical research: overview, applications, challenges, and future perspectives. Chin J Nat Med 2015; 13: 3–21. [DOI] [PubMed] [Google Scholar]
- 9. Watkins RR. Using precision medicine for the diagnosis and treatment of viral pneumonia. Adv Ther 2022. 10.1007/s12325-022-02180-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bodkin N, Ross M, McClain MT et al. Systematic comparison of published host gene expression signatures for bacterial/viral discrimination. Genome Med 2022; 14: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Magill SS, Edwards JR, Beldavs ZG et al. Prevalence of antimicrobial use in US acute care hospitals, May-September 2011. JAMA 2014; 312: 1438–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ko ER, Henao R, Frankey K et al. Prospective validation of a rapid host gene expression test to discriminate bacterial from viral respiratory infection. JAMA Netw Open 2022; 5: e227299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mahle RE, Suchindran S, Henao R et al. Validation of a host gene expression test for bacterial/viral discrimination in immunocompromised hosts. Clin Infect Dis 2021; 73: 605–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rello J, van Engelen TSR, Alp E et al. Towards precision medicine in sepsis: a position paper from the European Society of Clinical Microbiology and Infectious Diseases. Clin Microbiol Infect 2018; 24: 1264–72. [DOI] [PubMed] [Google Scholar]
- 15. Self WH, Balk RA, Grijalva CG et al. Procalcitonin as a marker of etiology in adults hospitalized with community-acquired pneumonia. Clin Infect Dis 2017; 65: 183–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schuetz P, Wirz Y, Sager R et al. Procalcitonin to initiate or discontinue antibiotics in acute respiratory tract infections. Cochrane Database Syst Rev 2017; 10: CD007498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kamat IS, Ramachandran V, Eswaran H et al. Procalcitonin to distinguish viral from bacterial pneumonia: a systematic review and meta-analysis. Clin Infect Dis 2020; 70: 538–42. [DOI] [PubMed] [Google Scholar]
- 18. Huang DT, Yealy DM, Filbin MR et al. Procalcitonin-guided use of antibiotics for lower respiratory tract infection. N Engl J Med 2018; 379: 236–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Metlay JP, Waterer GW, Long AC et al. Diagnosis and treatment of adults with community-acquired pneumonia. an official clinical practice guideline of the American Thoracic Society and Infectious Diseases Society of America. Am J Respir Crit Care Med 2019; 200: e45–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roy A, Powers HR, Craver EC et al. Antibiotic stewardship: early discontinuation of antibiotics based on procalcitonin level in COVID-19 pneumonia. J Clin Pharm Ther 2022; 47: 243–47. [DOI] [PubMed] [Google Scholar]
- 21. Calderon M, Li A, Bazo-Alvarez JC et al. Evaluation of procalcitonin-guided antimicrobial stewardship in patients admitted to hospital with COVID-19 pneumonia. JAC Antimicrob Resist 2021; 3: dlab133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Watkins RR, Lemonovich TL. Serum procalcitonin in the diagnosis and management of intra-abdominal infections. Expert Rev Anti Infect Ther 2012; 10: 197–205. [DOI] [PubMed] [Google Scholar]
- 23. Hellyer TP, McAuley DF, Walsh TS et al. Biomarker-guided antibiotic stewardship in suspected ventilator-associated pneumonia (VAPrapid2): a randomised controlled trial and process evaluation. Lancet Respir Med 2020; 8: 182–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Borges I, Carneiro R, Bergo R et al. Duration of antibiotic therapy in critically ill patients: a randomized controlled trial of a clinical and C-reactive protein-based protocol versus an evidence-based best practice strategy without biomarkers. Crit Care 2020; 24: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kooistra EJ, van Berkel M, van Kempen NF et al. Dexamethasone and tocilizumab treatment considerably reduces the value of C-reactive protein and procalcitonin to detect secondary bacterial infections in COVID-19 patients. Crit Care 2021; 25: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xiao H, Wang G, Wang Y et al. Potential value of presepsin guidance in shortening antibiotic therapy in septic patients: a multicenter, prospective cohort trial. Shock 2022; 57: 63–71. [DOI] [PubMed] [Google Scholar]
- 27. Brown M, Hahn W, Bailey B et al. Development and evaluation of a sensitive bacteriophage-based MRSA diagnostic screen. Viruses 2020; 12: 631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meile S, Kilcher S, Loessner MJ et al. Reporter phage-based detection of bacterial pathogens: design guidelines and recent developments. Viruses 2020; 12: 944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Price TK, Hilt EE, Thomas-White K et al. The urobiome of continent adult women: a cross-sectional study. BJOG 2020; 127: 193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Finucane TE. ‘Urinary tract infection’ and the microbiome. Am J Med 2017; 130: e97–8. [DOI] [PubMed] [Google Scholar]
- 31. Saatchi A, Yoo JW, Marra F. Outpatient prescribing and prophylactic antibiotic use for recurrent urinary tract infections in British Columbia, Canada. Can Urol Assoc J 2021; 15: 397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Watson KJ, Trautner B, Russo H et al. Using clinical decision support to improve urine culture diagnostic stewardship, antimicrobial stewardship, and financial cost: a multicenter experience. Infect Control Hosp Epidemiol 2020; 41: 564–70. [DOI] [PubMed] [Google Scholar]
- 33. Anahtar MN, Yang JH, Kanjilal S. Applications of machine learning to the problem of antimicrobial resistance: an emerging model for translational research. J Clin Microbiol 2021; 59: e0126020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Khaledi A, Weimann A, Schniederjans M et al. Predicting antimicrobial resistance in Pseudomonas aeruginosa with machine learning-enabled molecular diagnostics. EMBO Mol Med 2020; 12: e10264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Su M, Satola SW, Read TD. Genome-based prediction of bacterial antibiotic resistance. J Clin Microbiol 2019; 57: e01405-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Feretzakis G, Loupelis E, Sakagianni A et al. Using machine learning algorithms to predict antimicrobial resistance and assist empirical treatment. Stud Health Technol Inform 2020; 272: 75–8. [DOI] [PubMed] [Google Scholar]
- 37. Moehring RW, Phelan M, Lofgren E et al. Development of a machine learning model using electronic health record data to identify antibiotic use among hospitalized patients. JAMA Netw Open 2021; 4: e213460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kanjilal S, Oberst M, Boominathan S et al. A decision algorithm to promote outpatient antimicrobial stewardship for uncomplicated urinary tract infection. Sci Transl Med 2020; 12: eaay5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Minejima E, Wang J, Boettcher S et al. Distance between home and the admitting hospital and its effect on survival of low socioeconomic status population with Staphylococcus aureus bacteremia. Public Health Rep 2022; 137: 110–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Baur D, Gladstone BP, Burkert F et al. Effect of antibiotic stewardship on the incidence of infection and colonisation with antibiotic-resistant bacteria and Clostridium difficile infection: a systematic review and meta-analysis. Lancet Infect Dis 2017; 17: 990–1001. [DOI] [PubMed] [Google Scholar]
- 41. Rawson TM, Wilson RC, O'’Hare D et al. Optimizing antimicrobial use: challenges, advances and opportunities. Nat Rev Microbiol 2021; 19: 747–58. [DOI] [PubMed] [Google Scholar]
- 42. Neely MN, Kato L, Youn G et al. Prospective trial on the use of trough concentration versus area under the curve to determine therapeutic vancomycin dosing. Antimicrob Agents Chemother 2018; 62: e02042-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Paharik AE, Schreiber HL 4th, Spaulding CN et al. Narrowing the spectrum: the new frontier of precision antimicrobials. Genome Med 2017; 9: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang B, Fang D, Lv Q et al. Targeted therapeutic strategies in the battle against pathogenic bacteria. Front Pharmacol 2021; 12: 673239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Duan C, Cao H, Zhang LH et al. Harnessing the CRISPR-Cas systems to combat antimicrobial resistance. Front Microbiol 2021; 12: 716064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Murugaiyan J, Kumar PA, Rao GS et al. Progress in alternative strategies to combat antimicrobial resistance: focus on antibiotics. Antibiotics (Basel) 2022; 11: 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Araya D P, Palmer KL, Duerkop BA. CRISPR-based antimicrobials to obstruct antibiotic-resistant and pathogenic bacteria. PLoS Pathog 2021; 17: e1009672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gluck E, Nguyen HB, Yalamanchili K et al. Real-world use of procalcitonin and other biomarkers among sepsis hospitalizations in the United States: a retrospective, observational study. PLoS One 2018; 13: e0205924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Broyles MR. Impact of procalcitonin-guided antibiotic management on antibiotic exposure and outcomes: real-world evidence. Open Forum Infect Dis 2017; 4: ofx213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Langford BJ, Beriault D, Schwartz KL et al. A real-world assessment of procalcitonin combined with antimicrobial stewardship in a community ICU. J Crit Care 2020; 57: 130–33. [DOI] [PubMed] [Google Scholar]
- 51. Sathitakorn O, Jantarathaneewat K, Weber DJ et al. The feasibility of procalcitonin and CPIS score to reduce inappropriate antibiotics use among severe-critically ill COVID-19 pneumonia patients: a pilot study. Am J Infect Control 2022; 50: 581–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Branch-Elliman W, O’Brien W, Strymish J et al. Association of duration and type of surgical prophylaxis with antimicrobial-associated adverse events. JAMA Surg 2019; 154: 590–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shrayteh ZM, Rahal MK, Malaeb DN. Practice of switch from intravenous to oral antibiotics. Springerplus 2014; 3: 717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Heil EL, Bork JT, Abbo LM et al. Optimizing the management of uncomplicated Gram-negative bloodstream infections: consensus guidance using a modified Delphi process. Open Forum Infect Dis 2021; 8: ofab434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bebell LM, Muiru AN. Antibiotic use and emerging resistance: how can resource-limited countries turn the tide? Glob Heart 2014; 9: 347–58. [DOI] [PMC free article] [PubMed] [Google Scholar]