Abstract
The phylogenetic relationships of bacterial symbionts from three gall-bearing species in the marine red algal genus Prionitis (Rhodophyta) were inferred from 16S rDNA sequence analysis and compared to host phylogeny also inferred from sequence comparisons (nuclear ribosomal internal-transcribed-spacer region). Gall formation has been described previously on two species of Prionitis, P. lanceolata (from central California) and P. decipiens (from Peru). This investigation reports gall formation on a third related host, Prionitis filiformis. Phylogenetic analyses based on sequence comparisons place the bacteria as a single lineage within the Roseobacter grouping of the α subclass of the division Proteobacteria (99.4 to 98.25% sequence identity among phylotypes). Comparison of symbiont and host molecular phylogenies confirms the presence of three gall-bearing algal lineages and is consistent with the hypothesis that these red seaweeds and their bacterial symbionts are coevolving. The species specificity of these associations was investigated in nature by whole-cell hybridization of gall bacteria and in the laboratory by using cross-inoculation trials. Whole-cell in situ hybridization confirmed that a single bacterial symbiont phylotype is present in galls on each host. In laboratory trials, bacterial symbionts were incapable of inducing galls on alternate hosts (including two non-gall-bearing species). Symbiont-host specificity in Prionitis gall formation indicates an effective ecological separation between these closely related symbiont phylotypes and provides an example of a biological context in which to consider the organismic significance of 16S rDNA sequence variation.
Marine bacteria are associated with gall formation (tumorigenesis) on a number of species of red algae, although only two reports detail the specific causation of an algal gall by an identified bacterium (5, 10). This probably reflects the general situation encountered when attempting to cultivate, or isolate in pure culture, symbiotic microbes (10, 33, 44). In the red algal genus Prionitis (Rhodophyta, Halymeniaceae, Gigartinales) gall formation is known from at least four species world wide and is, in the case of Prionitis lanceolata (from central California), associated with the presence of a specific microorganism (5). Despite attempts by several authors, this bacterium has yet to be cultivated or isolated in pure culture (3–5). No physiological function of these bacterially induced Prionitis galls has been determined, nor is it apparent what, if any, benefit is derived by algal host or bacterial invader. This organismic relationship is termed a symbiosis sensu DeBary as used by Smith, meaning simply the living together of differently named organisms (43). The 16S rDNA phylotype of this eubacterium has been determined from its complete small-subunit ribosomal DNA sequence and whole-cell hybridization used to confirm the inductive role of this symbiont in gall formation (5).
The purpose of this investigation was to determine if bacterial gall formation on different members of the genus Prionitis is species specific and to investigate the possibility of coevolution occurring between these bacterial symbionts and their red algal hosts. To address this, the molecular phylogenetic relationships of the eubacterial symbionts from P. lanceolata, Prionitis filiformis, and Prionitis decipiens galls were compared to that of their algal hosts and bacterial phylotype-specificity was investigated both in nature and in the laboratory. Bacterial phylogeny was inferred from 16S sequence comparisons, and host phylogeny was inferred from comparisons of the algal nuclear internal-transcribed-spacer regions (ITS) of P. lanceolata, P. filiformis, P. decipiens, Prionitis lyalli, and Prionitis sp. (from Mission Bay). The relative branching orders of both host and symbiont lineages were compared.
Recent work in molecular microbial ecology has focused on the delineation of molecular phylotypes based on differences in the small subunit of rRNA (SSU) sequence variation (9, 14, 25–27, 30, 37, 44). This has raised the following question in a number of different ecological contexts: to what extent does SSU rDNA sequence variation (especially among closely related organisms) reflect underlying biological differences (natural history, physiology, ecology, etc.). A recent investigation of marine photosynthetic bacterioplankton has revealed ecotypic variation not apparent by direct comparison of rDNA sequences and illustrates how minor variations in the sequences of these genes can mask wholesale physiological differences (35). Our investigation of host-symbiont specificity in gall-bearing species of Prionitis was carried out using whole-cell hybridization of rRNA in bacteria from galls in nature and by laboratory cross-inoculation trials. The intercellular symbionts from P. lanceolata, P. decipiens, and P. filiformis galls were assayed from field-collected material using the species-specific oligodeoxynucleotide probes, S-S-P.l.sym-0949-a-A-25 and S-S-P.f/d.sym-0949-a-A-25 (7). Laboratory cross-inoculation trials using bacteria from galls on P. lanceolata and P. filiformis were then carried out as an assay for the potential of symbionts from a given host species to induce new gall formation on alternate gall- and non-gall-bearing host species.
The evidence presented here from molecular phylogeny, laboratory cross-inoculation trials, and whole-cell hybridization experiments with gall bacteria from nature shows that bacterial gall formation in the genus Prionitis is species specific. In particular, the results of molecular phylogenetic analyses are confirmed by laboratory cross-inoculation trials that show an effective biological separation of symbiont molecular phylotypes. The basis of this separation, including the mechanisms of symbiont recognition that must operate in these species-specific associations, remains unknown (43). The effective biological separation of symbiont phylotypes suggests that these lineages, at least in terms of their symbiotic ecology, may be considered different species. The combination of molecular phylogenetic analyses and host-symbiont specificity studies serves to illustrate that, in this system, extremely low levels of 16S rDNA sequence variation (ca. 1%) can reflect significant biological differences between organisms.
MATERIALS AND METHODS
Field collection of Prionitis sp.
Gall-bearing thalli of P. lanceolata, P. filiformis, and P. decipiens were collected from 11 intertidal and 2 subtidal locations along the Pacific coasts of North and South America (Table 1). Non-gall-bearing thalli of Prionitis lyalli and P. filiformis were collected from the Pacific Coast of central California (Table 1). Algae were transported in damp newspaper on ice and processed within several hours of collection. P. lanceolata and P. filiformis were maintained in robust condition for up to 5 months in high flow indoor tanks with ca. 1% ambient light levels and vigorous aeration. Living P. decipiens (from Peru) was obtained directly from the type locality via 2-day air express (Table 1) and was processed immediately upon arrival.
TABLE 1.
The distribution of Prionitis sp. utilized in this investigationa
| Prionitis sp. | Location of collection | Gall formation |
|---|---|---|
| P. lanceolata | Tomales Point, Marin Co., Calif. | + |
| P. lanceolata | Pigeon Point, San Mateo Co., Calif. | + |
| P. lanceolata | Santa Cruz Point, Santa Cruz Co., Calif. | + |
| P. lanceolata | Hopkins Marine Life Refuge, Stanford University, Pacific Grove, Calif.a | + |
| P. lanceolata | Carmel State Beach, Carmel Co., Calif. | + |
| P. lanceolata | Piedras Blancas, San Luis Obispo Co., Calif. | + |
| P. lanceolata | San Nicholas Island, Channel Islands, Calif. | + |
| P. lanceolata | Anacapa Island, Channel Islands, Calif. | + |
| P. lanceolata | Sycamore State Beach, Malibu Co., Calif. | + |
| P. lanceolata | Royal Palms Beach, Los Angeles Co., Calif. | + |
| P. filiformis | Cape Perpetua, Oreg. | + |
| P. filiformis | Lufenholtz Beach, Humbolt Co., Calif. | + |
| P. decipiens | Port Paita, Peru (type locality) | + |
| P. filiformis | Carmel State Beach, Carmel Co., Calif.b | − |
| Prionitis sp. | Mission Bay, San Diego Co., Calif.b | − |
| P. lyalli | Pigeon Point, San Mateo Co., Calif. | − |
The distribution and occurrence of gall-bearing Prionitis collected in nature.
Subtidal collections.
Phylogenetic analysis of Prionitis gall symbionts. DNA extraction-amplification-cloning and sequencing.
Total DNA was isolated from galls on P. filiformis (Lufenholtz population) and P. decipiens and from the thalli of P. filiformis (Lufenholtz population), P. filiformis (Carmel population), P. decipiens, P. lyalli, and Prionitis sp. (Mission Bay) by using Chelex-100 extraction (Bio-Rad, Richmond, Calif.) as described previously (6, 24). Amplification of 16S rDNA gene sequences and algal ITS sequences by PCR, cloning, and initial screening of amplification products was also carried out as described previously (6, 24). Plasmid DNA was prepared for sequencing by Wizard miniprep following the manufacturer's recommendations (Promega, Madison, Wis.). Cloned 16S rDNA genes (one amplicon from each of three PCRs) were sequenced entirely along both strands by using an ABI 373 automated sequencer with dye termination chemistry (ABI; Perkin-Elmer).
Phylogenetic analyses were performed as follows. Selected full-length 16S rDNA sequences from the Proteobacteria were obtained as a subalignment from the Ribosomal Database Project by using the SUBALIGN command and a user-defined ID LIST (36). Prionitis symbiont sequences and additional sequences of environmental clones (obtained as nucleotide sequences from GenBank by using the Entrez browser) related to the α subclass of the division Proteobacteria were manually aligned to this subalignment in the DNA sequence editor GDE 2.2 (provided by Steve Smith). Cloned symbiont amplicons from P. filiformis and P. decipiens were shown to be nonchimeric by comparison to the previously determined P. lanceolata symbiont sequence (7). A sequence mask was applied in McClade 3.0 that excluded gaps and all positions of uncertain alignment with final analyses performed on a 16S data set consisting of 20 full-length 16S sequences (1,038 positions included, 677 positions excluded).
Tree topologies were evaluated and branch lengths were calculated by using PAUP version 4.0.0d64 with maximum parsimony and distance or maximum likelihood optimality criteria (David Swofford, personal communication). Evolutionary distances were calculated by using either the Kimura two-parameter or the Hasegawa-Kishino-Yano (1985) (HKY85) (with rate heterogeneity) model of nucleotide change. Transition-transversion ratio, proportion of invariable positions, and gamma shape parameter were estimated from the data set by maximum likelihood. Phylogenetic tree topologies were reconstructed in PAUP by heuristic search using the branch-swapping algorithm, tree-bisection-reconnection, and starting trees generated by replicate stepwise random addition. Decay indices were calculated, and the significance of alternative evolutionary hypotheses was examined by statistical permutation test for mono- and nonmonophyly (T-PTP test) (17, 46). Host ITS sequences were edited, aligned, and analyzed as described above by using GDE 2.2 and PAUP. The relative consistencies of all resulting trees were estimated by bootstrap analyses consisting of 1,000 replicates each of both the 16S and ITS data sets, with the following exception. The bootstrap analysis of the 16S data set using maximum likelihood was performed on 100 replicates (19).
Accession numbers for the P. filiformis and P. decipiens symbiont 16S sequences are AF107209 and AF107210, respectively. Accession numbers for P. lanceolata, P. filiformis (Lufenholtz population), P. filiformis (Carmel population), P. lyalli, P. decipiens, and Prionitis sp. (Mission Bay) ITS sequences are AF274237, AF274234, AF274233, AF274235, AF274236, and AF274238, respectively.
Laboratory cross-inoculation trials of symbiont-host specificity.
Host-specificity in the Prionitis symbiosis was investigated by cross-inoculation trials performed in the laboratory using gall-bearing P. lanceolata and P. filiformis (Lufenholtz population) and non-gall-bearing P. filiformis (Carmel population) and P. lyalli. Individual thalli of P. decipiens (Peru) were held for 7 days but could not be maintained under laboratory conditions for sufficient time to carry out cross-inoculation experiments. Cross-inoculations were carried out as described previously in induction studies of the P. lanceolata symbiosis (5). Galls collected in nature were washed, homogenized in sterile seawater (passed through a 0.22-μm-pore-size filter) and used to inoculate, by wounding, the uninfected thalli of P. lanceolata, P. filiformis (Lufenholtz and Carmel populations) and the related P. lyalli (4, 6). Inoculation trials matched gall homogenates from P. lanceolata and P. filiformis (Lufenholtz population) with the alternate gall-bearing host and two non-gall-bearing hosts (P. filiformis from the Carmel population and P. lyalli). Thalli used in these experiments had been maintained in the laboratory before inoculation for 2 to 3 months and showed no evidence of gall initiation or formation.
Whole-cell hybridization of P. lanceolata gall bacterial rRNA.
Identification of the 16S rDNA eubacterial phylotype associated with P. lanceolata and P. filiformis galls was carried out by dual whole-cell hybridization by using the following monofluorescently labeled rRNA-targeted oligodeoxynucleotide probes: S-D-Bact-0338-a-A-18 (universal eubacterial probe), S-S-P.f/d.sym-0949-a-A-25 (P. filiformis and P. decipiens specific probe), and S-S-P.l.sym-0949-a-A-25 (P. lanceolata symbiont-specific probe) (6, 15). Probes were 5′ end labeled with either fluorescein isothiocyanate (green fluorescence) or TAMRA (red fluorescence) (see above for specific labeling). Symbionts from galls on P. lanceolata, P. filiformis, and P. decipiens were prepared and hybridized as described previously (6). Hybridization was carried out at 45°C followed by three high-stringency washes (0.2 × SET) of the same temperature. Slides were mounted in 3:1 Citifluor-DAPI (0.5 μg · ml−1 4′,6′-diamidino-2-phenylindole [DAPI]), sealed with nail polish and observed on an Olympus IMT2 inverted photoscope by using a Bio-Rad 600 laser confocal imaging system and an Olympus S-Plan-apo 60× oil objective. Fluorescence images generated by 560 and 510 nm excitation (corresponding to TAMRA and fluorescein, respectively) were captured separately, saved as PICT files, merged in Adobe Photoshop 4.0, and colored to match the original signal. No other image alterations were made.
RESULTS
Phylogenetic analysis.
The complete 16S rDNA sequences of three Prionitis eubacterial symbionts and the complete sequences of the ITS regions from six Prionitis sp. algal hosts were determined. Bacterial symbiont sequences ranged in length from 1,411 to 1,416 bp (excluding the universal eubacterial primer sequences used for amplification). No microheterogeneity was detected between three completely sequenced amplicons from each symbiont. Algal ITS sequences ranged in length from to 773 to 797 bp, also excluding primer sequences. A low level of microheterogeneity was detected between clones from different PCR amplifications: P. lanceolata and Prionitis sp. (Mission Bay) varied at three positions, P. decipiens varied at two positions, and P. lyalli varied at a single position. No microheterogeneity was detected in the ITS sequences of either P. filiformis (Lufenholtz population) or P. filiformis (Carmel population). Sequence alignments were unambiguous along the entire lengths of both the bacterial and the algal data sets.
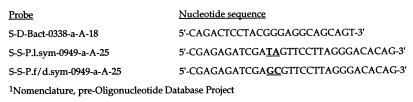
Phylogenetic trees inferred from sequence comparisons of both the 16S and ITS data sets are generally consistent under maximum parsimony, distance, and maximum likelihood optimality criteria (Fig. 1 and 2). Sequence comparisons of the 16S data set show a 99.4 to 98.25% unweighted nucleotide similarity between the bacterial symbionts of P. lanceolata, P. filiformis, and P. decipiens for the following pairwise comparisons. The symbionts of P. lanceolata and P. filiformis differ by 24 nucleotides, P. lanceolata and P. decipiens differ by 18 nucleotides, and P. filiformis and P. decipiens differ by eight nucleotides. The symbionts of P. decipiens (Peru) and P. filiformis (Lufenholtz population) also share an identical two-nucleotide substitution within the 24-bp species-specific rDNA probe target region (S-S-P.l.sym-0949-a-A-25) (Fig. 3).
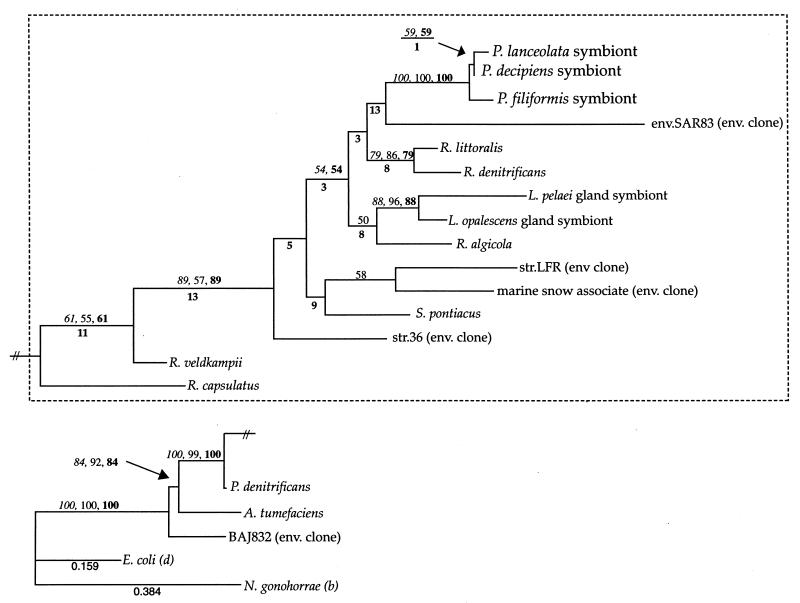
FIG. 1.
Phylogram (maximum likelihood/−ln likelihood value = 5563.81538) of the Prionitis gall symbionts and selected members of the Proteobacteria inferred from 16S rDNA sequence comparisons. Branch lengths were calculated by using maximum likelihood analysis with the following parameters: HYK85 model with an estimated transition/transversion ratio of 1.35048, an estimated proportion of invariable sites of 0.37302, and an estimated gamma shape parameter of 0.53896. Bootstrap values (above branches at relevant nodes) are reported when greater than 50% and represent results of parsimony (italic type), distance (regular type), and maximum likelihood (boldface type) analyses. Decay indices are given below relevant branch nodes. The likelihoods of several branches are included for scale. The branch lengths in the region of the tree set off by the marquee are increased 600% relative to the scale. In addition to the Prionitis bacterial symbionts, the following taxa are included (Ribosomal Database Project short identification): env.SAR83 (env SAR83), Roseobacter littoralis (Ros.litora); R. denitrificans (Ros.denitr); Roseobacter algicola (Ros.algico); str.36 (str.36); str.LFR (str.LFR); Paracoccus denitrificans (Par.denitr); Rhodobacter capsulatus (Rb.capsula); Rhodobacter veldkampii (Rb.veldkam); A. tumefaciens (Ag.tumefac); Escherichia coli (E.colirrnH); and Nisseria gonohorrae (Nis.gonorr). Additional taxa identified from GenBank accession numbers are the Loligo opalescens (AFO26462) and Loligo pealei (AF022392) endosymbionts, an environmental clone of a marine-snow associated bacterium (AF030780), Sulfitobacter pontiacus (SPY13155), and an environmental clone from Lake Baikal (AJ222832).
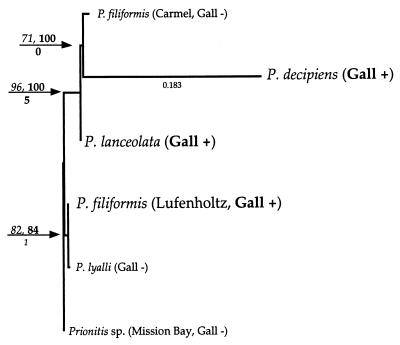
FIG. 2.
Phylogram of the Prionitis sp. hosts inferred from ITS sequence comparisons. Branch lengths were calculated by using maximum likelihood with the following parameters: HYK85 model with an estimated t/v ratio of 0.785 and an estimated proportion of invariable sites of 0.725. The gamma shape parameter of this data set was estimated at infinity. The most likely tree was constructed by random stepwise addition and was evaluated by exhaustive search using 10 replicates (random). Results of bootstrap analysis using maximum likelihood (1,000 replicates, full heruistic) are given at relevant branch nodes; decay indices are indicated below branches. Bootstrap values in boldface type indicate analysis after removal of the most divergent sequence (longest branch, P. decipiens). The likelihood of a single branch is given for scale. Based solely on morphological considerations, Prionitis sp. (Mission Bay) can be considered Prionitis angusta.
FIG. 3.
Oligonucleotide probes utilized in this investigation. Universal eubacterial probe, S-D-Bact-0338-a-A-18; P. lanceolata symbiont specific probe, S-S-P.l.sym-0949-a-A-25; P. filiformis-P. decipiens symbiont, S-S-P.f/d.sym-0949-a-A-25.
Phylogenetic analysis groups all three Prionitis gall bacterial symbionts in a monophyletic clade within the Roseobacter radiation (Rhodobacter grouping) of the α subclass of the division Proteobacteria (α-Proteobacteria) (Fig. 1). This placement of the gall symbionts is recovered by all phylogenetic analysis methods and is supported in 100% of bootstrap analyses (Fig. 1). The relative branching orders among bacterial gall symbionts were also resolved consistently by parsimony, distance, and maximum likelihood. These results indicate that the bacterial symbionts of P. lanceolata and P. decipiens galls are closest relatives to the exclusion of the P. filiformis microbe (Fig. 1). Bootstrap support for this topology, however, is not strong (Fig. 1). Maximum likelihood, distance, and parsimony tree scores were also calculated for constraint trees where alternative relative branching orders of the Prionitis sp. bacterial symbionts were forced; i.e., the grouping of the symbionts from P. lanceolata and P. filiformis to the exclusion of P. decipiens. In all cases, the most likely tree topology maintained the grouping of P. lanceolata with P. decipiens (−ln likelihood value less than 5,564 versus greater than 5,589).
Phylogenetic analyses based on sequence comparisons of host ITS regions are also consistent under the conditions of parsimony or either distance or maximum likelihood optimality criteria (Fig. 2). A single inferred tree topology is well supported by bootstrap resampling (Fig. 2). This reconstruction is also consistent with the evolutionary relationships inferred from the 16S data set in the placement of P. lanceolata with P. decipiens relative to the gall-bearing P. filiformis (Lufenholtz population). An important finding of our analyses is that gall-bearing thalli of P. filiformis (Lufenholtz population) are not the closest molecular relatives of morphologically assigned, non-gall-bearing con-specific P. filiformis (Carmel population) (Fig. 2). Instead, the closest molecular relative of the gall-bearing P. filiformis is an alga of extremely dissimilar morphology, P. lyalli (Fig. 2). This probably reflects the difficulties that may be encountered when using sequence data to reexamine the phylogenetic relationships underlying problematic, morphologically defined, marine algal taxa (see Discussion). Irrespective of this, the proposed molecular phylogenetic relationships among the three gall-bearing Prionitis hosts are consistent with the inferred phylogeny of their gall-inducing bacterial symbionts.
Laboratory cross-inoculation trials.
Laboratory cross-inoculation trials showed that the symbionts from P. lanceolata were able to induce gall formation on P. lanceolata but never on P. filiformis (Lufenholtz or Carmel populations) or P. lyalli (Table 2). Similarly, homogenates of P. filiformis (Lufenholtz population) galls were able to induce gall formation on P. filiformis (Lufenholtz population) but not on P. filiformis (Carmel population), P. lanceolata, or P. lyalli (Table 2). The timing of gall maturation differed between P. lanceolata (which required ca. 12 weeks for eruption of the gall from the algal thallus) and P. filiformis (Lufenholtz population) (which required 3 to 4 months to reach the erumpent gall stage).
TABLE 2.
Laboratory cross-inoculation trials showing the percentage of galls successfully induced by symbiont self and cross-inoculations
| Algal thalli used | % Gall formationb
|
|
|---|---|---|
| P. lanceolata | P. filiformis (Lufenholtz population) | |
| P. lanceolataa | 89 (48) | (45) |
| 89 (49) | (49) | |
| P. filiformis (Lufenholtz population) | 0 (43) | 36 (42) |
| 0 (46) | 40 (46) | |
| P. filiformis (Carmel population) | 0 (47) | 0 (46) |
| 0 (47) | 0 (47) | |
| P. lyalli | 0 (52) | 0 (48) |
| 0 (59) | 0 (44) | |
These results are consistent with a previous investigation of gall induction on P. lanceolata (5).
Number of gall inoculations.
Laboratory gall induction of P. lanceolata was successful ca. 90% of the time (Table 2). This was consistent with the results of an earlier study of gall causation on P. lanceolata (5). In the case of P. filiformis, the percentage of successful gall formation was lower, with inoculation sites forming tumors ca. 40% of the time (Table 2).
Hybridization of whole-cell rRNA in nature.
Whole-cell hybridization of the rRNA of bacteria from P. lanceolata and P. filiformis galls in nature confirmed the pattern of symbiont-host specificity observed in phylogenetic analyses and laboratory cross-inoculations. Both S-S-P.l.sym-0949-a-A-25 and S-D-Bact-0338-a-A-18 but never S-S-P.f.sym-0949-a-A-25 hybridized with rRNA in eubacteria recovered from galls on 10 geographically separated isolates of P. lanceolata. Similarly, both S-S-P.f.sym-0949-a-A-25 and S-D-Bact-0338-a-A-18 but never S-S-P.l.sym-0949-a-A-25 hybridized with the rRNA in eubacteria assayed from three geographically separated isolates of P. filiformis. The rRNA in bacteria in P. decipiens galls was not hybridized by either of the oligodeoxynucleotide probes utilized in this investigation.
Dual whole-cell hybridization studies of the P. lanceolata and the P. filiformis (Lufenholtz population) symbionts from the same preparation confirmed the specificity of the oligonucleotides used (Fig. 4). The rRNA in Agrobacterium tumefaciens and Roseobacter denitrificans cells used as controls for oligodeoxynucleotide specificity were hybridized by the universal eubacterial probe S-D-Bact-0338-a-A-18 but neither of the symbiont specific probes S-S-P.l.sym-0949-a-A-25 or S-S-P.f/d.sym-0949-a-A-25.
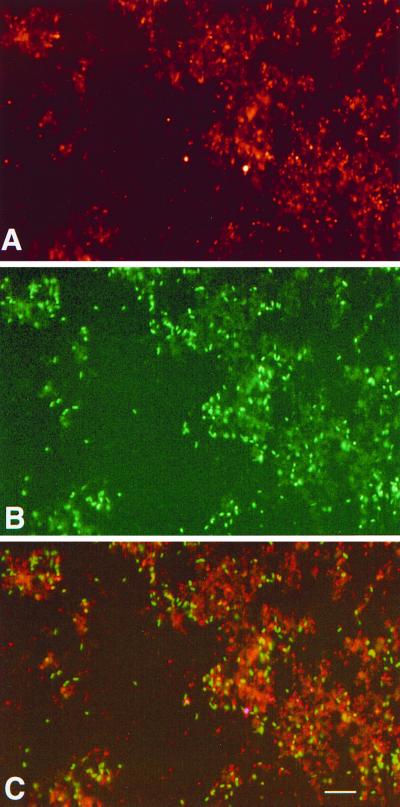
FIG. 4.
Whole-cell hybridization of the gall symbionts from P. lanceolata and P. filiformis galls. (A) P. lanceolata symbionts hybridized by S-S-P.l.sym-0949-a-A-25 (fluorescein under 510 nm excitation). (B) P. filiformis (Lufenholtz) symbionts hybridized by S-S-P.f/d.sym-0949-a-A-25 (TAMRA under 560 nm excitation). (C) Merged images of dual whole-cell hybridization, P. lanceolata and P. filiformis symbionts (scale = 15 μm).
DISCUSSION
The closest relatives of the Prionitis gall symbionts revealed by phylogenetic analysis are all members of a group of marine α-Proteobacteria that can be termed the Roseobacter group. Molecular investigation of marine microbial ecology has revealed a large group of coastal marine bacteria related to the Rhodobacter grouping of the α-Proteobacteria. This group includes, in addition to numerous environmental clones of uncertain biological significance, the associates, epiphyhtes, and symbionts of marine animals (8, 11, 15, 20, 25, 44). Examples of algal-associated members of this group that have been cultured include R. denitrificans, isolated from the surface of the marine alga Ulva (Chlorophyta), and R. algicola, isolated from the phycosphere of the toxic dinoflagellate Prorocentrum lima (32, 38). Close relatives of the Prionitis gall symbionts include the cultured members of the genus Roseobacter, several marine environmental clones, and a clade of squid endosymbionts (Fig. 1) (8, 28, 32, 42). The closest relative, however, of the Prionitis bacterial symbiont clade is not consistently resolved in our analyses (Fig. 1).
The overall level of variation in ITS sequences among Prionitis sp. was 4 to 7%, a level of variation similar to those observed among other algal ITS sequences used successfully for phylogenetic analysis (13, 24, 29). A limitation of the analysis of ITS sequence variation is the difficulty in defining suitable outgroups (relatives close enough that confident alignment is possible) for phylogenetic analyses. ITS sequences for closely related taxa are frequently not available (may not have been sequenced or identified) for inclusion in the analysis. The ability to unambiguously align the six algal host ITS sequences used in this study implies, based on the low level of sequence divergence among taxa, that a relatively recent radiation of these algal taxa has occurred. The ITS region is transcribed but then spliced out of mature rRNA during processing, leading to higher rates of evolution in this region relative to the rDNA (7, 24). This increase in sequence variability limits phylogenetically informative comparisons of the ITS to closely related taxa (intergeneric comparisons or very closely related genera) (7, 24).
The phylogenetic relationships inferred from molecular analyses of both the bacterial SSU and algal ITS data sets are consistent with laboratory and field evidence for species specificity. Additionally, the data are consistent with the interpretation that members of the red algal genus Prionitis and their symbiotic bacteria are coevolving. Bacteria from host species in central California and Peru (P. lanceolata and P. decipiens) appear more closely related to each other than either is to the gall bacteria from the northern California and Oregon populations of P. filiformis (Fig. 1). These relationships are weakly supported by bootstrap analyses and may reflect the radiation of these bacteria with their algal hosts. An alternative evolutionary hypothesis which groups the symbionts from P. decipiens and P. filiformis to the exclusion of the P. lanceolata symbiont is rejected by T-PTP test for nonmonophyly (P = 0.87). Although the evidence is not conclusive, coevolution in the Prionitis gall-forming symbioses is a consistent and parsimonious interpretation of the data.
The molecular phylogenetic relationships inferred for the members of the genus Prionitis examined in this study do not mirror the geographic location where these specimens were collected nor are they in agreement with traditional species assignations based on morphology or the field habit of gall formation (1). Based on comparisons of ITS sequences, gall-bearing P. filiformis from Lufenholtz Beach, northern California, and non-gall-bearing P. filiformis from Carmel State Beach, central California, are not closest relatives. The differences inferred from phylogenetic analyses are mirrored by variability in species susceptibility to gall induction and by the occurrence of galls in nature on these morphologically similar algae. In the field, P. filiformis (Lufenholtz population) thalli were found bearing galls ca. 30% of the time (J. B. Ashen, unpublished data). P. filiformis (Carmel population), however, despite the examination of numerous thalli (>200) collected over 3 years, was never found bearing galls (J. B. Ashen, unpublished data).
Field observations of the apparent geographic discontinuity of gall formation on P. filiformis are supported by examination of Prionitis herbaria sheets housed at the Jepson Herbarium, University of California, Berkeley. This collection contains 38 specimens of P. filiformis (collected from San Francisco, California, to Trinidad Head, Humbolt County, California), 40% of which bear small (1- to 5-mm) conspicuous galls (specimen dates, 1898 to 1971). No gall-bearing thalli were found among the 20 specimens of P. filiformis collected south of San Francisco, including several specimens from Carmel State Beach (specimen dates, 1898 to 1957).
Laboratory cross-inoculation data are also in agreement with observations of the gall-bearing habit made in the field and from herbaria specimens. Bacteria from galls on P. filiformis (Lufenholtz population) were capable of reinfection of their host thalli but were not observed to form galls on the related (morphological species) P. filiformis (Carmel population). The evidence from molecular phylogeny, the distribution of the gall-bearing habit in nature, and the species specificity of gall induction support the hypothesis that the P. filiformis morphospecies hides a cryptic species not readily distinguishable in the field or by traditional microscopic techniques.
Galls on P. filiformis (Lufenholtz population) were similar in gross appearance to those on the related P. lanceolata. P. filiformis (Lufenholtz population) galls were smaller in diameter (1 to 5 mm) and whitish to cream colored throughout their development, in contrast to the larger (5- to 10-mm) red-pink-colored galls on P. lanceolata (6). Microscopic examination of P. filiformis galls revealed aggregations of intercellular bacteria proliferating between abnormally growing host cells. This is very similar to the situation observed in cross sections of P. lanceolata galls, although the bacterial symbionts in P. filiformis were smaller (0.75 by 1.75 μm) than their P. lanceolata counterparts (6, 34). Gall formation on P. filiformis (Lufenholtz population) (as on Lobocolax deformans Howe) has not been reported in the literature, although tumorization of this alga is evident on herbarium specimens from 99 years ago (31) (J. B. Ashen, unpublished data).
Microscopic examination of galls from P. decipiens also revealed hypertrophied host cells harboring dense assemblages of intercellular symbionts and appeared identical to galls on P. lanceolata (4, 6). The oligodeoxynucleotide probes utilized in this investigation, however, did not hybridize the P. decipiens symbionts. Variability in the intensity of whole-cell fluorescence upon hybridization may be linked to the physiological state of the hybridized cell through the absolute number of ribosomes (3). The failure to detect whole cells of the P. decipiens gall symbionts was probably due to the physiological state of these bacteria after shipping; e.g., the thalli of this alga were maintained ca. 1 week before the onset of severe decomposition precluded further analyses.
The molecular data from this study indicate that the evolutionary relationships of the examined Prionitis taxa mirror the phylogenetic relationships inferred for their bacterial symbionts (Fig. 2 and 3). Bootstrap support for the proposed branching order among the algal taxa is very robust (Fig. 2). The removal of the most divergent sequence (P. decipiens) from this data set has little to no effect on the inferred tree topology or bootstrap values (Fig. 2). In addition, an alternative evolutionary hypothesis, one that groups P. decipiens and P. filiformis to the exclusion of P. lanceolata, is not supported by a T-PTP test for nonmonophyly (P = 1.0). This evidence suggests that coevolution has occurred between these lineages of red algae and their symbiotic α-Proteobacteria. An alternative explanation of the inferred phylogenetic relationships would require that the congruence of branching orders (phylogenetic relationships) between Prionitis hosts and their bacterial symbionts evolved independently, either from a single lineage of symbiotic bacteria and three different algal hosts or from three related bacteria and three different hosts. A more parsimonious interpretation of the data would be that gall formation evolved on a single species of Prionitis with subsequent radiation of that symbiosis leading to speciation of algal host and bacterial symbiont.
Bacterial interactions with algae in nature remain virtually unstudied, despite evidence from a number of laboratories that bacteria can have important consequences for algal growth and morphogenesis in culture (22, 23, 36, 39–41, 45). Axenic cultures of marine macroalgae do not grow normally, a condition which can be alleviated by the readdition of any number of uncharacterized bacterial epiphytes or the maintenance of appropriate ratios of phytohormones (36, 39–41, 45).
The role of bacteria in the physiology of Prionitis gall formation is not understood. In P. lanceolata galls, however, the overproduction of the plant phytohormone indole-3-acetic acid does occur relative the uninfected thallus (4a). The biosynthetic pathways involved are unknown, but, by analogy to terrestrial plant-bacterial gall symbioses, may be microbial (38, 47). Alternatively, the overproduction observed in galls relative to the uninfected algal thallus may be due to an unknown bacterial inductive signal acting on a native algal pathway.
The technological advances that allow the identification and monitoring of uncultivated (or uncultivable) bacteria have provided the means with which to reexamine this and other, similar, eukaryotic-eubacterial associations (2, 12, 16, 18, 21, 47). Our work on microbial symbioses in the genus Prionitis has furnished a biological context within which to view SSU rDNA sequence differences among these gall-forming microbes. Together with the relatively recent work by Moore et al., our work illustrates what has long been suspected to occur in the microbiological community. Small variations in rDNA sequence do not indicate, a priori, similarity of physiological capability or ecological niche (35).
The inability of the bacterial symbionts tested to infect alternate hosts (and their host restriction in the field as observed with whole-cell hybridization) suggests underlying differences in functional gene sequences or gene compliments. This indicates that, for this group of symbionts and hosts, a ca. 1% 16S rDNA sequence difference has significant ecological implications, particularly in terms of host-symbiont specificity. The possibility that coevolution has occurred between Prionitis sp. algal hosts and their bacterial symbionts is also supported, although this evidence remains open to alternative interpretation.
ACKNOWLEDGMENTS
Funds for this research and publication were provided by the Office of Naval Research-Assert (grant no. N00014-92-J-1462) and by the National Science Foundation (Systematic Biology Program no. BSR 940411) to L.J.G. and by grants from Friends of Long Marine Lab, Sigma Xi, and the UCSC Department of Biology to J.B.A.
We thank the following people for their help, suggestions, and support: Bill Sullivan; Ed Delong; Craig Cary; Debra Moon; Giuseppe Zuccarello; Kevin Redslob; Constance Gramlich; Betsy Steele; and the Long Marine Laboratories staff, Lynn J. Rothschild, Bob Rassmussen, Paul Silva, and Richard Moe.
REFERENCES
- 1.Abbott I A, Hollenberg G J. Marine algae of California. Palo Alto, Ca: Stanford University Press; 1976. [Google Scholar]
- 2.Amann R, Kuhl M. In situ methods for assessment of microorganisms and their activities. Curr Opin Microbiol. 1998;1:352–358. doi: 10.1016/s1369-5274(98)80041-6. [DOI] [PubMed] [Google Scholar]
- 3.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Apt K, Gibor A. Development and induction of bacteria-associated galls on Prionitis lanceolata (Rhodophyta) Dis Aquat Org. 1989;6:151–156. [Google Scholar]
- 4a.Ashen J B, Cohen J D, Goff L J. GC-SIM-MS detection and quantification of free indole-3-acetic acid in bacterial galls on the marine alga Prionitis lanceolata (Rhodophyta) J Phycol. 1999;35:493–500. [Google Scholar]
- 5.Ashen J B, Goff L J. Galls on the marine red alga Prionitis lanceolata (Halymeniaceae): specific induction and subsequent development of an algal-bacterial symbiosis. Am J Bot. 1998;85:1710–1721. [PubMed] [Google Scholar]
- 6.Ashen J B, Goff L J. Molecular identification of a bacterium associated with gall formation in the marine red alga, Prionitis lanceolata. J Phycol. 1996;32:286–297. [Google Scholar]
- 7.Baldwin B G. Phylogenetic utility of the internal transcribed spacers of nuclear ribosomal DNA in plants: an example from the Compositae. Mol Phylogenet Evol. 1992;1:3–16. doi: 10.1016/1055-7903(92)90030-k. [DOI] [PubMed] [Google Scholar]
- 8.Barbieri E, Gulledge J, Moser D, Chien C C. New evidence for bacterial diversity in the accessory nidamental gland of the squid (Loligo Pealei) Biol Bull (Woods Hole) 1996;191:316–317. doi: 10.1086/BBLv191n2p316. [DOI] [PubMed] [Google Scholar]
- 9.Barns S M, Delwiche C F, Palmer J D, Pace N R. Perspectives on archeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc Natl Acad Sci USA. 1996;93:9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breznak J A. Biochemical aspects of symbiosis between termites and their intestinal microbiota. In: Andersen J M, editor. Invertebrate-microbial interactions. Cambridge, England: Cambridge University Press; 1984. pp. 173–204. [Google Scholar]
- 11.Britschgi T B, Giovannoni S J. Phylogenetic analysis of a natural marine bacterioplankton population by rRNA gene cloning and sequencing. Appl Environ Microbiol. 1991;57:1313–1318. doi: 10.1128/aem.57.6.1707-1713.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cary S C. Vertical transmission of a chemoautotrophic symbiont in the protobranch bivalve, Solemya reidi. Mol Mar Biol Biotechnol. 1994;3:121–130. [PubMed] [Google Scholar]
- 13.Coleman A W, Suarez A, Goff L J. Molecular delineation of species and syngens in Volvocacean green algae (Chlorophyta) J Phycol. 1994;27:463–473. [Google Scholar]
- 14.DeLong E F. Marine microbial diversity: the tip of the iceberg. Trends Biotechnol. 1997;15:203–207. doi: 10.1016/S0167-7799(97)01044-5. [DOI] [PubMed] [Google Scholar]
- 15.DeLong E F. Single-cell identification using fluorescently labeled ribosomal RNA-specific probes. In: e. a. Kemp P, editor. Handbook of methods in aquatic microbial ecology. Boca Raton, Fla: Lewis Publishers; 1993. pp. 285–293. [Google Scholar]
- 16.Dubilier N, Giere O, Distel D L, Cavanaugh C M. Characterization of chemoautotrophic bacterial symbionts in a gutless marine worm (Oligochaeta, Annelida) by phylogenetic 16S rRNA analysis and in situ hybridization. Appl Environ Microbiol. 1995;61:2346–2350. doi: 10.1128/aem.61.6.2346-2350.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faith D P. Cladistic permutation tests for monophyly and nonmonophyly. Syst Zool. 1991;40:366–375. [Google Scholar]
- 18.Feldman R A, Black M B, Cary C S, Lutz R A, Vrijenhoek R C. Molecular phylogenetics of bacterial endosymbionts and their vestimentiferan hosts. Mol Mar Biol Biotechnol. 1997;6:268–277. [PubMed] [Google Scholar]
- 19.Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 20.Field K G, Gordon D, Wright T, Rappe M, Urbach E, Vergin K, Giovannoni S J. Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl Environ Microbiol. 1997;63:63–70. doi: 10.1128/aem.63.1.63-70.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer K, Hahn D, Amann R I, Daniel O, Zeyer J. In situ analysis of the bacterial community in the gut of the earthworm Lumbricus terrestris L. by whole-cell hybridization. Can J Microbiol. 1995;41:666–673. [Google Scholar]
- 22.Fries L. Growth regulating effects of phenylacetic acid and p-hydroxy phenylacetic acid on Fucus spiralis L. (Phaeophyceae, Fucales) in axenic culture. Phycologia. 1977;16:451–455. [Google Scholar]
- 23.Fries L. Some observations on the morphology of Enteromorpha linza (L.) J. Ag. and Enteromorpha compressa (L.) Grev. in axenic culture. Bot Mar. 1975;18:251–253. [Google Scholar]
- 24.Goff L J, Moon D A, Coleman A W. Molecular delineation of species and species relationships in the red algal agarophytes Gracilariopsis and Gracilaria (Gracilariales) J Phycol. 1994;30:521–537. [Google Scholar]
- 25.González J M, Moran M A. Numerical dominance of a group of marine bacteria in the α-subclass of the class Proteobacteria in coastal seawater. Appl Environ Microbiol. 1997;63:4237–4242. doi: 10.1128/aem.63.11.4237-4242.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gray J P, Herwig R P. Phylogenetic analysis of bacterial communities in marine sediments. Appl Environ Microbiol. 1996;62:4049–4059. doi: 10.1128/aem.62.11.4049-4059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hugenholtz P, Pitulle C, Hershberger K L, Pace N R. Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaufman M R, Ikeda Y, Patton C, VanDykhuizen G, Epel D. Bacterial symbionts colonize the accessory nidamental gland of the squid Loligo opalescens via horizontal transmission. Biol Bull (Woods Hole) 1998;194:36–43. doi: 10.2307/1542511. [DOI] [PubMed] [Google Scholar]
- 29.Kooistra W H C F, Stam W T, Olsen J L, van den Hoek C. Biography of Cladophoropsis membranacea (Chlorophyta) based on comparisons of nuclear rDNA ITS sequences. J Phycol. 1992;28:660–668. [Google Scholar]
- 30.Kuske C R, Barns S M, Busch J D. Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl Environ Microbiol. 1997;63:3614–3621. doi: 10.1128/aem.63.9.3614-3621.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kylin, H. 1941. Californische Rhodophyceen. Lunds Universitets Årsskrift, Ny Följd, Andra Afdelningen 37:51.
- 32.Lafay B, Ruimy R, Rausch de Traubenberg C, Breittmayer V, Gauthier M J, Christen R. Roseobacter algicola sp. nov., a new marine bacterium isolated from the phycosphere of the toxin-producing dinoflagellate Prorocentrum lima. Int J Syst Bacteriol. 1995;45:290–296. doi: 10.1099/00207713-45-2-290. [DOI] [PubMed] [Google Scholar]
- 33.Margulis L, Fester R. Symbiosis as a source of evolutionary inovation: speciation and morphogenesis. Bellagio, Italy: Massachusetts Institute of Technology Press; 1991. [PubMed] [Google Scholar]
- 34.McBride D P, Kugrens P, West J. Light and electron microscopic observations on red algal galls. Protoplasma. 1974;79:249–264. doi: 10.1007/BF01276605. [DOI] [PubMed] [Google Scholar]
- 35.Moore L R, Rocap G, Chisholm S W. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature. 1998;393:464–467. doi: 10.1038/30965. [DOI] [PubMed] [Google Scholar]
- 36.Nakanishi K, Nishijima M, Nishimura M, Kuwano K, Saga N. Bacteria that induce morphogenesis in Ulva pertusa (Chlorophyta) grown under axenic conditions. J Phycol. 1996;32:479–482. [Google Scholar]
- 37.Pace N R. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 38.Patten C L, Glick B R. Bacterial biosynthesis of indole-3-acetic acid. Can J Microbiol. 1996;42:207–220. doi: 10.1139/m96-032. [DOI] [PubMed] [Google Scholar]
- 39.Provasoli L. Effect of plant hormones on Ulva. Biol Bull (Woods Hole) 1958;114:375–384. [Google Scholar]
- 40.Provasoli L, Pintner I J. Bacteria induced polymorphism in an axenic laboratory strain of Ulva lactuca (Chlorophyceae) J Phycol. 1980;16:196–201. [Google Scholar]
- 41.Provasoli L, Pintner I J. Symbiotic relationships between microorganisms and seaweeds. Am J Bot. 1964;51:681. [Google Scholar]
- 42.Shiba T. The genus Roseobacter. In: Balows A, et al., editors. The prokaryotes: a handbook on the biology of bacteria: ecophysiology, isolation, identification, applications. 2nd ed. New York, N.Y: Springer-Verlag; 1992. pp. 202–206. [Google Scholar]
- 43.Smith D C, Douglas A E. The biology of symbiosis. 1st ed. London, England: Edward Arnold Ltd.; 1987. [Google Scholar]
- 44.Suzuki M T, Rappe M S, Haimberger Z W, Winfield H, Adair N, Strobel J, Giovannoni S J. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl Environ Microbiol. 1997;63:983–989. doi: 10.1128/aem.63.3.983-989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tatewaki M, Provasoli L, Pintner I J. Morphogenesis of Monostroma oxyspermum (Kutz.) Doty (Chlorophyceae) in axenic culture, especially in bialgal culture. J Phycol. 1983;19:409–416. [Google Scholar]
- 46.Trueman J W. Sponges, plants and T-PTP. Syst Biol. 1995;44:429–434. [Google Scholar]
- 47.Yamada T. The role of auxin in plant-disease development. Ann Rev Phytopathol. 1993;31:253–273. doi: 10.1146/annurev.py.31.090193.001345. [DOI] [PubMed] [Google Scholar]