Abstract
Small ubiquitin-like modifiers, SUMOs, are proteins that are conjugated to target substrates and regulate their functions in a post-translational modification called SUMOylation. In addition to its physiological roles, SUMOylation has been implicated in several neurodegenerative diseases, such as Alzheimer’s, Parkinson’s, and Huntington’s diseases (HD). HD is a neurodegenerative monogenetic autosomal dominant disorder caused by a mutation in the CAG repeat of the huntingtin (htt) gene, which expresses a mutant Htt protein more susceptible to aggregation and toxicity. Besides Htt, other SUMO ligases, enzymes, mitochondrial and autophagic components are also important for the progression of the disease. Here we review the main aspects of Htt SUMOylation and its role in cellular processes involved in the pathogenesis of HD.
Keywords: Huntingtin, Huntington’s disease, Neurodegeneration, Post-translational modification, SUMO, SUMOylation
1. Introduction
To allow for their numerous biological effects, proteins can be modified in many ways by processing events known as post-translational modifications (PTMs) (Ramazi et al., 2020). PTMs can add from simple chemicals (e.g. phosphate) to complex groups (e.g. carbohydrates) into proteins, altering their functions, destinations, structures and, thus, increasing the diversity of the proteome (Conibear, 2020). SUMOylation is among the top 10 studied PTMs, together with the best-characterized phosphorylation and ubiquitylation (Ramazi and Zahiri, 2021). In addition to its involvement in cardiovascular diseases (Shetty et al., 2020) and cancer (Kroonen and Vertegaal, 2020), SUMOylation has been implicated in neurodegenerative diseases (Princz and Tavernarakis, 2020). To date, several studies have focused on the role and functional consequences of SUMOylation in the two most prevalent neurodegenerative diseases, Alzheimer’s and Parkinson’s diseases (Guerra De Souza et al., 2016, Marcelli et al., 2018, Martins et al., 2016). It is important to point out that currently there is no cure for any neurodegenerative disease and that millions of people worldwide are affected, making the discovery of new treatments the focus of intense scientific research. A better understanding of the mechanisms underlying SUMOylation and how to manipulate them can provide novel therapeutic targets for neurodegeneration and have a clinical impact. Thus, the present review attempts to examine findings from in vitro and in vivo studies to provide a comprehensive picture of the involvement of SUMOylation in Huntington’s disease (HD).
2. SUMOylation
SUMOylation is the reversible attachment of SUMO (small ubiquitin-like modifier) proteins to lysine residues in a three-step enzymatic pathway (Chang and Yeh, 2020). Modification by SUMO may alter protein localization, stability, and activity, as well as gene expression, DNA repair, and RNA processing (Chen et al., 2021, Varejão et al., 2020).
Briefly, the first step in the SUMOylation pathway is the maturation of SUMO by the family of SUMO-specific proteases, SENPs. Before its ligation to target substrates, SUMO is activated by the SUMO-activating enzyme 1 and 2 complexes, SAE1/SAE2, and then conjugated by the SUMO-conjugating enzyme, Ubc9. Once attached, with or without the participation of E3 protein ligases (e.g. Rhes, PIAS, RanBP2, MAPL, Topors, ZATT), SUMO modifies the target protein and, after that, it is deconjugated, or deSUMOylated, by the same SENPs (Chang and Yeh, 2020).
Five SUMO isoforms are found in mammalians. SUMO-1 shares ~ 50% of its amino acid sequence with both SUMO-2 and SUMO-3 that differ by only three N-terminal amino acids. As the available antibodies are unable to distinguish between these two isoforms, they are often denoted as SUMO-2/3 (Jansen and Vertegaal, 2021). SUMO-1 and SUMO-2/3 are highly expressed in the brain and are primarily involved in physiological and pathological processes, respectively (Anderson et al., 2017, Guo et al., 2013, Luo et al., 2017). SUMOs 4 and 5 are the most recently described paralogs that have their expression limited to some tissues. SUMO-4 is found mainly in the lymph nodes, placenta, spleen, and kidneys and contributes to diabetes and stress responses. SUMO-5 is found in peripheral blood leukocytes and testes, where its main function is related to the regulation of promyelocytic leukemia (PML) nuclear bodies (Liang et al., 2016, Wilson, 2017).
3. Huntington’s disease
HD is a neurodegenerative monogenetic autosomal dominant disease that affects mainly the spiny projection neurons from the striatum, resulting in significant motor deficits (Barron et al., 2021, Schaffert and Carter, 2020). These motor deficits are marked by involuntary movement appearance and voluntary movement dysfunction (Lakke, 1981) that are accompanied by neuropsychiatric symptoms, such as depression (Paoli et al., 2017), and cognitive impairment (Papp et al., 2011). In addition, HD causes muscular and testicular atrophy, body weight loss, metabolic and endocrine disorders (van der Burg et al., 2009, van der Burg et al., 2017), as well as changes in the peripheral immune system (Valadão et al., 2020) and in the gut microbiota (Du et al., 2021). As a result, patients have a life expectancy of 10–20 years post-diagnosis (Hawton et al., 2019).
HD is caused by a mutation in the CAG repeat of the huntingtin (htt) gene that encodes the Htt protein, which is a 348 kDa protein and is expressed in both the cytoplasm and the nucleus of neurons (Princz and Tavernarakis, 2020). Htt is crucial for neural and embryonic development, and it also participates in RNA trafficking, vesicular transport, and transcription (Fields et al., 2021).
When an expansion in the CAG trinucleotides repeats (polyglutamine -polyQ- stretch) occurs in the htt gene, a mutant form of the Htt protein (mHtt) is encoded. This mHtt becomes more susceptible to aggregation in both cytoplasm and nucleus (Testa and Jankovic, 2019), and disturbs mitochondrial and synaptic function, axonal transport, transcription and translation processes, culminating in neuronal death (McColgan and Tabrizi, 2018, Rangel-Barajas and Rebec, 2018). However, it is important to note such inclusions do not always cause cellular damage. Actually, some studies suggest that mHtt inclusions are formed as a way to protect neurons from toxic mHtt oligomers (Arrasate et al., 2004, Pastore and Temussi, 2012, Tabrizi et al., 2020).
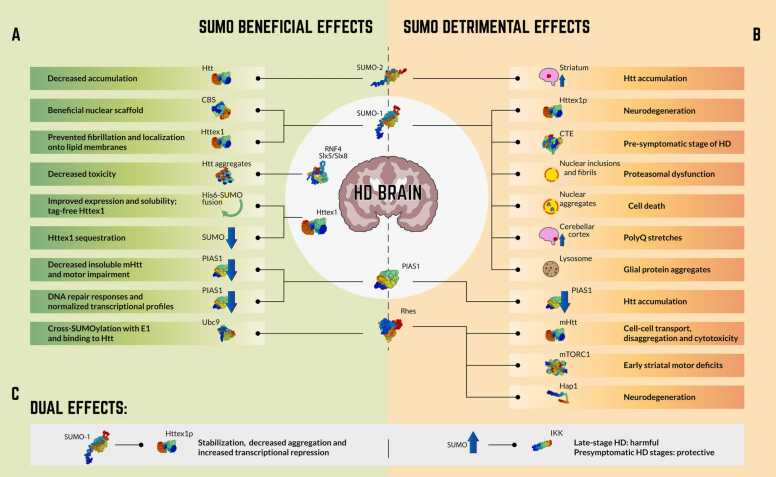
The main positive and negative effects of SUMOylation on HD are listed below and summarized in Fig. 1. These effects are triggered by the SUMO machinery and SUMO-target proteins, which are shown in Table 1, Table 2, respectively.
Fig. 1.
Effects of protein SUMOylation on Huntington’s disease (HD). The main beneficial (A), detrimental (B) and dual (C) effects of SUMOylation on SUMO machinery and HD-related proteins are highlighted. Proteins structure: SwissModel.expasy.org.
Table 1.
Summary of findings on SUMOylation pathway proteins in HD and polyQ disorders.
| SUMOylation pathway protein | Model | Event | Effect | Ref. |
|---|---|---|---|---|
| PIAS1 | zQ175 KI mice and HD patient-derived iPSCs | KD | DNA damage repair responses and normalized transcriptional profiles | Morozko et al. (2021) |
| R6/2 mice | KD and Overexpression | Reduction of insoluble protein and phenotypic benefit | Ochaba et al. (2016) | |
| Rhes | Striatal neuronal cells (STHdhQ7/Q7) or HD mutants (STHdhQ111/Q111) | Rhes-mediated mHtt transport | Facilitation of cell–cell transport of mHtt | Sharma and Subramaniam (2019) |
| HEK293 cells | Binding to mHtt | mHtt increased disaggregation and cytotoxicity | Subramaniam et al. (2009) | |
| Rhes KO mice | Binding to E1 and Ubc9 | Ubc9 binding to Htt and cross-SUMOylation of E1 and Ubc9 | Subramaniam et al. (2010) | |
| Rhes KO N171-82Q mice | mTORC1 signaling dysregulating | Early striatal motor deficit | Swarnkar et al. (2015) | |
| STUbL Slx5 | Yeast plasmids expressing 25Q and 103Q Htt | Expression and transcriptional properties of aggregation-prone Htt constructs | Reduced toxicity and abnormal transcriptional activity | Ohkuni et al. (2018) |
| SUMO-1 | NIID brain tissue and PC12 cells | Localization into fibrils of tissues and nuclear inclusions | Recruitment of SUMO-1 modified proteins into insoluble nuclear inclusions and proteasomal dysfunction | Pountney et al. (2003) |
| PC12 cells | SUMO-1 and atrophin-1co-transfection | Accelerated aggregate formation and cell death | Terashima et al. (2002) | |
| Brain tissue (HD, DRPLA, SCA1 and MJD) and B05 transgenic mouse | Increased SUMOylated proteins in cerebellar cortex | SUMO-1 activation contributes to polyQ diseases | Ueda et al. (2002) | |
| Brain tissue (MSA) | Cytoplasmic accumulation of α-synuclein, tau or HttQ74-GFP | SUMO-1 colocalization with lysosomes related to glial protein aggregates and protein aggregation | Wong et al. (2013) | |
| SUMO-2 | HeLa cells | Htt SUMO-2 modification | Decreased accumulation | O’Rourke et al. (2013) |
DRPLA: dentatorubral-pallidoluysian atrophy; E1: SUMO activating enzyme E1; GFP: green fluorescent protein; HD: Huntington’s disease; HEK: human embryonic kidney cells; Htt: huntingtin; iPSCs: induced pluripotent stem cells; KD: knockdown; KI: knock-in; KO: knockout; mHtt: mutant huntingtin; MJD: Machado-Joseph disease; MSA: multiple system atrophy; mTORC1: mechanistic target of rapamycin complex 1; NIID: neuronal intranuclear inclusion disease; PolyQ: polyglutamine; PIAS1: Protein Inhibitor of Activated STAT 1; Q: glutamine; Rhes: Ras homolog enriched in the striatum; SCA1: spinocerebellar ataxia type 1; STUbL: SUMO-targeted ubiquitin ligase; Ubc9: SUMO conjugating enzyme 9.
Table 2.
Summary of findings on SUMO target proteins in HD and polyQ disorders.
| SUMO target | Model | Event | Effect | Ref. |
|---|---|---|---|---|
| CBS | Yeast two-hybrid vector pGBDU-C2 | SUMO-1 modification | Beneficial nuclear scaffold | Kabil et al. (2006) |
| CTE | R6/2 mice | SUMO-1 conjugation | Prevented degradation | Gibb et al. (2007) |
| Hap1 | HD-KI and Hap1-KO mice | Blocking of Rhes’ binding, SUMOylating mHtt and Hap1 depletion | Neurodegeneration promoted by Rhes, Hap1 and cellular stress | Liu et al. (2020) |
| Httex1 | pTWIN1 vector (human Httex1 fused to His6-SUMO) | Site-specifically phosphorylated highly pure WT or mHttex1 proteins production | Improved expression and increased solubility | Chiki et al. (2021) |
| Neuroblastoma (Neuro2a) cells enriched with mHtt or WT Htt | Reduction of SUMOylation | Sequestration of Httex1 | Moily et al. (2017) | |
| Purification of Httex1 | Httex1 and SUMO (Smt3) fusion | Tag-free Httex1 | Reif et al. (2018) | |
| GST-Htt-exon1 (46Q) fusion proteins and Htt-exon1 (46Q) | Htt aggregation promoted by SUMOylation | Htt fibrillization and localization onto lipid membranes prevented by SUMOylation | Sedighi et al. (2020) | |
| Httex1p | Drosophila heterozygotes | Neurodegeneration promoted by SUMOylation | SUMOylation worsens HD | Steffan et al. (2004) |
| HeLa cells | Fusion of SUMO-1 to Httex1p | SUMOylation stabilizes Httex1p, reduces its ability to form aggregates, and promotes its capacity to repress transcription | Steffan et al. (2004) | |
| IKK | Hela, St12.7, ST14A, N548mu, WT STHdhQ7/HdhQ7 and homozygous mutant STHdhQ111/HdhQ111 cell lines | mHtt clearance | Increased SUMOylation: Late-stage HD: harmful; Presymptomatic stages: protective |
Thompson et al. (2009) |
CBS: Cystathionine β-synthase; CTE: COOH terminus of excitatory amino acid transporter 2; GST: glutathione S-transferase; Hap1: huntingtin-associated protein 1; HD: Huntington’s disease; Htt: huntingtin; Httex1: exon 1 of the huntingtin protein; Httex1p: pathogenic fragment of exon 1 of the huntingtin protein; IKK: IκB kinase; KI: knock-in; KO: knockout; mHTT: mutant huntingtin; mHtt: mutant huntingtin; mHttex1p: pathogenic fragment of exon 1 of the mutant huntingtin protein; Q: glutamine; Rhes: Ras homolog enriched in the striatum; WT: wild type.
4. Beneficial effects of SUMOylation on HD
PTMs are quite important in HD, especially because they modulate the clearance and toxicity of polyQ-expanded Htt, as well as the physiological functions of wild-type Htt (Wang et al., 2010). For instance, phosphorylation, acetylation, ubiquitylation, and SUMOylation in the first 17 amino acids of exon1 of Htt (Httex1), the smallest Htt fragment, have pivotal roles in modulating cellular processes and functions. SUMO-mHttex1 enhances Htt expression, increases the solubility of aggregates, and facilitates their handling and purification, as recently described by Chiki et al. (2021) that developed highly purified wild-type and mHttex1 proteins, which were site-specifically phosphorylated. Reif et al. (2018) also showed that these SUMO-Httex1 fusion proteins enable Htt manipulation and modification, contributing to a better understanding of how to prevent Htt aggregation.
Depending on which SUMO paralog is conjugated to Htt, different outcomes can be expected. Both SUMO-1 and SUMO-2/3 regulate HD through Htt modification and can represent new therapeutic targets for the development of neuroprotective treatments (Chen et al., 2021). Htt SUMOylation could facilitate cellular clearance when combined with ubiquitylation (Celen and Sahin, 2020). Nevertheless, SUMOylation and ubiquitylation pathways can be impaired in HD, disrupting cellular homeostasis.
Regarding mHtt inclusions, it is still unclear whether they are really neurotoxic in HD, with some pieces of evidence suggesting that they could be part of an endogenous neuroprotective event (Arrasate et al., 2004). Thus, SUMO proteins could participate in the formation of such inclusions in order to shield neurons from damage (Liebelt and Vertegaal, 2016). In fact, Sedighi et al. (2020), using GST-Htt-exon1 fusion proteins and Httex1, observed that SUMOylated Httex1 inhibited fibril formation and promoted aggregate species. These findings demonstrate that Htt aggregation by SUMO modifiers is distinct from other types of aggregates, since SUMOylation prevented Htt aggregation, binding, and accumulation. Another study using neuroblastoma (Neuro2a) cells enriched with mutant or wild-type Htt into different aggregation states suggested that a reduction in SUMOylation could be an adaptive response to help the sequestration of soluble Httex1, a therapeutic strategy to prevent Htt aggregation (Moily et al., 2017). In summary, Htt SUMOylation may represent a molecular mechanism essential for the maintenance of cellular function.
The SUMO E3 ligase PIAS1 was shown to modulate neurons and to change HD-related phenotypes in some studies using in vivo approaches. For example, PIAS1-directed miRNA injected into the striatum of R6/2 mice improved motor impairment, even in the presence of mHtt (Ochaba et al., 2016). When PIAS1 was reduced, mHtt did not accumulate, SUMOylation was reduced, synaptophysin levels were increased, and inflammatory markers were normalized, whereas PIAS1 overexpression led to Htt accumulation. These findings corroborate a previous study showing that PIAS1 regulates Htt accumulation and SUMOylation in cells (O’Rourke et al., 2013). The contribution of PIAS1 was also investigated in HD mice and HD patient-derived induced pluripotent stem cells (iPSCs), linking this E3 ligase to DNA damage repair pathways (Morozko et al., 2021). PIAS1 knockdown normalized HD transcriptional deregulation and improved DNA repair mechanisms in mice, supporting the conclusion that SUMOylation underlies DNA damage repair responses and transcriptional modulation.
Rhes (Ras homolog enriched in the striatum) is a small G protein that, besides being an unusual SUMO E3 ligase, since it has no structural similarity with other SUMO ligases (Liebelt and Vertegaal, 2016), plays different roles in the SUMOylation pathway. For instance, Rhes can enhance the capacity of Ubc9 to SUMOylate mHtt and the cross-SUMOylation between SAE1/2 and Ubc9 (Serra et al., 2021, Subramaniam et al., 2010). Also, Rhes regulates SUMOylation, which is significantly reduced in the striatum of Rhes-deleted mice (Subramaniam et al., 2010).
A significant role for Rhes in the selective toxicity found in HD patients was demonstrated by the loss of balance and ataxia-like features, with ectopic expression of Rhes in the cerebellum of mice (Swarnkar et al., 2015). Rhes deletion in HD mice (N171/82Q), expressing a mHtt with 82 glutamines, protected against motor deficits, induced anxiolytic responses, and prevented lateral ventricle dilatation. Harrison and LaHoste (2013) postulated that SUMO modification could decrease Htt aggregates and promote cell death, so Rhes could act as a physiological tissue-specific regulator of SUMOylation (Napolitano et al., 2018). Since Rhes binds to Htt and reduces its physiological functioning, increasing SUMOylation could represent a potential target for HD treatment.
As other neurodegenerative diseases, HD is also characterized by abnormal mitochondrial functioning (Han et al., 2011). The mHtt modifies the mitochondrial morphology machinery by increasing fission proteins, such as Drp1 (dynamin-related protein 1) and Fis1 (fission 1), and decreasing pro-fusion proteins, such as Mfn1/2 (mitofusins 1 and 2) and OPA1 (optic atrophy 1) (Han et al., 2011, Palmer et al., 2011). These GTPase family proteins are crucial for maintaining a fine mitochondrial fission and fusion balance (Reddy et al., 2011). The overexpression of Htt74Q, a mutant form of Htt, leads to mitochondrial fission and cell death, while the overexpression of Drp1 and Mfn2 can prevent these events (Wang and Dasso, 2009). Abnormal mitochondrial dynamics during HD progression was observed in patients, both through quantification of the levels of mitochondrial structural genes and localization of mitochondria-related proteins and mHtt (Shirendeb et al., 2012). SUMOylation, in turn, has an important role in the regulation of mitochondrial dynamics by modulating Drp1, SENP5, and SUMO-1 activities (Han et al., 2011). Therefore, considering that Htt overexpression can change the normal mitochondrial dynamics, SUMO proteins could restore the normal state of mitochondria during HD pathogenesis.
SUMO proteins are colocalized with polyQ protein aggregates and have become a key factor for neuronal inclusions observed in several polyQ disorders, besides HD (Dorval and Fraser, 2007, Terashima et al., 2002). Other well studied neurodegenerative polyQ diseases include spinal and bulbar muscular atrophy (SMBA), dentatorubro-pallidolysian atrophy (DRPLA), Machado-Joseph disease (MJD), spinocerebellar ataxias (SCAs) types 1, 2, 3, 6, 7 and 17, all characterized by polyQ expansions (Ehrnhoefer et al., 2011). All these diseases are hereditary, usually linked to some common enzymes, as the cystathionine β-synthase (CBS), a highly regulated enzyme responsible for the formation of cystathionine, due to some structural changes on CBS and its interaction with Htt. Oxidation products from homocysteine are also implicated in HD. Moreover, CBS is also a target for SUMOylation, as showed in in vivo and in vitro models, mainly in the cellular nucleus, associated with nuclear scaffold proteins (Kabil et al., 2006).
Both ubiquitin and SUMO proteins belong to the family of small ubiquitin-like modifiers (UBLs). SUMO-targeted ubiquitin ligases (STUbLs) include Slx5/Slx8 (heterodimer) found in yeast and its human ortholog RNF4 (homodimer). Besides being essential for SUMO conjugation, they also facilitate ubiquitylation. Ohkuni et al. (2018) analyzed how expanded glutamine residues affect cell viability by using WT, Slx5Δ and Slx8Δ yeast strains, i.e., cells with Slx5 and Slx8 deletion. They showed that an aggregation-prone Htt, with polyQ expansions, negatively affected Slx5Δ/Slx8Δ cells. However, when Slx5 was fused to the Gal4 DNA-binding domain, a positive regulator of gene expression, the activation of Htt constructs did not occur. In addition, RNF4 was also able to inhibit the transcriptional activity mediated by aggregation-prone Htt. Thus, STUbLs are able to prevent the toxicity caused by polyQ expanded Htt and might play a neuroprotective role in HD. It is also worth mentioning that some ubiquitin targets can become important SUMO targets to reduce HD pathology, like the neuronal chaperone HSJ1 that is capable of reducing Htt aggregation in R6/2 mice through lysine 63 (K63) when ubiquitylated (Labbadia et al., 2012). K63-linked polyubiquitin chains regulate proteasome-independent pathways and are also a SUMO substrate.
5. Detrimental effects of SUMOylation on HD
As already stated, inclusion body formation is not always a predictor of pathogenesis. However, SUMO-1 can increase Htt solubility and toxicity (Subramaniam et al., 2009) and SUMO-2 has been found to accumulate in the striatum, one of the most affected brain regions in HD, and it is related to the pathogenic accumulation of Htt (O’Rourke et al., 2013). In addition, SUMOylation can also indirectly affect Htt, as observed with the inflammatory kinase IKK that activates mHtt clearance and could be involved in its neurotoxicity (Steffan, 2010). Thompson et al. (2009) described that Htt is phosphorylated by IKK on serine 13, promoting Htt poly-SUMOylation, which modulates its clearance by both proteasome and lysosome pathways. Thus, as SUMOylation participates in IKK activation, its increase could be harmful to neurons.
A detrimental effect for SUMOylation was also observed in a SUMO target called CTE (caspase-cleaved fragment of excitatory amino acid transporter 2 (EAAT2) fragment). Using a R6/2 mouse model of HD, Gibb et al. (2007) observed that, when modified by SUMO-1, there was accumulation of CTE in the spinal cord of mice, characterizing a pre-symptomatic stage of HD. In a recent study with another SUMO target called Hap1 (huntingtin-associated protein 1), Liu et al. (2020) observed that Hap1 depletion led Rhes to bind to mHtt in adult HD knockout mice. Hap1 binds mHtt strongly and, therefore, there is more SUMOylated Htt, which, in turn, affects cell viability and contributes to HD development.
Despite being quite similar proteins, SUMO and ubiquitin can have divergent effects on the same target. The conflicting data related to Htt, for example, may be due to the competition between SUMOylation and ubiquitylation for the same 6, 9, and 15 lysine residues of the N-terminal fragment (Celen and Sahin, 2020, Sarge and Park-Sarge, 2011). Both SUMOylation and ubiquitylation are components of the normal cellular metabolism and can occur simultaneously and interfere with each other (Deger et al., 2015). Previous studies have linked the reduction in mHtt toxicity to ubiquitylation, whereas protein stabilization, decreased aggregation, and transcriptional imbalance have been linked to SUMOylation (Ehrnhoefer et al., 2011). Intriguingly, in an in vitro model SUMOylation has been shown to stabilize Httex1p, a pathogenic fragment of Htt, as well as decrease its aggregation and enhance transcriptional repression, whereas in a Drosophila model SUMOylation aggravated neurodegeneration, while Httex1p ubiquitylation ameliorated neuronal viability (Steffan et al., 2004). Nonetheless, it was also found that HD pathology is reduced due to the lysine mutations of Httex1p that prevent both SUMOylation and ubiquitylation. These results indicate that proteasome activation, ubiquitylation enhancement and SUMOylation promotion could be important to slow down or even prevent neurodegeneration (Huang et al., 2018). Therefore, it is undisputable that more studies are necessary to clarify the specific molecular mechanisms by which both these PTMs interfere with Htt.
We have discussed previously that Rhes has important beneficial effects in HD, because its ability to interfere with HD pathogenesis; however, it also has deleterious effects due to promoting SUMOylation. When attached to mHtt, SUMOylation increases and ubiquitylation decreases, which reduces the degradation of mHtt and increases its toxicity (Ross and Shoulson, 2009, Subramaniam et al., 2009). In an attempt to understand the link between Rhes/SUMO isoforms and mHtt through tunneling nanotubes (TNT)-like cellular protrusions, an important mechanism for intercellular communication, Sharma and Subramaniam (2019) described that SUMOylation-defective mHtt or CRISPR/Cas9-mediated depletion of the three main SUMO isoforms decreased Rhes-mediated mHtt transport. These data provide relevant evidence for the role of the SUMOylation machinery in the biogenesis of tunneling nanotubes (TNT)-like cellular protrusions and, even more remarkable, that Rhes contributes to the transport of mHtt from cell to cell. In addition, SUMOylation enhances cytotoxicity when it targets mHtt and leads to its disaggregation, but this could also happen due to the Rhes-mHtt interaction, causing striatal neurotoxicity.
The mammalian target of rapamycin (mTOR) protein kinase is an essential regulator of autophagy, a lysosomal degradation process that eliminates and recycles unwanted material from the cell, and it is well known that autophagy is deregulated in neurodegenerative diseases, such as HD (Croce and Yamamoto, 2019). Considering that Rhes-induced activation of mTOR alters protein synthesis (Subramaniam and Snyder, 2011) and that mTOR regulates autophagy, apoptosis and cell proliferation (Zou et al., 2020), mHtt-induced Rhes inhibition could decrease mTOR activation. Rhes could also enhance autophagy by interacting with beclin-1, an autophagy activator. Thus, Rhes inhibition may represent an effective way to attenuate mHtt deleterious effects (Carbo et al., 2019).
As much as positive effects are seen, negative effects for SUMOylation in polyQ disorders are also evident, especially related to SUMO-1. Its conjugation is increased in the cerebellar cortex of mice expressing mutant ataxin-1, indicating a role for SUMO-1 in polyQ disorders (Ueda et al., 2002). In this same study, SUMO-1 was also found in the affected brain regions from patients with HD, DRPLA, MJD and SCA1. Additionally, SUMO-1 colocalization with intranuclear inclusions was observed in both DRPLA brain tissue and in an in vitro model, where the aggregates were strongly SUMOylated (Terashima et al., 2002). Nuclear aggregate formation and apoptotic cellular death were also found in PC12 cells co-transfected with SUMO-1 and atrophin-1. SUMO-1 was observed again in three cases of neuronal intranuclear inclusion disease (NIID) related to proteasomal dysfunction (Pountney et al., 2003). The same was found in brain tissue from patients with multiple system atrophy (MSA), characterized by α-synuclein glial cytoplasmic inclusions, and progressive supranuclear palsy (PSP), marked by glial tau inclusions (Wong et al., 2013), which are a subset of neurodegenerative diseases called oligodendroglial inclusion bodies. SUMO-1 sub-domains were inside and surrounding these inclusion bodies, as well as the lysosomal marker, cathepsin D. A role for SUMO-1 in lysosome function was found by transfecting 1321N1 cells with an aggregation-prone mHttex1, HttQ74-GFP, that resulted in an association between SUMO-1-positive lysosomes and cytoplasmic accumulation of α-synuclein, tau or HttQ74-GFP.
6. Conclusions
Protein SUMOylation has a pivotal role in HD and other polyQ disorders. Here we reviewed several mechanisms by which SUMO proteins can modulate molecular aspects of HD-related proteins, pointing out its clinical relevance, especially considering that SUMOylation is highly associated with protein aggregation. As discussed above and summarized in Fig. 1, the data published so far are still controversial. In general, SUMOylation appears to have a dual effect, improving the cellular clearance and causing cytotoxicity, pointing to the need for more studies to best clarify the mechanisms underlying SUMOylation in HD and associated disorders. Based on the results from in vivo and in vitro studies to increase the modification of Htt by SUMO-2 instead of SUMO-1 could provide a promising treatment. Moreover, RNF4 overexpression protected against Htt aggregates and improved the transcriptional changes observed in HD by acting directly on the chromatin-associated Htt (Ohkuni et al., 2018). This finding could help to guide the design of an effective treatment for HD and other polyQ diseases, since RNF4-dependent degradation of Htt would be SUMO isoform insensitive. In addition to the previous effects mentioned, SUMO could still have a role in protein quality control, modulating misfolded protein, solubility, and stability (Gallagher et al., 2014), and could also prevent the formation of polyQ structures (Huang et al., 2018).
Conflicts of Interest
None.
Acknowledgements
This work was supported by grants from the Brazil National Council of Scientific and Technological Development (CNPq), in the form of PhD studentships (E.S.S.) and productivity in research scholarships (R.D.P., P.S.B. and H.I.C.). We are also grateful for grant support from the Newton Fund, ISN, and especially IBRO for Returning Home and PROLAB Grants (H.I.C.).
References
- Anderson D.B., Zanella C.A., Henley J.M., Cimarosti H. In: Advances in Experimental Medicine. Wilson V.G., editor. Springer International Publishing AG; 2017. SUMOylation: implications for neurodegenerative diseases; pp. 261–281. [DOI] [PubMed] [Google Scholar]
- Arrasate M., Mitra S., Schweitzer E.S., Segal M.R., Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- Barron J.C., Hurley E.P., Parsons M.P. Huntingtin and the synapse. Front. Cell. Neurosci. 2021;15:1–18. doi: 10.3389/fncel.2021.689332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbo M., Brandi V., Pascarella G., Staid D.S., Colotti G., Polticelli F., Ilari A., Morea V. Bioinformatics analysis of Ras homologue enriched in the striatum, a potential target for Huntington’s disease therapy. Int. J. Mol. Med. 2019;44:2223–2233. doi: 10.3892/ijmm.2019.4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celen A.B., Sahin U. Sumoylation on its 25th anniversary: mechanisms, pathology, and emerging concepts. FEBS J. 2020;287:3110–3140. doi: 10.1111/febs.15319. [DOI] [PubMed] [Google Scholar]
- Chang H.M., Yeh E.T.H. Sumo: from bench to bedside. Physiol. Rev. 2020;100:1599–1619. doi: 10.1152/physrev.00025.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Zhang Y., Wang Q., Qin Y., Yang X., Xing Z., Shen Y., Wu H., Qi Y. The function of SUMOylation and its crucial roles in the development of neurological diseases. FASEB J. 2021;35:1–17. doi: 10.1096/fj.202002702R. [DOI] [PubMed] [Google Scholar]
- Chiki A., Ricci J., Hegde R., Abriata L.A., Reif A., Boudeffa D., Lashuel H.A. Site-specific phosphorylation of huntingtin exon 1 recombinant proteins enabled by the discovery of novel kinases. ChemBioChem. 2021;22:217–231. doi: 10.1002/cbic.202000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear A.C. Deciphering protein post-translational modifications using chemical biology tools. Nat. Rev. Chem. 2020;4:674–695. doi: 10.1038/s41570-020-00223-8. [DOI] [PubMed] [Google Scholar]
- Croce K.R., Yamamoto A. A role for autophagy in Huntington’s disease. Neurobiol. Dis. 2019;122:16–22. doi: 10.1016/j.nbd.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deger J.M., Gerson J.E., Kayed R. The interrelationship of proteasome impairment and oligomeric intermediates in neurodegeneration. Aging Cell. 2015;14:715–724. doi: 10.1111/acel.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorval V., Fraser P.E. SUMO on the road to neurodegeneration. Biochim. Biophys. Acta - Mol. Cell Res. 2007;1773:694–706. doi: 10.1016/j.bbamcr.2007.03.017. [DOI] [PubMed] [Google Scholar]
- Du G., Dong W., Yang Q., Yu X., Ma J., Gu W., Huang Y. Altered gut microbiota related to inflammatory responses in patients with Huntington’s disease. Front. Immunol. 2021;11:1–12. doi: 10.3389/fimmu.2020.603594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrnhoefer D.E., Sutton L., Hayden M.R. Small changes, big impact: posttranslational modifications and function of huntingtin in huntington disease. Neuroscientist. 2011;17:475–492. doi: 10.1177/1073858410390378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields E., Vaughan E., Tripu D., Lim I., Shrout K., Conway J., Salib N., Lee Y., Dhamsania A., Jacobsen M., Woo A., Xue H., Cao K. Gene targeting techniques for Huntington’s disease. Ageing Res. Rev. 2021:70. doi: 10.1016/j.arr.2021.101385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher P.S., Oeser M.L., Abraham A.C., Kaganovich D., Gardner R.G. Cellular maintenance of nuclear protein homeostasis. Cell. Mol. Life Sci. 2014;71:1865–1879. doi: 10.1007/s00018-013-1530-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb S.L., Boston-Howes W., Lavina Z.S., Gustincich S., Brown R.H., Pasinelli P., Trotti D. A caspase-3-cleaved fragment of the glial glutamate transporter EAAT2 is sumoylated and targeted to promyelocytic leukemia nuclear bodies in mutant SOD1-linked amyotrophic lateral sclerosis. J. Biol. Chem. 2007;282:32480–32490. doi: 10.1074/jbc.M704314200. [DOI] [PubMed] [Google Scholar]
- Guerra De Souza A.C., Prediger R.D., Cimarosti H. SUMO-regulated mitochondrial function in Parkinson’s disease. J. Neurochem. 2016;137:673–686. doi: 10.1111/jnc.13599. [DOI] [PubMed] [Google Scholar]
- Guo C., Hildick K.L., Luo J., Dearden L., Wilkinson K.A., Henley J.M. SENP3-mediated deSUMOylation of dynamin-related protein 1 promotes cell death following ischaemia. EMBO J. 2013:1–15. doi: 10.1038/emboj.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X.J., Tomizawa K., Fujimura A., Ohmori I., Nishiki T., Matsushita M., Matsui H. Regulation of mitochondrial dynamics and neurodegenerative diseases. Acta Med. Okayama. 2011;65:1–10. doi: 10.18926/AMO/43824. [DOI] [PubMed] [Google Scholar]
- Harrison L.M., LaHoste G.J. The role of Rhes, Ras homolog enriched in striatum, in neurodegenerative processes. Exp. Cell Res. 2013;319:2310–2315. doi: 10.1016/j.yexcr.2013.03.033. [DOI] [PubMed] [Google Scholar]
- Hawton A., Green C., Goodwin E., Harrower T. Health state utility values (QALY weights) for Huntington’s disease: an analysis of data from the European Huntington’s Disease Network (EHDN) Eur. J. Heal. Econ. 2019;20:1335–1347. doi: 10.1007/s10198-019-01092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wu J., Xu L., Wang J., Chen Z., Yang R. Regulation of HSF1 protein stabilization: an updated review. Eur. J. Pharmacol. 2018;822:69–77. doi: 10.1016/j.ejphar.2018.01.005. [DOI] [PubMed] [Google Scholar]
- Jansen N.S., Vertegaal A.C.O. A chain of events: regulating target proteins by SUMO polymers. Trends Biochem. Sci. 2021;46:113–123. doi: 10.1016/j.tibs.2020.09.002. [DOI] [PubMed] [Google Scholar]
- Kabil O., Zhou Y., Banerjee R. Human cystathionine β-synthase is a target for sumoylation. Biochemistry. 2006;45:13528–13536. doi: 10.1021/bi0615644. [DOI] [PubMed] [Google Scholar]
- Kroonen J.S., Vertegaal A.C.O. Targeting SUMO signaling to wrestle cancer. Trends Cancer. 2020;7:496–510. doi: 10.1016/j.trecan.2020.11.009. [DOI] [PubMed] [Google Scholar]
- Labbadia J., Novoselov S.S., Bett J.S., Weiss A., Paganetti P., Bates G.P., Cheetham M.E. Supression of protein aggregation by chaperone modification of high molecular weight complexes. Brain. 2012;135:1180–1196. doi: 10.1093/brain/aws022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakke J.P.W.F. Classification of extrapyramidal disorders. J. Neurosci. 1981;51:311–327. doi: 10.1016/0022-510x(81)90109-x. [DOI] [PubMed] [Google Scholar]
- Liang Y.C., Lee C.C., Yao Y.L., Lai C.C., Schmitz M.L., Yang W.M. SUMO5, a novel poly-SUMO isoform, regulates PML nuclear bodies. Sci. Rep. 2016;6:1–15. doi: 10.1038/srep26509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebelt F., Vertegaal A.C.O. Ubiquitin-dependent and independent roles of SUMO in proteostasis. Am. J. 2016;311:284–296. doi: 10.1152/ajpcell.00091.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Cheng S., Yang H., Zhu L., Pan Y., Jing L., Tang B., Li S., Li X.J. Loss of Hap1 selectively promotes striatal degeneration in Huntington disease mice. Proc. Natl. Acad. Sci. USA. 2020;117:20265–20273. doi: 10.1073/PNAS.2002283117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Gurung S., Lee L., Henley J.M., Wilkinson K.A., Guo C. Increased SUMO-2/3-ylation mediated by SENP3 degradation is protective against cadmium-induced caspase 3–dependent cytotoxicity. J. Toxicol. Sci. 2017;42:529–538. doi: 10.2131/jts.42.529. [DOI] [PubMed] [Google Scholar]
- Marcelli S., Ficulle E., Piccolo L., Corbo M., Feligioni M. An overview of the possible therapeutic role of SUMOylation in the treatment of Alzheimer’s disease. Pharmacol. Res. 2018;130:420–437. doi: 10.1016/j.phrs.2017.12.023. [DOI] [PubMed] [Google Scholar]
- Martins W.C., Tasca C.I., Cimarosti H. Battling Alzheimer’s disease: targeting SUMOylation-mediated pathways. Neurochem. Res. 2016;41:568–578. doi: 10.1007/s11064-015-1681-3. [DOI] [PubMed] [Google Scholar]
- McColgan P., Tabrizi S.J. Huntington’s disease: a clinical review. Eur. J. Neurol. 2018;25:24–34. doi: 10.1111/ene.13413. [DOI] [PubMed] [Google Scholar]
- Moily N.S., Ormsby A.R., Stojilovic A., Ramdzan Y.M., Diesch J., Hannan R.D., Zajac M.S., Hannan A.J., Oshlack A., Hatters D.M. Transcriptional profiles for distinct aggregation states of mutant Huntingtin exon 1 protein unmask new Huntington’s disease pathways. Mol. Cell. Neurosci. 2017;83:103–112. doi: 10.1016/j.mcn.2017.07.004. [DOI] [PubMed] [Google Scholar]
- Morozko E.L., Smith-Geater C., Monteys A.M., Pradhan S., Lim R.G., Langfelder P., Kachemov M., Hill A., Stocksdale J.T., Cullis P.R., Wu J., Ochaba J., Miramontes R., Chakraborty A., Hazra T.K., Lau A., St-Cyr S., Orellana I., Kopan L., Wang K.Q., Yeung S., Leavitt B.R., Reidling J.C., William Yang X., Steffan J.S., Davidson B.L., Sarkar P.S., Thompson L.M. PIAS1 modulates striatal transcription, DNA damage repair, and SUMOylation with relevance to Huntington’s disease. Proc. Natl. Acad. Sci. USA. 2021;118:1–11. doi: 10.1073/pnas.2021836118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano F., D’Angelo L., de Girolamo P., Avallone L., de Lange P., Usiello A. The thyroid hormone-target gene Rhes a novel crossroad for neurological and psychiatric disorders: new insights from animal models. Neuroscience. 2018;384:419–428. doi: 10.1016/j.neuroscience.2018.05.027. [DOI] [PubMed] [Google Scholar]
- O’Rourke J.G., Gareau J.R., Ochaba J., Song W., Raskó T., Reverter D., Lee J., Monteys A.M., Pallos J., Mee L., Vashishtha M., Apostol B.L., Nicholson T.P., Illes K., Zhu Y.Z., Dasso M., Bates G.P., Difiglia M., Davidson B., Wanker E.E., Marsh J.L., Lima C.D., Steffan J.S., Thompson L.M. SUMO-2 and PIAS1 modulate insoluble mutant huntingtin protein accumulation. Cell Rep. 2013;4:362–375. doi: 10.1016/j.celrep.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochaba J., Monteys A.M., O’Rourke J.G., Reidling J.C., Steffan J.S., Davidson B.L., Thompson L.M. PIAS1 regulates mutant huntingtin accumulation and Huntington’s disease-associated phenotypes in vivo. Neuron. 2016;90:507–520. doi: 10.1016/j.neuron.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkuni K., Pasupala N., Peek J., Holloway G.L., Sclar G.D., Levy-Myers R., Baker R.E., Basrai M.A., Kerscher O. SUMO-targeted ubiquitin ligases (STUbLs) reduce the toxicity and abnormal transcriptional activity associated with a mutant, aggregation-prone fragment of huntingtin. Front. Genet. 2018;9:1–16. doi: 10.3389/fgene.2018.00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C.S., Osellame L.D., Stojanovski D., Ryan M.T. The regulation of mitochondrial morphology: intricate mechanisms and dynamic machinery. Cell Signal. 2011;23:1534–1545. doi: 10.1016/j.cellsig.2011.05.021. [DOI] [PubMed] [Google Scholar]
- Paoli R.A., Botturi A., Ciammola A., Silani V., Prunas C., Lucchiari C., Zugno E., Caletti E. Neuropsychiatric burden in Huntington’s disease. Brain Sci. 2017;7:1–15. doi: 10.3390/brainsci7060067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp K.V., Kaplan R.F., Snyder P.J. Biological markers of cognition in prodromal Huntington’s disease: a review. Brain Cogn. 2011;77:280–291. doi: 10.1016/j.bandc.2011.07.009. [DOI] [PubMed] [Google Scholar]
- Pastore A., Temussi P. Protein aggregation and misfolding: good or evil? J. Phys. Condens. Matter. 2012:24. doi: 10.1088/0953-8984/24/24/244101. [DOI] [PubMed] [Google Scholar]
- Pountney D.L., Huang Y., Burns R.J., Haan E., Thompson P.D., Blumbergs P.C., Gai W.P. SUMO-1 marks the nuclear inclusions in familial neuronal intranuclear inclusion disease. Exp. Neurol. 2003;184:436–446. doi: 10.1016/j.expneurol.2003.07.004. [DOI] [PubMed] [Google Scholar]
- Princz A., Tavernarakis N. SUMOylation in neurodegenerative diseases. Gerontology. 2020;66:122–130. doi: 10.1159/000502142. [DOI] [PubMed] [Google Scholar]
- Ramazi S., Allahverdi A., Zahiri J. Evaluation of post-translational modifications in histone proteins: a review on histone modification defects in developmental and neurological disorders. J. Biosci. 2020;45:3–9. doi: 10.1007/s12038-020-00099-2. [DOI] [PubMed] [Google Scholar]
- Ramazi S., Zahiri J. Post-translational modifications in proteins: resources, tools and prediction methods. Database. 2021;2021:1–20. doi: 10.1093/database/baab012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel-Barajas C., Rebec G.V. Overview of Huntington’s disease models: neuropathological, molecular, and behavioral differences. Curr. Protoc. Neurosci. 2018;83:1–21. doi: 10.1002/cpns.47. [DOI] [PubMed] [Google Scholar]
- Reddy P.H., Reddy T.P., Manczak M., Calkins M.J., Shirendeb U., Mao P. Dynamin-related protein 1 and mitochondrial fragmentation in neurodegenerative diseases. Brain Res. Rev. 2011;67:103–118. doi: 10.1016/j.brainresrev.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reif A., Chiki A., Ricci J., Lashuel H.A. Generation of native, untagged huntingtin exon1 monomer and fibrils using a SUMO fusion strategy. J. Vis. Exp. 2018;136:1–9. doi: 10.3791/57506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross C.A., Shoulson I. Huntington disease: pathogenesis, biomarkers, and approaches to experimental therapeutics. Park. Relat. Disord. 2009;15:S135–S138. doi: 10.1016/S1353-8020(09)70800-4. [DOI] [PubMed] [Google Scholar]
- Sarge K.D., Park-Sarge O.K. International Review of Cell and Molecular Biology. 1st ed. Elsevier Inc; 2011. SUMO and its role in human diseases. [DOI] [PubMed] [Google Scholar]
- Schaffert L.N., Carter W.G. Do post-translational modifications influence protein aggregation in neurodegenerative diseases: a systematic review. Brain Sci. 2020;10:1–37. doi: 10.3390/brainsci10040232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedighi F., Adegbuyiro A., Legleiter J. SUMOylation prevents huntingtin fibrillization and localization onto lipid membranes. ACS Chem. Neurosci. 2020;11:328–343. doi: 10.1021/acschemneuro.9b00509. [DOI] [PubMed] [Google Scholar]
- Serra M., Pinna A., Costa G., Usiello A., Pasqualetti M., Avallone L., Morelli M., Napolitano F. Involvement of the protein ras homolog enriched in the striatum, rhes, in dopaminergic neurons’ degeneration: link to Parkinson’s disease. Int. J. Mol. Sci. 2021;22:1–13. doi: 10.3390/ijms22105326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M., Subramaniam S. Rhes travels from cell to cell and transports Huntington disease protein via TNT-like protrusion. J. Cell Biol. 2019;218:1972–1993. doi: 10.1083/JCB.201807068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty P.M.V., Rangrez A.Y., Frey N. SUMO proteins in the cardiovascular system: friend or foe? J. Biomed. Sci. 2020;27:1–14. doi: 10.1186/s12929-020-00689-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirendeb U.P., Calkins M.J., Manczak M., Anekonda V., Dufour B., Mcbride J.L., Mao P., Reddy P.H. Mutant huntingtin’s interaction with mitochondrial protein Drp1 impairs mitochondrial biogenesis and causes defective axonal transport and synaptic degeneration in Huntington’ s disease. Hum. Mol. Genet. 2012;21:406–420. doi: 10.1093/hmg/ddr475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan J.S. Does huntingtin play a role in selective macroautophagy? Cell Cycle. 2010;9:3401–3413. doi: 10.4161/cc.9.17.12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan J.S., Agrawal N., Pallos J., Rockabrand E., Trotman L.C., Slepko N., Illes K., Lukacsovich T., Zhu Y.Z., Cattaneo E., Pandolfi P.P., Thompson L.M., Marsh J.L. SUMO modification of huntingtin and Huntington’s disease pathology. Science. 2004;304:100–104. doi: 10.1126/science.1092194. [DOI] [PubMed] [Google Scholar]
- Subramaniam S., Mealer R.G., Sixt K.M., Barrow R.K., Usiello A., Snyder S.H. Rhes, a physiologic regulator of sumoylation, enhances cross-sumoylation between the basic sumoylation enzymes E1 and Ubc9. J. Biol. Chem. 2010;285:20428–20432. doi: 10.1074/jbc.C110.127191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam S., Sixt K.M., Barrow R., Snyder S.H. Rhes, a striatal specific protein, mediates mutant-huntingtin cytotoxicity. Science. 2009;324:1327–1330. doi: 10.1126/science.1172871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam S., Snyder S.H. Huntington’s disease is a disorder of the corpus striatum: focus on Rhes (Ras homologue enriched in the striatum) Neuropharmacology. 2011;60:1187–1192. doi: 10.1016/j.neuropharm.2010.10.025. [DOI] [PubMed] [Google Scholar]
- Swarnkar S., Chen Y., Pryor W.M., Shahani N., Page D.T., Subramaniam S. Ectopic expression of the striatal-enriched GTPase Rhes elicits cerebellar degeneration and an ataxia phenotype in Huntington’s disease. Neurobiol. Dis. 2015;82:66–77. doi: 10.1016/j.nbd.2015.05.011. [DOI] [PubMed] [Google Scholar]
- Tabrizi S.J., Flower M.D., Ross C.A., Wild E.J. Huntington disease: new insights into molecular pathogenesis and therapeutic opportunities. Nat. Rev. Neurol. 2020;16:529–546. doi: 10.1038/s41582-020-0389-4. [DOI] [PubMed] [Google Scholar]
- Terashima T., Kawai H., Fujitani M., Maeda K., Yasuda H. SUMO-1 co-localized with mutant atrophin-I with expanded polyglutamines accelerates intranuclear aggregation and cell death. Neuroreport. 2002;13:2359–2364. doi: 10.1097/00001756-200212030-00038. [DOI] [PubMed] [Google Scholar]
- Testa C.M., Jankovic J. Huntington disease: a quarter century of progress since the gene discovery. J. Neurol. Sci. 2019;396:52–68. doi: 10.1016/j.jns.2018.09.022. [DOI] [PubMed] [Google Scholar]
- Thompson L.M., Aiken C.T., Kaltenbach L.S., Agrawal N., Illes K., Khoshnan A., Martinez-Vincente M., Arrasate M., O’Rourke J.G., Khashwji H., Lukacsovich T., Zhu Y.Z., Lau A.L., Massey A., Hayden M.R., Zeitlin S.O., Finkbeiner S., Green K.N., LaFerla F.M., Bates G., Huang L., Patterson P.H., Lo D.C., Cuervo A.M., Marsh J.L., Steffan J.S. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. J. Cell Biol. 2009;187:1083–1099. doi: 10.1083/jcb.200909067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H., Goto J., Hashida H., Lin X., Oyanagi K., Kawano H., Zoghbi H.Y., Kanazawa I., Okazawa H. Enhanced SUMOylation in polyglutamine diseases. Biochem. Biophys. Res. Commun. 2002;293:307–313. doi: 10.1016/S0006-291X(02)00211-5. [DOI] [PubMed] [Google Scholar]
- Valadão P.A.C., Santos K.B.S., Ferreira e Vieira T.H., Macedo e Cordeiro T., Teixeira A.L., Guatimosim C., de Miranda A.S. Inflammation in Huntington’s disease: a few new twists on an old tale. J. Neuroimmunol. 2020;348:1–11. doi: 10.1016/j.jneuroim.2020.577380. [DOI] [PubMed] [Google Scholar]
- van der Burg J.M., Björkqvist M., Brundin P. Beyond the brain: widespread pathology in Huntington’s disease. Lancet Neurol. 2009;8:765–774. doi: 10.1016/S1474-4422(09)70178-4. [DOI] [PubMed] [Google Scholar]
- van der Burg J.M.M., Gardiner S.L., Ludolph A.C., Landwehrmeyer G.B., Roos R.A.C., Aziz N.A. Body weight is a robust predictor of clinical progression in Huntington disease. Ann. Neurol. 2017;82:479–483. doi: 10.1002/ana.25007. [DOI] [PubMed] [Google Scholar]
- Varejão N., Lascorz J., Li Y., Reverter D. Molecular mechanisms in SUMO conjugation. Biochem. Soc. Trans. 2020;48:123–135. doi: 10.1042/BST20190357. [DOI] [PubMed] [Google Scholar]
- Wang Y., Dasso M. SUMOylation and deSUMOylation at a glance. J. Cell Sci. 2009;122:4249–4252. doi: 10.1242/jcs.050542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Lin F., Qin Z.H. The role of post-translational modifications of huntingtin in the pathogenesis of Huntington’s disease. Neurosci. Bull. 2010;26:153–162. doi: 10.1007/s12264-010-1118-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson V.G. In: SUMO Regulation of Cellular Processes. Wilson V.G., editor. Springer International Publishing; 2017. Introduction to sumoylation; pp. 1–12. [DOI] [Google Scholar]
- Wong M.B., Goodwin J., Norazit A., Meedeniya A.C.B., Richter-Landsberg C., Gai W.P., Pountney D.L. SUMO-1 is associated with a subset of lysosomes in glial protein aggregate diseases. Neurotox. Res. 2013;23:1–21. doi: 10.1007/s12640-012-9358-z. [DOI] [PubMed] [Google Scholar]
- Zou Z., Tao T., Li H., Zhu X. mTOR signaling pathway and mTOR inhibitors in cancer: progress and challenges. Cell Biosci. 2020:1–11. doi: 10.1186/s13578-020-00396-1. [DOI] [PMC free article] [PubMed] [Google Scholar]