Abstract
Posttraumatic stress disorder (PTSD) is a psychiatric disorder that may arise in response to severe traumatic event and is diagnosed based on three main symptom clusters (reexperiencing, avoidance, and hyperarousal) per the Diagnostic Manual of Mental Disorders (version DSM-IV-TR). In this study, we characterized the biological heterogeneity of PTSD symptom clusters by performing a multi-omics investigation integrating genetically regulated gene, splicing, and protein expression in dorsolateral prefrontal cortex tissue within a sample of US veterans enrolled in the Million Veteran Program (N total=186,689). We identified 30 genes in 19 regions across the three PTSD symptom clusters. We found nine genes to have cell-type specific expression, and over-representation of miRNA-families – miR-148, 30, and 8. Gene-drug target prioritization approach highlighted cyclooxygenase and acetylcholine compounds. Next, we tested molecular-profile based phenome-wide impact of identified genes with respect to 1678 phenotypes derived from the Electronic Health Records of the Vanderbilt University biorepository (N=70,439). Lastly, we tested for local genetic correlation across PTSD symptom clusters which highlighted colocalization and local genetic correlation with metabolic (e.g., obesity, diabetes, vascular health) and laboratory traits (e.g., neutrophil, eosinophil, tau protein, creatinine kinase). Overall, this study finds comprehensive genomic evidence including clinical and regulatory profiles between PTSD, hematologic and cardiometabolic traits, that support comorbidities observed in epidemiologic studies of PTSD.
Keywords: PTSD, brain, transcriptomics, metabolic, blood, expression
3. Introduction
Post-traumatic stress disorder (PTSD) is a psychiatric disorder that may arise in response to severely stressful events. The lifetime prevalence of PTSD in the United States is estimated at approximately 7%1,2, and more than 12% in the active US veteran population3. Twin-based studies estimated a 24–72% heritability of PTSD with strong variation depending on the characteristics of the cohort investigated4. Recent large-scale efforts from the Psychiatric Genomics Consortium and the Million Veteran Program (MVP) have identified several genetic risk loci associated with PTSD1. However, the understanding of processes underlying translation of genetic associations into pathophysiological processes remains limited due to symptomatic heterogeneity of PTSD patients and the multiplicity of implicated regulatory elements. As per the Diagnostic and Statistical Manual for Mental Disorders—Fourth Edition, Revised (DSM-IV), 17 symptoms of PTSD comprise three main clusters: reexperiencing i.e. recollection of traumatic memories, avoidance i.e avoiding thoughts of traumatic events, and hyperarousal i.e. irritability, reactivity, or high vigilance5. Furthermore, PTSD is highly polygenic and since genetic variants can affect multiple regulatory elements, it is challenging to prioritize targets for personalized therapeutics. Neuroimaging studies indicate PTSD is associated with structural and functional changes in the dorsolateral prefrontal cortex (dlPFC)6, which may impair learning and memory, executive cognitive functions, and emotional regulation7, and is therefore an important tissue to be investigated in the context of PTSD pathogenesis.
In the current study, we performed a characterization of PTSD symptom clusters by integrating mRNA, alternative splicing, and proteome expression of the dlPFC (N=1,280) with genome-wide association data of reexperiencing, avoidance, and hyperarousal symptoms from the MVP (N=186,689)1. We identified several novel dlPFC-specific genetically regulated loci through gene/splicing/protein-wide transcriptome-wide association studies (TWAS/SPWAS/PWAS). These loci were further investigated for their biological, and phenome-wide impact. To elucidate biological impact, we analyzed the identified genes for their role in brain cell-type specific expression, cortex-specific gene-co-expression, miRNAs, and drug targets. To assess the phenome-wide impact of identified loci, we applied two molecular-evidence based approaches. First, we performed a phenome-wide association study (PheWAS) using genetically-regulated expression (GReX) of identified genes in the electronic health records and laboratory measurements from the Vanderbilt Biobank repository (BioVU; n=70,439). Second, we performed phenome-wide colocalization analysis of the identified genes (i.e. exhibit a high probability of sharing the same causal variant between expression and phenotype). We further investigated these colocalizing traits with PTSD symptom clusters using locus-specific genetic correlation. Our findings also contribute new information on several molecular mechanisms associated with PTSD symptoms and highlight potential therapeutic targets in the context of PTSD comorbidities (Figure 1A).
Figure 1:
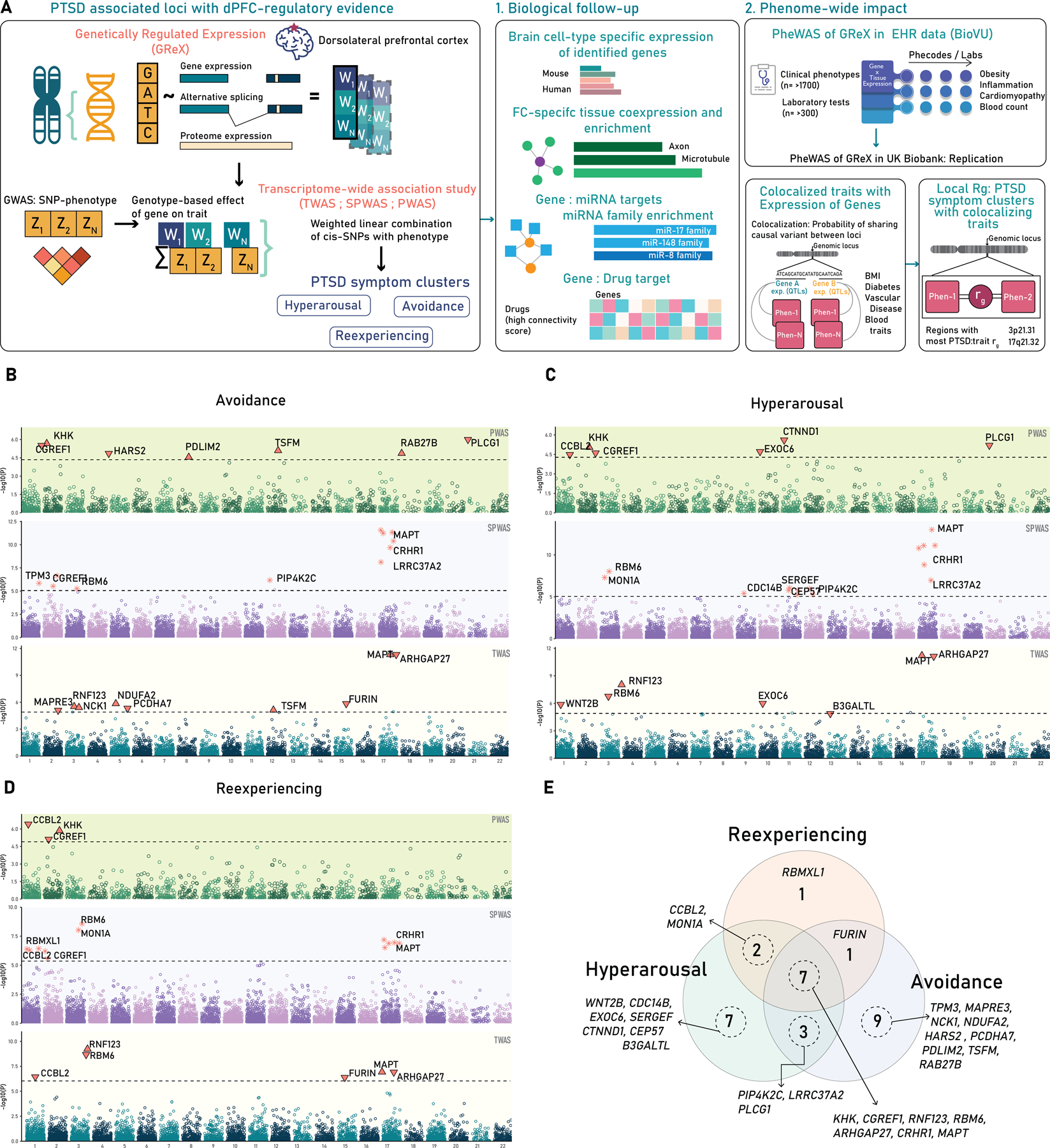
A. Study Design. A schematic of the study design showing the concept of genetically regulated expression (GReX) and applied to three PTSD symptom cluster. B-D. Three-tiered Manhattan plots of three PTSD symptoms B-Avoidance, C-Hyperarousal, D-Reexperiencing, the lowest layer is TWAS, middle is SPWAS, and top is PWAS (See text for more details). E. The Venn diagram of overlapping genes across the three PTSD symptom clusters.
4. Subjects and Methods
4.1. GWAS of PTSD symptom clusters
We obtained authorized access to Database of Genotype and Phenotype (dbGaP) for the genome-wide statistics of PTSD symptom clusters: reexperiencing, avoidance, and hyperarousal from the MVP (dbGaP Study Accession: phs001672.v6.p1)1. This GWAS of PTSD and its quantitative symptom clusters included 186,689 US military veterans from the MVP. Genome-wide association statistics reflect results from individuals of European ancestry, inverse-variance meta-analyzed across two tranches, and reported with variant-level beta, standard errors, and p values1. We performed strict quality control using the munging procedure provided by LD-score regression (LDSC)8. Our quality control procedure resulted in a final count of 1,191,504 high-quality HapMap autosomal genetic variants.
4.2. Gene expression weights for dlPFC and TWAS
We obtained pretrained models of gene and splicing expression of the dlPFC fitted in the data from the Common-Mind Consortium (CMC; N=452; 5,420 expression transcript and 7,772 splicing transcripts) stored in the FUSION repository (See URLs). The quality control of RNA-seq, RNA-seq splicing, and genotype from CMC data has been previously described in detail 9,10. The proteome expression weights for the dlPFC (n=376; features - 1467) were leveraged from the Religious Orders Study and Memory and Aging Project (ROSMAP) published by Wingo et. al11 (See URLs). Briefly, the expression weights were generated using FUSION’s recommended pipeline described previously for the CMC data9. The transcriptome-wide association study using gene, splicing, and proteome expression models was performed using FUSION9, and genome-wide transcriptomic correlation for each of TWAS/SPWAS/PWAS across three PTSD symptom clusters was tested using RhoGE12. We applied Bonferroni correction (0.05/#genes tested) for multiple testing correction. The genes identified through T/SP/PWAS approach as hereon referred to as PTSD-associated genes.
4.3. Biological characterization
4.3.1. Brain cell-types expression
We looked up the identified PTSD-associated genes, in the brain cell-type-specific lists from McKenzie et. al13. Briefly, the McKenzie et. al identified top genes for six brain-cell types by combining three mouse brain cell expression datasets from the cerebral cortex, somatosensory cortex and hippocampus, and primary visual cortex, and two human brain cell expression datasets from the temporal lobe.
4.3.2. Frontal cortex-based gene co-expression and gene ontology enrichment
To identify genes that are co-expressed with the identified genes in the frontal cortex, we used the GTEx v8 data stored within TCSBN (tissue and cancer specific biological network), a database of tissue-specific biological networks14. We extracted co-expression modules for frontal cortex, and created the minimum network (i.e., shortest path between query genes and co-expressed genes) to retain the most essential or closest genes using the igraph package in R. For all the genes in the network we performed an enrichment test for gene ontology of molecular function, biological processes, and cellular components. We applied false discovery rate (FDR q <0.05) for multiple testing correction.
4.3.3. miRNA-gene network and miRNA family enrichment
We mapped the PTSD-associated genes to miRNAs from the miRTarBase (hsa) using the Official Gene Id and performed enrichment of miRNA families with the miRNetR package15. FDR multiple testing correction (FDR q<0.05) was applied on enrichment p-values.
4.3.4. Gene-drug assessment
We used the Connectivity Map (CMap) Drug database to identify candidate drug targets for the PTSD-associated genes. The CMap database was accessed via the CLUE interface (See URLs), which compared the query gene list against the database of differentially expressed genes associated with various experimental perturbagens including knockdowns, overexpression, and chemical compounds. The median connectivity score ranges from −100 to 100 indexes the similarity between the CMap-reference expression pattern and the list of query genes.
4.4. Phenome-wide Assessment
4.4.1. PheWAS of genetically-predicted expression in electronic health records
To understand the phenotypic consequences of dysregulated mRNA expression across our 30 genes of interest, we performed a phenome-wide association study (PheWAS)16 using the Vanderbilt EHR and linked biobank, BioVU. Phenotypes in BioVU are represented as dichotomous traits called phecodes and they reflect a hierarchical clustering of the International Classification of Diseases 9th and 10th (ICD9/ICD10) codes. For each phenotype we required a minimum number of 100 cases which resulted in the inclusion of 1379 phecodes in 70,439 individuals of European ancestry. We used the PheWAS package in R to perform logistic regression to identify the phecodes that are significantly associated with genetically imputed gene expression of brain tissues after adjusting for sex, age, and the top ten within-ancestry principal components from genetic data to control for population stratification (Denny et al. 2010, 2013). Multiple testing correction (FDR q < 0.05) was applied per tissue.
4.4.2. BioVU Biomarker LabWAS
The lab-wide association scan (LabWAS)17 permitted screening of clinical lab tests from the Vanderbilt University Medical Center EHR. For each of the 30 genes identified in the analyses, we tested the association between its predicted gene expression pattern in brain tissues and the available set of clinical labs. We applied the QualityLab cleaning pipeline 17 with settings to yield median age-adjusted (residual taken after regressing the cubic splines of age with 4 knots) inverse normal quantile transformed lab values (to control for skewness and non-normality). We included labs with measurements in at least 100 individuals, which resulted in 299 labs in 70,439 individuals of European ancestry. The lab tests are divided into 12 sub-categories; blood, metabolic, endocrine, kidney, immune, liver, urinary, OB/GYN, toxicology, cardiovascular, and cancer. Our analyses included the covariates age, sex, and top ten within-ancestry principal components from genetic data to adjust for genetic ancestry. Multiple testing correction (FDR q< 0.05) was applied per tissue.
We replicated the genetically predicted PheWAS findings in an external cohort of the UK Biobank (UKBB) using PhenomeXcan18 which uses complementary TWAS method, S-MultiXcan, and gene expression weights from Genotype-Tissue Expression (GTEx) consortia 19. For the traits identified in BioVU cohort, we extracted phenotype associations for the same tissue and gene pairs identified in BioVU’s PheWAS and LabWAS. We selected – ‘Brain Cortex’, ‘Brain Cerebellum’, and ‘Brain Frontal Cortex BA9’, and four genes - LRRC37A2, ARHGAP27, RNF123, and WNT2B. Two traits from BioVU (‘Protein serum plasma’ and ‘Tau protein’) were not available in PhenomeXcan/UKBB cohort, and hence were not tested. An absolute z-score of 3 (or p=0.0013; 0.05/8 phenotypes reflecting Bonferroni correction) or higher was considered as evidence of replication.
4.4.3. Colocalization of identified loci with traits
We investigated all genes identified in this study for colocalization against all the phenotypes available in data resource-OpenTarget Genetics20. The colocalization was performed using the coloc-R package as described previously21, and the traits were considered to be colocalized when posterior probability (PP) of the hypothesis (H4) that two loci are shared under a single causal variant was H4>= 80%. We performed enrichment of phenotype categories as listed within OpenTarget Genetics using hypergeometric test in two ways; i) an ‘all-tissue: expression’ and ii) filtered tissues using the keyword “brain”. The hypergeometric test was performed for each phenotype category as ratio for colocalized/not-colocalized per category.
4.4.4. Local genetic correlation
We mapped 147 out of 172 colocalizing traits using the available StudyID from OpenTargets with UKBB phenotype ID (from Neal lab v2) to download their munged summary statistics (See URLs). To estimate local genetic correlation (rg) at 19 identified regions for 147 traits and the three PTSD symptom traits, we first estimated the heritability of the regions as recommended by Local Analysis of [co]Variant Association (LAVA) developers22. We applied an FDR multiple testing correction on heritability estimates, and subsequently performed local rg for region:trait pairs that survived multiple testing threshold (FDR q<0.05).
5. Results
5.1. Integrative analyses of dlPFC identifies PTSD symptom cluster-associated genes
We tested genetically regulated gene, splicing, and proteome expression of the dlPFC across the three PTSD symptoms clusters: reexperiencing, hyperarousal, and avoidance. We identified associations across our three regulatory models – TWAS, SPWAS, and PWAS, resulting in 11 unique genes for reexperiencing (6-gene expression; 7-splicing expression; 3-proteome expression), 19 unique genes for hyperarousal (7-gene expression; 9-splicing expression; 6-proteome expression), and 20 unique genes for avoidance (9-gene expression; 7-splicing expression; 7-proteome expression) (Figure 1B–D; Table S1). In total, we identified 30 unique gene associations at 19 independent genomic regions across the three PTSD symptom clusters. Seven genes (KHK, CGREF1, RBM6, MAPT, CRHR1, RNF123, ARHGAP27) were common to all the three PTSD symptoms, while one (RBMX1), seven (EXOC6, CDC14B, CTNND1, SERGEF, CEP57, WNT2B, B3GALTL) and nine (HARS2, PDLIM2, TSFM, RAB27B, MAPRE3, NDUFA2, PCDHA7, TPM3, NCK1) were distinct to Reexperiencing, Hyperarousal and Avoidance respectively (Figure 1E). We compared the genes identified from the GWAS of PTSD1, and the genes found through our TWAS/SPWAS/PWAS approach. Genes outside a 2MB window around the lead gene reported by the GWAS were considered novel. We found 17 genes that were novel relative to the GWAS of PTSD symptom clusters (Table S2). The transcriptomic correlation (RhoGE) for each of the TWAS/SPWAS/PWAS of nominally significant genes across three PTSD symptoms was >90% (p<2.92E-92; Table S3), which suggests shared regulatory architectures underlying susceptibility for each cluster.
5.2. Biological characterization of identified genes
We performed a biological evaluation of the 30 genes for gene-brain cell types, frontal-cortex based gene co-expression, miRNA family enrichment and drug targets.
5.2.1. Brain cell-type specific expression
To identify if any of the observed genes were enriched for brain cell types, we explored the mouse and human single-cell datasets from McKenzie et. al13. We found nine genes that showed specificity for five cell-types; astrocytes (CCBL2), endothelia (ARHGAP27, EXOC6), microglia (ARHGAP27, KHK, TPM3), neuron (MAPT, RAB27B), and oligodendrocytes (EXOC6, MAPT, PDLIM2, PIP4K2C) (Supplementary Figure 1; Table S4).
5.2.2. Prefrontal-cortex specific gene co-expression and gene ontology enrichment
To understand the effect of co-expressed genes, we created a network of genes co-expressed in the frontal cortex with the identified genes and retained 22 co-expressed genes with 12 TWAS-derived genes. We tested all the genes in the network for gene ontology and identified two cellular function processes; axon (p-value=4.04e-05) and microtubule (p-value=1.78e-04) that were FDR significant. Other nominally significant processes were neuron projection, cell projection part, and microtube cytoskeleton among others (Figure 2A–B; Table S5).
Figure 2:
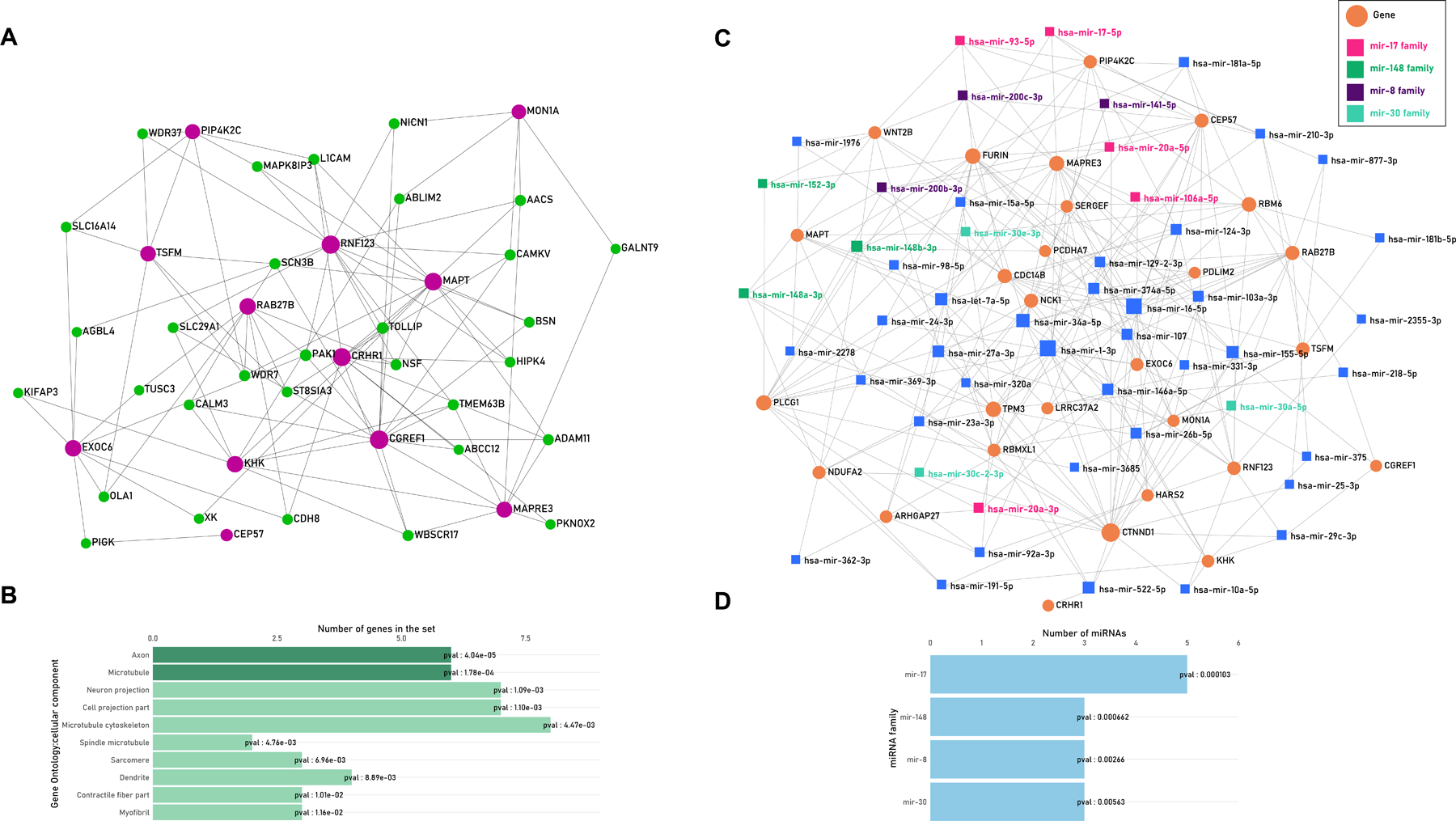
A. Tissue Co-expression Network. The network of TWAS genes (purple) with prefrontal-cortex based co-expressed genes (green) B. Bar plot showing top ten gene ontology enrichment processes of all genes in the network wherein dark green processes survived FDR testing. C. MiRNA-Gene network. The network of miRNAs (orange) which map to TWAS genes (orange circles); the miRNAs belonging to overrepresented miRNA-families are color coded in the legend. D. The details of the miRNA-family enrichment are shown as bar plot, all the four miRNA families survived FDR q<0.05.
5.2.3. Gene-miRNA mapping and enrichment
MiRNAs are short non-coding RNA sequences that regulate gene expression. Based on structure and function, miRNAs can be grouped into families. Using 28 genes of interest that were available in the database, we identified 51 miRNAs that interacted with > 2 genes. Enrichment analysis identified four miRNA families surviving FDR < 0.05, including 5 members within the miRNA-17 family, 3 members within the miRNA-148 family, 3 members within the miRNA-8 family, and 3 members within the miRNA-30 family. (Figure 2C–D; Table S6).
5.2.4. Gene-drug prioritization
To prioritize drug targets, we assessed perturbed drug-gene signatures similar to the transcriptional signatures of the identified genes, by performing a gene-drug target assessment using CMap. We found data pertaining to 11/30 genes and used the absolute median connectivity score of 90 or higher as prioritized interacting gene-drug targets. We found a total of 10 drugs with overall median connectivity score >90. These top drugs primarily include cyclooxygenase or COX-2 inhibitors, and acetylcholine receptor antagonists (Supplementary Figure 2).
5.3. Phenome-wide assessment of identified loci
5.3.1. PheWAS and LabWAS of GReX in BioVU and UKBB cohorts
We characterized the phenome-wide impact of the identified loci by analyzing clinical phenotypes in 70,439 individuals of European ancestry from BioVU cohort. We investigated GReX associations of 6 brain tissues for 1,379 clinical phenotypes and 299 laboratory measures. The PheWAS approach identified 5 phenotypes including bariatric surgery, obesity, hypertrophic cardiomyopathy, myoclonus, inflammatory bowel disease (Table S7; Figure 3A). The enrichment of phenotype categories shows a 9.3-fold increase (p-value=0.0045) for digestive processes (Figure 3B; Table S8). The LabWAS identified 12 blood traits – mean corpuscular hemoglobin concentration (MCHC), Eosinophils, tau protein, and protein-serum plasma volume (Table S9). The statistically overrepresented phenotype category was blood-related/hematological traits (7.13 log-fold enrichment; p-value=0.00027; Table S10).
Figure 3:
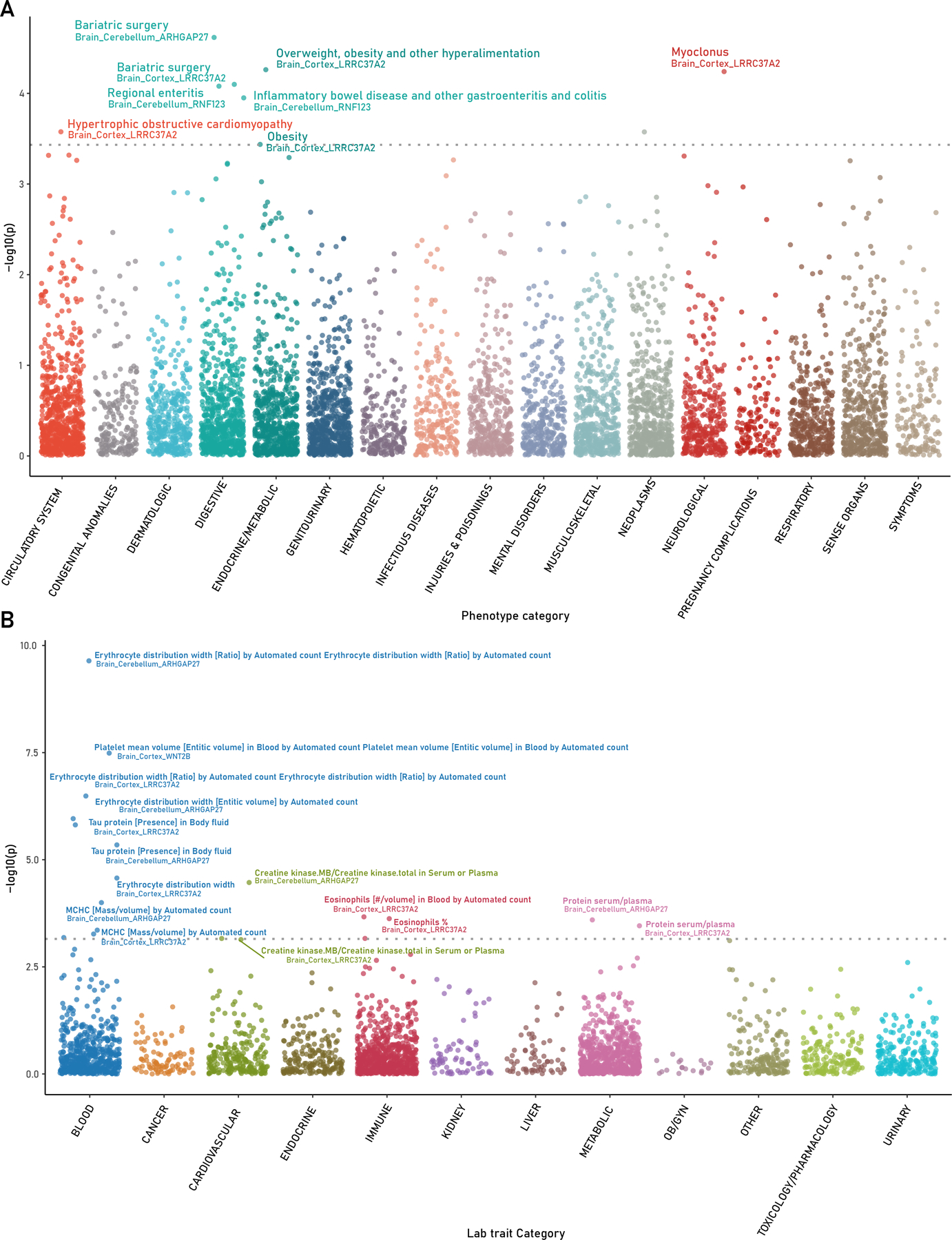
PheWAS and LabWAS of TWAS identified genes in BioVU cohort. A. PheWAS of 1379 clinical phenotypes and B. LabWAS of 299 laboratory measures using genetically regulated expression of identified TWAS genes and brain tissues.
We replicated these phenotypes in the UKBB cohort (N~100,000–300,000) using brain cortex, prefrontal cortex, and cerebellum-based GReX associations as per the gene:tissue pairs identified in BioVU (Table S11). Two traits from BioVU (‘Protein serum plasma’ and ‘Tau protein’) were not available in UKBB cohort. For hypertrophic cardiomyopathy identified in BioVU cohort, we found Illness of father, mother, and siblings- heart disease (LRRC37A2; zscore-3.1~4.1) in the UKBB cohort. For hematological traits - Erythrocyte distribution width, mean corpuscular hemoglobin concentration, platelet mean volume, identified in BioVU cohort, we found ‘Red blood cell erythrocyte count’ (ARHGAP27; z-score= 4.7~17.2), ‘Raw-mean corpuscular hemoglobin’ (ARHGAP27, LRRC37A2; z-score= −8.5 ~ −6), ‘Mean platelet thrombocyte volume’/ ‘Platelet count’ (WNT2B; z-score= −8.7~11.15) in UKBB cohort respectively. Metabolic traits – obesity, and bariatric surgery in BioVU cohort were replicated with phenotypes – Trunk fat percentage, body fat percentage, and trunk fat mass (ARHGAP27, LRRC37A2; z-score= −5.3 ~ −3.6) in UKBB cohort. Together, these replications highlight that GReX of the identified genes with PTSD is also involved in peripheral/non-psychiatric traits.
5.3.2. Colocalization of gene-QTL with human phenome
We employed the colocalization approach to assesses the probability that the association signal responsible for the predicted differences in expression and that the association signal for the trait are being caused by the same variant across a larger set of nearby cis-variants 23. Colocalization is a complementary approach to GReX- based methods that aggregates several cis-variants in the region. We identified 2,457 phenotype:gene:tissue pairs with H4 probability >= 80% across 20 genes and 74 tissues (Figure 4A). To prioritize phenotypes, we performed an enrichment test using the OpenTarget-assigned categories and found 12 over-represented categories that survived multiple testing correction (Figure 4B). We then selected tissues that contained the term ‘Brain’ in their description, we found 489 phenotypes:tissue:gene pairs across four genes and 12 brain tissues. Here, we found four enriched categories that were the same categories as seen in all tissue analysis (Figure 4C). Overall, we identified 172 unique phenotypes and we were able to map 147 phenotypes available in UKBB to test their pleiotropic relationship with PTSD symptom clusters using local rg (Table S12).
Figure 4:

A. Colocalization plot showing probability of having shared causal variant between gene expression (top-x axis), and phenotype (left axis), grouped by categories (right axis). B. Enrichment of Phenotype categories that survive multiple testing, comparing colocalized vs non-colocalized gene:phenotype pairs for all tissues. C. Enrichment of Phenotype categories comparing colocalized vs non-colocalized gene:phenotype pairs that survive multiple testing for brain tissues only.
5.3.3. Local genetic correlation of PTSD symptom clusters and colocalizing phenotypes
We sought to confirm the locus-specific relationship between expression-colocalizing traits from UKBB, and PTSD symptom clusters from the MVP, we performed a local rg analysis. We tested three PTSD symptom clusters across all 19 regions for heritability, and regions with significant heritability were tested further for local rg within and between three PTSD symptom clusters and 147 colocalizing traits, for a total of 450 PTSD:trait pairs being investigated. We found that the highest number of correlated traits were observed in 3p21.31 (231/450), followed by 17q21.32 (107/450), and 1p13.2 (103/450) locus (Figure 5; Table S13). Using the same phenotype categories as colocalization analyses, we performed an enrichment of phenotype categories for each of the three loci (Table S14). Across the 16 phenotype categories, the 3p21.31 locus which houses three TWAS genes (RBM6, RNF123, MON1A) was significantly enriched for anthropometric traits (3.39 log fold enrichment; p-value: 1.95 ×10−5), and uncategorized category (2.30 log fold enrichment; p-value: 5.29 ×10−3), which contains miscellaneous phenotypes such as lifestyle factors, medication use for vascular health, and body composition traits such as impendence of leg and arm. At the 3p21.31 locus, there is a positive rg between PTSD symptom clusters and several metabolic and blood traits (i.e., high blood pressure, cholesterol-lowering medication, BMI, Illness of father, diabetes; white blood cell count; neutrophil count). The 17q21.32 locus (ARHGAP2, CRHR1, MAPT, LRRC37A2) was enriched for hematological measurement (7.25 log fold enrichment; p-value: 3.20 ×10− 8), PTSD symptom clusters (8.13 log fold enrichment; p-value: 4.95 ×10− 3), and biological processes (11.4 log fold enrichment; p-value: 1.25 ×10−2). The 1p13.2 locus (WNT2B) was enriched for anthropometric traits (5.76 log fold enrichment; p-value: 1.55 ×10−11), and bone measurements (4.77 log fold enrichment; p-value: 7.53 ×10−5).
Figure 5:
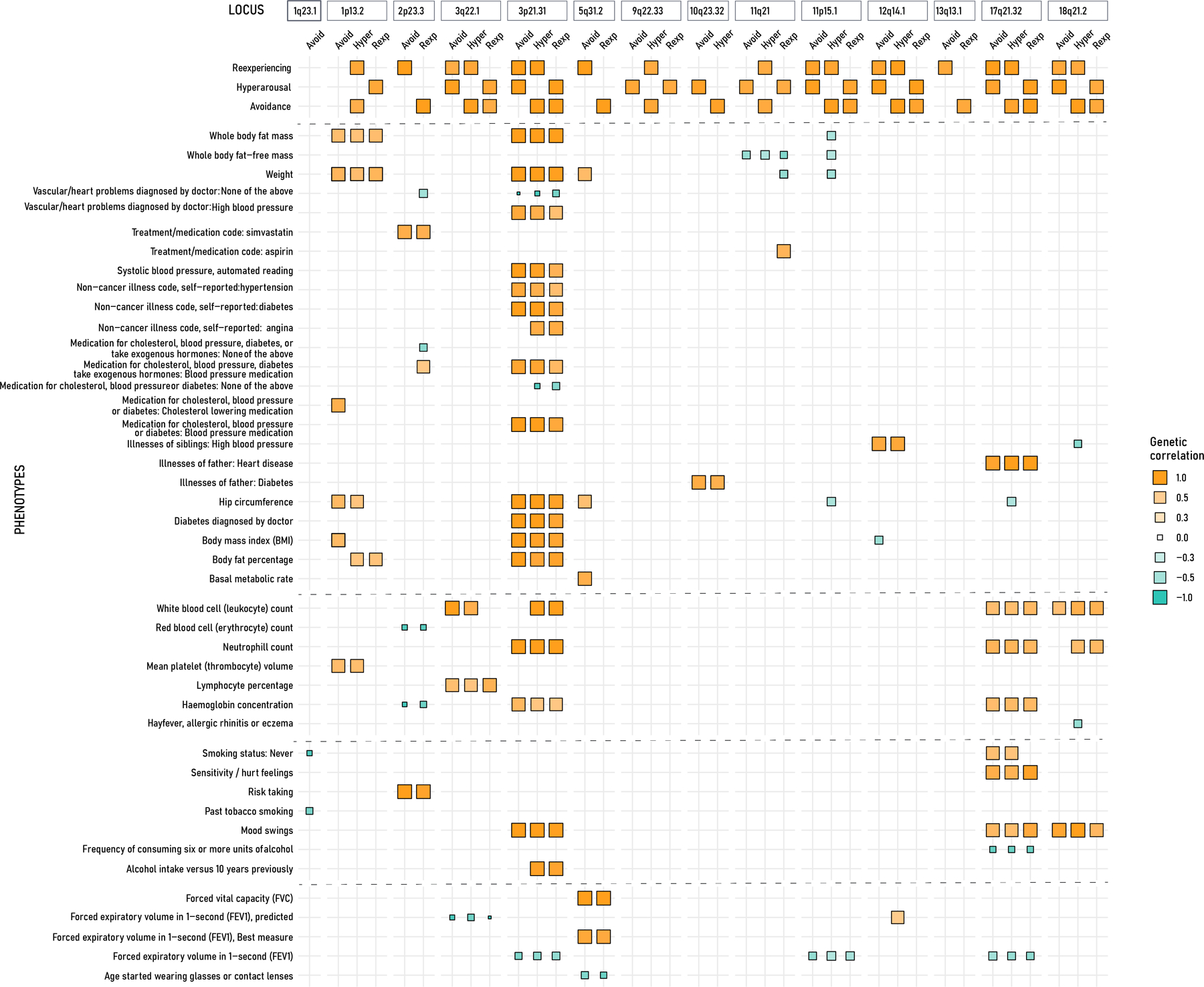
Matrix plot of local genetic correlation between three PTSD symptom clusters and colocalizing traits (few presented here on the left y-axis). The x-axis shows PTSD symptoms grouped for each locus as identified in the TWAS analysis. The positive correlation is denoted in orange, and blue is negative correlation, the size of the squares is relative to genetic correlation value.
6. Discussion
PTSD is characterized by symptoms related to hyperarousal (hypervigilance, irritability, sleep difficulties, heightened startle response), avoidance of situations and people that serve as reminders of the trauma, and reexperiencing the traumatic event, as for example in the form of nightmares, flashbacks and/or psychosomatic episodes5. To disentangle the molecular basis of PTSD symptom clusters, we integrated the multi-tiered genetically regulated expression of gene, splicing, and protein levels, identifying 30 genes in 19 genomic regions associated with the three main PTSD symptom clusters. We identified 17 novel genes that were not identified in the previous GWAS of PTSD symptom clusters1. Four of these genes (e.g., CCBL2, FURIN, KHK, and PDLIM2) have been previously identified in gene expression studies of PTSD24–27, highlighting the importance of genetically-regulated expression in PTSD pathogenesis. CCBL2 (also known as KYAT3) showed evidence of genetically predicted transcript expression, splicing, and protein expression and has previously shown specificity to astrocytes. This gene encodes a transaminase in the L-tryptophan catabolic pathway, converting kynurenine to kynurenic acid, which has antagonistic effects on glutamate receptors; dysregulation of this metabolic pathway has been implicated in a wide variety of neuropsychiatric disorders. KHK and PDLIM2 genes were specific for microglia and oligodendrocyte, respectively. Variants in FURIN were reported to exhibit gene-environment interaction effect with lifetime trauma burden28. We identified 14 miRNAs in four miRNA families that target multiple PTSD associated genes; among these, miRNA-20a (miR-17 family), miRNA-200c (miR-8 family), and miRNA-30c (miR-30 family) were significantly dysregulated in PTSD among military combat veterans29,30. These miRNAs along with miRNA-148a (miRNA-148 family) were also identified in rodent models of PTSD31,32. The present observation of genetically regulated expression of the genes regulated by these miRNAs highlights the possibility that genetic variants could affect target binding for miRNAs which also serve as compensatory mechanisms.
To understand the impact of GReX across the clinical and laboratory phenome, we performed PheWAS and LabWAS in BioVU. The brain-tissue based GReX of the TWAS genes highlighted obesity (and related bariatric surgery), cardiomyopathy, inflammation (e.g. inflammatory bowel disease and regional enteritis), neurological (myoclonus-twitching of muscles), and several hematological traits (erythrocytes, eosinophils, mean corpuscle hemoglobin) and these findings were replicated in the larger UKBB cohort. We observed an overrepresentation of similar phenotypes using colocalization as a complementary approach (i.e., anthropometric, metabolic traits including diabetes, and cardiovascular traits including, vascular health, blood pressure medication, and hematological traits). Our local genetic correlation findings provided further support for pleiotropic associations between PTSD, metabolic and hematological traits localize in 3p21.31 and 17q21.32, respectively. Our findings also highlight some loci and genes that may exert pleiotropic effects across multiple comorbid domains.
Several epidemiologic studies identify associations between PTSD and metabolic dysfunction like diabetes, hypertension, and abnormal lipid profiles33,34, which are risk factors to cardiovascular dysfunction and failure35,36. Our hypothesis-free phenome-wide approach provides convergent evidence and highlights loci and genes that appear to mediate risk for both PTSD symptoms and multiple associated metabolic disorder. The genes, RBM6, RNF123, MON1A (3p21.31), and WNTB2 (1p13.2) have shared effects in PTSD and metabolic-traits, which have been demonstrated to have highly pleiotropic effects traits 37. RBM6 and RNF123 were reported in GWAS and expression studies of PTSD, and cardiovascular diseases38, which may be due to association with cardiovascular-risk factors such as diabetes, and lipid metabolism 39,40. WNTB2 is associated with stress-induced neuroinflammation in PTSD41 and depression42. ARHGAP2, CRHR1, MAPT, and LRRC37A2 (17q21.32) are in a high LD region. These genes are known to be associated with PTSD1, but they also have functional links with metabolic traits hypothesized to guide diet-based signals via dysregulated expression in brain tissues43. Additionally, LRRC37A2 was found to be dysregulated in CD133+ cells which are involved in vascular injury response44. While directly measured gene expression is highly influenced by environmental and technical factors, genetically regulated expression is anchored in the inherited genetic variation.
Inflammation is a well-documented common denominator between PTSD and metabolic dysfunction45. Hematological measurements such as white blood cell and platelet count are considered inflammatory markers and are often altered in PTSD patients46,47. Several studies have reported short and long-term changes in inflammatory levels as a cause and consequence of PTSD 48. The drug-gene target approach further highlighted several cyclooxygenase inhibitors, including some used to treat IBD and other inflammatory disorders. Recent study using EHR data further identified role of non-psychotropic medications such as anti-inflammatory drugs with improved PTSD symptoms49. In addition, a study showed success with anticholinergics in treating reexperiencing-related traits such as PTSD-related nightmares50 and an animal model showed evidence for reductions in behavioral and stress hormone responses to fear conditioning51. Metabolic syndrome also contributes to proinflammatory response52, and on the other hand, PTSD has been associated with later development of obesity53. The underlying mechanisms indicate putative role of hypothalamic/pituitary hormone signaling, which might result in uncontrolled appetite and increased cortisol contributing to central obesity54.
The drug repurposing analysis identified several drugs implicated in the treatment of stress-related response. Pterostilbene is a cyclooxygenase and FAAH inhibitor that prevents catabolism of anandamide, and endocannabinoid55–57. WH-4023 is a SRC inhibitor with an ability to reduce NMDA receptor function which exhibit ketamine-like function, and shown to reduce craving-related activation of reward circuits58,59. We also observe Carbetocin, an analogue of oxytocin which has shown involvement in reward-related functional connectivity in PTSD-affected individuals60–62. Taken together, the findings highlight complex associations between PTSD, metabolic conditions, and inflammation that have been frequently identified within the epidemiologic literature.
While our study has taken multiple investigative steps to identify biological properties, and peripheral traits associated with genes involved with PTSD symptom clusters, our study has limitations. The MVP is a military cohort, and prevalence of combat exposure that may not generalize outside this cohort. To overcome this limitation, we replicated our associations in other cohorts such as UKBB and BioVU. Additionally, the MVP is primarily consisted of male participants, to circumvent this, we checked the identified genes against the sex-biased eQTLs from the GTEx63, and didn’t find any overlap. The study was performed using summary statistics from the European ancestry, and may not translate to other ancestries, future studies with diverse ancestries and improved sample sizes are needed for better understanding. Additionally, it is possible for some of the observed effects to be mediated by unmeasured mediating variables or confounding variable effects (e.g., health-related behaviors) that are causal for our target phenotypes and strongly associated with our loci of interest. Future work may involve causal inference methods to try and elucidate specific pleiotropic model. Some of the genes identified are in high LD region and will require functional experiments to dissect gene-specific effects for possible drug development and repurposing.
In summary, our study reports comprehensive evidence including clinical and regulatory data that highlight the molecular basis of the epidemiologic associations among PTSD, inflammation, and metabolic dysfunction.
Supplementary Material
9. Acknowledgements
The authors thank publicly available resources from Neale Lab (UK Biobank GWAS statistics), the Million Veteran Program (GWAS statistics via dbGaP), and Common-Mind Consortium (pretrained dlPFC models). The authors acknowledge support from the National Institutes of Health (R21DC018098, R33DA047527, F32MH122058, U54MD010722-04, R01MH113362, R01MH118223, R56MH120736, T32HG008341) and One Mind. The BioVU projects at Vanderbilt University Medical Center are supported by numerous sources: institutional funding, private agencies, and federal grants. These include the NIH-funded Shared Instrumentation Grant S10OD017985 and S10RR025141; CTSA grants UL1TR002243, UL1TR000445, and UL1RR024975 from the National Center for Advancing Translational Sciences. Its contents are solely the responsibility of the authors and do not necessarily represent official views of the National Center for Advancing Translational Sciences or the National Institutes of Health. Genomic data are also supported by investigator-led projects that include U01HG004798, R01NS032830, RC2GM092618, P50GM115305, U01HG006378, U19HL065962, R01HD074711; and additional funding sources listed at https://victr.vumc.org/biovu-funding/.
Footnotes
Data Availability and URLs
- GWAS summary statistics: https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001672.v6.p1
- TWAS method: http://gusevlab.org/projects/fusion/
- Splicing and Gene Expression Weights : http://gusevlab.org/projects/fusion/
- Proteome weights: https://www.synapse.org/#!Synapse:syn23627957
- Open Targets: https://genetics.opentargets.org/
- UKBB Summary Statistics : http://www.nealelab.is/uk-biobank
- CLUE: https://clue.io/
Conflict of Interest
RP and JG are paid for their editorial work on the journal Complex Psychiatry. JHK has served as a scientific consultant (Individual Consultant Agreements less than $5000 per year) to Amgen, AstraZeneca Pharmaceuticals, Bigen, Idec, MA, Biomedisyn Corporation, Forum Pharmaceuticals, Janssen Research & Development, Otsuka America Pharmaceutical, Sunovion Pharmaceuticals, Takeda Industries, and Taisho Pharmaceutical Co; is on the Scientific Advisory Board for Biohaven Pharmaceuticals, Blackthorn Therapeutics, Lohocla Research Corporation, Luc Therapeutics, Pfizer Pharmaceuticals, Tand RImaran Pharma; holds stock in Biohaven Pharmaceuticals Medical Sciences and stock options in Blackthorn Therapeutics and Luc Therapeutics; and is editor of Biological Psychiatry (income greater than $10,000 per year). JG is named as an inventor on PCT patent application no. 15/878,640 entitled “Genotype-guided dosing of opioid agonists,” filed January 24, 2018. RHP is a scientific consultant to Cogstate for work that bears no relationship to the current study. The other authors report no biomedical financial interests or potential conflicts of interest.
11 References
- 1.Stein MB, Levey DF, Cheng Z, Wendt FR, Harrington K, Pathak GA et al. Genome-wide association analyses of post-traumatic stress disorder and its symptom subdomains in the Million Veteran Program. Nat Genet 2021; 53: 174–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith SM, Goldstein RB, Grant BF. The association between post-traumatic stress disorder and lifetime DSM-5 psychiatric disorders among veterans: Data from the National Epidemiologic Survey on Alcohol and Related Conditions-III (NESARC-III). J Psychiatr Res 2016; 82: 16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steele M, Germain A, Campbell JS. Mediation and Moderation of the Relationship Between Combat Experiences and Post-Traumatic Stress Symptoms in Active Duty Military Personnel. Mil Med 2017; 182: e1632–e1639. [DOI] [PubMed] [Google Scholar]
- 4.Duncan LE, Ratanatharathorn A, Aiello AE, Almli LM, Amstadter AB, Ashley-Koch AE et al. Largest GWAS of PTSD (N=20 070) yields genetic overlap with schizophrenia and sex differences in heritability. Mol Psychiatry 2018; 23: 666–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pai A, Suris AM, North CS. Posttraumatic Stress Disorder in the DSM-5: Controversy, Change, and Conceptual Considerations. Behav Sci (Basel) 2017; 7. doi: 10.3390/bs7010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nutt DJ, Malizia AL. Structural and functional brain changes in posttraumatic stress disorder. J Clin Psychiatry 2004; 65 Suppl 1: 11–17. [PubMed] [Google Scholar]
- 7.Arnsten AFT, Raskind MA, Taylor FB, Connor DF. The Effects of Stress Exposure on Prefrontal Cortex: Translating Basic Research into Successful Treatments for Post-Traumatic Stress Disorder. Neurobiol Stress 2015; 1: 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh P-R et al. An atlas of genetic correlations across human diseases and traits. Nat Genet 2015; 47: 1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BWJH et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet 2016; 48: 245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gusev A, Mancuso N, Won H, Kousi M, Finucane HK, Reshef Y et al. Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat Genet 2018; 50: 538–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wingo AP, Liu Y, Gerasimov ES, Gockley J, Logsdon BA, Duong DM et al. Integrating human brain proteomes with genome-wide association data implicates new proteins in Alzheimer’s disease pathogenesis. Nat Genet 2021; 53: 143–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mancuso N, Shi H, Goddard P, Kichaev G, Gusev A, Pasaniuc B. Integrating Gene Expression with Summary Association Statistics to Identify Genes Associated with 30 Complex Traits. Am J Hum Genet 2017; 100: 473–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKenzie AT, Wang M, Hauberg ME, Fullard JF, Kozlenkov A, Keenan A et al. Brain Cell Type Specific Gene Expression and Co-expression Network Architectures. Sci Rep 2018; 8: 8868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee S, Zhang C, Arif M, Liu Z, Benfeitas R, Bidkhori G et al. TCSBN: a database of tissue and cancer specific biological networks. Nucleic Acids Res 2018; 46: D595–D600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang L, Zhou G, Soufan O, Xia J. miRNet 2.0: network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res 2020; 48: W244–W251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics 2010; 26: 1205–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dennis JK, Sealock JM, Straub P, Hucks D, Actkins K, Faucon A et al. Lab-wide association scan of polygenic scores identifies biomarkers of complex disease. medRxiv 2020. doi: 10.1101/2020.01.24.20018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pividori M, Rajagopal PS, Barbeira A, Liang Y, Melia O, Bastarache L et al. PhenomeXcan: Mapping the genome to the phenome through the transcriptome. Sci Adv 2020; 6. doi: 10.1126/sciadv.aba2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aguet F, Barbeira AN, Bonazzola R, Brown A, Castel SE, Jo B et al. The GTEx Consortium atlas of genetic regulatory effects across human tissues. BioRxiv 2019. doi: 10.1101/787903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghoussaini M, Mountjoy E, Carmona M, Peat G, Schmidt EM, Hercules A et al. Open Targets Genetics: systematic identification of trait-associated genes using large-scale genetics and functional genomics. Nucleic Acids Res 2021; 49: D1311–D1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet 2014; 10: e1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Werme J, van der Sluis S, Posthuma D, de Leeuw C. LAVA: An integrated framework for local genetic correlation analysis. BioRxiv 2021. doi: 10.1101/2020.12.31.424652. [DOI] [PubMed] [Google Scholar]
- 23.Giambartolomei C, Zhenli Liu J, Zhang W, Hauberg M, Shi H, Boocock J et al. A Bayesian framework for multiple trait colocalization from summary association statistics. Bioinformatics 2018; 34: 2538–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stein MB, Chen C-Y, Jain S, Jensen KP, He F, Heeringa SG et al. Genetic risk variants for social anxiety. Am J Med Genet B, Neuropsychiatr Genet 2017; 174: 120–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchese S, Cancelmo L, Diab O, Cahn L, Aaronson C, Daskalakis NP et al. Altered gene expression and PTSD symptom dimensions in World Trade Center responders. medRxiv 2021. doi: 10.1101/2021.03.05.21252989. [DOI] [PubMed] [Google Scholar]
- 26.Muhie S, Gautam A, Meyerhoff J, Chakraborty N, Hammamieh R, Jett M. Brain transcriptome profiles in mouse model simulating features of post-traumatic stress disorder. Mol Brain 2015; 8: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tylee DS, Chandler SD, Nievergelt CM, Liu X, Pazol J, Woelk CH et al. Blood-based gene-expression biomarkers of post-traumatic stress disorder among deployed marines: A pilot study. Psychoneuroendocrinology 2015; 51: 472–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamman AJF, Wendt FR, Pathak GA, Krystal JH, Southwick SM, Sippel LM et al. Attachment style moderates polygenic risk for incident posttraumatic stress in U.S. military veterans: A 7-year, nationally representative, prospective cohort study. Biol Psychiatry 2021. doi: 10.1016/j.biopsych.2021.09.025. [DOI] [PubMed] [Google Scholar]
- 29.Martin CG, Kim H, Yun S, Livingston W, Fetta J, Mysliwiec V et al. Circulating miRNA associated with posttraumatic stress disorder in a cohort of military combat veterans. Psychiatry Res 2017; 251: 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou J, Nagarkatti P, Zhong Y, Ginsberg JP, Singh NP, Zhang J et al. Dysregulation in microRNA expression is associated with alterations in immune functions in combat veterans with post-traumatic stress disorder. PLoS One 2014; 9: e94075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balakathiresan NS, Chandran R, Bhomia M, Jia M, Li H, Maheshwari RK. Serum and amygdala microRNA signatures of posttraumatic stress: fear correlation and biomarker potential. J Psychiatr Res 2014; 57: 65–73. [DOI] [PubMed] [Google Scholar]
- 32.Muiños-Gimeno M, Espinosa-Parrilla Y, Guidi M, Kagerbauer B, Sipilä T, Maron E et al. Human microRNAs miR-22, miR-138–2, miR-148a, and miR-488 are associated with panic disorder and regulate several anxiety candidate genes and related pathways. Biol Psychiatry 2011; 69: 526–533. [DOI] [PubMed] [Google Scholar]
- 33.Michopoulos V, Vester A, Neigh G. Posttraumatic stress disorder: A metabolic disorder in disguise? Exp Neurol 2016; 284: 220–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palmer BW, Shir C, Chang H, Mulvaney M, Hall JMH, Shu I-W et al. Increased prevalence of metabolic syndrome in Veterans with PTSD untreated with antipsychotic medications. Int J Ment Health 2021; : 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Donnell CJ, Schwartz Longacre L, Cohen BE, Fayad ZA, Gillespie CF, Liberzon I et al. Posttraumatic stress disorder and cardiovascular disease: state of the science, knowledge gaps, and research opportunities. JAMA Cardiol 2021; 6: 1207–1216. [DOI] [PubMed] [Google Scholar]
- 36.Edmondson D, von Känel R. Post-traumatic stress disorder and cardiovascular disease. Lancet Psychiatry 2017; 4: 320–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y, He XN, Li C, Gong L, Liu M. Identification of candidate genes and micrornas for acute myocardial infarction by weighted gene coexpression network analysis. Biomed Res Int 2019; 2019: 5742608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Civelek M, Wu Y, Pan C, Raulerson CK, Ko A, He A et al. Genetic Regulation of Adipose Gene Expression and Cardio-Metabolic Traits. Am J Hum Genet 2017; 100: 428–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fadason T, Ekblad C, Ingram JR, Schierding WS, O’Sullivan JM. Physical interactions and expression quantitative traits loci identify regulatory connections for obesity and type 2 diabetes associated snps. Front Genet 2017; 8: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rowlands DS, Page RA, Sukala WR, Giri M, Ghimbovschi SD, Hayat I et al. Multi-omic integrated networks connect DNA methylation and miRNA with skeletal muscle plasticity to chronic exercise in Type 2 diabetic obesity. Physiol Genomics 2014; 46: 747–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muhie S, Gautam A, Chakraborty N, Hoke A, Meyerhoff J, Hammamieh R et al. Molecular indicators of stress-induced neuroinflammation in a mouse model simulating features of post-traumatic stress disorder. Transl Psychiatry 2017; 7: e1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mariani N, Cattane N, Pariante C, Cattaneo A. Gene expression studies in Depression development and treatment: an overview of the underlying molecular mechanisms and biological processes to identify biomarkers. Transl Psychiatry 2021; 11: 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Merino J, Dashti HS, Sarnowski C, Lane JM, Todorov PV, Udler MS et al. Genetic analysis of dietary intake identifies new loci and functional links with metabolic traits. Nat Hum Behav 2021. doi: 10.1038/s41562-021-01182-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu D, Glaser AP, Patibandla S, Blum A, Munson PJ, McCoy JP et al. Transcriptional profiling of CD133(+) cells in coronary artery disease and effects of exercise on gene expression. Cytotherapy 2011; 13: 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson MA, Liberzon I, Lindsey ML, Lokshina Y, Risbrough VB, Sah R et al. Common pathways and communication between the brain and heart: connecting post-traumatic stress disorder and heart failure. Stress 2019; 22: 530–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koraishy FM, Salas J, Neylan TC, Cohen BE, Schnurr PP, Clouston S et al. association of severity of posttraumatic stress disorder with inflammation: using total white blood cell count as a marker”. Chronic Stress (Thousand Oaks) 2019; 3. doi: 10.1177/2470547019877651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lindqvist D, Mellon SH, Dhabhar FS, Yehuda R, Grenon SM, Flory JD et al. Increased circulating blood cell counts in combat-related PTSD: Associations with inflammation and PTSD severity. Psychiatry Res 2017; 258: 330–336. [DOI] [PubMed] [Google Scholar]
- 48.Hori H, Kim Y. Inflammation and post-traumatic stress disorder. Psychiatry Clin Neurosci 2019; 73: 143–153. [DOI] [PubMed] [Google Scholar]
- 49.Shiner B, Forehand JA, Rozema L, Kulldorff M, Watts BV, Trefethen M et al. Mining clinical data for novel PTSD medications. Biol Psychiatry 2021. doi: 10.1016/j.biopsych.2021.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sogo K, Sogo M, Okawa Y. Centrally acting anticholinergic drug trihexyphenidyl is highly effective in reducing nightmares associated with post-traumatic stress disorder. Brain Behav 2021; 11: e02147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaur H, Kaur R, Jaggi AS, Bali A. Beneficial role of central anticholinergic agent in preventing the development of symptoms in mouse model of post-traumatic stress disorder. J Basic Clin Physiol Pharmacol 2020; 31. doi: 10.1515/jbcpp-2019-0196. [DOI] [PubMed] [Google Scholar]
- 52.Hotamisligil GS. Inflammation, metaflammation and immunometabolic disorders. Nature 2017; 542: 177–185. [DOI] [PubMed] [Google Scholar]
- 53.Masodkar K, Johnson J, Peterson MJ. A review of posttraumatic stress disorder and obesity: exploring the link. Prim Care Companion CNS Disord 2016; 18. doi: 10.4088/PCC.15r01848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aaseth J, Roer GE, Lien L, Bjørklund G. Is there a relationship between PTSD and complicated obesity? A review of the literature. Biomed Pharmacother 2019; 117: 108834. [DOI] [PubMed] [Google Scholar]
- 55.Mayo LM, Rabinak CA, Hill MN, Heilig M. Targeting the Endocannabinoid System in the Treatment of Posttraumatic Stress Disorder: A Promising Case of Preclinical-Clinical Translation? Biol Psychiatry 2021. doi: 10.1016/j.biopsych.2021.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mayo LM, Asratian A, Lindé J, Morena M, Haataja R, Hammar V et al. Elevated anandamide, enhanced recall of fear extinction, and attenuated stress responses following inhibition of fatty acid amide hydrolase: A randomized, controlled experimental medicine trial. Biol Psychiatry 2020; 87: 538–547. [DOI] [PubMed] [Google Scholar]
- 57.Mayo LM, Asratian A, Lindé J, Holm L, Nätt D, Augier G et al. Protective effects of elevated anandamide on stress and fear-related behaviors: translational evidence from humans and mice. Mol Psychiatry 2020; 25: 993–1005. [DOI] [PubMed] [Google Scholar]
- 58.Thompson SL, Gianessi CA, O’Malley SS, Cavallo DA, Shi JM, Tetrault JM et al. Saracatinib Fails to Reduce Alcohol-Seeking and Consumption in Mice and Human Participants. Front Psychiatry 2021; 12: 709559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Patel KT, Stevens MC, Dunlap A, Gallagher A, O’Malley SS, DeMartini K et al. Effects of the Fyn kinase inhibitor saracatinib on ventral striatal activity during performance of an fMRI monetary incentive delay task in individuals family history positive or negative for alcohol use disorder. A pilot randomised trial. Neuropsychopharmacology 2021. doi: 10.1038/s41386-021-01157-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baldi E, Costa A, Rani B, Passani MB, Blandina P, Romano A et al. Oxytocin and fear memory extinction: possible implications for the therapy of fear disorders? Int J Mol Sci 2021; 22. doi: 10.3390/ijms221810000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sippel LM, Flanagan JC, Holtzheimer PE, Moran-Santa-Maria MM, Brady KT, Joseph JE. Effects of intranasal oxytocin on threat- and reward-related functional connectivity in men and women with and without childhood abuse-related PTSD. Psychiatry Res Neuroimaging 2021; 317: 111368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Melkonian AJ, Flanagan JC, Calhoun CD, Hogan JN, Back SE. Craving Moderates the Effects of Intranasal Oxytocin on Anger in Response to Social Stress Among Veterans With Co-Occurring Posttraumatic Stress Disorder and Alcohol Use Disorder. J Clin Psychopharmacol 2021; 41: 465–469. [DOI] [PubMed] [Google Scholar]
- 63.Oliva M, Muñoz-Aguirre M, Kim-Hellmuth S, Wucher V, Gewirtz ADH, Cotter DJ et al. The impact of sex on gene expression across human tissues. Science 2020; 369. doi: 10.1126/science.aba3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


