Abstract
Numerous drug treatments that have recently entered the clinic or clinical trials have their genesis in zebrafish. Zebrafish are well established for their contribution to developmental biology and have now emerged as a powerful preclinical model for human disease, as their disease characteristics, aetiology and progression, and molecular mechanisms are clinically relevant and highly conserved. Zebrafish respond to small molecules and drug treatments at physiologically relevant dose ranges and, when combined with cell-specific or tissue-specific reporters and gene editing technologies, drug activity can be studied at single-cell resolution within the complexity of a whole animal, across tissues and over an extended timescale. These features enable high-throughput and high-content phenotypic drug screening, repurposing of available drugs for personalized and compassionate use, and even the development of new drug classes. Often, drugs and drug leads explored in zebrafish have an inter-organ mechanism of action and would otherwise not be identified through targeted screening approaches. Here, we discuss how zebrafish is an important model for drug discovery, the process of how these discoveries emerge and future opportunities for maximizing zebrafish potential in medical discoveries.
RELATED LINKS
Chemical Probes Portal: https://www.chemicalprobes.org
Mechanism-of-action box: https://github.com/Novartis/MoaBox
Probes & Drug: https://www.probes-drugs.org
Probe Miner: https://probeminer.icr.ac.uk
The structural Genomics Consortium: https://www.thesgc.org
well-defined Donated Chemical Probes: https://www.sgc-ffm.uni-frankfurt.de
Zebrafish disease models society: https://www.zdmsociety.org
Zebrafish (Danio rerio), named for their stripes, are small (2–5 cm) tropical fish that live naturally in rivers in southern Asia. More than 40 years ago, zebrafish entered the laboratory setting as a model organism for the study of developmental biology. Zebrafish are ideal for this purpose because they are fertilized ex vivo, can generate hundreds of embryos from a breeding pair and are transparent, enabling the development of a vertebrate to be captured from a single cell through organogenesis to a swimming zebrafish. Zebrafish share 70% of their genome with humans, and more than 80% of disease proteins are conserved1. Complex gene editing coupled with live high-resolution imaging has meant that development — as well as diseases — can be modelled and captured in unprecedented molecular detail and resolution2. Crucially, the diseases modelled in zebrafish often represent bona fide models of human disease, capturing the disease aetiology, progression and resolution processes2,3. Precision gene editing in zebrafish can generate models of human disease alleles, and these models recapitulate some of the aetiology found in patients4,5. Moreover, genetic and transgenic models often have the same alterations in conserved underlying molecular pathways as those that drive human disease2,3; cancer cell states and transcriptional signatures, for example, are often conserved between zebrafish models and human patients6,7. Zebrafish are vertebrates and therefore have similar tissues and developmental biology to humans, and zebrafish models have been generated for a wide array of cancers8, liver disease9, blood disorders10, heart disease11 and behavioural disorders12,13, among others.
Disease proteins and processes are conserved between humans and zebrafish, which means that drugs that are active in humans often have the same target in zebrafish, especially drugs that interact with the active region of the target protein. This is especially notable in drugs designed to target cancer pathways, including targeted therapies such as MEK inhibitors14; drugs commonly used on a daily basis, such as acetaminophen15; and serotonin (5-HT) modulators16. There are some examples of drugs that work in humans but not in zebrafish, and vice versa, but evidence from more than 20 years of drug screening in zebrafish indicates that, on the whole, molecules that are active in zebrafish are similarly active in mouse and human systems with similar pharmacokinetic properties17.
In this Review, we discuss recent advances in disease modelling using the zebrafish that have highlighted the potential of this model organism for discovering drugs and exploring drug mechanisms in vivo, as well as its utility as a whole-animal model for preclinical drug repurposing and personalized medicine. More recently, small-molecule screens have been carried out on adult fish, enabling the examination of pathways involved in physiology or cancer. The ability to screen in vivo coupled with the relevance of phenotypes to human diseases have contributed to the emergence of zebrafish as a leading model organism for whole-animal chemical genetics and drug discovery18 (FIG. 1).
Fig. 1 |. The power of zebrafish to model disease and therapy.

The advantages of zebrafish as a model system can be applied to model human disease and drug treatments in the embryonic and adult stages. Hundreds of transparent zebrafish embryos are fertilized ex vivo and are easily accessible for gene editing, transgenics and small-molecule screening. During these embryonic stages, the biology and function of organs and tissues can be followed in living animals at single-cell resolution to study diverse and complex processes including developmental biology, developmental disease models, stem cells and regeneration, and neural circuitry and behaviours. By 3 months, zebrafish are breeding adults and generate large numbers of embryos for small-molecule and genetic screens, and are themselves studied as models for disease, including cancer, cardiovascular and other organ defects, behaviour, addiction, metabolism, and adult tissue regeneration and physiology.
Zebrafish phenotypic drug screens
One of the fundamental challenges in drug discovery is showing efficacy and understanding the mechanism of action within a living whole-animal system. Although many compounds and drug leads have activity in vitro or in cell systems, it is crucial to understand how drugs work in the context of disease in an organ, tissue and distal sites in the body. A major goal is to monitor these responses over time, during disease progression and resolution, and to evaluate drug-induced adverse effects8.
Zebrafish for phenotypic profiling.
The same characteristics that made zebrafish a tractable model organism for developmental biology and disease modelling have advanced zebrafish as a model system for phenotypic drug discovery17. In phenotypic drug screening, drug leads are selected by their potential to generate phenotypic outcomes, independently of known and validated targets19–22. Phenotypic drug screens were the historic basis for drug discovery and have re-emerged alongside targeted drug discovery on the basis of their success in identifying first-in-class drugs. Inherent features of the phenotypic screening process include: drugs or drug leads can be identified even if they generate the desired phenotypic outcome by targeting multiple targets (for example, kinase inhibitors can generate therapeutic benefit through targeting multiple kinases)23; there is the potential for a broad range of phenotypic outcomes; and counter-screening against compounds can be used to understand additional toxicity or unwanted phenotypes. The small size of zebrafish embryos means that thousands of embryos can be screened for phenotypic effects in the context of a living whole animal. Although phenotypic zebrafish screens are not as high throughput as those in cell culture, yeast, fruitflies or in vitro activity assays, zebrafish embryos can be screened on a scale beyond what is currently available in other vertebrate systems. Small molecules can be actively absorbed from the water by the embryo, absorbed through the gills in older fish or administered by oral gavage24.
The goal of most high-throughput drug screens in vitro is to identify a highly selective drug with a single target. However, many drugs act on multiple targets to affect multiple biological pathways. The capacity of a drug or small molecule to bind to multiple targets — polypharmacology — is being incorporated into pharmaceutical designs to purposefully develop drugs that target synergistic combinations of factors25,26. In a small-molecule screen, AD57, a compound with poly-pharmacological properties, was found to increase survival in Drosophila melanogaster models of multiple endocrine neoplasia (MEN) type 2 caused by activating mutations in the gene encoding the receptor tyrosine kinase Ret. The effects of AD57 on survival were mediated by the inhibition of Ret plus three other kinases, Raf, Src and S6K, and toxicity was caused by inhibition of Tor. Through rational chemical design, drug leads were designed that did not alter Tor activity and had improved efficacy and reduced toxicity for MEN in flies and human cancer cells27. In zebrafish, multi-dimensional phenotyping means that chemically induced phenotypes can assist in the identification of multi-target effects of chemical compounds. For example, the commonly used MEK inhibitor U0126 was shown to be both a copper chelator and a MEK inhibitor in vivo, based on shared zebrafish phenotypes and yeast chemical–genetic profiles with other copper chelators and MEK inhibitors28. Zebrafish screens are also beginning to incorporate the mechanisms of multi-target drugs, and this has been especially effective in large-scale behaviour-based phenotypic screens29. For example, one group analysed structurally diverse compounds and associated behaviour profiles to identify compounds with complex polypharmacology mechanisms, such as haloperidol, which they demonstrated provides anti-psychotic effects through multi-target mechanisms30.
For some drugs, zebrafish recapitulate effects seen in humans better than mouse models do. This is the case for thalidomide, which was prescribed to expectant mothers for nausea and caused thousands of birth defects in unborn children. In mouse, thalidomide did not cause any defects; however, in zebrafish, thalidomide causes the same morphological limb defects as it did in humans. In both species the drug binds to and activates the E3 ubiquitin ligase CUL4–RBX1–DDB1–CRBN (known as CRL4CRBN)31. Understanding the targets of thalidomide explains the effects of the drug on the developing fetus and has enabled the appropriate and therapeutic use of thalidomide in novel contexts, such as myeloma32,33. Target identification for this drug and its analogues has been crucial to understanding how subtle differences in target engagement can lead to differences in activity in vivo34.
Whole-animal integrated physiology.
With some exceptions, zebrafish have the same tissue development and homeostasis as humans, including the development of blood, muscle, heart, liver, pancreas, spleen, intestine, kidney, bone, fat, and neuronal and neural crest lineages. Drugs can be screened for on-target activity in tissues and simultaneously provide information about the compound absorption, distribution, metabolism, excretion and toxicity (ADME-tox) properties17,35. Establishing the compound activity profile in the target tissue while simultaneously testing the impact on other tissues can be integrated into iterative chemistry for compound development and specificity36. This approach provides important information about common side effects of drugs or drug combinations, including life-saving drugs that are dose-limited owing to severe toxicities in patients. For example, doxorubicin is a topoisomerase II inhibitor and is highly effective at killing cancer cells but comes with a substantial risk of heart toxicity and failure; these same cardiotoxicities are also observed in zebrafish treated with this drug. By screening for adjuvant therapies that can prevent the doxorubin-induced cardiomyopathies, the natural product visnagin, which can protect against cardiomyopathies without altering the cytotoxicity of doxorubicin towards cancer cells, was found37. Recently, this potential for integrated physiology has been developed further, using zebrafish to identify compounds that protect against the dose-limiting toxicities of cisplatin, a common and broad-spectrum chemotherapy that causes kidney damage and deafness. By employing a two-pronged toxicity counter-screening assay for kidney and lateral line cells, dopamine and its associated regulators were found to be otoprotective and nephroprotective, without affecting the potency of cisplatin to induce cell death in neuroblastoma or oral squamous cell carcinoma cell lines38.
Although molecular targets are often shared between zebrafish and humans, this does not always translate to shared pharmacology39. For example, there are differences in ligand regulation of the oestrogen receptor (ER)40–42. Oestrogen-dependent gene activity is regulated by ligand binding to ERα and ERβ in humans, and ERα, ERβ1 and ERβ2 in zebrafish. Although oestrogens and anti-oestrogens stimulate similar responses in human and zebrafish systems, there were inter-species differences between ERα and ERβ ligand specificity and active concentrations40–42. For example, propyl pyrazole triol (PPT) is selective for ERα in humans and ERβ in zebrafish, and conversely, diarylpropionitrile (DPN) is selective for ERβ in human cells and ERα in zebrafish40,41. Thus, subtle differences in ER sequences and regulation between species prevent direct translation of compounds that regulate the ER response from zebrafish to human.
Screening assays.
Some of the earliest zebrafish drug screens were based simply on the developmental phenotype of the zebrafish embryo, demonstrating that small molecules can generate phenotypes similar to genetic mutations20. This powerful approach identified the first bone morphogenetic protein (BMP) inhibitor, which phenocopied genetic mutations in BMP signalling; derivatives of this inhibitor are now in clinical trials43 (TABLE 1). In contrast to most genetic mutations, chemicals can conditionally control developmental pathways in a temporal manner, and the phenotypes can be dose-dependent. The ability to add small molecules at various stages in development is crucial for screening and selectively testing compounds within a specific developmental or disease time-window. Furthermore, observing how drugs alter cell and tissue physiology over time, and within the context of the whole animal, provides insights into possible inter-organ physiological, off-target or toxic effects of molecules. Modular variations and flexible screening designs can incorporate ranges of small-molecule concentrations and treatment timings within the context of sensitized genetic backgrounds to boost the hit rate from small-molecule screens and reduce the number of false negatives, as has been recently illustrated by screens for segmentation disorders44.
Table 1 |.
Drug discovery routes from zebrafish to the clinic
| Drug | Zebrafish screening assay | Indication | Status; clinical trial identifier |
|---|---|---|---|
| New applications for known targets | |||
| MEK inhibitor | Prevent lymphatic disease in ARAF-mutant transgenic model | Lymphatic disease | Compassionate use |
| MAPK pathway inhibitors | Rescue blood flow in BRAF-mutant transgenic model | Arteriovenous malformation | Compassionate use, planned clinical trials |
| Leflunomide | Inhibit neural crest development | Melanoma | Phase I (on hold); NCT01611675 |
| All-trans retinoic acid | Pluripotent zebrafish blastomere culture | Adenoid cystic carcinoma | Phase II; NCT03999684 |
| Olaparib plus temozolomide | Adult PDX and xenograft models | Rhabdomyosarcoma | Phase I; NCT01858168 |
| New target or new application | |||
| ProHema (PGE2 derivative) | HSC expansion in development | Leukaemia, graft versus host disease | Phase II; NCT01627314, NCT00890500 |
| Clemizole (EPX-100) and clemizole derivatives (EPX-101, EPX-102, EPX-103) | Rescue of seizures in scn1lab mutant | Dravet syndrome | Phase II; NCT04462770 |
| Trifluoperazine | Rescue of anaemia in rsp19 mutants | Diamond Blackfan anaemia | Phase I; NCT03966053 |
| New chemical entity | |||
| ORC-13661 | Protection of lateral line hair cells from cisplatin and aminoglycoside | Hearing loss following antibiotic treatment in patients with CF | Phase II |
| KER-047 | Dorsalization of developing embryo | Fibrodysplasia ossificans progressiva; iron deficiency anaemia; iron-refractory iron deficiency anaemia | Phase I; ACTRN12619000319178 |
| PP2A activator | Selective toxicity for MYC-overexpressing thymocytes | T cell acute lymphoblastic leukaemia | Preclinical |
CF, cystic fibrosis; HSC, haematopoietic stem cell; PDX, patient-derived xenograft; PGE2, prostaglandin E2; PP2A, protein phosphatase 2A.
Whole-animal zebrafish screening assays enable deep phenotyping of drug activity on organs and tissues at single-cell resolution. A wide range of screens have used assays that capture the dynamics of developmental biology, cell proliferation, cancer, infection and immunity, behaviour and toxicity, among others17 (FIG. 2). Screens can follow protein expression at the single-cell level in the developing embryos using antibodies (such as phosphohistone-H3 antibodies)45, or follow gene expression in single cells or patterns, such as using whole-mount in situ hybridization (such as crestin RNA expression)46 on fixed tissues. Screens can also take advantage of the remarkable ability of zebrafish to regenerate tissues from stem cell populations, including the regeneration of hair cells47, pigment cells48,49, bone50 and spinal cord51. Single cells and tissues can also be examined using fluorescent reporter lines in live embryos in drug screens. For example, a screen for regulators of pancreatic β-cells using gene expression fluorescence as a readout identified a subgroup of histone deacetylase (HDAC) inhibitors that could reduce glucose levels in zebrafish, even in the context of hyperglycaemia, and these inhibitors were shown to have conserved function in mouse and human β-cell models52.
Fig. 2 |. Examples of phenotypes that can be assessed using drug screening.

Fluorescent in situ hybridization (FISH), immunohistochemistry (IHC) or tissue-specific fluorescent transgene expression can be used to identify drugs that ablate specific cell types, including cancer cells, in embryonic, larval and adult zebrafish. Sensitized screens use genetically mutant animals and reversion of phenotypes following drug administration. In the example provided, small molecules can be identified that sensitize p53-deficient embryos to radiation-induced cell killing when assessed by whole-mount terminal deoxynucleotidyl transferase dUTP nick end labelling (TUNEL). The lightning bolt indicates DNA damage, such as irradiation. Transgenic approaches can be used in which a fluorescent protein is expressed under the control of a gene promoter that is activated by a specific molecular pathway. In the example provided, DUSP6 is a well-known target of RAS pathway activity and can be used to identify inhibitory drugs in that pathway.
Screens using assays with fluorescent reporter lines can be performed at high throughput using low-resolution microscopes or plate readers; however, this limits the cellular resolution of the screening assay. This problem has been partly solved with an automated, high-resolution image analysis platform for drug screening in live embryos; this platform was used to screen for small molecules that regulate myelinating oligodendrocytes, which are crucial for axon insulation and which support the central nervous system (CNS)53. In this set-up, zebrafish embryos were automatically taken from multi-well plates in liquid and pulled through rotating thin-wall glass capillary tubes in which they were scanned by a spinning disc confocal microscope. This approach enabled high-throughput, complex, deep phenotyping of the mechanisms of drugs in vivo without damaging the embryos through handling and mounting53.
Screening complex visible phenotypes can also be useful for cardiovascular biology and diseases. Zebrafish hearts are clearly visible and beat with active blood circulation by 48 h of development. Zebrafish larvae can present with key indicators of cardiovascular diseases, including cardiomyopathy, arrhythmic disorders and altered angiogenesis11. These features enabled the first small-molecule screen for whole-organ function in a vertebrate to be performed in zebrafish54. Recently, in a quest for large-scale, accurate quantification, an automated platform was developed to quantify heart rate and rhythm in non-anaesthetized zebrafish embryos, allowing large-scale scoring of more than 500 embryos per 96-well plate in 20–60 min, in which dose-dependent effects were observed55. To better define and screen the metrics used to monitor cardiac function, another group employed light-sheet microscopy coupled with deep-learning image analysis to identify the earliest indicators of heart deficits caused by drugs, well before current standard methods could detect potential problems56. Cardiovascular disease is one of the leading causes of death globally and there is a current downward trend of cardiovascular drug discovery — zebrafish drug screens have the potential to reveal new, unanticipated drug leads for cardiovascular biology and disease11.
Zebrafish liver disease is similar to human disease, even in embryonic stages, and the transparent nature of the zebrafish embryo enables a dynamic readout of liver function9,57–59. Using a fluorescent reporter line for the fasting response in zebrafish embryos, new chemical regulators of the feeding-to-fasting metabolic state were identified; these compounds could also effectively improve the health of diet-induced obese mice60,61. Zebrafish liver disease models extend to toxicity caused by acetaminophen, one of the most common causes of drug-induced liver toxicity. Through small-molecule screening on zebrafish embryos, prostaglandin E2 (PGE2), together with N-acetylcysteine, was found to reduce toxicity in both embryo and adult acetaminophen liver toxicity models15. Subsequent chemical screens for modulators of liver development identified regulators of nitric oxide and S-nitrosothiol signalling that could promote liver growth, protect against acetaminophen liver injury and stimulate liver repair in zebrafish and mice62,63. Zebrafish liver assays can also be integrated into the drug discovery pipeline to identify toxicities early in the drug development process, as well as to identify chemical toxins in the environment39. Zebrafish liver assays in embryos have been used to demonstrate that sub-toxic levels of inorganic arsenic, commonly found in drinking water and a worldwide public health concern, can interact with low levels of ethanol to promote fatty liver disease64. This provides clear evidence in animals that environmental toxins can amplify the risks from common factors that promote liver disease64.
Zebrafish embryos begin to swim by 3 days of development, and the rapid, simultaneous development of the hundreds of small, free-swimming zebrafish embryos provides an opportunity to follow complex zebrafish behaviours. Zebrafish develop stereotypical behaviours, with evolutionarily conserved motivation and decision-making neuronal networks, that can be captured at scale with high-speed cameras65. More than 100 embryonic and adult behaviours have been defined12, and because the neuropharmacology of drugs applied to zebrafish is similar to that in humans, zebrafish can be used as a drug screening platform for a wide range of CNS psychiatric disorders including sleep–wake cycles, depression and anxiety, epilepsy and neurodegenerative disorders30,66–68.
Phenotypic behaviours can be coupled with fluorescent molecular readouts of the animal without perturbation. A recent screen for molecular regulators of the circadian rhythm using a luminescent reporter in zebrafish embryos identified drugs, such as nonsteroidal anti-inflammatory drugs (NSAIDs) and glucocorticoids, that regulate the inflammatory state to alter the circadian clock69. This could have implications for how inflammatory drugs alter the circadian rhythms that control daily physiology and metabolic health. Notably, the effects of anti-inflammatory and pro-inflammatory compounds were absent in cell cultures, so whole-animal zebrafish embryos were needed to identify non-cell autonomous drug targets in this complex biological process69.
Identifying mechanisms and targets.
The selection of drugs used for screening or testing depends heavily upon subsequent follow-up and the necessity for target identification. One of the advantages of phenotypic screening is that the molecular target of the drug does not need to be known or have a hypothesized function in the disease70. For a compound with unknown targets, one technique to identify its mechanism of action is to compare the drug-induced phenotype with known genetically induced phenotypes. For example, the chemical dorsomorphin induces the same phenotype as mutations in the BMP signalling pathway43. Similarly, comparison with known chemical–phenotypic profiles has been used to identify new target pathways for drug leads — new psychoactive target pathways for poorly characterized small molecules71, and more recently, the specific activity of tolterodine towards muscarinic M3 receptors over M2 receptors in heart development72. Chemoinformatic target identification, the comparison of a compound with drug structures with known targets, can also assist in candidate target identification73. Unbiased screening coupled with barcoding of drug-induced behavioural phenotypes in zebrafish embryos has been used to identify new targets for previously uncharacterized drugs29,30,68. Similar chemoinformatic target identification approaches were used to identify the target pathway for NSC10627, a compound found to strongly inhibit the zebrafish neural crest but with no known target74. Through structural similarity to a dihydroorotate dehydrogenase (DHODH) inhibitor, NSC10627 was tested and found to inhibit DHODH; at this stage, the clinically active DHODH inhibitor leflunomide was selected for preclinical and eventual clinical trials74.
Biochemical methods to identify targets, such as through pull-down interaction assays, coupled with in silico drug modelling, can reveal new targets for drugs or targets for first-in-kind molecules75. This approach was used to identify aldehyde dehydrogenase (ALDH) enzymes as a new class of enzymes that can bioactivate the 5-nitrofuran antibiotic class of prodrugs76. Drug targets can also be validated using genetics to knock down targets or through overexpression assays28,77–79. In an example using zebrafish genetics, an unanticipated target of clemastine was found in macrophages in a drug screen for anti-mycobacterial activity in a zebrafish tuberculosis model80. Clemastine is well known as an antihistamine, but in this context, clemastine stimulated P2X purinoceptor 7 (P2RX7) channels in host macrophages to clear the bacterial infection80.
Library and drug selection.
The choice of chemical library for a zebrafish small-molecule screen strongly influences the phenotypic outcomes and future decisions about whether the compound could be developed as a probe (tool) compound, a drug lead or for repurposing. This is especially important because the high-content complexity of zebrafish screening assays means that most libraries are screened in low to medium throughput. Many groups use commercially available libraries of compounds or clinically active drugs (libraries of FDA-approved drugs) that are widely available and well established for drug repurposing. These drugs have demonstrated activity but are often non-selective, complicating subsequent data deconvolution and target identification81. The reliance on these libraries, which represent only a fraction of chemical diversity, in the zebrafish community may partly explain why common targets and pathways, such as NSAIDs, often emerge from zebrafish drug screens.
As an alternative to screening libraries of known drugs, using sets of chemical probes with well-defined mechanisms of action provides hypotheses of target function. These hypotheses can be experimentally validated in vivo72, for example, by using zebrafish genetics to alter the chemical target or to explore the timing and active dose in a living system17. Probes also provide insight into the capacity of the target to be modulated pharmacologically for drug-lead development82. Recent efforts by the chemical biology community have generated publicly available, highly annotated chemical probe resources81–83 (BOX 1).
Box 1 |. Chemical libraries.
Zebrafish investigators have the opportunity to use libraries of selective probes of well-defined pharmacological agents82, such as the mechanism-of-action box (Novartis Institutes for Biomedical Research; see Related links), which is a dynamic chemogenetic library that covers more than 2,100 mammalian targets and includes information about known target engagement and appropriate control compounds83. The Structural Genomics Consortium (SGC; see Related links) and the well-defined donated chemical probes81 (see Related links) offer libraries of high-quality probes, including associated data (off-target and on-target profiling), control compounds, and recommendations on usage and established guidelines. For zebrafish investigators who require specific small molecules to functionally interrogate the biology of a specific molecular pathway, the Chemical Probes Portal (see Related links) is a web-based resource that provides investigators with a comprehensive overview of >350 tool compounds and recommendations for selection and use from their scientific advisory board. Probes & Drug (see Related links) is a resource that allows investigators to explore integrated target and chemistry data from more than 800 probes and 5,700 drugs, and Probe miner (see Related links) is a resource for probe selection based on medicinal chemistry data from more than 1.8 million small molecules on more than 2,200 human targets. These resources can assist zebrafish researchers to use high-quality probes (selective, active and chemically stable) that can be best matched to assay, and avoid poor quality probes (non-selective, reactive or unstable) that cause unreliable data and interpretation. For more detailed information see REF.82.
Because phenotypic screening has been central to the clinical success of first-in-class drugs22, screening libraries that explore novel chemical space provides an opportunity for the zebrafish field to discover probes and new drug leads with novel mechanisms of action that can be translated to the clinic (TABLE 1). Libraries of compounds that maximize structural novelty and diversity provide a starting point when no known bioactive compounds exist to modulate a specific biological function or to identify a novel mechanism of action82. To capture this diversity, libraries that use diversity-oriented synthesis are considered more like natural products and have been the source of chemical probes and drug leads for a number of diseases, including cancer, heritable diseases and infectious diseases82,84.
Natural products are another rich source of novel chemical structures with novel mechanisms of action84,85. Natural product libraries can comprise single compounds, but more often extracts contain tens to hundreds of compounds per sample. Natural products can be challenging as a starting point for screening because the active metabolite or molecule must be identified from the extract, the compounds can be difficult to synthesize and/or rare, and selective pressures may mean that natural products often target a limited number of essential targets of protein–protein networks86. Despite these challenges, more than half of the currently available drugs for cancer come from nature84,85. For example, using natural product chemical libraries in zebrafish screens, a new fungal filtrate mycotoxin library, comprised of secreted secondary metabolites from more than 10,000 fungal species, was generated87. Secreted secondary metabolites have already brought us antibiotics such as penicillin and cephalosporin, and the immunosuppressant cyclosporine. The mycotoxin filtrates were screened to identify those that caused specific developmental phenotypes (such as defects in the notochord, pigmentation, fin and heart) and those that caused overall toxicity in zebrafish embryos. To identify the active mycotoxin that elicited developmentally specific phenotypes, activity-guided purification was performed using zebrafish phenotypes as a readout, followed by identification of the active molecules using high-resolution mass spectrometry and NMR spectrometry88. Using this approach, among the compounds they identified was cercosporamide, which they demonstrated was a potent inhibitor of BMP receptor (BMPR) type I kinase88. Cercosporamide treatment pheno copied zebrafish BMP loss-of-function genetic mutants and other BMP inhibitors; however, the structure is distinct from known BMP inhibitors and, based on phenotypic analysis, cercosporamide primarily inhibits ALK2, one of the BMP receptors, in zebrafish. Cercosporamide represents a new class of BMP inhibitors for diseases that have overactive BMP signalling, especially those with activated ALK2, such as fibrodysplasia ossificans progressiva and diffuse intrinsic pontine glioma.
From zebrafish to human disease
Many small molecules discovered to have disease-rescuing activity in the zebrafish have made it to clinical trials (TABLE 1). Below, we illustrate this success with a selection of examples based on screens using embryos, adults and tumour xenografts (FIG. 3).
Fig. 3 |. Zebrafish drug discovery and optimization for clinical development.
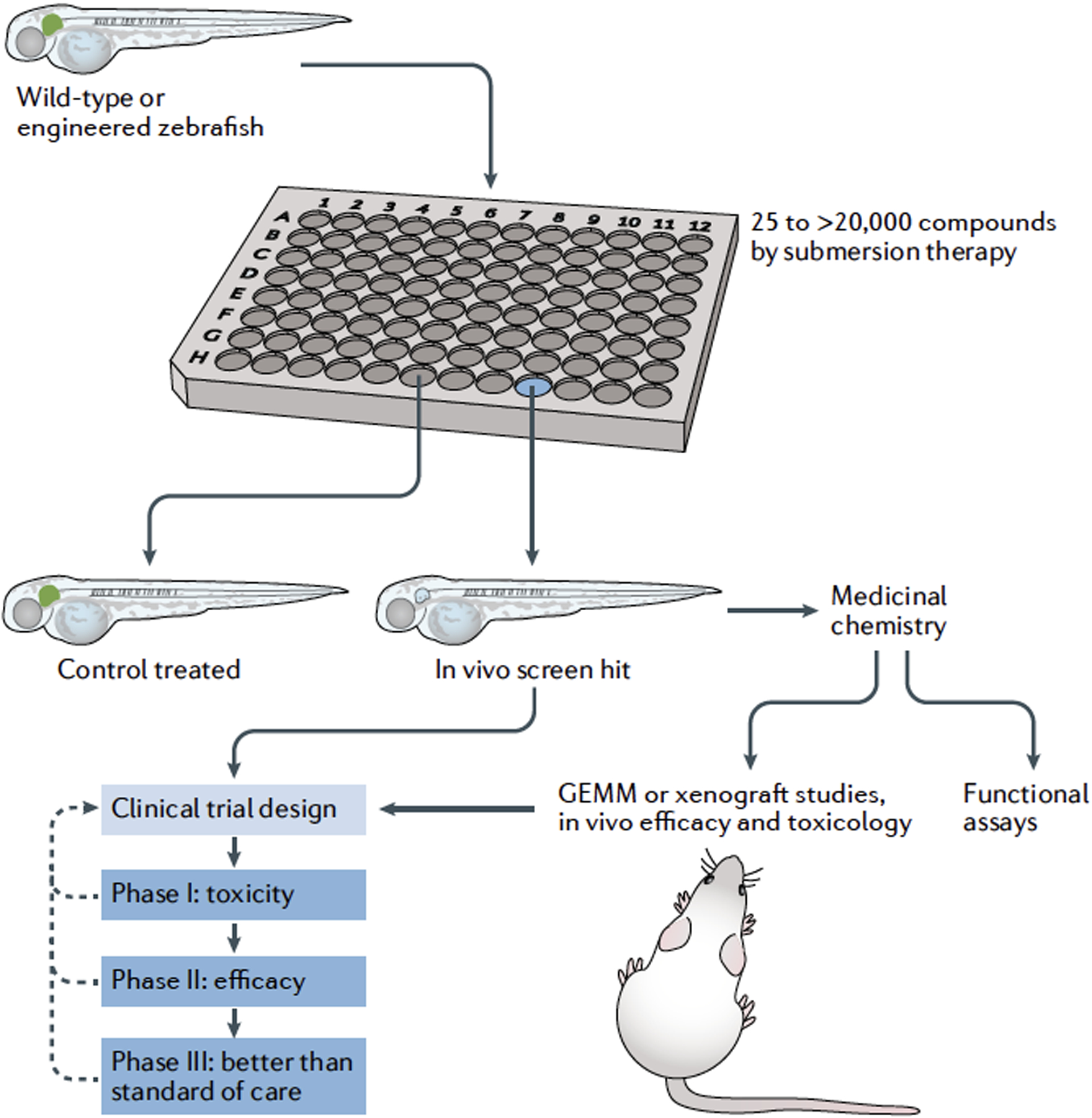
Zebrafish are innovative drivers of drug discovery and drug-lead optimization in the drug development pipeline. Phenotypic drug screens in zebrafish involve arraying zebrafish embryos into multi-well plates (for example, a 96-well plate) in which each well contains a different drug, drug combination or extract. Zebrafish embryos absorb small molecules at physiological drug concentrations. Zebrafish can be screened for altered cell or tissue development marked by fluorescent reporters, or by other phenotypic assays. Once a screening hit is identified, the phenotypic readout coupled with drug-induced toxicity can be evaluated, and chemical compound optimization rapidly assessed through structure–activity relationship (SAR) profiling using zebrafish embryos. These novel drug leads are then further processed through absorption, distribution, metabolism, excretion and toxicity (ADME-tox) studies, additional functional assays and in vivo models before testing in the stages of human clinical trials. In some situations, screening hits can be taken directly from zebrafish to inform clinical trial design. GEMM, genetically engineered mouse model.
Prevention of ototoxicity.
The zebrafish has provided key insights into ear development and a model for deafness. For instance, the congenital Usher syndrome, which is associated with deafness, has been accurately modelled using the zebrafish89,90. A zebrafish screen was used to identify chemicals that prevented toxicity of the mechanosensory lateral line hair cells, cells that show functional similarities to mammalian inner ear hair cells, from aminoglycosides, a commonly used antibiotic class91. The identified molecule, PROTO-1, was used as a starting point for medicinal chemistry to improve its efficacy, physiochemical, pharmacokinetic and toxicological properties. These modifications resulted in ORC-13661, which prevented aminoglycoside-induced ototoxicity in the mature rat without interfering with the antimicrobial efficacy of the aminoglycoside92. Subsequent studies using zebrafish, rat and organotypic mouse cochlear cultures showed that ORC-13661 protected sensory hair cells from ototoxicity induced by both aminoglycosides and cisplatin by high-affinity blockade of the mechanoelectrical transducer channel in outer hair cells, which is used for cisplatin entry93.
A phase I double-blind, placebo-controlled study of ORC-13661 was completed in 2019. Single oral doses of up to 400 mg of ORC-13661 and multiple daily oral doses of up to 80 mg of ORC-13661 appeared generally safe and well tolerated by the healthy volunteers. None of the treatment-related adverse effects were serious, dose-related or significantly different from those in the placebo group93. Based on these results, a phase II study will begin shortly to assess whether ORC-13661 can protect against hearing loss in patients with cystic fibrosis who can suffer from ototoxicity and hearing loss when administered high-dose aminoglycoside antibiotics to treat their respiratory infections. Another group of patients who might benefit from ORC-13661 are those with non-tuberculous mycobacterial infections, who are often treated with the aminoglycoside antibiotic amikacin, which can cause hearing loss.
PGE2 for stem cell transplantation.
A chemical screen designed to find inducers of haematopoietic stem cells (HSCs) in the developing aorta of the zebrafish showed that PGE2 could directly stimulate HSC production and engraftment. This principle was also shown in mouse bone marrow transplants. This discovery that dimethyl prostaglandin E2 (dmPGE2) treatment in zebrafish increases the number of stem cells94 was followed by mechanistic studies of stem cell activity95, and PGE2 was then tested in a phase I clinical trial. PGE2 activates the WNT pathway, which drives self-renewal, and upregulates CXC-chemokine receptor 4 (CXCR4), which enhances HSC homing to the marrow95–98. In the mouse, a 2 h pulse of dmPGE2 increased engraftment rates fourfold, and inhibition of prostaglandin dehydrogenase also enhanced HSC activity99. Using a competitive transplantation experiment akin to the mouse competitive repopulation assay, a phase I clinical trial was designed in which patients with leukaemia, but no matched donor marrow, received two cord blood units as an alternative source of stem cells (single cords have too few stem cells to ensure engraftment): one unit was treated with dmPGE2, the other was untreated100. Both were transplanted into patients. DNA polymorphisms in white blood cells revealed preferential engraftment and earlier return of neutrophils and platelets from dmPGE2-treated cord bloods in 10 out of 12 patients100. This work progressed to a larger phase II clinical trial of more than 150 patients who have received dmPGE2-treated cord blood or mobilized peripheral blood stem cells. As was suggested by phase I studies, patients who received dmPGE2-treated blood had enhanced blood engraftment and did not subsequently develop graft versus host disease. As PGE2 was subsequently shown to enhance gene transfer, this treatment has become a popular method for treating blood stem cells during gene therapy trials because it improves viral transduction and stem cell effects93.
Clemizole for genetic epilepsy.
Dravet syndrome is a medically refractory catastrophic epileptic encephalopathy in children that is caused by a mutation in SCN1A. As with many genetic forms of epilepsy, Dravet syndrome is an orphan indication with no definitive or convincing treatment. Zebrafish with a loss-of-function mutation in scn1lab have spontaneous electrographic seizures and convulsive-like swim behaviours101. These models capture the clinical features of Dravet syndrome in children, and as in patients, the zebrafish Dravet syndrome model was resistant to antiepileptic drugs101. A screen of more than 3,000 commercially available and FDA-approved drugs identified the 5-HT receptor agonist clemizole as a chemical suppressor of spontaneous seizures in scn1lab mutants, inhibiting both convulsive-like behaviour and electrographic events101–103. Clemizole binds to serotonin receptors, and the FDA-approved serotonin receptor agonist lorcaserin also had antiepileptic activity in zebrafish models16. On the basis of these findings, the investigators evaluated lorcaserin for compassionate use in five children with Dravet syndrome, and all children had fewer total seizures16. This was the first ‘aquarium-to-bedside’ example of evaluating drugs to treat epilepsy13. EpyGenix Therapeutics is now developing clemizole (EPX-100) and clemizole derivatives (EPX-101, EPX-102 and EPX-103) for clinical use. Phase I studies confirmed that EPX-100 is safe and well-tolerated by children (n = 24), and EPX-100 has now moved to phase II clinical trials, with the first patient dosed in November 2020. Approximately 85% of known human single-gene epilepsy mutations have homologues in zebrafish, providing a framework for modelling additional rare genetic forms of epilepsy in zebrafish for drug screens.
Drug leads for melanoma.
Zebrafish models of melanoma have been central to understanding the genomics and aetiology of the disease, and for identifying new therapeutic strategies104. Crucially, zebrafish melanomas share molecular and histopathological features with human melanomas, indicating that these models are relevant for melanoma cancer biology and drug discovery. The reactivation of melanocyte developmental pathways and the dependence of tumours on these pathways in melanoma has positioned the zebrafish as a model for small-molecule screens in the developing embryo that can then be tested in adult systems8.
Melanoma arises from the melanocyte lineage — pigmented cells that emerge from the neural crest during development105. A neural crest transcriptional signature is activated in melanoma, and a small-molecule screen to find compounds that selectively target the neural crest identified DHODH inhibitors, including the anti-arthritis drug, leflunomide74. Leflunomide is an FDA-approved drug, so this discovery led to preclinical trials in mouse xenografts and clinical trials that examined leflunomide in combination with BRAF inhibitors. Despite the strong preclinical rationale and potent effects in mouse xenografts, patient recruitment has been difficult because there is strong competition between clinical trials to enrol patients with melanoma, largely because of the wide array of immuno-oncology and combination targeted therapy approaches being investigated in this cancer type.
In addition to providing a novel drug treatment approach to target the neural crest, zebrafish models have been central to understanding the contribution of reactivation of the neural crest transcriptional signature to melanoma initiation106 and providing a detailed mechanistic understanding of how nucleotide metabolism and RNA polymerase II transcriptional elongation regulates melanocyte stem cells and melanoma states46,49,107. These studies revealed new vulnerabilities in melanoma and suggested that some subtypes of melanoma may be sensitive to inhibitors of transcriptional elongation49,108. Screening for compounds that affect neural crest and melanocytes in development has provided a rich source of new melanoma drug leads. Through a small-molecule screen on the zebrafish neural crest, the natural compound caffeic acid phenethyl ester (CAPE) was shown to be a novel regulator of the neural crest through phosphatidylinositol 3-kinase (PI3K) and SOX10 activity109, and the antifungal clotrimazole was shown to inhibit ectopic melanocytes in a KIT transgenic zebrafish model and to have activity in melanoma cells110. Furthermore, through a zebrafish melanocyte cytotoxicity screen, the 5-nitrofuran prodrugs were found to be activated by ALDHs76, and later shown to selectively target ALDH-high cancer stem-like cells in human cancers111.
Zebrafish blastomeres.
Adenoid cystic carcinoma (ACC) is a rare and malignant form of head and neck cancer that arises from the salivary glands. There is no standard chemotherapy treatment for recurrent or metastatic ACC, and most patients die within 2 years of diagnosis. Most ACCs have recurrent MYB translocations, and ACC is characterized at the molecular level by overexpression of the master transcription factor, MYB, and its target genes that control proliferation and differentiation112. Few experimental models are available; there is only one human cell line (recently developed), and culturing primary ACC cells has been challenging113. To identify chemical regulators of MYB expression, one group used a zebrafish bacterial artificial chromosome (BAC) reporter system with GFP at the ATG of myb, and then disassociated the cells at the sphere stage113. This approach generated a cell culture system that enabled high-throughput screening for regulators of myb–GFP-expressing cells that would not otherwise be possible in a whole-embryo screening approach. Screening 3,840 bioactive small molecules in duplicate identified all-trans retinoic acid (ATRA), a retinoic acid agonist, as a potent and selective suppressor of myb–GFP expression. To test the potential for ATRA as a therapeutic for ACC, MYB-translocation positive, patient-derived xenograft (PDX) mouse models were treated with ATRA, which strongly inhibited growth; ATRA reduced the MYB occupancy on DNA. A single-institution, phase II study of ATRA is treating 18 patients with advanced, incurable ACC (NCT03999684), and although full trial results are pending, early reports, presented at the 2021 Annual Meeting of the American Association for Cancer Research, suggest some stabilization effect among certain patients with excellent overall tolerability.
MAPK pathway inhibitors in RASopathy.
Mutations in the RAS–MAPK pathway that occur de novo in development lead to a range of rare genetic conditions called the RASopathies114. The RASopathies include Noonan syndrome, cardio-facio-cutaneous (CFC) syndrome, Costello syndrome and Legius syndrome, Noonan with multiple lentigines and neurofibromatosis type 1 (NF1), which together make RASopathies one of the most common genetic conditions. Although these are distinct syndromes, individuals share common clinical features including characteristic facial features, developmental and growth delay, heart defects, neurocognitive delays and gastrointestinal issues. RASopathies are progressive syndromes, with clinical phenotypes developing in utero and postnatally, the latter of which may be preventable or even reversible and are the focus of current clinical trials115–117. Despite functional predictions based on sequence analysis, early expression studies of BRAF and MEK CFC-syndrome alleles in zebrafish indicated that they are gain-of-function in vivo118. At the molecular level, in zebrafish and Drosophila models, RASopathy mutations can lead to both increased and attenuated levels of RAS signalling, depending on the cellular context119,120. In other zebrafish models, RASopathy mutations caused defective convergence and extension cell movements in gastrulation, leading to later craniofacial, pigmentation, heart and growth defects121–123. These zebrafish assays provide a means to quantitatively rank the severity of the MEK1 mutations, and zebrafish phenotypes correlate with severity of clinical features in patients121.
As with patients with RASopathy, adult zebrafish with Costello RAS alleles or mutations in NF1, the gene associated with NF1, are cancer prone124. Mutations in RABL3, which encodes a RAS-like gene, lead to an increased risk of pancreatic cancer, and RABL3 mutations are now considered to be another RASopathy123. In zebrafish, rabl3 mutants show features of RASopathies, including craniofacial and growth defects, and are cancer prone as adult animals. Activated MAPK pathway activity also leads to increased anxiety in adult zebrafish, a common mental health issue for individuals with RASopathies125.
These consistent zebrafish phenotypes in early embryonic stages, as well as adult phenotypes, have enabled preclinical testing of BRAF and MEK inhibitors in RASopathy models in two important and unexpected ways. First, MEK inhibitors restored normal development in the most common CFC syndrome zebrafish embryos using a continuous concentration of MEK inhibitors at a lower dose than is used for cancer treatment126. Similarly, low-dose semi-continual treatment reduced RASopathy phenotypes in rabl3 mutant zebrafish123, and alleviated anxiety in adult zebrafish with activated MAPK signalling123. These studies reflect the need to restore, rather than abolish, disease-associated activated MAPK signalling in patients with RASopathy. Second, unlike cancer treatments, which require high drug concentrations to inhibit the MAPK pathway throughout treatment, zebrafish studies identified developmental periods that were especially sensitive to MAPK pathway activation and defined treatment windows118. Within these windows, inhibiting the activity of the RASopathy mutation required mutation-specific drug doses, such that mutations with higher MAPK pathway signalling require higher doses of drug121. Thus, sophisticated modelling of a diverse spectrum of RASopathy alleles in zebrafish enables innovative preclinical drug testing and shows how to fine-tune therapy to match the disease mutation activity within individual patients.
MEK inhibitors for lymphatic anomaly.
Zebrafish preclinical models have recently been central to informing drug treatment. Whole-exome sequencing was used to identify somatic, gain-of-function mutations in ARAF kinase in two patients with central conducting lymphatic anomaly (CCLA), and zebrafish models were then generated to test a new treatment strategy14 (FIG. 4). One patient, an otherwise healthy and active child of 12 years of age, had been frequently hospitalized for 2 years owing to unexplained swellings in the groin and legs that were unresponsive to standard sirolimus treatment. A second, adult patient also presented with severely advanced lymphatic disease and died from complications of the illness. The mutation, at a conserved residue (S214), inhibited binding by the 14-3-3 proteins and thus increased the localization and activation by RAS at the membrane. In endothelial cells, expression of ARAF-S214P promoted vessel sprouting, and these vessels were sensitive to the MEK inhibitor trametinib. To develop a preclinical model, the researchers then expressed the ARAF mutation specifically in the developing lymphatic vessels of zebrafish and showed that the mutant ARAF induced dilation of the lymphatic vessels and thoracic duct, similar to clinical features seen in CCLA. Treatment of these zebrafish with another MEK inhibitor, cobimetinib, prevented the vessel phenotypes in zebrafish without causing additional toxicity. On the basis of the ability of MEK inhibitors to prevent the ARAF-induced vessel phenotypes in cells and in zebrafish, the team administered trametinib to the child, who had clinical improvements in lymphoedema by 3–6 months. By 12 months of treatment, there were dramatic improvements in pulmonary function with reduced pleural effusions around the lungs coupled with near complete regression of the massively dilated lymphatic ducts. Even more strikingly, lymph vessels underwent remodelling, and new and normal-appearing lymphatic networks developed. After a year of treatment, the child was able to resume schooling and physical activity. Thus, the rapid and physiological relevance of the zebrafish model can directly support clinical decisions to move forward with life-saving treatments.
Fig. 4 |. Compassionate drug treatment: from zebrafish to child.

Preclinical modelling of human genetic disease in zebrafish facilitates compassionate use of drugs in patients. A zebrafish model of the ARAFS214P mutation identified as a somatic heterozygous event in a child with lymphatic anomaly supported compassionate administration of MEK inhibitors. a | Images of a transgenic zebrafish embryo expressing ARAFS214P mutation (red) in the lymphatic endothelial cells (green), and lymphangiograms of the 12-year-old child with ARAFS214P before treatment. The white box in the top panel indicates the region shown at higher magnification in the images below. In zebrafish, lymphatic vessels become dilated with ARAFS214P expression and, in the child, there are inadequate ducts in the thighs and dilation of the lymphatic ducts, resulting in significant swelling in the legs. Treatment with MEK inhibitors rescues the zebrafish phenotype and resolves abnormal dilated ducts and normalizes lymphatic networks in the child. b | Standard growth chart, with measurements from the patient indicated as dots plotted on a graph that shows growth centiles by age. The improved lymphoedema is clearly seen in the patient’s legs (inset photos from the indicated times) and weight. Treatment was initiated just before age 13. PCV, posterior cardinal vein; TD, thoracic duct. Reprinted from REF.14, Springer Nature Limited. Figure design based on REF.17.
BRAF and arteriovenous malformation.
Arteriovenous malformation (AVM) is the abnormal growth and tangle of arteries and veins, leading to bleeding, pain, disfigurement and associated serious medical issues, including stroke. Treatment options have been limited and are especially problematic. Molecular understanding of the mechanism of disease indicates that somatic, activating mutations in the MAPK signalling pathway, including mutations in KRAS, BRAF or MEK, promote AVM127–129. Some of the AVM mutations overlap with cancer mutations, such as BRAFV600E, suggesting that they may be sensitive to BRAF inhibitors. To examine the activity of these mutations in an animal model, and to address the potential for therapies, BRAF and MEK mutations were expressed under a vessel-specific promoter such that their expression in zebrafish, as in patients, was mosaic129. Zebrafish expressing BRAF and MEK mutations developed vessel phenotypes that recapitulated the clinical features of the patients, including tangled and disordered vessel development, and severely impeded blood flow. Critically, for some AVM zebrafish, the BRAF inhibitor vemurafenib could resolve the vessel phenotypes once they were already established, and for almost all AVM zebrafish blood flow was improved upon treatment. BRAF inhibitors are now in compassionate use for BRAF-mutant AVM, and clinical trials with trametinib for MAPK pathway mutations in AVM are in preparation (NCT04258046).
Genetically engineered zebrafish
Although most screens have been performed in embryos, larval and adult zebrafish cancer models have also contributed to our understanding of how compounds kill cancer. Importantly, genetically engineered zebrafish models that incorporate tumour-suppressor inactivation and/or oncogene activation by transgenesis have, by and large, proved to be accurate models of human disease. These models often share common initiating cell types, underlying pathway activations that drive cancer and conserved vulnerabilities to chemotherapies and targeted pathway inhibitors. To date, a plethora of genetic zebrafish models of cancer have been generated, many of which have been used for drug discovery.
T cell acute lymphoblastic leukaemia.
A panel of 4,880 bioactive small molecules, including a wide array of FDA-approved compounds, was screened for the ability to kill fluorescently labelled T lymphocytes in larval transgenic zebrafish. The lead compound, phenothiazine (PPZ), also killed T cells in MYC-induced T cell acute lymphoblastic leukaemia (T-ALL) transgenic zebrafish models and human T-ALL cell lines. PPZ bound to and activated phosphatase PP2A, leading to the rapid dephosphorylation of its targets. Despite the activity of PPZ in killing T-ALL cells, this drug had severe in vivo toxicity due to inhibition of the dopamine receptor D2 (DRD2) within the basal ganglia, which led to extra pyramidal movement disorders130,131. Medicinal chemistry approaches were then used to develop a PP2A-activating drug that does not inhibit DRD2, obviating the effects associated with neurotoxicity19. This small molecule, improved heterocyclic activators of PP2A-1 (iHAP1), led to activation of a specific set of PP2A complexes and dephosphorylation of Ser241 of MYB-related protein B (MYBL2), a transcription factor, resulting in irreversible cell cycle arrest and cell death. iHAP1 killed T-ALL cells in both transgenic zebrafish transplant models and human T-ALL xenografts grown in NOD-scid IL2RGnull (NSG) mice, and the neurological effects of the parent compound, PPZ, were not detected. This example provides a roadmap of how findings made in zebrafish larval transgenic screens can identify new drug targets for cancer and then can aid in the development of novel small-molecule classes.
In a similar screen of 26,000 compounds, drugs that killed fluorescently labelled T cells in larval zebrafish were subsequently analysed in zebrafish transgenic models of T-ALL132. Lenaldekar was thus identified and elicited tumour cell killing of both zebrafish T-ALL and human T-ALL grown in mouse xenografts with overall negligible toxicity132. Lenaldekar also killed additional leukaemia subtypes, including therapy-refractory B-ALL and chronic myelogenous leukaemia, suggesting additional therapeutic opportunities. In another screen, inducible tg(hsp:AML–ETO) zebrafish were used to screen for chemical modifiers of this fusion oncogene, which causes acute myeloid leukaemia (AML). This screen identified the cyclooxygenase 2 (COX2) inhibitor, nimesulide, as a suppressor of AML1–ETO using early haematopoi etic differentiation as a larval readout133. Mechanistically, this work uncovered the COX2–β-catenin axis as a potent regulator of self-renewal, and showed that nimesulide treatment reduced xenograft tumour growth of human leukaemia cells grown in zebrafish134.
Hepatocellular carcinoma.
Using an AlphaScreen-based assay to identify inhibitors of the histone acetyl-transferases KAT6A and KAT6B, 243,000 diverse small-molecule compounds were screened135. Medicinal chemistry approaches were used to optimize the parent compound and produced WM-8014, which had an IC50 of 8 nM and induced a 60-fold increase in KAT6A inhibitory activity. Importantly, in a larval zebrafish transgenic model of KRAS-G12V-induced hepatocellular carcinoma, WM-8014 only elicited anti-proliferative effects on KRAS-G12V-expressing cells, not control cells, suggesting a large therapeutic window136. WM-8014 inhibited cancer growth in other models, including genetically engineered mouse lymphoma models, suggesting that this compound class could be broadly used in the treatment of cancer136. New medicinal chemistry derivations of WM-8014 have now been developed that have better oral bioavailability and are even more potent KAT6A inhibitors137, raising hopes that this novel drug class will soon progress to preclinical modelling and eventual clinical evaluation.
RAS pathway inhibitors for cancer.
Transgenic zebrafish cancer models can be used to identify drug pathways that lie downstream of specific oncogenic drivers. For example, the RAS pathway was first identified as a crucial driver of rhabdomyosarcoma (RMS) using transgenic zebrafish models and cross-species gene expression comparisons with human RMS138,139. Moreover, drug screening in embryos and then in transgenic models of this same disease found that combining mTOR/AKT inhibitors and MEK inhibitors killed RMS cells, a strikingly similar approach to what is now being evaluated clinically in this disease140. Using a transgenic NRAS model of zebrafish primitive neuroectodermal tumours of the CNS (CNS-PNETs), investigators showed that these tumours are arrested at an oligoneural precursor cell stage and have activated RAS–MAPK signalling141. An orthotopic zebrafish CNS-PNET transplant model was then used to show that MEK inhibitors selectively eliminate cancer cells in vivo without affecting the normal brain. This work provided crucial preclinical validation for the use of MEK inhibitors for the treatment of childhood oligoneural neuroblastoma with FOXR2 activation (NB-FOXR2) CNS-PNETs.
Ablain et al.142 recently pioneered approaches in melanoma modelling to uncover new pathways that drive tumour progression and can be targeted therapeutically. In this work, potential driver and tumour-suppressor mutations were identified by targeted sequencing of primary and metastatic human mucosal melanoma; these mutations were then expressed (for oncogenes) or the genes were inactivated (for tumour suppressors) simultaneously within developing melanomas using transgenics and CRISPR–Cas9 genome editing in F0 mosaic injected zebrafish. This platform confirmed a tumour-suppressor role for sprouty-related, EVH1 domain-containing protein (SPRED1), a negative regulator of the RAS–MAPK pathway, in early onset and aggressive melanomas, specifically in KIT-driven disease. Preclinical drug modelling using adult transgenic zebrafish uncovered that dasitinib, a multi-kinase inhibitor, potently killed KIT-driven melanomas, but not KIT-active melanomas that harboured SPRED inactivation. These results suggest that prioritizing treatment based on SPRED1 mutations will provide a much-needed therapeutic window for the treatment of KIT-active melanomas. Finally, this work suggests that KIT-active, SPRED1-deleted melanomas have elevated MAPK activity and that MEK inhibitors, such as trametinib, could be beneficial.
Transplant models in drug discovery
Allograft and xenograft transplant assays using larval and adult fish provide opportunities to assess drug effects in live animals and directly within human cancers (FIG. 5).
Fig. 5 |. Adult zebrafish xenograft transplantation approaches using FDA-approved drugs to clinical trials.

Zebrafish xenograft transplantation models enable preclinical testing of drug synergies in cancer. Drug synergy combinations are identified in cell models and then directly tested in adult xenograft cancer models. The drug effects are monitored by imaging fluorescent tumours in the fish, and through high-resolution imaging of single-cell end points, such as cell cycle or drug target pathways. Drug combinations can be tested further in other animal models and cell lines for toxicity and efficacy. The simultaneous monitoring for toxicity in the adult zebrafish enables the zebrafish results to be directly incorporated into the clinical trial design. *, statistically significant difference. GEMM, genetically engineered mouse model.
Allogeneic transplantation models.
Cell transplantation approaches have provided a novel means to expand the number of animals available for study, and thus have yielded new insights into pathways that drive cancer. For example, Chen et al.143 initially performed a drug screen of approximately 40,000 compounds in human RMS cell lines to identify drugs that cause cell death and/or differentiation using a high-content imaging screen. Top screen hits defined five distinct classes of inhibitors. Well-vetted experimental and FDA-approved drugs from these inhibitor classes were then re-tested in a hit-expansion screen to ensure reproducible and robust effects on the differentiation and viability of RMS cells. In total, 95 additional hit-expansion drugs were selected from the top classes of inhibitory drugs identified in the screen and then assessed for effects on KRAS-G12D-induced RMS cells grown in 6- to 8-week-old syngeneic zebrafish and human cell line models. This work uncovered an unexpected role for the WNT–β-catenin pathway in reducing self-renewal in embryonal RMS in vivo. Other drug classes, including RAS pathway inhibitors and HDAC inhibitors, also curbed growth and differentiation in zebrafish and human RMS models; both of these classes have progressed into clinical trials in RMS.
In addition to using allogeneic transplant models for drug screening, these same models can be used to assess new biology and uncover drug responses that often translate to the clinic. For example, Blackburn et al.144 also used the zebrafish T-ALL model to identify druggable pathways that regulate clonal evolution, elevate intratumoural heterogeneity and drive cancer progression. Using single-cell transplantation into syngeneic zebra fish, functional differences between single T-ALL clones could be assessed, including differences in latency, stem cell number and therapy responses. This work uncovered important roles for acquired AKT pathway activation during leukaemia evolution: AKT pathway activation increased the number of leukaemia-propagating cells (via mTORC1 activation), stabilized MYC to increase cell proliferation and rendered cells resistant to dexamethasone. This work also showed that evolution can create subclones that stochastically acquire mutations that render them insensitive to dexamethasone treatment, even in the absence of therapy. The work went on to show that combining dexamethasone and AKT pathway inhibition is a potent therapy in a subset of relapsed T-ALLs144. Other groups have used similar allogeneic transplantation assays to assess the preclinical responses of T-ALLs and B-ALLs to dexamethasone and cytotoxic therapies145,146, melanomas to BRAF inhibitors, and CNS-PNETs to MEK inhibitors141.
Xenograft transplantation into larvae.
The first cancer xenograft studies were performed in 2005 using larval zebrafish and involved the adoptive transfer of human melanoma cells into developing embryos147. Importantly, human melanoma cells engrafted into the zebrafish embryo survive, migrate and divide. Melanoma cells also unexpectedly secrete the embryonic morphogen Nodal and consequently cause ectopic embryonic axis formation148. Nodal signalling was found to regulate melanoma cell plasticity and tumorigenicity. Independent of the important biological insights gleaned from this work, this was the first description of human cancers grown in larval zebrafish and was performed at a developmental stage earlier than the establishment of the acquired immune system; thus, tumour cells were not rejected by engrafted animals. These studies led to a wide array of engraftment studies that included many types of human cancer, HSCs149 and induced pluripotent stem cell-derived neural precursors150.
Subsequent refined xenograft models capitalized on real-time imaging of cancer at single-cell resolution, facilitated by labelling cells with fluorescent dyes and transgenic reporters. For example, in xenografted transgenic VEGFR2:G-RCFP zebrafish, prototypic anti-angiogenic inhibitors potently suppressed tumour-induced angiogenesis, as observed in live animals151. Further work discovered why VEGF signalling suppresses local tumour growth but accelerates micrometastatic disease: VEGF signalling and myeloid cells cooperate in vivo to promote metastasis152. Individual human prostate cancer cells can be visualized in vivo and monitored for migration and metastasis in these xenograft models153. Using this model, investigators have found that inhibiting the nuclear factor-κB (NF-κB)–activin A pathway decreases the pool of prostate cancer stem cells and decreases invasion, metastatic growth and bone lesion formation using zebrafish and mice xenografts154. Xenograft studies of brain cancers have uncovered the therapeutic efficacy of combining the HSP90 inhibitor onalespib and the chemotherapeutic agent temozolomide in malignant gliomas155, and identified protein arginine N-methyltransferase 5 (PRMT5) as a drug target in glioblastoma156, again suggesting the broad utility of larval xenograft models for assessing preclinical therapies. When coupled with automated xenograft injection and quantitative bio-imaging platforms, xenotransplantation assays can now be further stream-lined to efficiently deliver cancer cells into zebrafish embryos, distribute engrafted animals to 96-well plates, deliver drugs and then automatically image animals after treatment. This automated pipeline has the potential to greatly accelerate target discovery and demonstrate the efficacy of new drugs in preclinical xenograft models157.
Larval xenograft models are also now being used to assess therapy responses of patients, providing new insights into how tumours become refractory to therapy and predicting which patients will respond to therapy. For example, Bentley et al.158 used zebrafish xenografts to match targeted compounds with molecular aberrations in human T-ALL. These investigators could identify xenografted tumours that harboured activating mutations in NOTCH1 by their sensitivity to γ-secretase inhibitors. Patient-derived larval xenografts of colorectal cancer have also been assessed for differential sensitivities to two standard therapies using a wide array of tumour cell behaviours that included proliferation, metastasis and angiogenesis159. Relative sensitivities identified in the zebrafish xenograft model were predictive of responses seen in patients and were similar to those observed in tumours grown in immune-deficient mice. Building on these observations, larval xenograft assays were developed to predict individual responses to bevacizumab, an anti-VEGF therapy, in triple-negative breast cancer and colorectal cancer160; and sensitivity to the poly(ADP-ribose) polymerase (PARP) inhibitor olaparib in triple-negative breast cancer, independent of BRCA mutational status161.
Xenografts in immune-deficient fish.
Recent advances in genome engineering have provided opportunities to create immune-deficient adult zebrafish that lack T, B and natural killer (NK) cells. Advantages of these models include being able to raise fish at 37 °C, dose drugs in a clinically achievable manner using oral gavage instead of submersion therapy, and implanting enough cells that they form tumours with histology and long-term growth kinetics that are similar to those found in patients162,163. Building on previous prkdc−/− and il2rga−/− fish models164,165, Yan et al.163 created compound mutant animals that lack T, B and NK cells but still survive to adulthood. These animals stably engraft a wide array of human cancers, including melanoma, triple-negative breast cancer and RMS. Oral gavage of zebrafish engrafted with RMS cells identified that the PARP inhibitor olaparib, in combination with the DNA-damaging agent temozolomide, could be a new therapy for RMS163. These findings were extended to a zebrafish xenograft model that allowed for real-time imaging of therapeutic effects on the cell cycle at single-cell resolution and identified that the combination therapy induced a G2 cell cycle arrest followed by apoptosis in human RMS cells. Importantly, this work used oral gavage and dosing schedule that mimicked the frequency and blood levels of drugs that could be achieved clinically. Based on these findings, and their extension in PDX mouse models, this drug combination has now progressed to clinical evaluation in RMS (NCT01858168).
Although this is the first description of using xenografts grown in adult immune-deficient fish for drug discovery, these models are expected to complement the high-throughput assays available in larval zebrafish xenografts and also provide advantages similar to those of the NSG mouse models for studying drug responses of human xenografted cancers. Future studies will likely develop refined models that express human cytokines to sustain xenograft growth, akin to those pioneered for larval xenografts166.
Routes to the clinic
In the past 15 years, zebrafish chemical biology has moved forward rapidly. Many of the initial practical complexities of drug screens, including methodology and access to libraries, have been addressed through community efforts such as the Zebrafish Disease Models Society (see Related links). These efforts continue to provide connections, know-how and the advisory networks needed for discovery scientists to interact with clinical scientists, medicinal chemists and the pharmaceutical industry to move compounds from the lab to the clinic. We identify three routes to progress a small-molecule phenotype in zebrafish to the clinical setting. Drugs that have emerged via these three routes are highlighted in TABLE 1 and the phenotypic screening approaches are discussed in BOX 2.
Box 2 |. Zebrafish phenotypic profiling in drug development.
Phenotypic profiling of the activity of a chemical compound, including discovery-based screens and target-based drug testing on preclinical models, is at the heart of zebrafish drug development and discovery. Here, we discuss some of the distinct chemical–genetic approaches that were used for the drugs in TABLE 1.
Matching a chemical phenotype with a genetic phenotype
Screening for compounds that induce a phenotype similar to a known genetically caused phenotype can identify highly selective compounds that target a specific pathway. To discover and generate specific AlK2 inhibitors, a phenotypic screen of 7,500 small molecules for compounds that caused dorsalization of zebrafish embryos identified a lead compound, dorsomorphin, which is a bone morphogenic protein (BMP) inhibitor43. Based on the phenotypes caused by genetic mutations in alk8 and a dominant mutation in BMP type I receptor, these investigators knew that loss of BMP signalling caused dorsalization phenotypes in zebrafish embryos and incorporated this knowledge into screening for small molecules.
Unbiased chemical screening for specific phenotypes
Screening for chemical compounds that give rise to a specific phenotype without a priori knowledge of the molecular targets can point to new biology and targetable pathways. In an unbiased screening approach, prostaglandin E2 (PGE2) was found to increase haematopoietic stem cell number in zebrafish embryos94, and all-trans retinoic acid (ATRA) was found to reduce MYB gene expression in zebrafish blastomere cultures113. Similarly, the 5-nitrofurans were identified in an unbiased small-molecule screen for compounds that killed melanocytes, leading to the identification of aldehyde dehydrogenase as a new target class for nifuroxazide pro-drug antibiotics in melanoma76,111. In each of these examples, the screening assay readout directly reflected the biology that the investigators wanted to chemically control, and the molecular targets and mechanism were identified subsequently.
Rescuing a disease model, or pseudonormalization, can be incorporated into unbiased screens to identify drug leads. Unbiased screens that rescue disease models identified that DB-041 prevented hair cell toxicity in a model for hearing loss following antibiotics47,93, that trifluoperazine rescued anaemia in a rsp19 zebrafish mutant model of Diamond Blackfan anaemia171–173 and that clemizole (EPX-100) rescued spontaneous seizures in a scn1lab mutant model of Dravet syndrome101. In these screens, there was no a priori knowledge about how to reverse a disease-related phenotype, and the chemical leads identified new information about the mechanism of the disease and therapy.
Target-based suppressor of disease phenotypes
Examining the capacity of compounds to rescue a disease phenotype can test drugs in a new disease context and provide the preclinical studies to move the drug towards clinical use. MEK inhibitors are now in compassionate use in lymphatic anomalies based on cell culture and zebrafish models14, and targeted MAPK pathway inhibitors were shown to restore blood flow and pseudonormalize the vasculature in a zebrafish model of BRAF-mutant arteriovenous malformation129. Preclinical drug trials in adult zebrafish xenografts identified a new drug combination — olaparib plus temozolomide — for the treatment of rhabdomyosarcoma; this combination had similar pharmacokinetic properties in zebrafish, mice and humans163,174. In each of these target-based modelling approaches, the molecular pathways that underlie the disease phenotypes were known and the small molecules restored normal function or reduced disease burden.
New applications for known targets.
Developing new applications for therapies with known targets is arguably the most direct route to the clinic. In this instance, there is molecular evidence that the target in the disease is the known target of the drug. Examples of this route include using MAPK pathway inhibitors for patients with mosaic MEK mutations in lymphatic anomaly14 and mosaic BRAF or MEK mutations in AVM129, and the combination of olaparib and temozolomide for RMS163. In these cases, the drugs were already in clinical use and the targets were well described; the zebrafish model generated the functional link between the molecular and clinical features in the model, and demonstrated the efficacy of drugs on the disease outcome. In combination with cell-based assays, these models can provide the preclinical animal model evidence required for compassionate use or for early-stage clinical trials. Furthermore, they provide a foundation for evaluating the in vivo biology of disease resolution and predicting drug resistance.
New target, new application.
Drug repurposing can identify a new molecular target or disease for a known drug. In this situation, the pharmacological and ADME-tox properties of the drug may be known, but new information about the dose and metabolites may be needed to use the drug in a new context or for a new purpose. Biomarkers will need to be developed to show on-target efficacy of the drug for the new target. Most likely, these studies will require further validation in another animal model, such as mice or primates96, although some have moved directly from zebrafish to patients16. This route led to the development of PGE2 to stimulate stem cell production and engraftment in cord blood96,100.
New chemical entity.
New drugs have been developed from novel composition of matter or new chemical entities (NCEs)19,93,167. This is the most challenging of the three screening routes to progress to the clinical setting. Zebrafish assays can support compound optimization through structure–activity relationship profiling, selecting against compounds that cause toxicity, and demonstrating multispecies efficacy, which improves confidence in the drug lead. This route is costly, as it requires medicinal chemistry to optimize the compounds, the generation of biomarkers and assays to assess drug efficacy, studies on pharmacokinetics and pharmacodynamics, and metabolite and toxicity studies in multiple species. One advantage to NCEs is that composition of matter patents, instead of re-use patents, can be granted to provide ownership.
Future directions and conclusions
Drug discovery remains an expensive and laborious process, and large scientific gaps between the laboratory and clinical settings hinder drug development. Given the shared drug responses between zebrafish and humans, zebrafish may be as powerful as a predictive preclinical model as mammalian models such as the mouse. The shared pharmacology between zebrafish and humans makes zebrafish an important partner in drug repurposing as well as a tool for the discovery of NCEs that opens the door to new drug classes. Gene editing and transgenic technologies are already being used to engineer humanized zebrafish with specific genes, which paves the way for engineering humanized zebrafish to investigate drug metabolism and in vivo pharmacokinetics. Zebrafish are emerging as avatars for human cancers, and future studies may further involve cross-species xenotransplantation between other tissues and organs for drug studies. Most drugs are absorbed by zebrafish through the water or by oral gavage; however, zebrafish are also a model on which to innovate novel drug administration routes, such as timed and tissue-specific drug release from beads or nanoparticles168, and to visualize drug–target interactions in vivo.
To better translate new drug uses from zebrafish to humans, further efforts are needed to understand pharmacokinetics in zebrafish, including the internal drug and metabolite concentrations. To this end, pharmacokinetic models have been developed in zebrafish larvae35,169. Zebrafish metabolic enzymes are conserved and, using micro-blood sampling and metabolic profiling in zebrafish, these models have been used to analyse the pharmacokinetics of paracetamol, which correlates well with the pharmacokinetics in humans. In adult xenografts, olaparib and temozolomide, administered by oral gavage, had similar pharmacokinetic properties in adult zebrafish to those in both mice and humans, suggesting that such approaches will be useful for future studies163. Using zebrafish to explore drug exposure–response relationships for both on-target and adverse effects (for example, hepatotoxicity), as well as internal exposure (ADME-tox studies), suggests that drug metabolism in zebrafish may be scalable to other vertebrates, including humans. Efforts to develop zebrafish pharmacology will support zebrafish within the pharmaceutical industry by ultimately improving drug screening protocols, providing mechanistic insight into drug metabolism and identifying adverse events.
Looking towards future zebrafish drug screens, incorporating multiple, simultaneous screening parameters into whole-animal screens will help us understand chemical targets and responses to drug treatments, including on-target and off-target drug effects at distal sites. These can be further enriched with high-content and high-speed live imaging of fluorescent reporters to capture large datasets of cell responses over timescales of days. Single-cell omic technologies, such as single-cell RNA sequencing and whole-organism lineage tracing, will enable the impact of drugs to be assessed across large cell populations, providing unprecedented detail of the cellular manifestation of disease and how treatments resolve disease and restore health170. Incorporating large-scale genome-wide editing with chemical screens will assist in target identification and will be an important step towards personalized gene–drug interactions, including gene–drug contraindicated interactions, synthetic lethality drug mechanisms for cancer treatments and drug efficacy within specific genetic populations.
The uses of the zebrafish system span the length of the drug discovery pipeline from discovery-stage high-content, high-throughput screening through to preclinical models for therapy175. We predict that as more zebrafish models are evaluated in the context of therapy, zebrafish will emerge as a preclinical platform for drug discovery and drug applications that lead directly to the clinic. Indeed, compelling human genetic data coupled with zebrafish models have already supported the direct use of MAPK pathway inhibitors in patients with lymphatic anomaly14 and arterial venous malformations129 on compassionate grounds, and clemizole and lorcaserin for patients with Dravet syndrome13,16. Xenografts in zebrafish have directly informed the clinical trial design for RMS163 without additional animal model prerequisites. The exciting and diverse range of zebrafish small-molecule screens and disease models has already firmly established zebrafish in the drug discovery and development pipeline and positions zebrafish to generate even more significant contributions to future therapeutics.
Acknowledgements
The authors thank R. Peterson, D. Raible, S. Baraban, C. MacRae and G. Hanna for helpful discussions, and their funders for support for this work. E.E.P. is funded by a MRC HGU Programme (MC_UU_00007/9), the European Research Council (ZF-MEL-CHEMBIO-648489) and Melanoma Research Alliance (687306). L.I.Z. is funded by Cancer Biology R01 CA103846, NIH Melanoma PPG, P01CA63222, Melanoma Research Alliance, Starr Cancer Consortium grant. D.M.L. is funded by NIH grants R01CA211734, R01CA154923, R01CA215118, RO1CA226926, R24OD016761 and the MGH Research Scholar Award.
Footnotes
Competing interests
L.I.Z. is a founder and stockholder of Fate Therapeutics, CAMP4 Therapeutics, Amagma Therapeutics and Scholar Rock. He is a consultant for Celularity and Cellarity. D.M.L. has a sponsored research agreement with NextCure and patents issued/pending on immune-deficient zebrafish. E.E.P. declares no competing interests.
References
- 1.Howe K et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 496, 498–503 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phillips JB & Westerfield M Zebrafish models in translational research: tipping the scales toward advancements in human health. Dis. Model. Mech 7, 739–743 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford YM et al. Zebrafish models of human disease: gaining insight into human disease at ZFIN. ILAR J. 58, 4–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prykhozhij SV & Berman JN Zebrafish knock-ins swim into the mainstream. Dis. Model. Mech 11, dmm037515 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu K, Petree C, Requena T, Varshney P & Varshney GK Expanding the CRISPR toolbox in zebrafish for studying development and disease. Front. Cell Dev. Biol 7, 13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baron M et al. The stress-like cancer cell state is a consistent component of tumorigenesis. Cell Syst. 11, 536–546 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Travnickova J et al. Zebrafish MITF-low melanoma subtype models reveal transcriptional subclusters and MITF-independent residual disease. Cancer Res. 79, 5769–5784 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cagan RL, Zon LI & White RM Modeling cancer with flies and fish. Dev. Cell 49, 317–324 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goessling W & Sadler KC Zebrafish: an important tool for liver disease research. Gastroenterology 149, 1361–1377 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rissone A & Burgess SM Rare genetic blood disease modeling in zebrafish. Front. Genet 9, 348 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Y, Zhang K, Sips P & MacRae CA Screening drugs for myocardial disease in vivo with zebrafish: an expert update. Expert Opin. Drug Discov 14, 343–353 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Kalueff AV et al. Towards a comprehensive catalog of zebrafish behavior 1.0 and beyond. Zebrafish 10, 70–86 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffin A et al. Preclinical animal models for dravet syndrome: seizure phenotypes, comorbidities and drug screening. Front. Pharmacol 9, 573 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li D et al. ARAF recurrent mutation causes central conducting lymphatic anomaly treatable with a MEK inhibitor. Nat. Med 25, 1116–1122 (2019). [DOI] [PubMed] [Google Scholar]
- 15.North TE et al. PGE2-regulated wnt signaling and N-acetylcysteine are synergistically hepatoprotective in zebrafish acetaminophen injury. Proc. Natl Acad. Sci. USA 107, 17315–17320 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffin A et al. Clemizole and modulators of serotonin signalling suppress seizures in Dravet syndrome. Brain 140, 669–683 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacRae CA & Peterson RT Zebrafish as tools for drug discovery. Nat. Rev. Drug Discov 14, 721–731 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Cully M Zebrafish earn their drug discovery stripes. Nat. Rev. Drug Discov 18, 811–813 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Morita K et al. Allosteric activators of protein phosphatase 2A display broad antitumor activity mediated by dephosphorylation of MYBL2. Cell 181, 702–715.e20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Peterson RT, Link BA, Dowling JE & Schreiber SL Small molecule developmental screens reveal the logic and timing of vertebrate development. Proc. Natl Acad. Sci. USA 97, 12965–12969 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peterson RT et al. Chemical suppression of a genetic mutation in a zebrafish model of aortic coarctation. Nat. Biotechnol 22, 595–599 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Swinney DC & Anthony J How were new medicines discovered? Nat. Rev. Drug Discov 10, 507–519 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Sonoshita M et al. A whole-animal platform to advance a clinical kinase inhibitor into new disease space. Nat. Chem. Biol 14, 291–298 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dang M, Henderson RE, Garraway LA & Zon LI Long-term drug administration in the adult zebrafish using oral gavage for cancer preclinical studies. Dis. Model. Mech 9, 811–820 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moya-Garcia A et al. Structural and functional view of polypharmacology. Sci. Rep 7, 10102 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Antolin AA, Workman P, Mestres J & Al-Lazikani B Polypharmacology in precision oncology: current applications and future prospects. Curr. Pharm. Des 22, 6935–6945 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dar AC, Das TK, Shokat KM & Cagan RL Chemical genetic discovery of targets and anti-targets for cancer polypharmacology. Nature 486, 80–84 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishizaki H et al. Combined zebrafish-yeast chemical-genetic screens reveal gene-copper-nutrition interactions that modulate melanocyte pigmentation. Dis. Model. Mech 3, 639–651 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCarroll MN, Gendelev L, Keiser MJ & Kokel D Leveraging large-scale behavioral profiling in zebrafish to explore neuroactive polypharmacology. ACS Chem. Biol 11, 842–849 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bruni G et al. Zebrafish behavioral profiling identifies multitarget antipsychotic-like compounds. Nat. Chem. Biol 12, 559–566 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito T et al. Identification of a primary target of thalidomide teratogenicity. Science 327, 1345–1350 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Lu G et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science 343, 305–309 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kronke J et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 343, 301–305 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sperling AS et al. Patterns of substrate affinity, competition, and degradation kinetics underlie biological activity of thalidomide analogs. Blood 134, 160–170 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Wijk RC et al. Impact of post-hatching maturation on the pharmacokinetics of paracetamol in zebrafish larvae. Sci. Rep 9, 2149 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fraser C et al. Rapid discovery and structure-activity relationships of pyrazolopyrimidines that potently suppress breast cancer cell growth via SRC kinase inhibition with exceptional selectivity over ABL kinase. J. Med. Chem 59, 4697–4710 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y et al. Visnagin protects against doxorubicin-induced cardiomyopathy through modulation of mitochondrial malate dehydrogenase. Sci. Transl Med 6, 266ra170 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wertman JN et al. The identification of dual protective agents against cisplatin-induced oto- and nephrotoxicity using the zebrafish model. eLife 9, e56235 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cassar S et al. Use of zebrafish in drug discovery toxicology. Chem. Res. Toxicol 33, 95–118 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gorelick DA, Pinto CL, Hao R & Bondesson M Use of reporter genes to analyze estrogen response: the transgenic zebrafish model. Methods Mol. Biol 1366, 315–325 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Pinto C et al. Selectivity of natural, synthetic and environmental estrogens for zebrafish estrogen receptors. Toxicol. Appl. Pharmacol 280, 60–69 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gorelick DA, Iwanowicz LR, Hung AL, Blazer VS & Halpern ME Transgenic zebrafish reveal tissue-specific differences in estrogen signaling in response to environmental water samples. Env. Health Perspect 122, 356–362 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu PB et al. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat. Chem. Biol 4, 33–41 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richter S, Schulze U, Tomancak P & Oates AC Small molecule screen in embryonic zebrafish using modular variations to target segmentation. Nat. Commun 8, 1901 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stern HM et al. Small molecules that delay S phase suppress a zebrafish bmyb mutant. Nat. Chem. Biol 1, 366–370 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Santoriello C et al. RNA helicase DDX21 mediates nucleotide stress responses in neural crest and melanoma cells. Nat. Cell Biol 22, 372–379 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Namdaran P, Reinhart KE, Owens KN, Raible DW & Rubel EW Identification of modulators of hair cell regeneration in the zebrafish lateral line. J. Neurosci 32, 3516–3528 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Allen JR, Skeath JB & Johnson SL Maintenance of melanocyte stem cell quiescence by gaba-a signaling in larval zebrafish. Genetics 213, 555–566 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johansson JA et al. PRL3-DDX21 transcriptional control of endolysosomal genes restricts melanocyte stem cell differentiation. Dev. Cell 54, 317–332.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bergen DJM, Kague E & Hammond CL Zebrafish as an emerging model for osteoporosis: a primary testing platform for screening new osteo-active compounds. Front. Endocrinol 10, 6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reimer MM et al. Dopamine from the brain promotes spinal motor neuron generation during development and adult regeneration. Dev. Cell 25, 478–491 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Helker CSM et al. A whole organism small molecule screen identifies novel regulators of pancreatic endocrine development. Development 146, dev172569 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Early JJ et al. An automated high-resolution in vivo screen in zebrafish to identify chemical regulators of myelination. eLife 7, e35136 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burns CG et al. High-throughput assay for small molecules that modulate zebrafish embryonic heart rate. Nat. Chem. Biol 1, 263–264 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Martin WK et al. High-throughput video processing of heart rate responses in multiple wild-type embryonic zebrafish per imaging field. Sci. Rep 9, 145 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Akerberg AA, Burns CE, Burns CG & Nguyen C Deep learning enables automated volumetric assessments of cardiac function in zebrafish. Dis. Model. Mech 12, dmm040188 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miyares RL, de Rezende VB & Farber SA Zebrafish yolk lipid processing: a tractable tool for the study of vertebrate lipid transport and metabolism. Dis. Model. Mech 7, 915–927 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salmi TM, Tan VWT & Cox AG Dissecting metabolism using zebrafish models of disease. Biochem. Soc. Trans 47, 305–315 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Wrighton PJ, Oderberg IM & Goessling W There is something fishy about liver cancer: zebrafish models of hepatocellular carcinoma. Cell Mol. Gastroenterol. Hepatol 8, 347–363 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gut P et al. Whole-organism screening for gluconeogenesis identifies activators of fasting metabolism. Nat. Chem. Biol 9, 97–104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gut P & Stainier DY Whole-organism screening for modulators of fasting metabolism using transgenic zebrafish. Methods Mol. Biol 1263, 157–165 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Garnaas MK et al. Rargb regulates organ laterality in a zebrafish model of right atrial isomerism. Dev. Biol 372, 178–189 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cox AG et al. S-nitrosothiol signaling regulates liver development and improves outcome following toxic liver injury. Cell Rep. 6, 56–69 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bambino K et al. Inorganic arsenic causes fatty liver and interacts with ethanol to cause alcoholic liver disease in zebrafish. Dis. Model. Mech 11, dmm031575 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marques JC, Li M, Schaak D, Robson DN & Li JM Internal state dynamics shape brainwide activity and foraging behaviour. Nature 577, 239–243 (2020). [DOI] [PubMed] [Google Scholar]
- 66.Khan KM et al. Zebrafish models in neuropsychopharmacology and CNS drug discovery. Br. J. Pharmacol 174, 1925–1944 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McCammon JM & Sive H Challenges in understanding psychiatric disorders and developing therapeutics: a role for zebrafish. Dis. Model. Mech 8, 647–656 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McCarroll MN et al. Zebrafish behavioural profiling identifies GABA and serotonin receptor ligands related to sedation and paradoxical excitation. Nat. Commun 10, 4078 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mosser EA et al. Identification of pathways that regulate circadian rhythms using a larval zebrafish small molecule screen. Sci. Rep 9, 12405 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moffat JG, Vincent F, Lee JA, Eder J & Prunotto M Opportunities and challenges in phenotypic drug discovery: an industry perspective. Nat. Rev. Drug Discov 16, 531–543 (2017). [DOI] [PubMed] [Google Scholar]
- 71.Rihel J et al. Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science 327, 348–351 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Burczyk MS et al. Muscarinic receptors promote pacemaker fate at the expense of secondary conduction system tissue in zebrafish. JCI Insight 4, e121971 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Laggner C et al. Chemical informatics and target identification in a zebrafish phenotypic screen. Nat. Chem. Biol 8, 144–146 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.White RM et al. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature 471, 518–522 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wagner BK & Schreiber SL The power of sophisticated phenotypic screening and modern mechanism-of-action methods. Cell Chem. Biol 23, 3–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou L et al. ALDH2 mediates 5-nitrofuran activity in multiple species. Chem. Biol 19, 883–892 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fellmann C, Gowen BG, Lin PC, Doudna JA & Corn JE Cornerstones of CRISPR–Cas in drug discovery and therapy. Nat. Rev. Drug Discov 16, 89–100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Minikel EV et al. Evaluating drug targets through human loss-of-function genetic variation. Nature 581, 459–464 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Neggers JE et al. Target identification of small molecules using large-scale CRISPR-Cas mutagenesis scanning of essential genes. Nat. Commun 9, 502 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Matty MA et al. Potentiation of P2RX7 as a host-directed strategy for control of mycobacterial infection. eLife 8, e39123 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Muller S et al. Donated chemical probes for open science. eLife 7, e34311 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gerry CJ & Schreiber SL Chemical probes and drug leads from advances in synthetic planning and methodology. Nat. Rev. Drug Discov 17, 333–352 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Canham SM et al. Systematic chemogenetic library assembly. Cell Chem. Biol 27, 1124–1129 (2020). [DOI] [PubMed] [Google Scholar]
- 84.Newman DJ & Cragg GM Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod 79, 629–661 (2016). [DOI] [PubMed] [Google Scholar]
- 85.Harvey AL, Edrada-Ebel R & Quinn RJ The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov 14, 111–129 (2015). [DOI] [PubMed] [Google Scholar]
- 86.Dancik V, Seiler KP, Young DW, Schreiber SL & Clemons PA Distinct biological network properties between the targets of natural products and disease genes. J. Am. Chem. Soc 132, 9259–9261 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hoeksma J et al. A new perspective on fungal metabolites: identification of bioactive compounds from fungi using zebrafish embryogenesis as read-out. Sci. Rep 9, 17546 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hoeksma J, van der Zon GCM, Ten Dijke P & den Hertog J Cercosporamide inhibits bone morphogenetic protein receptor type I kinase activity in zebrafish. Dis. Model Mech 13, dmm045971 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pickett SB & Raible DW Water waves to sound waves: using zebrafish to explore hair cell biology. J. Assoc. Res. Otolaryngol 20, 1–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Blanco-Sanchez B et al. Grxcr1 promotes hair bundle development by destabilizing the physical interaction between harmonin and sans Usher syndrome proteins. Cell Rep. 25, 1281–1291.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Owens KN et al. Identification of genetic and chemical modulators of zebrafish mechanosensory hair cell death. PLoS Genet. 4, e1000020 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chowdhury S et al. Phenotypic optimization of urea-thiophene carboxamides to yield potent, well tolerated, and orally active protective agents against aminoglycoside-induced hearing loss. J. Med. Chem 61, 84–97 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kitcher SR et al. ORC-13661 protects sensory hair cells from aminoglycoside and cisplatin ototoxicity. JCI Insight 4, e126764 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.North TE et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature 447, 1007–1011 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Goessling W et al. Genetic interaction of PGE2 and Wnt signaling regulates developmental specification of stem cells and regeneration. Cell 136, 1136–1147 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Goessling W et al. Prostaglandin E2 enhances human cord blood stem cell xenotransplants and shows long-term safety in preclinical nonhuman primate transplant models. Cell Stem Cell 8, 445–458 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hoggatt J, Mohammad KS, Singh P & Pelus LM Prostaglandin E2 enhances long-term repopulation but does not permanently alter inherent stem cell competitiveness. Blood 122, 2997–3000 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hoggatt J, Singh P, Sampath J & Pelus LM Prostaglandin E2 enhances hematopoietic stem cell homing, survival, and proliferation. Blood 113, 5444–5455 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang Y et al. Inhibition of the prostaglandin-degrading enzyme 15-PGDH potentiates tissue regeneration. Science 348, aaa2340 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cutler C et al. Prostaglandin-modulated umbilical cord blood hematopoietic stem cell transplantation. Blood 122, 3074–3081 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Baraban SC, Dinday MT & Hortopan GA Drug screening in Scn1a zebrafish mutant identifies clemizole as a potential Dravet syndrome treatment. Nat. Commun 4, 2410 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Griffin AL et al. Zebrafish studies identify serotonin receptors mediating antiepileptic activity in Dravet syndrome. Brain Commun. 1, fcz008 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dinday MT & Baraban SC Large-scale phenotype-based antiepileptic drug screening in a zebrafish model of dravet syndrome. eNeuro 2, ENEURO.0068–15.2015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Frantz WT & Ceol CJ From tank to treatment: modeling melanoma in zebrafish. Cells 9, 1289 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mort RL, Jackson IJ & Patton EE The melanocyte lineage in development and disease. Development 142, 620–632 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kaufman CK et al. A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation. Science 351, aad2197 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tan JL et al. Stress from nucleotide depletion activates the transcriptional regulator HEXIM1 to suppress melanoma. Mol. Cell 62, 34–46 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liang K et al. Targeting processive transcription elongation via SEC disruption for MYC-induced cancer therapy. Cell 175, 766–779.e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ciarlo C et al. A chemical screen in zebrafish embryonic cells establishes that Akt activation is required for neural crest development. eLife 6, e29145 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Precazzini F et al. Automated in vivo screen in zebrafish identifies clotrimazole as targeting a metabolic vulnerability in a melanoma model. Dev. Biol 457, 215–225 (2020). [DOI] [PubMed] [Google Scholar]
- 111.Sarvi S et al. ALDH1 bio-activates nifuroxazide to eradicate ALDHHigh melanoma-initiating cells. Cell Chem. Biol 25, 1456–1469.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Persson M et al. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc. Natl Acad. Sci. USA 106, 18740–18744 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mandelbaum J et al. Zebrafish blastomere screen identifies retinoic acid suppression of MYB in adenoid cystic carcinoma. J. Exp. Med 215, 2673–2685 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jindal GA, Goyal Y, Burdine RD, Rauen KA & Shvartsman SY RASopathies: unraveling mechanisms with animal models. Dis. Model. Mech 8, 769–782 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gripp KW et al. The sixth international RASopathies symposium: precision medicine — from promise to practice. Am. J. Med. Genet. A 182, 597–606 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gross AM et al. Advancing RAS/RASopathy therapies: an NCI-sponsored intramural and extramural collaboration for the study of RASopathies. Am. J. Med. Genet. A 182, 866–876 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.MacRae CA et al. A clinical trial of MEK inhibition in Noonan syndrome with hypertrophic cardiomyopathy. RASopathies Network. https://rasopathiesnet.org/clinical-trial-mek-inhibition-noonan-syndrome-hypertrophic-cardiomyopathy/ [Google Scholar]
- 118.Anastasaki C, Estep AL, Marais R, Rauen KA & Patton EE Kinase-activating and kinase-impaired cardio-facio-cutaneous syndrome alleles have activity during zebrafish development and are sensitive to small molecule inhibitors. Hum. Mol. Genet 18, 2543–2554 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Goyal Y et al. Divergent effects of intrinsically active MEK variants on developmental Ras signaling. Nat. Genet 49, 465–469 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Goyal Y et al. Parallel imaging of Drosophila embryos for quantitative analysis of genetic perturbations of the Ras pathway. Dis. Model. Mech 10, 923–929 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jindal GA et al. In vivo severity ranking of Ras pathway mutations associated with developmental disorders. Proc. Natl Acad. Sci. USA 114, 510–515 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Niihori T et al. Germline-activating RRAS2 mutations cause Noonan syndrome. Am. J. Hum. Genet 104, 1233–1240 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nissim S et al. Mutations in RABL3 alter KRAS prenylation and are associated with hereditary pancreatic cancer. Nat. Genet 51, 1308–1314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Castel P, Rauen KA & McCormick F The duality of human oncoproteins: drivers of cancer and congenital disorders. Nat. Rev. Cancer 20, 383–397 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lundegaard PR et al. MEK inhibitors reverse cAMP-mediated anxiety in zebrafish. Chem. Biol 22, 1335–1346 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Anastasaki C, Rauen KA & Patton EE Continual low-level MEK inhibition ameliorates cardio-facio-cutaneous phenotypes in zebrafish. Dis. Model. Mech 5, 546–552 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nikolaev SI et al. Somatic activating KRAS mutations in arteriovenous malformations of the brain. N. Engl. J. Med 378, 250–261 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Couto JA et al. Somatic MAP2K1 mutations are associated with extracranial arteriovenous malformation. Am. J. Hum. Genet 100, 546–554 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Al-Olabi L et al. Mosaic RAS/MAPK variants cause sporadic vascular malformations which respond to targeted therapy. J. Clin. Invest 128, 1496–1508 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Miller R Mechanisms of action of antipsychotic drugs of different classes, refractoriness to therapeutic effects of classical neuroleptics, and individual variation in sensitivity to their actions: part II. Curr. Neuropharmacol 7, 315–330 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Miller R Mechanisms of action of antipsychotic drugs of different classes, refractoriness to therapeutic effects of classical neuroleptics, and individual variation in sensitivity to their actions: part I. Curr. Neuropharmacol 7, 302–314 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ridges S et al. Zebrafish screen identifies novel compound with selective toxicity against leukemia. Blood 119, 5621–5631 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yeh JR et al. Discovering chemical modifiers of oncogene-regulated hematopoietic differentiation. Nat. Chem. Biol 5, 236–243 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang Y et al. AML1-ETO mediates hematopoietic self-renewal and leukemogenesis through a COX/beta-catenin signaling pathway. Blood 121, 4906–4916 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Falk H et al. An efficient high-throughput screening method for MYST family acetyltransferases, a new class of epigenetic drug targets. J. Biomol. Screen 16, 1196–1205 (2011). [DOI] [PubMed] [Google Scholar]
- 136.Baell JB et al. Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth. Nature 560, 253–257 (2018). [DOI] [PubMed] [Google Scholar]
- 137.Priebbenow DL et al. Discovery of acylsulfonohydrazide-derived inhibitors of the lysine acetyltransferase, KAT6A, as potent senescence-inducing anti-cancer agents. J. Med. Chem 63, 4655–4684 (2020). [DOI] [PubMed] [Google Scholar]
- 138.Langenau DM et al. Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev. 21, 1382–1395 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yohe ME et al. Insights into pediatric rhabdomyosarcoma research: Challenges and goals. Pediatr. Blood Cancer 66, e27869 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Le X et al. A novel chemical screening strategy in zebrafish identifies common pathways in embryogenesis and rhabdomyosarcoma development. Development 140, 2354–2364 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Modzelewska K et al. MEK inhibitors reverse growth of embryonal brain tumors derived from oligoneural precursor cells. Cell Rep. 17, 1255–1264 (2016). [DOI] [PubMed] [Google Scholar]
- 142.Ablain J et al. Human tumor genomics and zebrafish modeling identify SPRED1 loss as a driver of mucosal melanoma. Science 362, 1055–1060 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chen EY et al. Glycogen synthase kinase 3 inhibitors induce the canonical WNT/beta-catenin pathway to suppress growth and self-renewal in embryonal rhabdomyosarcoma. Proc. Natl Acad. Sci. USA 111, 5349–5354 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Blackburn JS et al. Clonal evolution enhances leukemia-propagating cell frequency in T cell acute lymphoblastic leukemia through Akt/mTORC1 pathway activation. Cancer Cell 25, 366–378 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Park G et al. Zebrafish B cell acute lymphoblastic leukemia: new findings in an old model. Oncotarget 11, 1292–1305 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mizgirev IV & Revskoy S A new zebrafish model for experimental leukemia therapy. Cancer Biol. Ther 9, 895–902 (2010). [DOI] [PubMed] [Google Scholar]
- 147.Lee LM, Seftor EA, Bonde G, Cornell RA & Hendrix MJ The fate of human malignant melanoma cells transplanted into zebrafish embryos: assessment of migration and cell division in the absence of tumor formation. Dev. Dyn 233, 1560–1570, (2005). [DOI] [PubMed] [Google Scholar]
- 148.Topczewska JM et al. Embryonic and tumorigenic pathways converge via Nodal signaling: role in melanoma aggressiveness. Nat. Med 12, 925–932 (2006). [DOI] [PubMed] [Google Scholar]
- 149.Staal FJ, Spaink HP & Fibbe WE Visualizing human hematopoietic stem cell trafficking in vivo using a zebrafish xenograft model. Stem Cell Dev. 25, 360–365 (2016). [DOI] [PubMed] [Google Scholar]
- 150.Strnadel J et al. Transplantation of human-induced pluripotent stem cell-derived neural precursors into early-stage zebrafish embryos. J. Mol. Neurosci 65, 351–358 (2018). [DOI] [PubMed] [Google Scholar]
- 151.Nicoli S, Ribatti D, Cotelli F & Presta M Mammalian tumor xenografts induce neovascularization in zebrafish embryos. Cancer Res. 67, 2927–2931 (2007). [DOI] [PubMed] [Google Scholar]
- 152.He S et al. Neutrophil-mediated experimental metastasis is enhanced by VEGFR inhibition in a zebrafish xenograft model. J. Pathol 227, 431–445 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chen L, Boleslaw Olszewski M, Kruithof-de Julio M & Snaar-Jagalska BE Zebrafish microenvironment elevates EMT and CSC-like phenotype of engrafted prostate cancer cells. Cells 9, 797 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chen L et al. A NF-kB-activin A signaling axis enhances prostate cancer metastasis. Oncogene 39, 1634–1651 (2020). [DOI] [PubMed] [Google Scholar]
- 155.Canella A et al. Efficacy of onalespib, a long-acting second-generation HSP90 inhibitor, as a single agent and in combination with temozolomide against malignant gliomas. Clin. Cancer Res 23, 6215–6226 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Banasavadi-Siddegowda YK et al. PRMT5 as a druggable target for glioblastoma therapy. Neuro Oncol. 20, 753–763 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ghotra VP et al. Automated whole animal bio-imaging assay for human cancer dissemination. PLoS ONE 7, e31281 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bentley VL et al. Focused chemical genomics using zebrafish xenotransplantation as a pre-clinical therapeutic platform for T-cell acute lymphoblastic leukemia. Haematologica 100, 70–76 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Fior R et al. Single-cell functional and chemosensitive profiling of combinatorial colorectal therapy in zebrafish xenografts. Proc. Natl Acad. Sci. USA 114, E8234–E8243 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Rebelo de Almeida C et al. Zebrafish xenografts as a fast screening platform for bevacizumab cancer therapy. Commun. Biol 3, 299 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Varanda AB, Martins-Logrado A, Ferreira MG & Fior R Zebrafish xenografts unveil sensitivity to olaparib beyond BRCA status. Cancers 12, 1769 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Fazio M, Ablain J, Chuan Y, Langenau DM & Zon LI Zebrafish patient avatars in cancer biology and precision cancer therapy. Nat. Rev. Cancer 20, 263–273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yan C et al. Visualizing engrafted human cancer and therapy responses in immunodeficient zebrafish. Cell 177, 1903–1914.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tang Q et al. Dissecting hematopoietic and renal cell heterogeneity in adult zebrafish at single-cell resolution using RNA sequencing. J. Exp. Med 214, 2875–2887 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Moore JC et al. Single-cell imaging of normal and malignant cell engraftment into optically clear prkdc-null SCID zebrafish. J. Exp. Med 213, 2575–2589 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Rajan V et al. Humanized zebrafish enhance human hematopoietic stem cell survival and promote acute myeloid leukemia clonal diversity. Haematologica 105, 2391–2399 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yu PB et al. BMP type I receptor inhibition reduces heterotopic ossification. Nat. Med 14, 1363–1369 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Weiss JT et al. Extracellular palladium-catalysed dealkylation of 5-fluoro-1-propargyl-uracil as a bioorthogonally activated prodrug approach. Nat. Commun 5, 3277 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Van Wijk RC et al. Mechanistic and quantitative understanding of pharmacokinetics in zebrafish larvae through nanoscale blood sampling and metabolite modeling of paracetamol. J. Pharmacol. Exp. Ther 371, 15–24 (2019). [DOI] [PubMed] [Google Scholar]
- 170.Travnickova J & Patton EE Deciphering melanoma cell states and plasticity with zebrafish models. J. Invest. Dermatol 141, 1389–1394 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Taylor AM, Raiser DM, Humphries JM, Ebert JL & Zon LI Calmodulin inhibition rescues the effects of ribosomal protein deficiency by modulating p53 activity in models of Diamond Blackfan anemia [abstract]. Blood 120, 512 (2012). [Google Scholar]
- 172.Macari ER et al. Calmodulin inhibition rescues DBA models with ribosomal protein deficiency through reduction of RSK signaling. Blood 128, 332 (2016). [Google Scholar]
- 173.Uechi T & Kenmochi N Zebrafish models of Diamond-Blackfan anemia: a tool for understanding the disease pathogenesis and drug discovery. Pharmaceuticals 12, 151 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Yan C, Yang Q, Do D, Brunson DC & Langenau DM Adult immune compromised zebrafish for xenograft cell transplantation studies. EBioMedicine 47, 24–26 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Patton E et al. Melanoma models for the next generation of therapies. Cancer Cell 10.1016/j.ccell.2021.01.011 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]


