Abstract
We present here the Arkansas AI-Campus solution method for the 2019 Kidney Tumor Segmentation Challenge (KiTS19). Our Arkansas AI-Campus team participated the KiTS19 Challenge for four months, from March to July of 2019. This paper provides a summary of our methods, training, testing and validation results for this grand challenge in biomedical imaging analysis. Our deep learning model is an ensemble of U-Net models developed after testing many model variations. Our model has consistent performance on the local test dataset and the final competition independent test dataset. The model achieved local test Dice scores of 0.949 for kidney and tumor segmentation, and 0.601 for tumor segmentation, and the final competition test earned Dice scores 0.9470 and 0.6099 respectively. The Arkansas AI-Campus team solution with a composite DICE score of 0.7784 has achieved a final ranking of top fifty worldwide, and top five among the United States teams in the KiTS19 Competition.
Keywords: Kidney tumor segmentation, CT images, U-Net model, Biomedical imaging
1. Introduction
The 2019 Kidney Tumor Segmentation Challenge (KiTS19) provides a good platform for encouraging computational approach development for automatic kidney tumor segmentation with patient computed tomography (CT) scans. In this paper we provide our method to address the challenge question. Our method is based on neural network models and trained by the dataset provided by the KiTS19 Challenge [1].
1.1. Medical Relevance and Significance
A machine-learning algorithm for segmenting kidneys and kidney tumors would be valuable for the medical community. Tumor characteristics, such as size and shape, are routinely used both for patient prognosis and surgical planning.
One of the most important factors influencing patient survival in renal cell carcinoma is the TNM stage of the tumor [5]. The TNM staging system is widely used in oncology and is determined by the size and shape of the primary tumor, number of lymph nodes involved, and the presence or absence of distant metastasis [6]. A CT scan of the primary tumor provides enough information to ascertain the tumor portion of the cancer’s TNM stage [5]. A segmentation algorithm will help automate the determination of the cancer’s T-stage, providing valuable prognostic information to the physician and patient.
A segmentation algorithm would also help in surgical planning. The most common treatment for solid renal masses is surgery [7]. Until recently, radical nephrectomy was the standard of care, but with more advanced imaging and surgical techniques, partial nephrectomy is now more feasible [7]. Partial nephrectomy is equally effective at achieving cancer remission, but with less morbidity than a radical nephrectomy [7]. However, not all patients are suitable for partial nephrectomy. A segmentation tool will assist surgeons in determining who is a candidate for partial nephrectomy and who would be better treated with a radical nephrectomy. Such a tool would allow the surgeon to see the tumor’s size and shape, and its relationship to nearby vital structures, such as the aorta, vena cava, collecting system, etc. These tumor aspects have been shown to influence the complication rate in partial nephrectomies [8].
2. Materials and methods
In this section, we first describe the dataset used and then present our model for the biomedical imaging problem based on neural networks.
2.1. Dataset
The data were provided by the KiTS19 Challenge organization [1]. The whole dataset consisted of 300 individual patient CT scans. 210 scans were made available to the competition teams as a training set, and the remaining 90 scans were retained for testing predictions; no segmentation information was provided for these.
We used the 210 patient CT scans and corresponding ground truth provided by the KiTS19 Challenge organizers [1] for our training and validation. A validation group was set aside before training began by selecting 20% (N=42) of available patients at random. The same validation group was isolated and used for validation on all models that were tested. The remaining 168 patients were used as the training group for all models. We chose to hold out a validation set instead of using cross-validation, because using cross-validation would force us to multiply the computational time by the number of folds we had, and an independent ensemble of neural networks would have to be trained for each fold. We chose 20% to achieve a balance between having adequate training data and being able to predict our error.
2.2. Methods
We investigated several model architectures as possible solutions to this challenge. Primarily, we looked at two high-level configurations: Mask-RCNN [2] and U-Net [3]. We decided on an ensemble of U-Net models as our final configuration after testing many variations. We discuss our experience with Mask-RCNN further in the Discussion section, as well as our rationale for ultimately choosing U-Net.
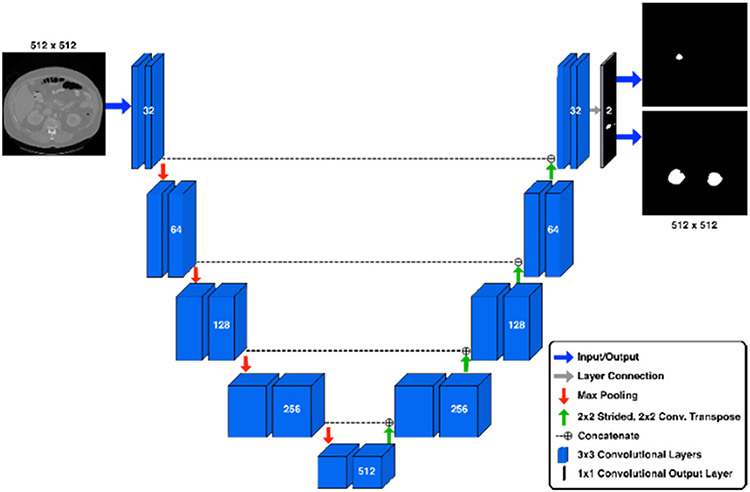
Our final ensemble consists of two U-Net models working in tandem, followed by a post-processing “cleanup” phase to minimize prediction artifacts. All of our U-Nets share the same structural architecture shown in Figure 1. The input layer accepts images of dimension 512x512 pixels. The network consists of four “downsampling” blocks, a feature representation block, four “upsampling” blocks, and one output convolutional layer. Each block contains two identical 3x3 convolutional layers. Each “downsampling” block is followed by a 2x2 max pooling operation. Each “upsampling” block is preceeded by a 2x2 2-D convolutional transpose layer with a stride of 2x2 and a concatenation with features from the corresponding downsampling block (see Figure 1). The final upsampling block connects directly to the output layer, which is a 1x1 2-D CNN layer with 2 output channels of shape 512x512 pixels. All intermediate CNN layers utilize a ReLU activation. The output layer utilizes a sigmoid activation representing the probability that any pixel location should belong to the region of interest. Feature dimension sizes for all blocks are shown in Figure 1. Both models in our ensemble were trained on axial slices, differing in the number of epochs trained and the interpretation of the output masks from each. One model was tasked with predicting the kidney and tumor masks separately in its two output channels. We will refer to this as the “K/T” model. The other model was trained to predict the combined kidney+tumor mask on the first output channel, and the tumor portion on the second output channel. We will refer to this as the “KT/T” model. The output from the two models was combined such that both models voted equally for the inclusion of any individual mask voxel, and voxels receiving a vote from either model were included in the result sent to the post-processing stage.
Fig. 1.
U-Net architecture. The U-Net operates by compressing the input image into a low-level feature representation at the apex of the U. Following this, the model expands this low-level representation into the predicted segmentations for the image.
This ensemble is unique as the two models are slightly different from each other, viewing the problem in slightly different ways. The KT/T model views the kidney and tumor as belonging together as a single unit, helping prevent errors in which the tumor voxels are predicted in locations far removed from the kidney. The K/T model is more flexible and not bound by this restriction, as it searches for the kidney independent from the tumor.
Finally, we post-processed the proposed mask by 1) filling gaps of width ≤ 2 in the tumor mask along each of the three axes, 2) computing and filling the convex hull of each connected region in the tumor mask, 3) removing any segmentations that occupy only a single ‘slice’ along each of the three axes, 4) retaining only the largest five connected regions in the tumor mask, 5) computing the two largest connected regions that intersect with a kidney segmentation in the union of the tumor and kidney masks and using those two largest connected regions to filter all proposals, removing any proposed segmentation voxels outside these two regions.
This post-processing stage removed spurious predictions as well as filling in any missing interior regions in the tumor prediction.
Pre-processing.
We found that loading the NiFTi-format files for each patient created a bottleneck in the training process, so we pre-processed the images and saved the pre-processed versions in a format that could be read directly by the Numpy [4] package. For our axial models, we saved each axial ‘slice’ in an individual Numpy file. This allowed us to load slices individually instead of loading an entire CT scan volume, further optimizing our loading times. For training with coronal and sagittal views, we saved the entire CT volume for each patient in a single Numpy file. We optimized training on these views such that all possible slices for a single patient were used preferentially before moving to a different patient, so that we could reduce the impact of the longer load times.
Our pre-processing also included a window normalization of the CT image data which imposed a threshold of the raw Hounsfield units to the range [−500, 500] and mapped the values to the numeric range [0, 1] according to the formula:
This step must also be performed prior to inference with all our models, so it is part of the input stage for the inference algorithm.
For inference, our algorithm reads the NiFTi file directly; it is not necessary to cache the image in Numpy format at this stage. The window normalization step is required as a pre-processing step during inference.
Training.
Our training data consisted of 168 scans that included a ground-truth segmentation. For each of our models, we proceeded as follows using the Keras[Keras] deep learning framework in Python with the Tensorflow[Tensorflow] back-end.
Starting weights were seeded at random and trained for eight epochs each. We continued this process until we found an initial model that seemed to be converging at a reasonable rate. Many starts did not converge in any meaningful way within the first eight epochs, and they were discarded. In general, good starting weights could be found in about five attempts.
All training for the axial models proceeded by dividing all available axial slices into two sets: ‘Positive’ slices contained at least one segmented voxel of either tumor or kidney, and ‘Negative’ slices contained no segmented voxels. We balanced our training set by randomly choosing enough slices from the positive and negative sets to create a 2:1 ratio of positive slices.
Image slices were augmented in the following ways (each augmentation had a 50% chance of being applied to any slice):
Randomly flipped vertically (this augmentation was disabled after ~135 epochs).
Randomly flipped horizontally.
Randomly shifted up to 15% in both the vertical and horizontal directions.
Randomly zoomed in/out up to 15% and recropped or padded with zeros to maintain image size (only used on epochs > 150 K/T and > 200 KT/T).
Models were trained using approximately 2000 slices per epoch. Training loss was a weighted cross-entropy loss where tumor segmentation errors were weighted ten times versus kidney segmentation errors. We also monitored a per-slice Dice metric to determine how training was proceeding.
After training the models until the training metrics indicated a performance plateau, we ranked the weights by training and validation Dice metric, and chose several top ranked checkpoints for further testing. For both axial models, we eventually trained in excess of 250 epochs, but the later checkpoints were not always best. Selected best weights were then used in an ensemble as described previously; we chose one checkpoint from the K/T and KT/T models for our final ensemble.
We provide detailed instructions for training both our K/T model and the KT/T model in the README.md file contained in our source repository on Github (https://github.com/jcausey-astate/ai_campus_kits19). The best K/T weights occurred at epoch 150 and the best KT/T weights occurred at epoch 205.
Implementation details.
We utilized both local and cloud-based Amazon Web Services (AWS) GPU instances to train our models. Our two local instances included a single NVIDIA Tesla P-40 GPU and a single NVIDIA Tesla V-100 GPU, respectively. We also utilized up to three concurrent AWS cloud instances using the Deep Learning AMI, with one NVIDIA Tesla P-100 on each instance.
3. Evaluation Metrics
Each model was evaluated and ranked by its average Sørensen–Dice score [9, 10, 11] across all CT scans in the validation set. The metric is defined by the following formula:
In this formula, TP is the number of correctly labeled voxels and FP is the number of voxels falsely labeled as belonging to the class, and FN is the number of voxels incorrectly labeled as belonging to the background. The Sørensen–Dice score is computed on a per-class basis. We ranked our models first by the Sørensen–Dice score on the union of the kidney and tumor classes and secondarily ranked them on the tumor class alone.
4. Results
4.1. Performance of our model
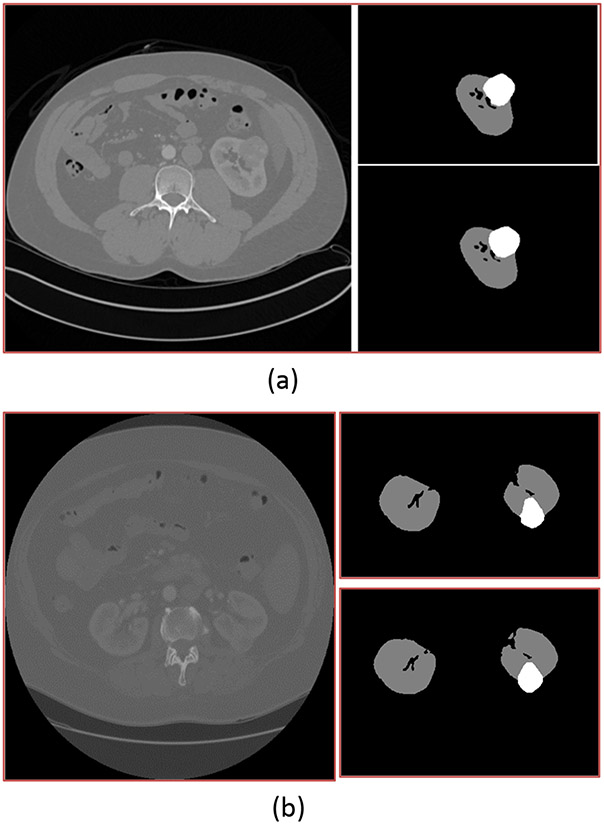
The table below shows the performance of our Arkansas AI-Campus models on our local validation group of 42 scans. Shown is the performance for each of the individual models in the ensemble, as well as the ensemble itself. The performance of the individual models does not include the described post-processing steps. Please refer to Figure 2 for exemplary prediction outputs from our ensemble model.
Fig. 2.
For both (a) and (b), the image on the left is a single slice from one patient's CT scan. The upper right image shows the outlines of the kidney and tumor as identified by radiologists, while the lower right image shows the same outline as identified by our ensemble model.
At the time of judging of the KiTS19 challenge, the Arkansas AI-Campus team solution placed 50th overall, and among the top five of teams from the United States. The evaluation of our model on the retained test set of 90 scans shows that our model has consistent performance, and it has achieved Dice scores 0.9470 for kidney and tumor segmentation, and 0.6099 for tumor segmentation respectively. The Arkansas AI-Campus model has a composite Dice score of 0.7784.
4.2. Discussion of other models
Mask-RCNN.
We attempted to adapt Mask-RCNN [2] to the segmentation problem. This model was selected for its state-of-the-art ability to perform segmentation tasks. However, we encountered challenges in adapting this model to the problem of segmenting the kidney and the tumor. Much like the U-Net model, Mask-RCNN is a 2D model and was trained with individual slices of CT scans. However, Mask-RCNN assumes that every training image will contain at least one object of interest. Training errors occur if this is not the case. To work around this problem, we added a ‘dummy’' mask consisting of a single pixel placed in a random position on each slice that did not contain kidney or tumor. The randomness was intended to prevent the model from perfectly learning the dummy mask's position.
After making these modifications, we were able to train Mask-RCNN models for the axial, coronal, and sagittal views. However, its performance was quickly outclassed by the U-Net, even in ensemble and with post-processing. We suspect this is because Mask-RCNN has a much higher model capacity (and complexity) and trained more slowly.
Multi-View U-Net.
While planning the U-Net ensemble, we planned to train models for all three views: axial, sagittal, and coronal. However, the coronal and sagittal models did not reach the same level of performance as the axial model, and ensembles containing these models underperformed ensembles with axial models only. We suspect that the reason was that these models were using the original images and not the interpolated dataset that allowed a common spacing in the Z-axis direction. This led to a much higher variance in the model's experience of anatomical structures for the non-axial views, reflected in the inability to reach adequate performance.
4.3. Further Work of Segmentation Algorithms
The development of automatic medical imaging segmentation algorithms, such as the work of the KiTS19 challenge, will contribute to the methods for imaging analysis. Also, the segmentation algorithms will be of clinical importance. Renal cell carcinomas are aggressive cancers, and further work for kidney tumor segmentation algorithms should be able to provide a tumor stage for the cancer as well as information about the tumor's size and location. Most kidney cancers are treated with surgery. Segmentation algorithms will assist surgeons in deciding whether or not a patient is a candidate for partial nephrectomy by helping decide if a sufficient margin of healthy tissue can be left behind to minimize risk of cancer recurrence.
5. Summary
The goal of the KiTS19 Challenge was to take standard human abdominal computed tomography (CT) images and build models that would scan the digital computed tomography (CT) scans and autonomously identify the kidneys and any solid tumors within the kidneys. The completion teams were initially given 210 scans to use as a training set from patients with kidney tumors identified by a team of radiologists at the University of Minnesota Medical Center. Using the training set, each team constructed models to identify tumors and were graded by how well their models performed on a separate testing set of 90 scans. The teams were scored by how closely the tumor and kidney tissue identified by their models matched tumor tissue identified by the experienced radiologists. After investigating several options, the Arkansas AI-Campus team developed an ensemble neural network algorithm that performed comparatively well and resulted in a composite Dice score of 0.7784 and 50th place globally. A limitation of the model is that it was developed iteratively during the competition, possibly leading to overfitting pressure from evaluation feedback. Additional validation on novel data would be required before deploying the model in a clinical setting.
Table 1.
The performance of our models on our local validation set of 42 CT scans.
| Model | K+T Dice, Std. Dev. |
T Dice, Std. Dev. |
|---|---|---|
| K/T axial | 0.927, 0.096 | 0.512, 0.293 |
| KT/T axial | 0.932, 0.072 | 0.517, 0.294 |
| Ensemble + post-processing | 0.949, 0.053 | 0.601, 0.292 |
Table 2.
The performance of our Mask-RCNN based models on our local validation set of 42 CT scans.
| Model | K+T Dice | T Dice |
|---|---|---|
| Mask-RCNN Axial + post-processing | 0.340 | 0.098 |
| Mask-RCNN Coronal + post-processing | 0.400 | 0.073 |
| Mask-RCNN Sagittal + post-processing | 0.463 | 0.079 |
| Mask-RCNN Ensemble + post-processing | 0.724 | 0.166 |
Acknowledgment
This research work was partially supported by National Institute of Health NCI grant U01CA187013, and National Science Foundation with grant number 1452211, 1553680, and 1723529, National Institute of Health grant R01LM012601, Arkansas Biosciences Institute grant #200144 "Develop Novel Informatics Algorithms for Lung Cancer Early Screening with CT Scans", as well as was partially supported by National Institute of Health grant from the National Institute of General Medical Sciences (P20GM103429).
Footnotes
Code Availibity
The code is available through Github (https://github.com/jcausey-astate/ai_campus_kits19), and some intermediate files we processed and generated with this study could be made available to an investigator upon request for academic, research, and noncommercial use.
Contributor Information
Jason Causey, Department of Computer Science and Molecular Bioscicneces Program, Center for No-Boundary Thinking (CNBT), Arkansas State University, Jonesboro, Arkansas 72467..
Jonathan Stubblefield, Department of Computer Science and Molecular Bioscicneces Program, Center for No-Boundary Thinking (CNBT), Arkansas State University, Jonesboro, Arkansas 72467..
Jake Qualls, Department of Computer Science and Molecular Bioscicneces Program, Center for No-Boundary Thinking (CNBT), Arkansas State University, Jonesboro, Arkansas 72467..
Jennifer Fowler, Department of Computer Science and Molecular Bioscicneces Program, Center for No-Boundary Thinking (CNBT), Arkansas State University, Jonesboro, Arkansas 72467..
Lingrui Cai, Ann Arbor Algorithm, Ann Arbor, Michigan 48103, United States of America.
Karl Walker, Department of Mathematics and Computer Science, University of Arkansas at Pine Bluff, Pine Bluff, Arkansas 55455.
Yuanfang Guan, Department of Computational Medicine & Bioinformatics, University of Michigan, Ann Arbor, Michigan 48109..
Xiuzhen Huang, Department of Computer Science and Molecular Bioscicneces Program, Center for No-Boundary Thinking (CNBT), Arkansas State University, Jonesboro, Arkansas 72467..
Data Availibity
The LIDC/IDRI data (https://luna16.grand-challenge.org/data/), LUNA16 data (https://wiki.cancerimagingarchive.net/display/Public/LIDC-IDRI) and DSB2017 Competition data (https://www.kaggle.com/c/data-science-bowl-2017/data) are publicly available through their individual websites and were previously used for biomedical imaging studies and computational approach development and testing by different research groups in the research field. The NLST data is NCI-controlled data; different research groups get their permission from NCI to use the NLST data for their study. Please refer to the NCI website for the information (https://biometry.nci.nih.gov/cdas/publications/?study=nlst).
References
- [1].Heller N, Sathianathen N, Kalapara A, Walczak E, Moore K, Kaluzniak H, Rosenberg J, Blake P, Rengel Z, Oestreich M, Dean J, Tradewell M, Shah A, Tejpaul R, Edgerton Z, Peterson M, Raza S, Regmi S, Papanikolopoulos N, and Weight C, “The KiTS19 Challenge Data: 300 Kidney Tumor Cases with Clinical Context, CT Semantic Segmentations, and Surgical Outcomes,” arXiv:1904.00445 [cs, q-bio, stat], Mar. 2019. [Google Scholar]
- [2].He K, Gkioxari G, Dollár P, and Girshick R, “Mask R-CNN,” arXiv:1703.06870 [cs], Mar. 2017. [DOI] [PubMed] [Google Scholar]
- [3].Ronneberger, Fischer P, and Brox T, “U-net: Convolutional networks for biomedical image segmentation,” in International Conference on Medical Image Computing and Computer-Assisted Intervention, 2015, pp. 234–241. [Google Scholar]
- [4].van der Walt S, Colbert SC, and Varoquaux G, “The NumPy Array: A Structure for Efficient Numerical Computation,” Computing in Science Engineering, vol. 13, no. 2, pp. 22–30, Mar. 2011. [Google Scholar]
- [5].Cohen HT and McGovern FJ, “Renal-Cell Carcinoma,” New England Journal of Medicine, vol. 353, no. 23, pp. 2477–2490, Dec. 2005. [DOI] [PubMed] [Google Scholar]
- [6].National Cancer Institute, “Cancer Staging,” https://www.cancer.gov/about-cancer/diagnosis-staging/staging.
- [7].Picken MM, Wang L, and Gupta GN, “Positive Surgical Margins in Renal Cell Carcinoma Translating Tumor Biology Into Clinical Outcomes,” American Journal of Clinical Pathology, vol. 143, no. 5, pp. 620–622, May 2015. [DOI] [PubMed] [Google Scholar]
- [8].Ficarra Vincenzo, et al. “Preoperative aspects and dimensions used for an anatomical (PADUA) classification of renal tumours in patients who are candidates for nephron-sparing surgery,” European urology 56.5 (2009): 786–793. [DOI] [PubMed] [Google Scholar]
- [9].Sørensen T (1948). "A method of establishing groups of equal amplitude in plant sociology based on similarity of species and its application to analyses of the vegetation on Danish commons". Kongelige Danske Videnskabernes Selskab. 5 (4): 1–34. [Google Scholar]
- [10].Dice Lee R. (1945). "Measures of the Amount of Ecologic Association Between Species". Ecology. 26 (3): 297–302. doi: 10.2307/1932409. JSTOR 1932409. [DOI] [Google Scholar]
- [11].Carass A; Roy S; Gherman A; Reinhold JC; Jesson A; et al. (2020). "Evaluating White Matter Lesion Segmentations with Refined Sørensen-Dice Analysis". Scientific Reports. 10 (1): 8242. Bibcode:2020NatSR..10.8242C. doi: 10.1038/s41598-020-64803-w. ISSN 2045-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Skourt BA, Hassani AE, & Majda A (2018). Lung CT Image Segmentation Using Deep Neural Networks. Procedia Computer Science, 127, 109–113. doi: 10.1016/j.procs.2018.01.104 [DOI] [Google Scholar]
- [13].Dong X, Lei Y, Wang T, Thomas M, Tang L, Curran WJ et al. , Automatic multiorgan segmentation in thorax CT images using U-net-GAN. Med Phys. 2019; 46: 2157–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The LIDC/IDRI data (https://luna16.grand-challenge.org/data/), LUNA16 data (https://wiki.cancerimagingarchive.net/display/Public/LIDC-IDRI) and DSB2017 Competition data (https://www.kaggle.com/c/data-science-bowl-2017/data) are publicly available through their individual websites and were previously used for biomedical imaging studies and computational approach development and testing by different research groups in the research field. The NLST data is NCI-controlled data; different research groups get their permission from NCI to use the NLST data for their study. Please refer to the NCI website for the information (https://biometry.nci.nih.gov/cdas/publications/?study=nlst).