Abstract
Many studies have highlighted the complex and diverse basis for heterosis in inbred crops. Despite the lack of a consensus model, it is vital that we turn our attention to understanding heterosis in undomesticated, heterozygous, and polyploid species, such as willow (Salix spp.). Shrub willow is a dedicated energy crop bred to be fast-growing and high yielding on marginal land without competing with food crops. A trend in willow breeding is the consistent pattern of heterosis in triploids produced from crosses between diploid and tetraploid species. Here, we test whether differentially expressed genes are associated with heterosis in triploid families derived from diploid Salix purpurea, diploid Salix viminalis, and tetraploid Salix miyabeana parents. Three biological replicates of shoot tips from all family progeny and parents were collected after 12 weeks in the greenhouse and RNA extracted for RNA-Seq analysis. This study provides evidence that nonadditive patterns of gene expression are correlated with nonadditive phenotypic expression in interspecific triploid hybrids of willow. Expression-level dominance was most correlated with heterosis for biomass yield traits and was highly enriched for processes involved in starch and sucrose metabolism. In addition, there was a global dosage effect of parent alleles in triploid hybrids, with expression proportional to copy number variation. Importantly, differentially expressed genes between family parents were most predictive of heterosis for both field and greenhouse collected traits. Altogether, these data will be used to progress models of heterosis to complement the growing genomic resources available for the improvement of heterozygous perennial bioenergy crops.
Keywords: allele-specific expression, copy number variation, differential gene expression, heterosis, hybrid vigor, polyploidy, regulatory divergence, ridge regression
Introduction
Numerous studies on heterosis have focused on hybrids derived from crossing inbred parents (Guo et al. 2004, 2006), few on those derived from outcrossing parents (Landry et al. 2005; Zhuang and Adams 2007), and even fewer on hybrids derived from outcrossing parents of different species or ploidy (Wittkopp and Kalay 2011). From early expression studies based on only a few dozen genes to recent research employing RNA-Seq, a common result in maize, wheat, and rice is that additive gene expression in hybrids makes up the greatest proportion of those differentially expressed between the parents (Guo et al. 2006; Stupar and Springer 2006; Stupar et al. 2008; Wei et al. 2009), yet genes with nonadditive expression display allele-specific expression (ASE) (Guo et al. 2004; Springer and Stupar 2007; Wei et al. 2009). This differential expression could be due to the presence of remote trans-factors, whereby a small number of key regulatory genes could play significant roles in heterosis (Ni et al. 2009; He et al. 2010; Goff 2011).
The heritability of gene expression has been attributed to both local cis-regulatory elements and distant trans-regulatory factors in the cell. Variation in these gene regulators can play dramatic roles in the evolution of gene expression. Cis-regulatory variation is thought to account for evolutionarily significant phenotypic differences, whereas trans-regulatory variation is thought to account more for adaptive differences (Wray 2007). For instance, cis-regulatory variation in promoter regions within-species should be minimal, compared to that among species. So, it is more likely that trans-effects should account for most of the regulatory variation in the intraspecific hybrid, and cis-effects in the interspecific hybrid (Wittkopp et al. 2008a). More simply, the greater phylogenetic distance between parents, the more likely it is that differential gene expression in the progeny will be due to gene localized polymorphism. However, it is uncertain whether nonadditive gene expression and regulatory divergence are more commonly observed in plants exhibiting hybrid vigor (heterosis).
A major ongoing topic in heterosis research is to what extent is nonadditive gene expression correlated with nonadditive phenotypic expression (Birchler et al. 2005), and how this informs combining ability or response to hybridization in the F1. In most crop plants, heterosis has been observed in hybrids bred from inbred parents of contrasting genetic backgrounds (East 1936; Birchler et al. 2003). In maize, high numbers of low-frequency alleles near conserved cis-regulatory regions in the genome have been thought to lead to gene misexpression and are implicated as having a deleterious impact on important component traits (Kremling et al. 2018). Dominance may help explain the phenomenon of heterosis in maize (McMullen et al. 2009), and there are efforts to purge these deleterious alleles from breeding material via targeted gene-editing technologies. Answers to questions regarding the genomic basis of heterosis are not only relevant in breeding and selection, but contribute to our understanding of the evolution of dioecious plant species that regularly undergo interspecific hybridization and polyploidization events.
Willow (Salix spp.) is exceptionally diverse, with over 350 species characterized across most of the temperate range, and ploidy levels ranging from diploid to dodecaploid (Kuzovkina et al. 2008), so the genomic basis of heterosis is likely to be different from that of conventional crop plants. Interspecific hybridization has been a key component in shrub willow improvement, as F1 hybrids often display heterosis for biomass yield (Kopp et al. 2001; Cameron et al. 2008; Serapiglia et al. 2014a; Fabio et al. 2016), especially those derived from diploid and tetraploid parents (Smart and Cameron 2008; Serapiglia et al. 2014b; Carlson and Smart 2016). What is promising for the biomass production industry, is that these high-yielding triploids outperform foundational commercial cultivars for dry weight biomass yield and other biomass-related morphological and physiological traits (Fabio et al. 2017). While there is good evidence of heterosis in triploid hybrids of willow (Serapiglia et al. 2014a; Carlson and Smart 2021), the genomic basis of this phenomenon is not well-characterized.
To support breeding efforts, an Illumina-based reference genome assembly of female S. purpurea 94006 was constructed using a F2 map-guided approach to orient scaffolds into pseudomolecules (Salix purpurea v1.0, DOE-JGI, https://phytozome-next.jgi.doe.gov/, last accessed 12/30/2021). Recently, sex-specific long-read genome assemblies of S. purpurea have been completed (S.purpurea v5.1, S. purpurea Fish Creek’ v.3.1, DOE-JGI, https://phytozome-next.jgi.doe.gov/, last accessed 12/30/2021). The genome size is an estimated 330 Mb and contains approximately 35,125 protein-coding genes (57,462 transcripts), and has proven useful in read alignment, variant discovery, and candidate gene selection (Hyden et al. 2021). There have been a handful of studies in shrub willow that focused on genetic mapping (Gunter et al. 2003; Berlin et al. 2010; Hanley and Karp 2014; Hällingbäck et al. 2016; Zhou et al. 2018; Carlson et al. 2019) of quantitative trait loci (QTL) associated with biomass yield traits to aid in marker-assisted selection (MAS), but most have been low-resolution. There have also been attempts at correlating cell wall biosynthesis genes with variation in biomass composition in Salix spp. (Serapiglia et al. 2012), as well as correlating sex dimorphism (Gouker et al. 2021) with gene expression and methylation patterns in F2S. purpurea (Hyden et al. 2021). Thus far, family-based ASE in Salix is restricted to a single study of F1 and F2 intraspecific S. purpurea (Carlson et al. 2017), where expression-level dominance comprised the greatest proportion of differentially expressed genes between the parents of both families. Overall, there were more genes with ASE in the F1 compared to F2, but both families displayed greater levels of cis- than trans-regulatory divergent expression patterns. In high-yielding, triploid hybrids of bioenergy willow, the heritability of gene expression and its broad influence on modulating heterosis for biomass yield and other traits important for biomass production has not been characterized.
Using willow as a model for understanding heterosis in heterozygous polyploid perennials, the objectives of this study were to: (1) describe the inheritance and regulatory divergence patterns influencing gene expression within and among 3 interspecific hybrid triploid families, (2) test for dosage effects on parent alleles in triploid progeny, and (3) determine which genes and gene sets are most predictive of heterosis for biomass growth and wood chemical composition traits important for bioenergy production.
Materials and methods
Plant material and growing conditions
Progeny individuals from 3 full-sib F1 triploid families included in this study were derived from the interspecific crosses: S. purpurea 94006 × S. miyabeana 01-200-003 (Family 415), S. viminalis 07-MGB-5027 × S. miyabeana 01-200-003 (Family 423), and S. miyabeana 01-200-006 × S. viminalis Jorr’ (Family 430). Herein, we refer to parents of the F1 families by their clone identifiers and discriminate the female and male parents as P1 and P2, respectively.
The field trial was established May 2014 at Cornell AgriTech (Geneva, NY). All parents and progeny were transplanted from nursery beds as stem cuttings (20 cm) in a randomized complete block design with 4 replicate blocks. The field perimeter was buffered using S. purpurea genotypes 94006 and ‘Fish Creek’ to avoid edge effects. Each plot consisted of 3 clones (within-row spacing: 0.4 m; between row spacing: 1.82 m), of which the middle plant was measured. The field trial was evaluated for 3 years.
Parent genotypes and randomly selected progeny were grown from stem cuttings (20 cm) in 12-L plastic pots with peat moss-based potting mix (Fafard, Agawam, MA) to evaluate growth traits under greenhouse conditions over the course of 12 weeks. Plot was defined as a single cutting planted in a pot, which were arranged in a randomized complete block design with 4 replicate blocks. Two blocks were located on benches in 1 greenhouse with the other 2 blocks in an adjacent greenhouse set for identical growing conditions. Supplemental greenhouse lighting was provided on a 14 h day:10 h night regimen with maximum daytime temperature of 26° and a nighttime temperature of 18°. Beyond weekly applications of beneficial insects and mites for pest management, no pesticides were required, as there were no symptoms of biotic or abiotic stress on any plant material throughout the length of the study. Liquid fertilizer (Peter’s 15-16-17 Peat-Lite Special, Scott’s, Marysville, OH, USA) was applied weekly beginning 4 weeks after planting.
For more information on the experimental design and phenotypes recorded in the field and greenhouse trial, see Carlson and Smart (2021).
Determination of ploidy level
The relative DNA content (pg 2C−1) of family parents and progeny was determined by flow cytometry using young leaf material harvested from actively growing shoots in greenhouse conditions. Analysis of 50 mg of mature leaf tissue from parental genotypes and selected progeny was performed at the Flow Cytometry and Imaging Core Laboratory at Virginia Mason Research Center in Seattle, WA, USA. A minimum of 4 replicates of all samples were independently assessed using the diploid female S. purpurea clone 94006 as an internal standard. Diploid parent clones from multiple runs were averaged and then divided by the 2C-value of the check for that run. This factor was then multiplied by each sample value within the same run as the check. When a clone was analyzed more than once, 2C-values were averaged.
Sample preparation and sequencing
A total of 3 biological replicate shoot tips (∼1 cm) of all triploid progeny individuals, as well as their parents, were excised from the primary stem and immediately flash-frozen in liquid N2 in the greenhouse, then placed in −80° storage. Shoot tips were defined as the shoot axis that is the most distal part of a shoot system, comprised of a shoot apical meristem and the youngest leaf primordia. This tissue was chosen based on previous work (Carlson et al. 2017) which consistently identified a greater number of genes were differentially expressed in the shoot tip transcriptome, compared to that of internode tissue. For each sample, a single shoot tip was removed from −80° storage, and ground to a fine powder (100–200 mg) prior to RNA isolation using the Spectrum Total Plant RNA Kit with DNase I digestion (Sigma, St. Louis, MO, USA). The only modification to manufacturer’s “Protocol B” was that prior to the tissue lysis step, the 2-ME/lysate mixture was incubated at 65° for 5 min, otherwise, the manufacturers’ procedures were followed. After elution, cold ethanol precipitations were performed by the addition of 10 μl acetic acid and 280 μl 100% cold ethanol to 100 μl eluate and placed in −80° for 3 h. Samples were centrifuged at 17,000 × g for 30 min at 4°, washed with 80% ethanol, then centrifuged at 17,000 × g for 20 min at 4°. After centrifugation, the supernatant was discarded, and the pellet resuspended in ribonuclease-free 10 mM Tris-HCl. Quantification of RNA sample quality and concentration was performed using the Experion “StdSens” kit (Bio-Rad Laboratories, Inc., Hercules, CA). Stranded RNA-Seq libraries were created and quantified by qPCR (2 × 76 or 2 × 151 bp) and sequenced on an Illumina Hi-Seq 2500 at J. Craig Venter Institute. Library sizes ranged from 8.3 to 53 million reads.
Read filtering, mapping, and variant discovery
Low-coverage paired-end genomic DNA sequencing of the parents of the F1 families was performed to validate variants from RNA-Seq data. Biallelic SNPs were used to quantify ASE within and among triploid progeny individuals. Parent DNA libraries were sequenced (Illumina HiSeq 2500, 2 × 101 bp) and aligned to the S. purpurea v1 reference genome using BWA mem (Li and Durbin 2009). Subsequent BAM files were sorted, marked for duplicates, and indexed in Picard (https://broadinstitute.github.io/picard, last accessed 12/30/2021). Indel realignment and variant calling was performed using HaplotypeCaller (emit_conf = 10, call_conf = 30) in the Genome Analysis Toolkit (GATK) (DePristo et al. 2011). Using BBDuk in the BBTools program (https://jgi.doe.gov/data-and-tools/bbtools/, last accessed 12/30/2021), raw reads were evaluated for artifact sequences by kmer matching (kmer = 25), allowing 1 mismatch and detected artifact was trimmed from the 3′ end of the reads. RNA spike-in reads, PhiX reads and reads containing any Ns were removed. Following quality trimming (phred = Q6), reads under the length threshold were removed (≥25 bp or 1/3 original read length). BWA mem was used for alignment of interleaved RNA-Seq reads to the reference. SAMtools was used to filter (-Shb -F 4 -f 0x2 -q 30), sort, and index resulting sequence alignment files. Duplicate reads were flagged using MarkDuplicates in Picard and GATK was used to flag and realign indels with RealignmentTargetCreator (minReads = 20) and IndelRealigner.
Gene expression inheritance classifications
To categorize inheritance of gene expression in the hybrid, Negative Binomial (NB) exact tests were performed with library-normalized count data in edgeR (Robinson et al. 2010) in R (R Core Team 2021), at a False Discovery Rate (FDR) of 0.005, for only genes with a minimum counts-per-million (CPM) ≥ 1. Prior to NB tests, dispersions were estimated using 3 biological replicates of each group to account for library-to-library variability. Tests for differential expression were for paired comparisons between (1) diploid and tetraploid parents (P2X and P4X), (2) diploid parent and the triploid hybrid (P2X and H), and (3) tetraploid parent and the triploid hybrid (P4X and H). Gene expression inheritance classifications were based on log2 fold-change > 1.2 and q-value < 0.005 resulting from exact tests between the parents and hybrid, according to Carlson et al. (2017).
Regulatory divergence classifications
To determine cis- and trans-effects on gene expression, separate binomial exact tests were performed using library-normalized read counts of diploid (P2X) and tetraploid (P4X) parent alleles in the parents (P2X and P4X) and the F1 triploid progeny individual (H2X and H4X) from the P2X × P4X cross. For a 2-sided binomial test, the null hypothesis is that the expected counts are in the same proportions as the library sizes, or that the binomial probability for the first library is n1/(n1 + n2). To test the null of independence of rows (P2X vs P4X and H2X vs H4X) and columns (P2X vs H2X and P4X vs H4X), Fisher’s exact test was performed on a 2 × 2 matrix comprised of P2X and P4X and H2X and H4X normalized read counts. For all tests, a fixed FDR was applied at a level of 0.005. Filtering parameters required ≥ 20 reads summed between the parents, and 2 alleles at a locus, such that each allele corresponds to either the diploid or tetraploid parent.
For each site, significant differences (FDR = 0.005) on the expression of parent alleles can occur either between the parents (P, binomial exact test), the hybrid (H, binomial exact test), or all (F, Fisher’s exact test). Categories of regulatory functions considered cis-only, trans-only, cis + trans, cis × trans, compensatory were assigned following previously described methods (Landry et al. 2005; McManus et al. 2010). Conservation of expression was attributed to cases where no significant differences could be observed. Ambiguous cases were observed when only one of the 3 tests (P, H, or F, described above) were deemed significant. While ambiguous cases could somewhat be resolved by lowering the significance threshold (e.g. FDR = 0.05), approximately equal proportions of ambiguous assignments were observed across regulatory divergence classes and triploid individuals. However, parent-only (P)-ambiguous genes were more common than the other ambiguous cases, of which, F-ambiguous genes were the least frequent.
Copy number variation
Copy number variation (CNV) was analyzed on a chromosome-wide scale, using median log2 (P2X/P4X) difference of logs in the parents and the median percentage of reads attributable to the P2X allele in the triploid hybrid (diploid %). Diploid % was calculated as HP2X/(HP2X + HP4X) × 100, where HP2X is a vector of library-normalized counts of the P2X allele in the hybrid and HP4X is that of the P4X allele in the hybrid. The expected CN of each homeolog in the hybrid was either determined to be deficient, normal, or replete, depending on these 2 parameters (Supplementary Fig. 1). To avoid over-estimating CNV in triploids, binned coverage of paired-end Illumina DNA-Seq reads of the parents was compared to validate RNA-Seq results.
Gene ontology analysis
Gene ontology (GO) term enrichment was performed in agriGO (Du et al. 2010) using the subset of the S. purpurea v1 transcriptome (reference set) that passed filtering, prior to tests of differential expression. Only significant ontologies (FDR = 0.05) were reported. Salix purpurea gene models and associated GO-terms which were annotated as hypothetical proteins were inferred using the best-hit (blastp e-value ≤ 0.01) to Populus trichocarpa (Phytozome v10.3) and Arabidopsis thaliana (TAIR10 and Araport11 annotations) proteomes.
Gene-trait correlations and prediction of heterosis using selected and random gene sets
Gene-trait correlations were performed for each family using scaled log2 (CPM + 1) library-normalized gene expression values (Supplementary File 1) and midparent heterosis values for field and greenhouse collected traits (Supplementary File 2), which were calculated as the % deviation of the F1 from the midparent value, as described in Carlson and Smart (2021). Ridge regression (α = 0) was used to predict midparent heterosis trait values with different gene sets using 10 replications of nested cross validation (10-fold inner and outer) with cv.glmnet in glmnet (Friedman et al. 2010). Gene sets were comprised of scaled log2 (CPM + 1) library normalized expression values of: (1) 5,000 randomly sampled genes, (2) the top 5,000 most highly expressed genes, (3) 4,986 genes which were differentially expressed between at least 1 pair of family parents, and (4) 379 genes that were commonly differentially expressed between all 3 family parent pairs. Prediction accuracy was assessed via linear regression of mean predicted and observed values.
Results
Transcriptome analysis indicates global additive inheritance of gene expression
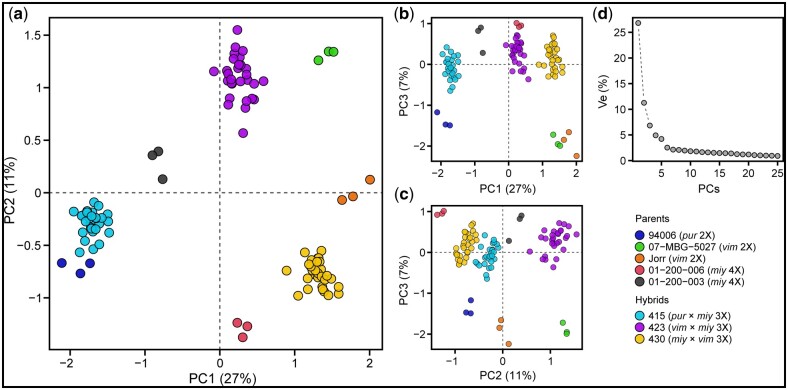
After quality filtering and alignment of triploid F1 progeny and parent paired-end RNA-Seq reads to the S. purpurea v1 reference, the library sizes ranged from 10 to 56 million. Of the 105 libraries sequenced, there were 2 identified as outliers (13X-430-035, greenhouse plot 59, biological replicate 1; 12X-415-074, greenhouse plot 278, biological replicate 3) and removed prior to downstream statistical analyses. In a multidimensional scaling plot of normalized transcriptome-wide gene expression of all triploid F1 progeny individuals and their parents (Fig. 1a), the first dimension represents sample distances based on species pedigree (Ve = 27%), with individuals containing S. viminalis in their background clustering to the right of the first dimension, S. miyabeana in the center, and S. purpurea to the left, such that the S. viminalis parents (07-MBG-5027 and ‘Jorr’) and S. purpurea 94006 are at extremes, or most distantly related. While family 415 and 423 individuals share the common tetraploid S. miyabeana parent 01-200-003, the proximity of family 423 and 430 clusters indicates that common parent species (S. viminalis and S. miyabeana) is a more important factor contributing to transcriptome-wide distances. The second and third dimensions further separate sample libraries by pedigree (Ve = 11%) and ploidy (Ve = 7%), respectively (Fig. 1c). For all 3 triploid families, the respective diploid and tetraploid parents flank clusters of family individuals and are relatively equidistant from the offspring cluster centers. Taking the first 2 dimensions into account, Euclidean distances approximated here implies transcriptome-wide gene expression inheritance is mostly conserved or additive.
Fig. 1.
Multidimensional scaling plot of library-normalized transcriptome-wide gene expression of all triploid F1 progeny individuals (families 415, 423, and 430) and their diploid (94006, 07-MBG-5027, and Jorr’) and tetraploid (01-200-006 and 01-200-003) parents. a) PC1 vs PC2, b) PC1 vs PC3, c) PC2 vs PC3, and d) % variance explained (% Ve) by the first 25 PCs. Euclidean distances on the 2-dimensional plot approximate leading log2 fold-changes between samples, using the top 500 genes with the largest standard deviations. Parents and progeny libraries are colored according to the legend.
Differential gene expression shows primarily additive and dominant inheritance patterns
Exact tests (FDR = 0.005) of differential gene expression between triploid family parent genotypes yielded similar numbers of differentially expressed genes, but the P1: P2 ratios differed (Table 1). The comparison of family 415 parents, S. purpurea 94006 (P1) vs. S. miyabeana 01-200-003 (P2), had 5,166 differentially expressed genes, with 2,661 genes greater in 94006 and 2,505 genes greater in 01-200-003 (P1: P2 = 1.06). The family 423 parents, S. viminalis 07-MBG-5027 (P1) vs S. miyabeana 01-200-003 (P2), had 5,523 differentially expressed genes, with 2,469 genes greater in 07-MBG-5027 and 3,054 genes higher expressed in 01-200-003 (P1: P2 = 0.81). The family 430 parent comparison, S. miyabeana 01-200-006 (P1) vs S. viminalis Jorr’ (P2), yielded 5,155 differentially expressed genes, with 2,467 genes greater in 01-200-006 and 2,688 genes greater in ‘Jorr’ (P1: P2 = 0.91). Globally, the parents of family 423 had a greater percentage of genes that were differentially expressed (22.1%), compared to the parents of families 415 (20.8%) and 430 (20.5%). A total of 379 genes were differentially expressed in common among all 3 family parent duos.
Table 1.
Number of differentially expressed genes between triploid family parents.
| Family | Female (P1) parent | Male (P2) parent | P1 > P2 (%) | P1 < P2 (%) | P1 = P2 (%) | Total |
|---|---|---|---|---|---|---|
| 415 | 94006 [2x] | 01-200-003 [4x] | 2,661 (10.7) | 2,505 (10.1) | 19,641 (79.2) | 24,807 |
| 423 | 07-MBG-5027 [2x] | 01-200-003 [4x] | 2,469 (9.86) | 3,054 (12.2) | 19,519 (77.9) | 25,042 |
| 430 | 01-200-006 [4x] | ‘Jorr’ [2x] | 2,467 (9.81) | 2,688 (10.7) | 19,993 (79.5) | 25,148 |
For those genes differentially expressed between parents, inheritance patterns were determined based on both the parent expression values and those observed in the hybrids (Table 2). The percentage of differentially expressed genes showing nonadditive inheritance ranged from 27% to 39% (mean = 33.5%) (Supplementary Fig. 2) in family 415, 40% to 56% (mean = 49.8%) in family 423, and 34% to 60% (mean = 50.3%) in family 430. Transgressively expressed genes (under- and overdominant) averaged just 0.7%, 1.1%, and 1.0%, for families 415, 423, and 430, respectively. The percentage of genes with underdominant expression out of total transgressively expressed genes was 98%, 74%, and 80% for families 423, 430, and 415, respectively. All individuals had a greater percentage of genes with expression level dominance in the direction of the tetraploid parent, ranging from 66% to 88% and a mean of 70% across all triploid families.
Table 2.
Number of genes assigned to inheritance classifications in triploid F1 progeny individuals and their averages by family.
| Family | P1-dominant | P2-dominant | Over-dominant | Under-dominant | Additive | Conserved |
|---|---|---|---|---|---|---|
| 415 (S. purpurea × S. miyabeana) | ||||||
| 415-018 | 236 | 960 | 0 | 28 | 3,010 | 20,573 |
| 415-020 | 376 | 1,109 | 0 | 31 | 2,786 | 20,505 |
| 415-023 | 336 | 1,024 | 0 | 23 | 2,842 | 20,582 |
| 415-031 | 343 | 1,209 | 1 | 40 | 2,760 | 20,454 |
| 415-038 | 308 | 1,048 | 0 | 35 | 2,887 | 20,529 |
| 415-054 | 414 | 945 | 0 | 20 | 2,882 | 20,546 |
| 415-073 | 326 | 1,038 | 0 | 34 | 2,864 | 20,545 |
| 415-074 | 280 | 869 | 0 | 18 | 3,034 | 20,606 |
| 415-082 | 334 | 1,208 | 2 | 29 | 2,702 | 20,532 |
| 415-257 | 331 | 1,298 | 2 | 44 | 2,633 | 20,499 |
| Mean | 328 | 1,071 | 1 | 30 | 2,840 | 20,537 |
| 423 (S. viminalis × S. miyabeana) | ||||||
| 423-004 | 447 | 1,219 | 14 | 51 | 1,544 | 21,767 |
| 423-034 | 268 | 895 | 7 | 12 | 1,759 | 22,101 |
| 423-043 | 302 | 1,282 | 5 | 31 | 1,513 | 21,909 |
| 423-048 | 325 | 1,428 | 7 | 27 | 1,447 | 21,808 |
| 423-051 | 286 | 1,055 | 8 | 19 | 1,681 | 21,993 |
| 423-063 | 422 | 1,116 | 8 | 21 | 1,593 | 21,882 |
| 423-066 | 278 | 1,021 | 7 | 21 | 1,707 | 22,008 |
| 423-067 | 317 | 1,247 | 10 | 22 | 1,543 | 21,903 |
| 423-070 | 491 | 1,337 | 26 | 29 | 1,466 | 21,693 |
| 423-072 | 324 | 1,332 | 3 | 34 | 1,495 | 21,854 |
| Mean | 346 | 1,193 | 10 | 27 | 1,575 | 21,892 |
| 430 (S. miyabeana × S. viminalis) | ||||||
| 430-004 | 1,498 | 370 | 5 | 41 | 1,330 | 21,904 |
| 430-005 | 1,517 | 340 | 4 | 29 | 1,249 | 22,009 |
| 430-006 | 867 | 274 | 5 | 22 | 1,609 | 22,371 |
| 430-016 | 1,067 | 285 | 7 | 16 | 1,492 | 22,281 |
| 430-018 | 1,162 | 291 | 6 | 14 | 1,410 | 22,265 |
| 430-025 | 1,175 | 283 | 2 | 25 | 1,380 | 22,283 |
| 430-031 | 1,026 | 279 | 6 | 20 | 1,482 | 22,335 |
| 430-033 | 1,007 | 458 | 6 | 31 | 1,496 | 22,150 |
| 430-034 | 1,519 | 351 | 14 | 21 | 1,295 | 21,948 |
| 430-035 | 704 | 207 | 3 | 16 | 1,780 | 22,438 |
| Mean | 1,154 | 314 | 6 | 24 | 1,452 | 22,198 |
There were fewer numbers of diploid parent dominant genes (15) (Supplementary Table 1) than tetraploid parent dominant genes (89) (Supplementary Table 2) that were common across all families and individuals. Due to the low number of common diploid parent dominant genes, there were no significant functional enrichments. Tetraploid dominant genes were enriched for GO molecular functions: beta-glucosidase activity and catalytic activity. In addition, tetraploid parent dominant genes were enriched for the KEGG pathways: phenylpropanoid biosynthesis, cyanoamino acid metabolism, biosynthesis of secondary metabolites, metabolic pathways, and starch and sucrose metabolism (Supplementary Table 3).
Allele-specific expression
To determine the extent of regulatory divergent expression, tests for ASE were conducted using expression data on biallelic sites that were first called with parent DNA-Seq and RNA-Seq libraries prior to calling parent alleles in the progeny. Family averages for the total number of genes assigned to at least 1 regulatory class were 15,391 (±114), 16,800 (±72), and 16,711 (±113), for families 415, 423, and 430, respectively (Table 3). On average, the percentage of genes assigned to nonconserved regulatory classes was 12%, 11%, and 10%, for families 415, 423, and 430, respectively (Supplementary Fig. 3). Family 415 had the greatest percentage of nonconserved genes with cis-regulation (65%), compared to families 423 (58%) and 430 (54%). The greatest mean percentage of genes with trans (24.6%), cis × trans (7.4%), and compensatory (10.8%) regulatory divergence patterns was for family 430, whereas family 415 had the greatest mean cis + trans (5.1%). Across all triploid individuals, a total of 49 genes were in common, having either cis, trans, cis + trans, cis × trans, or compensatory regulatory classifications (Supplementary Table 4). In addition, higher proportions of overdominant and underdominant expression coincided with higher proportions of cis × trans and compensatory regulatory classes. Further, a higher proportion of cis + trans divergence coincided with a lower proportion of underdominant expression, most notably for the comparison of families 423 and 430 with family 415.
Table 3.
Number of genes assigned to regulatory divergence classifications (FDR = 0.005) in triploid F1 individuals and their means by family.
| Family | cis | trans | cis + trans | cis × trans | Compensatory | Ambiguous | Conserved |
|---|---|---|---|---|---|---|---|
| 415 (S. purpurea × S. miyabeana) | |||||||
| 415-018 | 152 | 210 | 21 | 34 | 36 | 1,151 | 13,692 |
| 415-020 | 412 | 123 | 32 | 36 | 44 | 1,084 | 13,447 |
| 415-023 | 384 | 105 | 34 | 35 | 28 | 1,084 | 13,984 |
| 415-031 | 316 | 119 | 29 | 32 | 30 | 1,109 | 14,004 |
| 415-038 | 328 | 109 | 29 | 29 | 20 | 1,089 | 13,802 |
| 415-054 | 381 | 122 | 35 | 32 | 31 | 1,049 | 13,839 |
| 415-073 | 354 | 116 | 23 | 41 | 32 | 1,097 | 14,025 |
| 415-074 | 288 | 76 | 24 | 22 | 19 | 1,201 | 12,841 |
| 415-082 | 359 | 91 | 27 | 25 | 28 | 1,125 | 13,924 |
| 415-257 | 329 | 95 | 17 | 22 | 22 | 1,131 | 13,896 |
| Mean | 330 | 117 | 27 | 31 | 29 | 1,112 | 13,745 |
| 423 (S. viminalis × S. miyabeana) | |||||||
| 423-004 | 252 | 105 | 26 | 28 | 29 | 1,224 | 15,071 |
| 423-034 | 317 | 108 | 14 | 48 | 50 | 1,219 | 15,116 |
| 423-043 | 306 | 103 | 11 | 33 | 45 | 1,248 | 15,112 |
| 423-048 | 178 | 94 | 10 | 14 | 18 | 1,284 | 15,472 |
| 423-051 | 316 | 117 | 16 | 38 | 59 | 1,225 | 15,051 |
| 423-063 | 292 | 124 | 17 | 24 | 30 | 1,218 | 15,096 |
| 423-066 | 187 | 84 | 11 | 24 | 26 | 1,351 | 14,519 |
| 423-067 | 283 | 99 | 16 | 33 | 45 | 1,238 | 15,094 |
| 423-070 | 252 | 113 | 20 | 36 | 56 | 1,237 | 15,199 |
| 423-072 | 274 | 85 | 13 | 35 | 43 | 1,261 | 15,216 |
| Mean | 266 | 103 | 15 | 31 | 40 | 1,251 | 15,095 |
| 430 (S. miyabeana × S. viminalis) | |||||||
| 430-004 | 295 | 111 | 21 | 47 | 63 | 1,095 | 14,797 |
| 430-005 | 203 | 77 | 10 | 32 | 37 | 1,110 | 15,386 |
| 430-006 | 219 | 115 | 13 | 27 | 31 | 1,095 | 15,298 |
| 430-016 | 245 | 117 | 24 | 34 | 47 | 1,086 | 15,501 |
| 430-018 | 252 | 95 | 14 | 42 | 62 | 1,118 | 15,210 |
| 430-025 | 180 | 79 | 12 | 16 | 30 | 1,182 | 15,615 |
| 430-031 | 174 | 105 | 5 | 22 | 26 | 1,171 | 15,366 |
| 430-033 | 265 | 107 | 14 | 47 | 79 | 1,126 | 14,884 |
| 430-034 | 206 | 94 | 15 | 19 | 40 | 1,126 | 15,301 |
| 430-035 | 144 | 96 | 10 | 17 | 24 | 1,187 | 14,399 |
| Mean | 218 | 100 | 14 | 30 | 44 | 1,130 | 15,176 |
More genes are silenced than activated in F1 triploids
The presence (CPM > 1) or absence (CPM = 0) of transcripts in the parent and triploid hybrid was compared for each family. Overall, more genes were silenced than activated in the triploid hybrids, especially for families 415 and 430, in which nearly 5 times the number of genes were silenced than activated (Supplementary Fig. 4). Family 423 had a greater number of genes activated than the other 2 families, whereas family 430 had the greatest number of genes silenced. There were no GO-terms enriched for either activated or silenced gene-sets.
Deviations in expected dosage but global conformity of gene expression is apparent
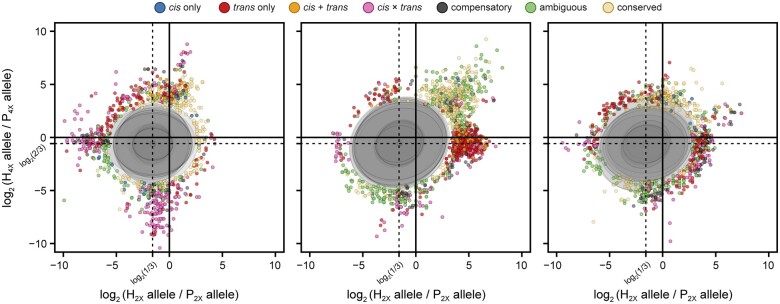
To test whether there was a dosage effect on parent alleles in triploid progeny, ASE ratios were compared within and among families. Only extreme deviations from expected dosage ratios (Pr = 1 × 10−5) were included in the analysis and considered to be dysregulated. Because it is expected that the triploid hybrid has inherited a single copy of the diploid parent allele and 2 copies of the tetraploid parent allele, if there was no deviation in expression of the parent alleles in the hybrid, all loci would be represented by a single point at the intersection of expected P2X/P4X difference of logs, log2 (P2X/P4X) (Fig. 2).
Fig. 2.
Superimposed dosage differential scatterplots of 10 individuals from each of the families 415, 423, and 430 (left to right, respectively). Each point depicts the log2 ratio of the diploid parent allele in the hybrid and the diploid parent allele in the parent against the log2 ratio of the tetraploid parent allele in the hybrid and the tetraploid parent allele in the parent. Points are colored according to their regulatory assignment. Ellipses mask most of the distribution of log2 dosage ratios (Pr = 1 × 10−5), such that points sitting outside ellipses are extreme outliers from expected dosage. Dotted lines at log2 (1/3) = −1.585 and log2 (2/3) = −0.585 represent distribution averages for diploid and tetraploid ratios, which is where the average distribution of dosage ratios is expected to occur.
While the dosage ratios were comparable within families, and all genome-wide family means fell within expected ranges, there were significant departures from expected dosage. For dosage ratio outliers, all 3 triploid families exhibited unique patterns. There was an abundance of genes showing up- and down-regulation of the tetraploid parent allele in family 415, a majority of which showed cis × trans regulatory divergence. Family 423 outliers featured up-regulation of the diploid parent allele in the hybrid, as well as high expression levels of both diploid and tetraploid parent alleles in the hybrid (i.e., trans). Many outliers with greater diploid and tetraploid ASE in the hybrid were classified ambiguous, and those ratios were not different for the tetraploid parent allele, but a greater diploid ratio had either trans or cis × trans regulatory patterns. All regulatory patterns were represented in family 430, which had the greatest number of unique genes among the families that had dosage ratio outliers from at least 1 individual. Unlike family 423, there were few genes with greater expression for both diploid and tetraploid parents in family 430. In general, common dosage dysregulated genes showed significant enrichment for response to stress, transcription, small molecule activity, and binding activity (Supplementary Table 5).
Genes with the greatest mean expression coincided with greater gene expression variance. Those genes with a μ/σ < 1 (n = 338) were significantly (q-value < 0.005) enriched for: GO biological functions of photosynthesis, translation, and response to stimulus; GO molecular functions of structural molecule activity, structural constituent of the ribosome, tetrapyrrole binding, and chlorophyll binding; GO cellular components of chloroplast, cytoplasm, chloroplast thylakoid, photosystem, and apoplast; KEGG pathways of photosynthesis, photosynthesis—antenna proteins, ribosome, metabolic pathways, and flavonoid biosynthesis. Over 50% of the top 50 most variable genes are either class I chaperonin heat shock proteins or ribosomal complex subunits, with the latter being most prominent. On the converse, the most highly expressed but least variable genes were enriched for the GO molecular function of ADP binding, of which most are annotated as stress-associated or disease resistance proteins (e.g. receptor-like kinases) and pentatricopeptide repeat-containing proteins.
Chromosomal copy number can be determined via allele-specific expression ratios and reads attributable to the diploid parent
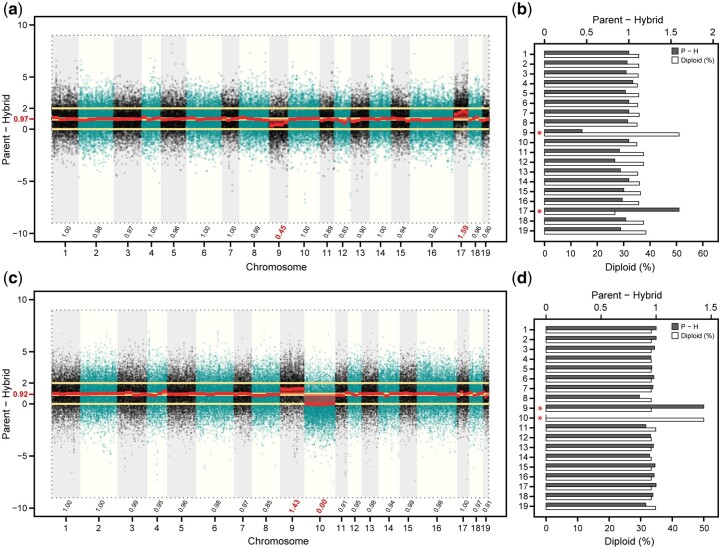
The difference in log2 (P2X/P4X) expression of parent alleles in the hybrid from respective diploid and tetraploid progenitors can help determine if major departures from expected dosage in the hybrid are a result of CNV in the tetraploid parent. For instance, it is expected that triploids inherit 1 chromosome copy from the diploid parent, and 2 copies from the tetraploid parent, such that the difference of the hybrid log2 (P2X/P4X) from the parent is equal to 1. Although these ratios are tetraploid parent informative, aneuploidy in the diploid parent cannot easily be determined, because at least 1 diploid parent copy must be present to infer chromosomal inheritance patterns in triploid progeny. Further, these ratios are not fully informative because any copy number in the tetraploid parent (1/4, 2/4, 3/4, 4/4) can potentially exist in 4 observable cases in the triploid (4/3, 3/3, 2/3, 1/3). However, the percentage of reads attributable to the diploid parent in the triploid hybrid (i.e., % diploid) can be utilized as a second parameter to rectify overlapping parent-hybrid ratios of different parent and hybrid combinations (Supplementary Fig. 1 and Fig. 3).
Fig. 3.
Manhattan plot showing a) chromosome-wide differences of log2 (P2X/P4X) expression (parent‒hybrid) between the family 415 parents (female diploid 94006 and male tetraploid 01-200-003) and the triploid hybrid 12X-415-031. Median parent‒hybrid values are shown above chromosome identifiers (x-axis). The barplot b) depicts the median parent‒hybrid difference (dark gray bars, scale top x-axis) and the % expression in the hybrid attributable to the diploid parent allele (white bars, scale lower x-axis) by chromosome (y-axis). The Manhattan plot in panel c) and barplot in panel (d), represent the same analyses, but between the family 423 parents (female diploid 07-MBG-5027 and male tetraploid 01-200-003) and the triploid hybrid 12X-423-070. Red text on x-axes in panels a) and d) correspond to red asterisks on y-axes in panels b) and c), which denote significant differences (Wilcoxon P-value < 1 × 10−16).
While chromosome-wide log2 (P2X/P4X) expression of the female diploid (S. purpurea 94006 and S. viminalis 07-MBG-5027) and male tetraploid (S. miyabeana 01-200-003) parents showed consistent median values approximately equal to 0, Salix chr09 significantly deviated from the expected (Wilcoxon p-value < 1 × 10−16), with a log2 (P2X/P4X) of 0.49. This suggests that only 3 copies of chr09 were present in S. miyabeana parent 01-200-003 (Supplementary Table 6). This was the case for both families 415 and 423, which share the male tetraploid parent 01-200-003. No significant deviations from the expected was observed for the parents of family 430 (S. miyabeana 01-200-006 × S. viminalis Jorr’).
For family 415, 5 triploid individuals had a median log2 (P2X/P4X) parent‒hybrid difference of 1.43 and approximately 34% of the reads which could be attributed to the diploid parent for chr09, which is expected, given the male parent was limited to 3 chr09 copies. The other 5 individuals had a log2 (P2X/P4X) difference of 0.45 for chr09 and were ∼50% diploid over all loci for the chromosome. Thus, the latter group in family 415 inherited 2 of the 3 tetraploid parent copies of chr09 and the former inherited only one. A total of 6 individuals in family 423 had a log2 (P2X/P4X) difference of 1.44 and were 33.3% diploid on average for chr09, which is expected if they inherited 2 copies from the tetraploid, because family 415 and 423 share the same male tetraploid S. miyabeana parent. The other 4 individuals in family 423 had a log2 (P2X/P4X) difference of 0.47 and were 50% diploid on average, so these individuals only inherited one of the 3 tetraploid parent copies of chr09.
It was not uncommon for individuals to possess an additional tetraploid copy of a chromosome and lack another. For instance, the family 415 individual, 12X-415-031, had a log2 (P2X/P4X) difference of 1.59 for chr17, but only 25% diploid, which suggests that 12X-415-031 inherited an additional copy of the male tetraploid parent 01-200-003 chr17. Stunningly, the same individual also lacked 1 copy of the male chr09 (log2 (P2X/P4X) = 0.45 and 50% diploid) (Fig. 3, a and b). Another example was for the family 423 individual 12X-423-070 (Fig. 3, c and d). While 12X-423-070 inherited 2 copies of chr09 from the tetraploid parent 01-200-003 (log2 [P2X/P4X] = 1.43 and 33.3% diploid), this individual lacked 1 copy of the tetraploid parent chr10 (log2 [P2X/P4X] = 0.0 and 50% diploid), which seems to be spurious, given there was no DNA-Seq or RNA-Seq coverage to indicate that 01-200-003 lacked a copy of chr10.
Gene-trait correlations
Since CNV in triploids may posit drastic phenotypic consequences, Pearson correlations (r) were made for mean genome-wide diploid (%) and heterosis for important biomass-related growth traits collected in the field and greenhouse (Supplementary Table 7). In general, diploid % was positively correlated with heterosis for foliar traits and inversely correlated with heterosis for biomass stem growth traits (Table 4 and Fig. 4) described in Carlson and Smart (2021). Diploid % was positively correlated with the field-collected leaf growth traits (length, perimeter, ratio, and specific leaf area) and inversely correlated with stem growth traits (height, basal diameter, area, and volume). For greenhouse-collected traits, diploid % was positively correlated with specific leaf area only, but inversely correlated with biomass yield, stem growth traits, and vegetative phenology. Field and greenhouse collected traits most positively correlated with diploid % were crown form (r = 0.65) and specific leaf area (r = 0.65), respectively, and inversely were plot height (r = −0.82) and root dry mass (r = −0.73), respectively. The only foliar field trait with an inverse relationship with diploid % was leaf shape factor (r = −0.51), which is a measure of leaf symmetry. Diploid % had a strong inverse relationship with the proportion of differentially expressed genes showing nonadditive inheritance (r = −0.71).
Table 4.
Pearson correlation coefficients (r) of heterosis for biomass-related traits and the mean percentage of each locus in triploid progeny attributable to the respective diploid parent (diploid %).
| Trait | Trait description | Trait class | Timea | Year 1b | Year 2 | ||
|---|---|---|---|---|---|---|---|
| Field study | |||||||
| HT | Plot height | Stem growth | 1, 2 | −0.82 | *** | −0.53 | ** |
| STN | Stem number | Stem growth | 1 | −0.41 | * | – | n.s. |
| MDIA | Mean stem diameter | Stem growth | 1, 2 | −0.47 | * | −0.40 | * |
| DIA | Stem diameter | Stem growth | 1, 2 | −0.54 | ** | −0.51 | ** |
| MSA | Mean stem area | Stem growth | 1 | −0.41 | * | – | n.s. |
| SA | Stem area | Stem growth | 1, 2 | −0.51 | ** | −0.47 | * |
| VOL | Stem volume | Stem growth | 1, 2 | −0.53 | ** | −0.52 | ** |
| LFL | Leaf length | Leaf growth | 2 | – | n.s. | 0.45 | * |
| LFP | Leaf perimeter | Leaf growth | 2 | – | n.s. | 0.5 | ** |
| LFR | Leaf ratio | Leaf growth | 2 | – | n.s. | 0.42 | * |
| LFF | Leaf shape factor | Leaf growth | 2 | – | n.s. | −0.51 | ** |
| SLA | Specific leaf area | Leaf growth | 1 | 0.58 | ** | n.s. | |
| CDIA | Basal crown diameter | Architecture | 1, 2 | −0.40 | * | −0.65 | *** |
| FORM | Crown form | Architecture | 1, 2 | 0.46 | * | 0.65 | *** |
| DEN | Wood density | Composition | 2 | – | n.s. | −0.41 | * |
| DVOL | Wood density × stem volume | Biomass | 1, 2 | −0.56 | ** | −0.58 | ** |
| Greenhouse study | |||||||
| SDW | Stem dry mass | Biomass | 70 | −0.52 | ** | ||
| LDW | Leaf dry mass | Biomass | 70 | −0.57 | ** | ||
| RDW | Root dry mass | Biomass | 70 | −0.73 | *** | ||
| AGB | Aboveground dry mass | Biomass | 70 | −0.56 | ** | ||
| TBM | Total dry mass | Biomass | 70 | −0.65 | *** | ||
| HT | Plot height | Stem growth | 42, 21–56 | −0.68 | *** | ||
| MSL | Mean stem length | Stem growth | 42, 21–56 | −0.60 | *** | ||
| TSL | Total stem length | Stem growth | 42, 21–56 | −0.59 | ** | ||
| SA | Stem area | Stem growth | 70 | −0.48 | ** | ||
| VOL | Stem volume | Stem growth | 70 | −0.60 | *** | ||
| SLA | Specific leaf area | Leaf growth | 70 | 0.31 | * | ||
| PHE | Vegetative phenology | Leaf growth | 11, 13 | −0.52 | ** | ||
| SPAD | Chlorophyll content | Leaf growth | 14 | −0.39 | * | ||
Time in years since coppice (field study) or days after planting (greenhouse study).
Asterisks ***, **, * denote significant at P-value < 0.001, P < 0.01, and P < 0.05, respectively.
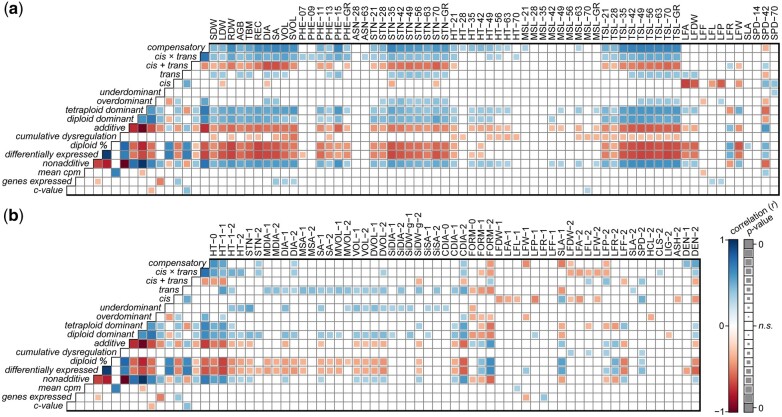
Fig. 4.
Correlations of nonadditive, regulatory divergent, and cumulative expression dysregulation class proportions in F1 progeny with heterosis values for a) greenhouse and b) field phenotypes. Pearson correlation coefficients (r), positive correlations are illustrated by filled blue squares and negative correlations by filled-red squares. Nonsignificant correlations (P-value > 0.01) were left blank. Significance levels (P-values) were used to scale the area of each square, such that smaller squares represent correlation coefficients with lower significance and larger squares represent those with greater significance.
Overall, there were stronger associations in the greenhouse trial than the field trial. The proportions of additive expression, and cis− and cis + trans divergence, were inversely correlated with heterosis for nearly all biomass traits in the greenhouse trial, as well as the total proportion of differentially expressed genes, cumulative expression dysregulation, and the proportion of differentially expressed genes with additive expression inheritance. For both the field and greenhouse trials, trans-divergence, differential expression, and the proportion of diploid- and tetraploid-parent dominant genes were positively correlated with heterosis for total stem volume. Heterosis for hemicellulose content was positively correlated with the proportion of cis-divergence and inversely with overdominance. Heterosis for cellulose and lignin content were positively and inversely correlated with the proportion of differentially expressed genes with diploid parent dominant expression. Chlorophyll content (SPAD) in both trials was inversely correlated with the proportion of nonadditive expression, trans, and compensatory regulatory divergence, and the proportion of differentially expressed genes with tetraploid parent dominant inheritance. Cis-divergence was positively correlated with the proportion of differentially expressed genes with additive inheritance, but inversely correlated with the proportion of diploid parent dominant and overdominant expression. The proportion of differentially expressed genes with trans-divergent expression was inversely correlated with the proportion of additive inheritance, but positively correlated with dominant and overdominant proportions. Finally, mean normalized expression levels (CPM) had the greatest positive association with cumulative dysregulation (r = 0.68); however, nonadditive inheritance and regulatory proportions lacked any significant associations with cumulative dysregulation.
Genes most commonly associated with heterosis for biomass growth traits were a peripheral-type benzodiazepine receptor (SapurV1A.0155s0220; r = 0.64–0.81) located on Salix chr02 and a squamosa promoter-binding-like protein (SapurV1A.0056s0240; r = 0.67 to 0.73) on chr03 (Supplementary File 3). Most common inverse gene associations with biomass growth traits were mediator of RNA polymerase II subunit 20 (SapurV1A.0616s0090; r = −0.74 to −0.81) on chr06 and a NLI interacting factor-like serine/threonine specific protein phosphatase (SapurV1A.0546s0050; r = −0.67 to −0.78) on chr15. Genes with a strong positive relationship with total biomass yield (r > 0.6, n = 189) were enriched for GO molecular functions: catalytic activity, transferase activity, and acyltransferase activity, and those inversely associated (r < −0.6, n = 94) were enriched for GO molecular functions: structural molecule activity and structural constituent of the ribosome (Supplementary Table 8).
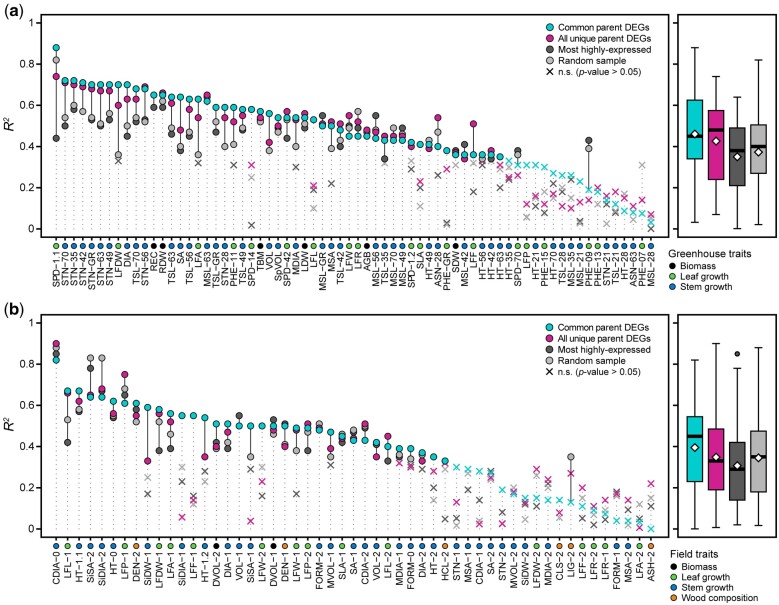
Prediction of heterosis with random and selected gene expression sets
For most traits, genes that were differentially expressed between the F1 parents were more predictive of midparent heterosis than those 5,000 genes either randomly sampled or most highly expressed (Fig. 5). Prediction accuracies of midparent heterosis using genes commonly differentially expressed between all 3 family parent pairs (n = 379) were akin to the larger set of genes differentially expressed between at least 1 pair (n = 4,986), yet there were cases in which 1 gene set performed substantially better, vice versa.
Fig. 5.
Prediction accuracies (R2) of heterosis values for a) greenhouse and b) field phenotypes (Supplementary Table 7) using selected and random gene expression sets. These 4 gene sets were: differentially expressed genes common among parent pairs (cyan, n = 379), differentially expressed genes in at least 1 parent pair (magenta, n = 4,978), the most highly expressed genes (dark gray, n = 5,000), and a random sampling of genes (light gray, n = 5,000). Boxplots to the right of each panel depict the distribution of prediction accuracies, with means represented as yellow diamond points. Traits are colored according to respective classes described in the lower left legend of each panel.
Overall, mean prediction accuracies were greater for greenhouse traits (Fig. 5a) than field traits (Fig. 5b), of which the common, overlapping differential expression gene set had better prediction accuracies on average. This can partly be explained by more frequent repeated measurements recorded in the greenhouse and the impact of pest damage on field phenotypes not present in the greenhouse. In addition, lower R2 values were observed for second year postcoppice measurements in the field compared to those taken the first year following coppice. Across all 4 gene sets, basal crown diameter had the highest prediction accuracies (R2 = 0.82–0.90), followed by leaf perimeter, chlorophyll content, primary stem number, and basal stem diameter.
Discussion
Differential gene expression is both additive and nonadditive
In interspecific Salix F1 triploid hybrid families described in this study, genes differentially expressed between parent species of contrasting ploidy exhibit both additive and nonadditive patterns of gene expression. Using microarrays of maize, Stupar and Springer (2006) determined that approximately 20% of the genes that were differentially expressed between inbred parents were nonadditively expressed in the hybrid, although very few were above the high parent (overdominant) or below the low parent (underdominant). Swanson and Wagner et al. (2006) found all inheritance categories in the hybrid represented among differentially expressed genes between 2 inbred parents of maize. In our study, there were very few genes that were differentially expressed between diploid and tetraploid parents and were outside the parental range in the triploid hybrid, especially overdominant genes, which were 3 times less-frequent on average than underdominant genes. In contrast, differentially expressed genes between heterozygous thistle (C. arvense) parents were more frequently overdominant than underdominant in intraspecific hybrids (Bell et al. 2013). Expression-level dominance was most-prominent in both shoot tip and stem internode tissues of F1 and F2 diploid S. purpurea families (Carlson et al. 2017) and was primarily biased in the direction of the female parent, especially in shoot-tip tissues. Very little additive gene expression was observed in S. purpurea, which is a unique result, compared with model crop plants (Guo et al. 2006; Stupar and Springer 2006; Song et al. 2013).
Among the triploid Salix families investigated in this study, expression-level dominance was prominent, as was established in diploid S. purpurea, but the percentage of differential expression attributed to dominance inheritance ranged from 28% to 60%. Progeny from reciprocal crosses between Salix Sections Vimen and Helix, showed the greatest percentage of dominant expression, which was 50% of those genes expressed differently between diploid and tetraploid parents. Cases of expression-level dominance in polyploid crops have been described in intraspecific thistle (Bell et al. 2013), interspecific coffee (Combes et al. 2015), as well as in allotetraploids of both rice (Xu et al. 2014) and Arabidopsis (Shi et al. 2012). This preferential expression is thought to be orchestrated by allelic interactions, which functions to silence one of the parent alleles in a parent-of-origin manner (Chen and Pikaard, 1997; Stupar et al. 2007; Donoghue et al. 2014; Baldauf et al. 2016).
Further analysis of ASE in triploid hybrids of willow indicated that gene expression variation was associated with both cis− and trans-regulatory divergence, and that cis‒trans compensatory interactions accounted for up to 25% of the variation. ASE has been extensively studied in model species, most notably, in interspecific hybrids and allopolyploids of Arabidopsis (Shi et al. 2012) and Drosophila (Landry et al. 2005; Wittkopp et al. 2008a, 2008b; McManus et al. 2010). There is a general trend that cis-regulatory divergence accounts for a greater proportion of expression variation in interspecific hybrids and that trans-regulatory divergence is more frequent in intraspecific hybrids (Wittkopp et al. 2004). In hybrids of inbred maize, cis-acting variation accounted for most of the divergent expression between parents and was largely attributed to additive expression patterns (Stupar and Springer 2006). Greater sequence divergence was proposed to promote the flexibility of trans-factors in their binding to interacting factors and cis-elements in A.thaliana and A. arenosa parent alleles (Shi et al. 2012). McManus et al. (2010) hypothesized that greater transgressive inheritance is associated with greater proportions of cis × trans divergence. Likewise, what was identified in triploid Salix hybrids, greater proportions of overdominant and underdominant (transgressive) expression inheritance did coincide with greater proportions of cis × trans and compensatory regulatory classes. Further, a greater proportion of cis + trans divergence coincided with a lower proportion of underdominant expression.
Global dosage balance with local sensitivities
Dosage in all 3 triploid families appeared to behave in an extraordinarily additive manner, irrespective the number of parent copies inherited. However, a handful of genes did depart from expected dosage in triploids, most notably, those coding for heat shock proteins. In this study, genes annotated as coding for heat shock proteins displayed greater expression in individuals with normal chr09 copies, whereas those null for a tetraploid parent copy had greater expression of stress- or senescence-associated genes. Overall, there were greater proportions of loci showing cis × trans and compensatory regulatory patterns in family 415 and 423 individuals that were aneuploid with only 1 tetraploid parent copy (e.g. chr2, 9, 10, and 17), or a greater average diploid %. While the quantity of a translation product (protein subunit) may impact the assembly of a particular complex, the mere involvement in a complex can also impact protein stability (Veitia et al. 2008). It may be that null mutations in metabolic functions are tolerated in a heterozygous state, but only weak, loss-of-function, dosage-sensitive genes can survive negative selection as heterozygotes (Birchler and Veitia 2010). The balance of regulatory hierarchies (dosage balance) (Birchler et al. 2005) are sensitive to gene dosage and changes in individual components can influence phenotype. In macromolecular complexes, dosage balance is essential, because partial aneuploidy of a dosage-sensitive gene can change the stoichiometry of the complexes and lead to fitness defects (Veitia et al. 2008). In maize, greater proportions of nonadditive expression were observed in triploid and tetraploid hybrids with genome dosage effects (Guo et al. 1996; Auger et al. 2005; Birchler et al. 2005; Riddle et al. 2010).
Previous gene expression studies in inbred and outcrossing species have regularly pooled F1 progeny libraries prior to sequencing. While this is not an issue for inbred crops, our results show that pooled RNA-Seq can underestimate factors contributing to the inheritance of gene expression in heterozygous species, especially for families derived from natural polyploids. For instance, without sequencing individual libraries, we would not have detected aneuploidy for chromosomes of polyploid progenitors in the F1 based on pooled RNA-Seq data alone, which could distort assumptions about the evolution of gene expression inferred from inheritance and regulatory assignments. Even if the expected ploidy in the hybrid is based on chromosome counts or DNA-Seq of the parents, there may not be an equal inheritance, and binomial tests for ASE between the parents and the hybrid would be incorrect if based on a fixed probability estimate. Thus, prior to tests for ASE, a simple adjustment could be made, which would first require that each chromosome (or scaffold) be tested independently. Utilizing median fold-changes in the parents and the percentage of reads in the hybrid attributable to the diploid parent, a probability of success under the null could be properly assigned.
Beyond the fact that the parents in this study were highly heterozygous, it is possible that CNV or aneuploidy can help explain some of the variation in heterosis observed within and among triploid families. In aneuploid studies, changing numerous chromosome segments can alter quantitative characters (Guo and Birchler 1994). Here, genome-wide averages of ASE attributable to the diploid parent (diploid %) in triploids were inversely correlated with heterosis for important stem growth traits (e.g. total harvestable biomass), but positively with foliar traits. The dosage balance hypothesis, outlined by Birchler (2005), may very well apply to slight deviations in the global inheritance of parent ASE or major differences in chromosomal copy number, as was observed for Salix chr09 aberrations in families 415 and 423. Genetic mapping in F2S. purpurea (Carlson et al. 2019) identified QTL on chr09 for leaf length, leaf perimeter, and specific leaf area, so positive correlations between diploid % and foliar traits could indicate a dosage sensitivity of genes controlling the variation for these traits.
A cluster of genes collocated on a 50 kb interval on chr10 contained genes highly-expressed in tetraploid S. miyabeana parents and triploid progeny, but with very low expression in diploid parents. These genes included duplicates, annotated as 3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransferase (DBTNBT, TAX10), which catalyzes the final step in biosynthesis of the anticancer compound Taxol (Walker et al. 2002). The constitutive high expression levels of DBTNBT in triploids and tetraploid S. miyabeana parents could suggest involvement in the synthesis of an important defensive compound in Salix. There are multiple copies of genes with this annotation in the Salix genome and a number of them collocate with QTL for variation in tremuloidin on chr08, 10, 15, and 16 (Keefover-Ring et al., in preparation). The abundance of tremuloidin or a related phenolic glycoside could be a source of broad-spectrum pest and/or disease resistance conferred to triploid hybrids by S. miyabeana parents, as all triploid genotypes in Carlson and Smart (2021) displayed field resistance to most willow pests and pathogens, but not intra- and inter-specific diploids. In a F2S. purpurea mapping population, Carlson et al. (2019) identified QTL associated with many important traits for biomass production. One QTL for willow leaf rust (Melampsora spp.) incidence was identified on chr10, and DBTNBT genes described here fall within that confidence interval, meriting further investigation.
Nonadditive gene expression correlates with nonadditive phenotypic expression
One of the major challenges in molecular genetics is disentangling the relationship of transcriptome-wide expression patterns to phenotypic effects (Birchler and Veitia 2007). Rather than concentrating on the terminologies of heterosis models (e.g. dominance, overdominance, or pseudo-overdominance), Birchler (2010) promoted a progression to a more nuanced quantitative and interactive or network-oriented framework for dissecting the phenomenon of heterosis. We utilized DNA-Seq and RNA-Seq to unravel the underlying regulatory architecture of differential expression and improve our understanding of heterosis in high-yielding triploid hybrids of willow. We showed that the proportion of genes differentially expressed between diploid and tetraploid parents attributable to nonadditive gene expression in the triploid hybrid (namely expression-level dominance) was positively correlated with heterosis for biomass yield as well as biomass-related growth traits collected in the greenhouse and in the field. In addition, we corroborate some of the key findings reported in Kremling et al. (2018), for example, that cumulative expression dysregulation is inversely correlated with heterosis for biomass; and that individuals with greater absolute expression tended to display greater levels of dysregulation.
Importantly, tetraploid parent dominant genes among triploid hybrids were enriched for the following pathways: phenylpropanoid biosynthesis, cyanoamino acid metabolism, biosynthesis of secondary metabolites, and starch and sucrose metabolism. Some of the most intriguing tetraploid parent dominant genes identified in this study were those annotated as uridine diphosphate (UDP) glycosyltransferases (UGTs). UGTs catalyze the transfer of sugars to a wide range of acceptor molecules, including plant hormones, and all classes of plant secondary metabolites (Ross et al. 2001). Both tetraploid parent dominant genes and those genes positively correlated with biomass yield and stem growth traits were enriched for the GO molecular function of catalytic activity. Further analysis of these candidate genes and gene sets, with regards to their relevance in overlapping support intervals from mapping experiments or regulatory patterns in other high-yielding triploid hybrid individuals, will prove useful in the genetic improvement of shrub willow as a bioenergy crop.
Parent differentially expressed genes are most predictive of heterosis in F1 hybrids
Here, we tested whether parent differentially expressed genes are predictive of heterosis in 3 interspecific triploid F1Salix crosses, by comparing prediction accuracies of those gene sets to a random sampling of genes as well as a selection of genes most highly expressed. While it would be assumed that the most highly expressed genes would also be the most variable, these genes had the lowest mean prediction accuracies for most traits and performed similarly as did a random sampling of genes of equal size. Differentially expressed genes were most predictive of heterosis, and often moreso using a reduced gene set of only common, overlapping differentially expressed genes among family parent pairs. However, there were a handful of traits where prediction accuracies of all gene sets were not considerably different from that of a random sample. This could mean that midparent heterosis values are attributable to population structure and/or highly quantitative in nature, such that a random gene sample is sufficient to illustrate the inherent transcriptome-wide differences between parent species. Thus, strong family-specific responses to hybridization and transgressive phenotypic expression would result in higher prediction accuracies for specific traits that have high among but not within family variances. Yet, this was not often the case. The phenotypes used in this study were from Carlson and Smart (2021), which reported both hybrid vigor and hybrid necrosis within all intra- and inter-specific F1 families in field and greenhouse conditions. Midparent heterosis values were normally distributed, besides traits with low variance, like later vegetative phenology dates.
Genes that were differentially expressed between the parents showed primarily additive and dominant inheritance patterns among F1 progeny, but segregated within families. Progeny individuals with a greater frequency of genes with dominant expression patterns were more apt to display heterosis for biomass growth. Further, genes that were differentially expressed between parents were more predictive of heterosis in F1 progeny compared to a random sampling of genes, irrespective of expression level. These gene sets could be used to aid in the selection of genotypes or breeding populations in the greenhouse by utilizing expression levels as an indicator of performance based on prior related datasets. While only 3 species were assayed in this study, the inclusion of additional, diverse parent species of varying heterozygosity would help determine if there is a core set of genes and/or transcriptional regulators that, when differentially expressed, comprise a network predictive of triploid heterosis in F1 crosses.
This work highlights regulatory factors influencing differential expression, as well as genes and gene sets predictive of heterosis for biomass growth, physiological, and wood chemical composition traits collected in the greenhouse and field. It is vital that we apply our ever-improving understanding of heterosis from studies of well-characterized diploid crop species, such as maize, tomato, and rice to the improvement of yield and biomass quality of undomesticated crops, including willow and poplar, which provide sustainable sources of lignocellulosic biomass for bioenergy, biofuels, and bioproducts. Additional characterization of the genomic basis of heterosis in related genera or more diverse Salix crosses will be valuable in understanding the broad evolutionary benefits of wide hybridization and incidence of polyploidy.
Data availability
The gene expression data (Supplementary File 1), heterosis values (Supplementary File 2), and gene-trait correlations (Supplementary File 3) used in this study, as well as Supplementary Tables 1–8 and Supplementary Figs. 1–4 are available online: https://www.github.com/Willowpedia/Carlson2021_TriploidHeterosis. Original sequence read files are accessible at NCBI SRA BioProject: PRJNA785270.
Supplemental material is available at G3 online.
Supplementary Material
Acknowledgments
We are grateful to Lauren Carlson and Dawn Fishback for their excellent technical assistance and to Dr. Fred Gouker and Dr. Eric Fabio for their assistance with harvest, processing, and tissue collection.
CHC wrote the manuscript, performed DNA and RNA isolations, phenotyping, bioinformatics, and statistics, YC and APC conducted sequencing and bioinformatics, and CDT and LBS devised the study and managed research programs. All authors participated in reviewing and editing the manuscript.
Funding
This research was supported by U.S. Department of Energy Office of Science, Office of Biological and Environmental Research, grant DE-SC0008375.
Conflicts of interest statement. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Literature cited
- Auger DL, Gray AD, Ream TS, Kato A, Coe EH, Birchler JA.. Nonadditive gene expression in diploid and triploid hybrids of maize. Genetics. 2005;169(1):389–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldauf JA, Marcon C, Paschold A, Hochholdinger F.. Nonsyntenic genes drive tissue-specific dynamics of differential, nonadditive, and allelic expression patterns in maize hybrids. Plant Phys. 2016;171:1144–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell GD, Kane NC, Rieseberg LH, Adams KL.. RNA-seq analysis of allele-specific expression, hybrid effects, and regulatory divergence in hybrids compared with their parents from natural populations. Genome Biol Evol. 2013;5(7):1309–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin S, Lagercrantz U, von Arnold S, Öst T, Rönnberg-Wästljung A.. High-density linkage mapping and evolution of paralogs and orthologs in Salix and Populus. BMC Genomics. 2010;11:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Auger DL, Riddle NC.. In search of the molecular basis of heterosis. Plant Cell. 2003;15(10):2236–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Riddle NC, Auger DL, Veitia RA.. Dosage balance in gene regulation: biological implications. Trends Genet. 2005;21(4):219–226. [DOI] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA.. The gene balance hypothesis: from classical genetics to modern genomics. Plant Cell. 2007;19(2):395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA.. The gene balance hypothesis: implications for gene regulation, quantitative traits and evolution. New Phytol. 2010;186(1):54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Yao H, Chudalayandi S, Vaiman D, Veitia RA.. Heterosis. Plant Cell. 2010;22(7):2105–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron KD, Phillips IS, Kopp RF, Volk TA, Maynard CA, Abrahamson LP, Smart LB.. Quantitative genetics of traits indicative of biomass production and heterosis in 34 full-sib F1Salix eriocephala families. Bioenerg Res. 2008;1(1):80–90. [Google Scholar]
- Carlson CH, Choi Y, Chan AP, Serapiglia MJ, Town CD, Smart LB.. Dominance and sexual dimorphism pervade the Salix purpurea L. transcriptome. Genome Biol Evol. 2017;9(9):2377–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CH, Gouker FE, Crowell CR, Evans L, DiFazio SP, Smart CD, Smart LB.. Joint linkage and association mapping of complex traits in shrub willow (Salix purpurea L.). Ann Bot. 2019;124(4):701–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CH, Smart LB.. Electrical capacitance as a predictor of root dry weight in shrub willow (Salix; Salicaceae) parents and progeny. Appl. Plant Sci. 2016;4(8):e1600031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CH, Smart LB.. Heterosis for biomass-related traits in interspecific triploid hybrids of willow (Salix spp.). BioEnergy Res. 2021. doi: 10.1007/s12155-021-10305-0. [DOI] [Google Scholar]
- Chen CZ, Pikaard CS.. Transcriptional analysis of nucleolar dominance in polyploid plants: biased expression/silencing of progenitor rRNA genes is developmentally regulated in Brassica. Proc Natl Acad Sci USA. 1997;94(7):3442–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combes M-C, Hueber Y, Dereeper A, Rialle S, Herrera J-C, Lashermes P.. Regulatory divergence between parental alleles determines gene expression patterns in hybrids. Genome Biol Evol. 2015;7(4):1110–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue MTA, Fort A, Clifton R, Zhang X, McKeown PC, Voigt-Zielinksi ML, Borevitz JO, Spillane C.. C(m)CGG methylation-independent parent-of-origin effects on genome-wide transcript levels in isogenic reciprocal F1 triploid plants. DNA Res. 2014;21(2):141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z.. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38:W64–W70. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East EM. Heterosis. Genetics. 1936;21(4):375–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabio ES, Kemanian AR, Montes F, Miller RO, Smart LB.. A mixed model approach for evaluating yield improvements in interspecific hybrids of shrub willow, a dedicated bioenergy crop. Ind Crop Prod. 2016;96:57–70. [Google Scholar]
- Fabio ES, Volk TA, Miller RO, Serapiglia MJ, Gauch HG, Van Rees KCJ, Hangs RD, Amichev BY, Kuzovkina YA, Labrecque M, et al. Genotype x environment interactions analysis of North American shrub willow yield trials confirms superior performance of triploid hybrids. Glob Change Biol Bioenergy. 2017;9(2):445–459. [Google Scholar]
- Friedman J, Hastie T, Tibshirani R.. Regularization paths for generalized linear models via coordinate descent. J Stat Soft. 2010;33(1):22. [PMC free article] [PubMed] [Google Scholar]
- Goff SA. A unifying theory for general multigenic heterosis: energy efficiency, protein metabolism, and implications for molecular breeding. New Phytol. 2011;189(4):923–937. [DOI] [PubMed] [Google Scholar]
- Gouker FE, Carlson CH, Zou J, Evans L, Crowell CR, Smart CD, DiFazio SP, Smart LB.. Sexual dimorphism in the dioecious willow Salix purpurea. Am J Bot. 2021;108(8):1374–1387. [DOI] [PubMed] [Google Scholar]
- Gunter LE, Roberts GT, Lee K, Larimer FW, Tuskan GA.. The development of two flanking SCAR markers linked to a sex determination locus in Salix viminalis L. J Hered. 2003;94(2):185–189. [DOI] [PubMed] [Google Scholar]
- Guo M, Birchler JA.. Trans-acting dosage effects on the expression of model gene systems in maize aneuploids. Science. 1994;266(5193):1999–2002. [DOI] [PubMed] [Google Scholar]
- Guo M, Davis D, Birchler JA.. Dosage effects on gene expression in a maize ploidy series. Genetics. 1996;142(4):1349–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Rupe MA, Yang X, Crasta O, Zinselmeier C, Smith OS, Bowen B.. Genome-wide transcript analysis of maize hybrids: allelic additive gene expression and yield heterosis. Theor Appl Genet. 2006;113(5):831–845. [DOI] [PubMed] [Google Scholar]
- Guo M, Rupe MA, Zinselmeier C, Habben J, Bowen BA, Smith OS.. Allelic variation of gene expression in maize hybrids. Plant Cell. 2004;16(7):1707–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallingbäck HR, Fogelqvist J, Powers SJ, Turrion‐Gomez J, Rossiter R, Amey J, Martin T, Weih M, Gyllenstrand N, Karp A, et al. Association mapping in Salix viminalis L. (Salicaceae)—identification of candidate genes associated with growth and phenology. Glob Change Biol Bioenergy. 2016;8(3):670–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley SJ, Karp A.. Genetic strategies for dissecting complex traits in biomass willows (Salix spp.). Tree Physiol. 2014;34(11):1167–1180. [DOI] [PubMed] [Google Scholar]
- He G, Zhu X, Elling AA, Chen L, Wang X, Guo L, Liang M, He H, Zhang H, Chen F, et al. Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell. 2010;22(1):17–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyden B, Carlson CH, Gouker FE, Schmutz J, Barry K, Lipzen A, Sharma A, Sandor L, Tuskan GA, Feng G, et al. Integrative genomics reveals paths to sex dimorphism in Salix purpurea L. Hort Res. 2021;8:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp RF, Smart LB, Maynard CA, Isebrands JG, Tuskan GA, Abrahamson LP.. The development of improved willow clones for eastern North America. Forest Chron. 2001;77(2):287–292. [Google Scholar]
- Kremling KAG, Chen S-Y, Su M-H, Lepak NK, Romay MC, Swarts KL, Lu F, Lorant A, Bradbury PJ, Buckler ES, et al. Dysregulation of expression correlates with rare allele burden and fitness loss in maize. Nature. 2018;555(7697):520–523. [DOI] [PubMed] [Google Scholar]
- Kuzovkina YA, Weih M, Romero MA, Charles JG, Hust S, McIvor I, Karp A, Trybush S, Labrecque M, Teodorescu TI, et al. Salix: botany and global horticulture. In: Jannick J, editor. Horticulture Reviews. New Jersey (NJ): John Wiley & Sons; 2008. p. 447–489. [Google Scholar]
- Landry CR, Wittkopp PJ, Taubes CH, Ranz JM, Clark AG, Hartl DL.. Compensatory cis-trans evolution and the dysregulation of gene expression in interspecific hybrids of Drosophila. Genetics. 2005;171:1813–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus CJ, Coolon JD, Duff MO, Eipper-Mains J, Graveley BR, Wittkopp PJ.. Regulatory divergence in Drosophila revealed by mRNA-seq. Genome Res. 2010;20(6):816–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen MD, Kresovich S, Villeda HS, Bradbury P, Li H, Sun Q, Flint-Garcia S, Thornsberry J, Acharya C, Bottoms C, et al. Genetic properties of the maize nested association mapping population. Science. 2009;325(5941):737–740. [DOI] [PubMed] [Google Scholar]
- Ni Z, Kim E-D, Ha M, Lackey E, Liu J, Zhang Y, Sun Q, Chen ZJ.. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature. 2009;457(7227):327–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Australia: R Foundation for Statistical Computing; 2021. [Google Scholar]
- Riddle NC, Jiang HM, An LL, Doerge RW, Birchler JA.. Gene expression analysis at the intersection of ploidy and hybridity in maize. Theor Appl Genet. 2010;120(2):341–353. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J, Li Y, Lim EK, Bowles DJ.. Higher plant glycosyltransferases. Genome Biol. 2001;2(2):REVIEWS3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serapiglia MJ, Cameron KD, Stipanovic AJ, Smart LB.. Correlations of expression of cell wall biosynthesis genes with variation in biomass composition in shrub willow (Salix spp.) biomass crops. Tree Genet. Genome. 2012;8(4):775–788. [Google Scholar]
- Serapiglia MJ, Gouker FE, Hart JF, Unda F, Mansfield SD, Stipanovic AJ, Smart LB.. Ploidy level affects important biomass traits of novel shrub willow (Salix) hybrids. Bioenerg Res. 2014a;8(1):259–269. [Google Scholar]
- Serapiglia MJ, Gouker FE, Smart LB.. Early selection of novel triploid hybrids of shrub willow with improved biomass yield relative to diploids. BMC Plant Biol. 2014b;14:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Ng DWK, Zhang C, Comai L, Ye W, Chen ZJ. Cis- and trans-regulatory divergence between progenitor species determines gene-expression novelty in Arabidopsis allopolyploids. Nat Commun. 2012;3:950. [DOI] [PubMed] [Google Scholar]
- Smart LB, Cameron KD.. Genetic improvement of willow (Salix spp.) as a dedicated bioenergy crop. In: Vermerris W, editor. Genetic Improvement of Bioenergy Crops. New York (NY): Springer Science; 2008. p. 377–396. [Google Scholar]
- Song G, Guo Z, Liu Z, Cheng Q, Qu X, Chen R, Jiang D, Liu C, Wang W, Sun Y, et al. Global RNA sequencing reveals that genotype-dependent allele-specific expression contributes to differential expression in rice F1 hybrids. BMC Plant Biol. 2013;13:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer NM, Stupar RM.. Allele-specific expression patterns reveal biases and embryo-specific parent-of-origin effects in hybrid maize. Plant Cell. 2007;19(8):2391–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar RM, Gardiner JM, Oldre AG, Haun WJ, Chandler VL, Springer NM.. Gene expression analyses in maize inbreds and hybrids with varying levels of heterosis. BMC Plant Biol. 2008;8:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar RM, Hermanson PJ, Springer NM.. Nonadditive expression and parent-of-origin effects identified by microarray and allele-specific expression profiling of maize endosperm. Plant Physiol. 2007;145(2):411–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar RM, Springer NM.. Cis-transcriptional variation in maize inbred lines B73 and Mo17 leads to additive expression patterns in the F1 hybrid. Genetics. 2006;173(4):2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson-Wagner RA, Jia Y, DeCook R, Borsuk LA, Nettleton D, Schnable PS.. All possible modes of gene action are observed in a global comparison of gene expression in a maize F1 hybrid and its inbred parents. Proc Natl Acad Sci USA. 2006;103(18):6805–6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitia RA, Bottani S, Birchler JA.. Cellular reactions to gene dosage balance: genomic, transcriptomic and proteomic effects. Trends Genet. 2008;24(8):390–397. [DOI] [PubMed] [Google Scholar]
- Walker K, Long R, Croteau R.. The final acylation step in Taxol biosynthesis: cloning of the taxoid C13-side-chain N-benzoyltransferase from Taxus. Proc Natl Acad Sci USA. 2002;99(14):9166–9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G, Tao Y, Liu G, Chen C, Luo R, Xia H, Gan Q, Zeng H, Lu Z, Han Y, et al. A transcriptomic analysis of superhybrid rice LYP9 and its parents. Proc Natl Acad Sci USA. 2009;106(19):7695–7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG.. Evolutionary changes in cis and trans gene regulation. Nature. 2004;430(6995):85–88. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG.. Regulatory changes underlying expression differences within and between Drosophila species. Nat Genet. 2008a;40(3):346–350. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG.. Independent effects of cis- and trans-regulatory variation on gene expression in Drosophila melanogaster. Genetics. 2008b;178(3):1831–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Kalay G.. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet. 2011;13(1):59–69. [DOI] [PubMed] [Google Scholar]
- Wray G. Evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8(3):206–216. [DOI] [PubMed] [Google Scholar]
- Xu C, Bai Y, Lin X, Zhao N, Hu L, Gong Z, Wendel JF, Liu B.. Genome-wide disruption of gene expression in allopolyploids but not hybrids of rice subspecies. Mol Biol Evol. 2014;31(5):1066–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Macaya-Sanz D, Rodgers-Melnick E, Carlson CH, Gouker FE, Evans LM, Schmutz J, Jenkins JW, Yan J, Tuskan GA, et al. Characterization of a large sexually dimorphic genome interval in Salix purpurea L. (Salicaceae). Mol Genet Genomics. 2018;293(6):1437–1452. [DOI] [PubMed] [Google Scholar]
- Zhuang Y, Adams KL.. Extensive allelic variation in gene expression in Populus F1 hybrids. Genetics. 2007;177(4):1987–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The gene expression data (Supplementary File 1), heterosis values (Supplementary File 2), and gene-trait correlations (Supplementary File 3) used in this study, as well as Supplementary Tables 1–8 and Supplementary Figs. 1–4 are available online: https://www.github.com/Willowpedia/Carlson2021_TriploidHeterosis. Original sequence read files are accessible at NCBI SRA BioProject: PRJNA785270.
Supplemental material is available at G3 online.