Abstract
rRNA-targeted oligonucleotide probes have become powerful tools for describing microbial communities, but their use in sediments remains difficult. Here we describe a simple technique involving homogenization, detergents, and dispersants that allows the quantitative extraction of cells from formalin-preserved salt marsh sediments. Resulting cell extracts are amenable to membrane blotting and hybridization protocols. Using this procedure, the efficiency of cell extraction was high (95.7% ± 3.7% [mean ± standard deviation]) relative to direct DAPI (4′,6′-diamidino-2-phenylindole) epifluorescence cell counts for a variety of salt marsh sediments. To test the hypothesis that cells were extracted without phylogenetic bias, the relative abundance (depth distribution) of five major divisions of the gram-negative mesophilic sulfate-reducing delta proteobacteria were determined in sediments maintained in a tidal mesocosm system. A suite of six 16S rRNA-targeted oligonucleotide probes were utilized. The apparent structure of sulfate-reducing bacteria communities determined from whole-cell and RNA extracts were consistent with each other (r2 = 0.60), indicating that the whole-cell extraction and RNA extraction hybridization approaches for describing sediment microbial communities are equally robust. However, the variability associated with both methods was high and appeared to be a result of the natural heterogeneity of sediment microbial communities and methodological artifacts. The relative distribution of sulfate-reducing bacteria was similar to that observed in natural marsh systems, providing preliminary evidence that the mesocosm systems accurately simulate native marsh systems.
Over the past decade, 16S rRNA-targeted specific oligonucleotide probes have become a powerful tool for describing the structure of microbial communities in a variety of natural environments (1, 35). In planktonic environments, a number of straightforward hybridization techniques are available for utilizing oligonucleotide probes, including whole-cell fluorescence in situ hybridization (1, 6, 16, 28, 29) and direct whole-cell blot hybridization (2, 5). These techniques simplify the use of 16S rRNA-targeted probes and therefore allow processing of larger numbers of samples required for conducting ecologically relevant studies. However, sediment and detritus often interfere with the enumeration of bacteria, hybridization, hybridization detection, and the extraction and purification of RNA (34). Thus, probe hybridization studies of sediment samples are more difficult and time-consuming than analogous studies in planktonic environments.
Despite the difficulty involved, many studies have demonstrated the utility of applying 16S rRNA probe hybridization strategies in sediment environments (3, 4, 7, 9, 10, 12, 17, 23, 24, 25, 30). In general, these studies have relied on hybridization of probes to RNA extracted, purified, and immobilized onto charged nylon membranes or fluorescence in situ hybridization (41). To our knowledge, no studies have utilized more straightforward whole-cell membrane hybridization techniques (5). One impediment to using whole-cell hybridization protocols in association with sediments has been the difficulty of quantitative extraction of cells from sediments. Extraction of cells from fine-grained highly organic sediments typical of salt marsh environments is particularly difficult.
Several problems must be overcome to facilitate the extraction of cells from sediments. First, since cells are often tenaciously attached to sediment particles, cells must be detached and separated from sediment particles and organic material that might interfere with the immobilization of cells to charged nylon or complicate microscopic visualization of cells. Second, cells must be extracted without regard to their phylogenetic identity or physiological status, since extraction biases would artifactually influence the observed microbial structure. Extraction bias is not thought to be a problem for total RNA extraction procedures, but this assumption has not been experimentally investigated.
Several studies have investigated the quantitative extraction of cells from sediments. Methods have typically involved ultrasonication, vigorous homogenization, and/or the addition of detergents and dispersants (13, 34, 36, 38). These methods have been successful with a variety of sediment types (11, 13, 14, 31, 32, 36, 38). In this study, we evaluated a modified protocol for extracting cells from salt marsh sediments that involved the use of homogenization, detergents, and dispersants, but did not require sonication. This technique allowed the rapid and quantitative extraction of cells from sediments that had been stored in formalin, while avoiding possible cell lysis by sonication. Resulting cell extracts were substantially reduced in sediment and detritus content and were therefore amenable to membrane blotting and hybridization protocols. The intent of this study was to determine if the whole-cell and RNA extraction techniques could be used to provide equivalent information regarding the composition of salt marsh sediment microbial communities by using a 16S rRNA-targeted oligonucleotide probe hybridization approach. To test the hypothesis that cells were extracted without phylogenetic biases, the microbial community structure (depth distribution) of five major divisions of the gram-negative mesophilic sulfate-reducing bacteria (SRB) were determined using a suite of SRB group-specific [32P]-labeled 16S rRNA-targeted oligonucleotide probes. The apparent structure of SRB communities was compared between RNA and whole-cell extracts from replicate salt marsh sediment samples.
MATERIALS AND METHODS
Collection of sediments.
Sediments used for developing the cell extraction protocol were collected from a pristine Spartina alterniflora-dominated salt marsh located adjacent to the Skidaway River, Savannah, Ga., on the campus of the Skidaway Institute of Oceanography. Intact sediment cores were collected using a custom manufactured surface-sterilized polycarbonate corer with a diameter of 5 cm and total length of 40 cm. Sediment from 2- to 4-cm intervals were extruded using a Teflon-tipped plunger into sterile disposable 50-ml tubes. Sediment (up to 4 g) was aliquoted into fresh sterile tubes, and an amount of 10 ml of artificial seawater (ASW) g of sediment−1 (27) that was 3.7% formalin (Sigma Chemical Co., St. Louis, Mo.) was added to the sediment and slurried by vortexing. Preserved sediments were stored at 4°C until use.
Salt marsh sediments used in probe hybridization studies were originally collected from an Environmental Protection Agency (EPA) Superfund site (LCP chemical site) located in Brunswick, Ga. These sediments were contaminated with mercury and polychlorinated biphenyls (ca. 10 ppt each) as a result of the operation of a chlor-alkali plant (18, 37). Contaminated sediments planted with S. alterniflora and unplanted are maintained in a salt marsh mesocosm system operated by the Skidaway Institute of Oceanography (21). These mesocosms consist of 2.88- by 1.44- by 1.44-m cells filled to a depth of 0.91 m (3.8 m3) with marsh sediments and operated on a 6.5-h tidal cycle that approximates the natural tidal cycle in coastal south Georgia. Replicate sediment cores (10-cm depth) were collected from randomly selected locations in the mesocosms by using a 50-ml custom-manufactured syringe coring device. For these studies, sediment cores were collected in June 1999, 6 months after sediments were initially placed in the mesocosm systems. Sediments were collected, extruded, and preserved frozen at −80°C until use.
Whole-cell extraction.
Formalin-fixed sediments (generally 1 g in 10 ml of ASW plus 3.7% formalin) were made 0.01 M (final concentration) with respect to sodium pyrophosphate (stock solution at 0.1 M; Sigma Chemical Co.) and 0.09% Tween 80 (stock solution at 100%; Sigma Chemical Co.) and were vigorously vortexed for 1 min. Sediments were incubated for 30 min at room temperature and were again vortexed for 15 s. The bulk of sediment particles were removed by centrifugation at 700 × g for 2 min. The supernatant containing the bacteria was removed to a fresh tube by aspiration. The sediment was washed two times with 10 ml of sterile ASW by vortexing for 1 min, and the wash supernatant was collected after centrifugation as described above. Both the original extract and wash supernatant (approximately 30 ml) were combined. Cells were collected by centrifugation at 12,000 × g for 10 min at 4°C and resuspended in 3 ml of fresh sterile ASW. In some cases, remaining fine sediment particles were removed by centrifugation (200 × g) for 1 min. Total cell abundance was determined by epifluorescence microscopy after staining cells with DAPI (4′,6′-diamidino-2-phenylindole) (40).
RNA extraction.
Total RNA was extracted and purified from salt marsh sediments essentially by following the procedure described by Moran et al. (25). The yield and quality of extracted RNA was improved when sediments were frozen prior to extraction (data not shown), and thus RNA was routinely extracted from sediment samples (5 g) that had been stored frozen −80°C for at least 2 days. Sediments were thawed on ice and equilibrated in 20 ml of ice-cold 120 mM sodium phosphate (pH 5.2) for 15 min with shaking. Following equilibration, the sediment was collected by centrifugation at 6,000 × g for 10 min at 4°C. The supernatant was discarded, and the sediment pellet was washed with another 20 ml of phosphate buffer. Following centrifugation at 6,000 × g for 10 min, the resulting pellet was resuspended in 7 ml of lysing buffer (50 mM Tris [pH 8.0], 0.25 mM EDTA [pH 8.0], 25% sucrose), to which 5 mg of lysozyme (Sigma Chemical Co.) ml−1 was freshly added. The sediment was incubated at room temperature for 15 min and centrifuged at 6,000 × g for 10 min at 4°C. The resulting supernatant was decanted, and the pellet was equilibrated with 7 ml of ACE buffer (10 mM sodium acetate, 10 mM NaCl, 3 mM EDTA [pH 5.1]). The pellet was again collected by centrifugation (6,000 × g, 10 min) and warmed to 60°C in a heated water bath for 10 to 15 min. ACE-buffered phenol (250 μl) and 20% sarcosyl (500 μl warmed to 60°C) were added to the pellet. The sediment was vortexed briefly and was incubated at room temperature. After 5 min, the solution was made 0.075 M NaCl by the addition of 300 μl of a 2 M NaCl solution that had been warmed to 60°C. The resulting sediment was extracted once with 6 ml of ACE-buffered phenol-chlorform-isoamyl alcohol (pH 5.1; 75:24:1). The aqueous extract (ca. 8 to 13 ml) was made 0.3 M sodium acetate by the addition of 1/10 volume of a 3 M sodium acetate solution. Following nucleic acid concentration, DNA was digested with 100 U of RNase-free DNase I (Sigma Chemical Co.) and purified using ACE-buffered phenol chlorform as previously described (25). Generally, the RNA-containing aqueous phase remained dark brown at this stage. Contaminating humic substances were removed by size filtration in 2.5-ml Sephadex G-75-120 spun columns constructed in standard 3-ml disposable syringes as described by Moran et al. (25). Sephadex was equilibrated in diethyl pyrocarbonate-treated distilled water prior to use. The quantity, purity, and quality of the extracted RNA were routinely determined by spectroscopy and visualization after agarose gel electrophoresis, respectively (33). Purified RNA was stored at −80°C until use.
Determination of microbial community structure.
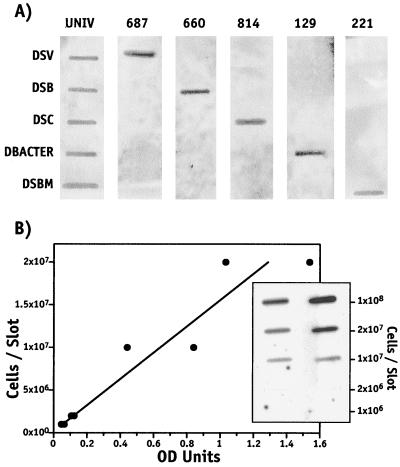
The relative abundance of the total eubacteria community and five phylogenetically distinct groups of SRB (Desulfobulbus, Desulfobacterium, Desulfobacter, Desulfovibrio, and Desulfococcus) was determined by hybridization of phylogenetic group-specific, 32P-labeled 16S rRNA-targeted oligonucleotide probes previously developed (8, 15). The Desulfovibrio group probe (SRB 687) also hybridizes with a number of the iron-reducing geobacters which are not SRB (22). All probes used in this study are shown in Table 1. The specificity of each of these probes was confirmed empirically at the indicated hybridization temperature by using purified RNA from cultured SRB strains (Fig. 1a). The sensitivity of each probe was also determined by dilution series studies using representative SRB cultures (Fig. 1b). Probe hybridization was conducted in a slot blot format using charged nylon (Zeta Probe, catalog no. 162-0165; Bio-Rad Laboratories, Hercules, Calif.). Initially, hybridization of concentration dilution series of RNA and sediment whole-cell extracts were performed to determine the optimal RNA and cell concentrations most appropriate for quantification by scanning densitometry. These studies indicated that under the conditions of this study, 2 to 5 ng of purified RNA/slot or ca. 106 cells/slot yielded hybridization densities well within the dynamic range of the scanning densitometer for quantification (data not shown). Therefore, in all studies reported here, RNA (ca. 2 to 5 ng/slot) and whole-cell extracts containing ca. 3 × 106 cells in 100 μl were immobilized on nylon membranes using a slot blot apparatus (Schleicher & Schuell, Inc., Keene, N.H.). Following blotting, RNA was cross-linked to the membrane by baking in vacuo at 80°C for 2 h or by UV cross-linking for 30 s at the 120,000-microjoule setting using a UV cross-linker (UV Stratalinker model 1800; Stratagene Cloning Systems, La Jolla, Calif.). Membranes were stored desiccated at −20°C until use.
TABLE 1.
16S rRNA-targeted oligonucleotide probes used in this study
| Probe | Probe sequence (5′ to 3′)a | THb | Specificity | Reference |
|---|---|---|---|---|
| Univ 342 | CTG CTG CSY CCC GTA G | 55°C | All eubacteria | 39 |
| SRB 687 | TAC GGA TTT CAC TCC T | 55°C | Desulfovibrio | 8 |
| SRB 660 | GAA TTC CAC TTT CCC CTC TG | 55°C | Desulfobulbus | 8 |
| SRB 221 | TGC GCG GAC TCA TCT TCA AA | 55°C | Desulfobacterium | 8 |
| SRB 129 | CAG GCT TGA AGG CAG ATT | 55°C | Desulfobacter | 8 |
| SRB 814 | ACC TAG TGA TCA ACG TTT | 55°C | Desulfococcus | 8 |
| Desulfosarcina | ||||
| Desulfobotulus |
FIG. 1.
Specificity of 16S rRNA-targeted probes used in this study and the detection sensitivity of a typical probe (UNIV 342) applied for the detection of membrane blotted cells. (A) The specificity of each of the probes used in this study was determined by hybridization at 37°C for the universal eubacterium-targeted probe UNIV and at 55°C for the SRB group-specific probes (SRB 687, 660, 814, 129, 221). RNA (2 ng/slot) purified from cultures of Desulfovibrio desulfuricans ATCC 13541 (DSV), Desulfobulbus propionicus ATCC 33891 (DSB), Desulfococcus multivorans ATCC 33890 (DSC), Desulfobacter sp. strain BG-8 (DBACTER [30]), and Desulfobacterium sp. strain BG-33 (DSBM [30]) were immobilized on replicate charged nylon membranes and hybridized with each probe used in this study. (B) Hybridization sensitivity of a 16S rRNA-targeted probe (UNIV 342) for the detection of immbolized D. desulfuricans cells. Hybridization signal was proportional (r2 = 0.90) to the number of cells. Hybridization detection is expressed in relative optical density (OD) units. Autoradiograph of replicate whole-cell hybridization used to construct the regression line is shown below the regression line.
Oligonucleotides were synthesized using an ABI DNA/RNA synthesizer (model 394) by the Molecular Genetics Facility at the University of Georgia. Oligonucleotides were end labeled with 60 μCi of [γ-32P]ATP (6,000 Ci/mmol; Du Pont/NEN, Boston, Mass.) using T4 polynucleotide kinase (Promega Corp., Madison, Wis.) as described previously (15).
Prehybridization, hybridization, and washes were performed by following published methods (15) at 55°C. Hybridization and wash temperatures were previously empirically determined for each probe (8, 19, 20; Table 1). The blots were prehybridized in a filter-sterilized (0.2 μm) hybridization solution containing 6× SSPE (1× SSPE is 180 mM NaCl, 10 mM NaH2PO4, and 1 mM Na2EDTA [pH 7.7]), 0.1% sodium dodecyl sulfate, and 1× Denhardt's solution (0.2% Ficoll, 0.02% polyvinylpyrrolidone, 0.02% bovine serum albumin). After 3 h, the prehybridization solution was replaced with fresh hybridization solution containing 20 pmol of 32P-labeled probe and was incubated overnight. Prehybridization and hybridization were conducted in 50- or 100-ml hybridization tubes (Bockel Scientific, Feasterville, Pa.) in a rotary hybridization oven (Robbins Scientific Model 2000 microhybridization incubator; Robbins Scientific, Corp., Sunnvale, Calif.). To avoid excessive nonspecific background hybridization, especially with the RNA blots, it was critical to perform prehybridization and hybridization of cell and RNA blots separately (data not shown). Following hybridization, the blots were washed for 1 h in three changes of 6× SSPE plus 0.1% SDS at the hybridization temperature. The blots were dried briefly under an infrared lamp and were attached to filter paper, and hybridization was detected by autoradiography using medical X-ray film (Fuji Medical Systems, USA, Inc., Stamford, Conn.). Generally, a clear hybridization signal was detected after 3 to 24 h for whole-cell blots and 24 to 72 h for RNA blots, depending on the probe used and the sample. Hybridization was quantified by scanning densitometry using the Quantity One version 4 software package and a GS-710 Calibrated Imaging Densitometer system (Bio-Rad Laboratories).
Statistics.
The observed relative distribution of SRB groups with respect to depth were compared between the whole-cell extraction and RNA extraction techniques for each sample and at each depth by two-way analysis of variance. The relative abundance of the five SRB groups in each sample was estimated by normalizing against the abundance of total eubacteria determined using the universal eubacterial probe UNIV 342 in the same sample. The overall comparison of the hybridization results between the whole-cell extraction technique and hybridization of purified RNA was made by linear regression. Cell concentrations of extracted sediments were compared to cell concentrations of unextracted samples by simple t tests. All comparisons were performed at the 95% confidence level. Statistical analyses were facilitated using the SigmaStat version 2.01 software package (SPSS, Inc., Chicago, Ill.).
RESULTS
Whole-cell extraction from salt marsh sediments.
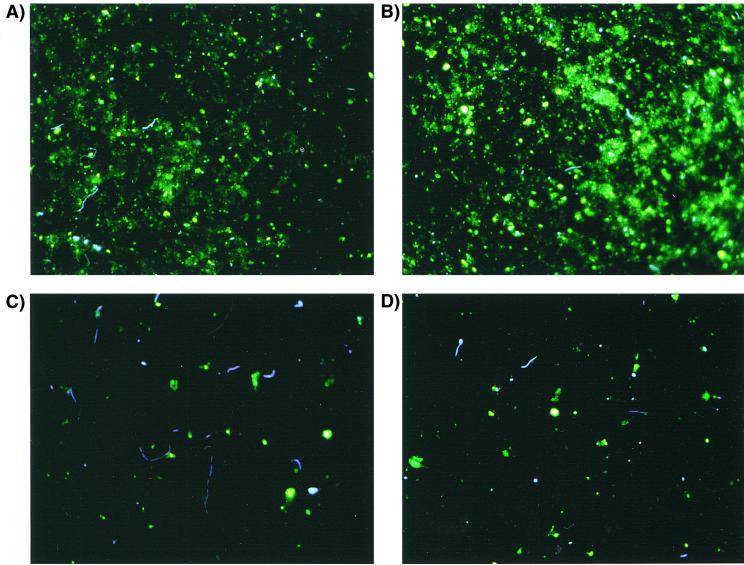
The efficiency of cell extraction from sediment was determined by comparing the concentrations of cells in sediment-free extracts to cell concentrations determined in the same sediments prior to extraction. Cell recoveries from salt marsh sediments sampled from various depths ranged from 91 to 102% relative to those from unextracted cell counts (Table 2). The average extraction recovery was 95.7% ± 3.7% (mean ± standard deviation) and was not significantly different from unextracted cell counts at the 95% confidence limit (P = 0.118). The amount of detrital material in sediment extracts appeared to be substantially reduced compared to that in unextracted samples (Fig. 2). In these micrographs, the irregularly shaped yellow stained material is believed to be detritus and sediment, while bacteria appear blue-white after DAPI staining (26). Cell extracts were sufficiently free of sediments so that 106 to 108 cells could be quantitatively blotted and cross-linked to charged nylon membranes and hybridized without interference from sediment particles. The cell extraction procedure typically required only 2 h, compared to nearly 2 days required for extraction and purification of RNA from sediments.
TABLE 2.
Cell extraction efficiency from salt marsh sediments
| Sediment (depth) | Unextracted sediment (log10 cells/g wet wt [SD]) | Extracted sediment (log10 cells/g wet wt [SD]) | % Recoverya |
|---|---|---|---|
| Surface (1–2 cm) | 8.07 (7.24) | 7.51 (6.67) | 93 |
| 8.00 (7.26) | 7.69 (6.99) | 96 | |
| Active rhizosphere (2–15 cm) | 7.50 (6.91) | 7.69 (7.12) | 102 |
| 7.89 (7.13) | 7.16 (6.44) | 91 | |
| Below active rhizosphere (15–60 cm) | 8.21 (7.76) | 7.91 (7.08) | 96 |
| 7.34 (6.71) | 7.05 (6.28) | 96 |
Average cell recovery was 95.7% ± 3.7% (mean ± standard deviation).
FIG. 2.
Photomicrographs of DAPI-stained bacteria before (A and B) and after (C and D) extraction using the whole-cell extraction procedure.
Blot hybridization; whole-cell versus RNA.
Slot blot hybridization with 32P-labeled oligonucleotide probes of whole cells and purified RNA are shown in Fig. 3. On average, 105 to 107 cells that originated from 0.015 to 0.03 g of sediment and 2 to 4 ng of RNA were blotted per individual slot. Generally, significantly stronger hybridization signals were obtained from whole-cell blots versus purified RNA relative to the amount of sediment initially extracted. Thus, in addition to the whole-cell extraction procedure being considerably simpler and less time-consuming than the RNA extraction procedure, hybridization sensitivity appears also to be greater using the whole-cell technique. These observations suggest that a greater fraction of the total sediment bacterial RNA is recovered when cells are extracted from the sediment prior to lysis rather than lysing cells in situ as is done when total RNA is extracted. Because stronger hybridization signals were routinely obtained from the whole-cell blots than from the RNA blots, the time required for autoradiographic quantification was also shorter for whole-cell blots relative to that for RNA blots. Typically, whole-cell blots with 105 to 107 cells · slot−1 rarely required more than 24 h of exposure, while RNA blots with 3 to 5 ng of RNA · slot−1 typically required several days of exposure for adequate quantification.
FIG. 3.
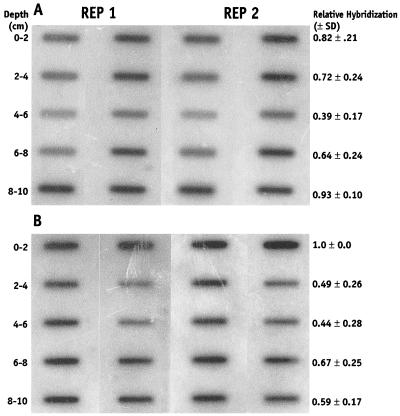
Hybridization of whole-cell (A) and RNA-extracted (B) sediments with 32P-labeled universal eubacterial 16S rRNA-targeted oligonucleotide probe UNIV 342. Fifty microliters of the whole-cell extract (ca. 105 to 106 cells) and 3 ng of extracted RNA was immobilized on the membrane for each depth sample (slot). Sediment depth and average relative hybridization density (optical density) for each depth is shown. Average hybridization signal is the result of replicate blots from replicate core samples indicated as REP 1 and REP 2.
Estimating phylogenetic diversity.
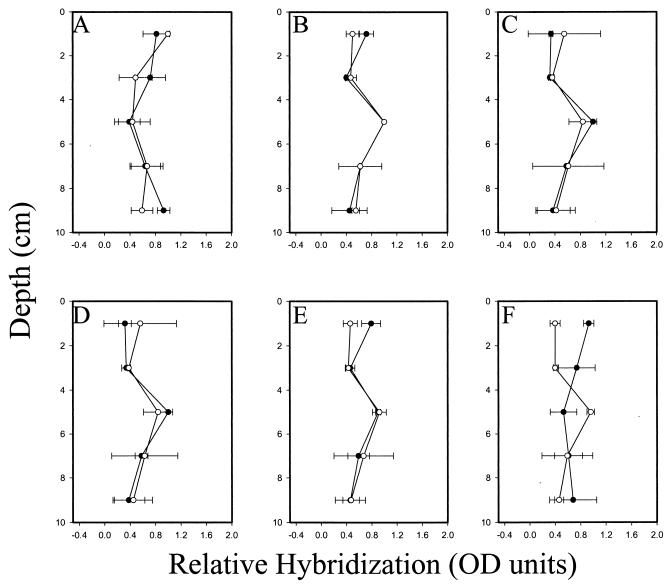
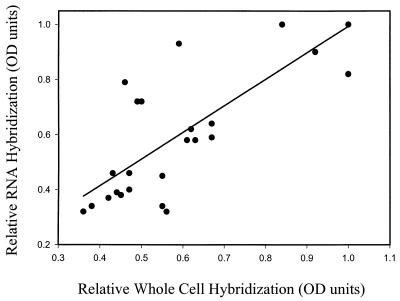
The distribution of total eubacteria and of five groups of gram-negative mesophilic SRB populations in salt marsh sediments that had recently been placed in a tidal mesocosm system was determined by hybridization with a suite of phylogenetic group-specific probes. The distribution in the top 10 cm of the sediment was determined using purified RNA and whole-cell extracts simultaneously. The relative distribution of each group is reported after normalization to the total eubacterial rRNA signal that was determined by quantifying the hybridization signal of replicate blots hybridized with the universal eubacterium-targeted oligonucleotide probe UNIV 342 (Table 1). With the exception of one group of the SRB (Desulfobulbus, probe SRB 660), the distribution of SRB populations and of total eubacteria was statistically identical whether the distribution was determined using the whole-cell or RNA extraction technique (Fig. 4). Within the top 10 cm of sediment the distribution of eubacteria was approximately homogeneous (Fig. 4A), while four of the five SRB groups examined exhibited a relative maximum between the depths of 4 and 6 cm (Fig. 4B to E). In the one instance in which the results from the whole-cell and RNA extraction techniques did not yield the same results (Desulfobulbus; P = 0.037), the whole-cell technique indicated a relative maximum of Desulfobulbus at 4 to 6 cm (Fig. 4F). Results obtained from RNA blots suggested that there were no differences in the relative abundance of this group with depth. Overall, the relative hybridization signals obtained with the whole-cell and RNA extraction techniques were similar. Relative hybridization of RNA blots could be reasonably predicted from relative whole-cell hybridization intensities by a simple first-order linear regression model (r2 = 0.60) when the results from the hybridization with the Desulfobulbus-specific probe were omitted (Fig. 5). The slope (0.96) and intercept (0.03) were not significantly different from 1 and 0, respectively, indicating a quantitatively direct relationship between the techniques.
FIG. 4.
Relative depth distribution of total eubacteria and five groups of gram-negative mesophilic SRB in mercury- and PCB (Aroclor 1268)-contaminated salt marsh sediment equilibrated in the BERM mesocosm system for 6 months. Relative normalized hybridization is reported as a fraction of the strongest hybridization signal at any depth in a given core. Profiles of sulfate-reducing groups were normalized to the total eubacterial signal. Distribution was determined after whole-cell extraction (○) and direct RNA extraction (●). Depth profiles of total eubacteria (probe UNIV 342) (A), Desulfobacterium (probe SRB 221) (B), Desulfobacter (probe SRB 129) (C), Desulfovibrio (probe SRB 687) (D), Desulfococcus/Desulfosarcina/Desulfobotulus group (probe SRB 814) (E), and Desulfobulbus (probe SRB 660) (F) are shown. Error bars indicate standard deviation of the mean from four independent hybridizations. Depth distributions for each probe determined after whole-cell and RNA extractions were compared by two-way analysis of variance.
FIG. 5.
Linear regression of relative hybridization of all 16S rRNA-targeted oligonucleotide probes used in this study determined after whole-cell extraction and RNA extraction in paired samples from the same cores and depths. Comparisons include all phylogenetic groups except the Desulfobulbus group (probe SRB 660). The regression coefficient is 0.60, the slope is 0.96, and the intercept is 0.03.
DISCUSSION
The objectives of this study were to develop a whole-cell extraction technique suitable for use with salt marsh sediments and to determine whether cell extracts were suitable for microbial diversity studies utilizing phylogenetic group-specific 16S rRNA-targeted oligonucleotide probes.
Direct comparisons of cell counts obtained from a variety of salt marsh sediments before and after the whole-cell extraction indicated that cells could be quantitatively recovered from salt marsh sediments by using the cell extraction procedure developed in this study. Furthermore, it was not possible to extract sufficient RNA for additional hybridization analysis from extracted sediments, strengthening the conclusion that cells were quantitatively removed from marsh sediments using the procedure described here (data not shown). Sonication, which in some studies has been shown to be required for dislodging sediment-attached bacteria but also to reduce cell recoveries (13), was not required to achieve quantitative cell recoveries using the technique developed in this study with salt marsh sediments. The cell extraction technique required substantially less time compared to the relatively laborious RNA extraction and purification techniques previously used in conjunction with molecular probe hybridization studies. Therefore, the whole-cell extraction technique is more amenable to the analysis of larger ecologically significant numbers of samples. Furthermore, because hybridization signals were consistently stronger when whole-cell blots were hybridized relative to blotted RNA, it can be speculated that the efficiency of RNA extraction was greater when the cells are extracted prior to cell lysis. Therefore, the sensitivity of the whole-cell extraction technique is greater than that achieved by RNA extraction. Thus, smaller sample sizes are required, further improving the ability to conduct ecologically relevant studies that typically can require the analysis of hundreds to thousands of individual samples. The greater efficiency of RNA extraction in the whole-cell extraction procedure is most likely due to a number of factors associated with lysing cells in the presence of native sediments. For example, it is likely that the degradation rate of free RNA is greater than cellular RNA in sediments and that sediments more readily bind free RNA rather than cellular RNA. Alternatively, if the specificity of the probes when hybridized to whole-cell extracts is different from that achieved when purified RNA was hybridized, the inferred greater hybridization sensitivity detected with whole-cell blots might be due to nonspecific probe hybridization. However, several reports in the literature suggest that the specificity of 16S rRNA-targeted oligonucleotide probes are maintained when used in whole-cell formats (1, 5) although the sensitivity of probes may be reduced in whole cells due to ribosomal higher order structure (15).
A priori there is reason to believe that the extraction of cells from sediments may lead to extraction bias in cell recovery. For example, some cell types may be more fastidiously attached to sediment particles and therefore may be less efficiently extracted. However, in the studies described here, the differences observed in the relative distribution of cell types in sediment cores between the RNA and whole-cell extraction methods were not significant. This observation was true whether essentially all eubacterial cells or specific groups of the delta proteobacteria were studied. Of six comparisons made (120 individual hybridization blots), in only one case (Desulfobulbus probe) were the relative hybridization signals different between the RNA and whole-cell extracts. These results suggest that the whole-cell extraction and RNA extraction hybridization approaches for describing sediment microbial communities are equally robust. Although the relative results obtained with the two techniques could be compared directly, since the absolute quantities of specific RNA targets or cell types were not determined, it was not possible to infer the diversity (species richness and abundance) of the sediment microbial communities from these studies.
Although the relative hybridization results obtained with the two methods were similar, the variability associated with replicates was high. In some cases, variability between replicate samples exceeded 2 standard deviations from the mean. This variability appears to stem both from natural heterogeneity of sediment microbial communities and from methodological artifacts, since there was an equally high degree of variability between replicate cores and between replicate extracts of single core samples (data not shown). Although the source of this variability remains largely unknown, it seems unlikely that the bulk of variability was associated with the inherent complexity of the target microbial consortia. If this were the case, we would have expected that the magnitude of hybridization variability associated with a particular probe would be inversely proportional to its phylogenetic specificity, since the broader the phylogenetic specificity the more complex the target community. This was not the case. For example, the average precision (standard deviation) associated with the mean relative hybridization of whole-cell extracts using the universal eubacterium-targeted probe (UNIV 342) was similar to the average precision associated with the five delta proteobacterium-specific probes. The standard deviation associated with the eubacterial probe was 0.2 compared with an average standard deviation of 0.2 ± 0.1 associated with the delta proteobacterial group-specific probes. However, the source of this variability remains unclear. No obvious correlation between sediment characteristics, such as density, porosity, or organic content, was noted in these studies, although the variability associated with sediments that were obtained from a 6- to 8-cm depth generally seemed to be the highest (Fig. 4). The high variability associated with direct RNA hybridization, regardless of whether the whole-cell extraction or RNA extraction technique is used, limits the resolution of these techniques to discern differences in the composition of sediment microbial community structures. Therefore, methodological and sampling variability should be an important consideration in the generation and interpretation of sediment rRNA hybridization data. Furthermore, these studies suggest that continued research is required to determine the source and nature of the variability associated with rRNA phylogenetic specific probe hybridization of sediment microbial communities.
Although only relative abundances of bacteria were determined in this study, the overall distribution pattern of gram-negative mesophilic SRB was similar to those reported in other studies (17, 30). SRB consortia were observed at all depths sampled, but were maximal between 4 and 6 cm. Similar distribution patterns have also been observed in natural S. alterniflora-dominated salt marshes in Georgia by using the same suite of rRNA-targeted oligonucleotide probes (19, 20).
Because the sediments used in this study originated from an experimental mesocosm system that had been established for 6 months, an evaluation of the distribution patterns of different microbial groups provides an opportunity to evaluate how accurately the mesocosm reflects native salt marsh sediments. Although the observations that are reported here must still be considered preliminary from this perspective, these results suggest that the mesocosm systems do accurately simulate the distribution of microbial consortia after a 6-month period of equilibration. The equilibration process of these mesocosms with respect to other parameters, including sediment physical characteristics, porewater chemistry, microbial activity, and plant growth, will be reported elsewhere. To our knowledge, the structure of microbial consortia has not been previously evaluated in these types of mesocosm systems and suggests that salt marsh mesocosm systems can be established that accurately simulate a native S. alterniflora salt marsh environment typical of southeastern United States coastal systems.
ACKNOWLEDGMENTS
This work was supported by grants from the National Science Foundation (grant number DEB-9706317), the Office of Naval Research (grant number N00014-97-1-0955), and the U.S. EPA (Hazardous Substance Research Council) (grant number R825513-01).
We thank J. K. King for technical assistance and the Skidaway Institute of Oceanography physical operations staff for construction and operation of the BERM tidal mesocosm facility. Anna Boyette helped prepare the figures and Dee Peterson prepared the manuscript.
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Betzl D, Ludwig W, Schleifer K-H. Identification of lactococci and enterococci by colony hybridization with 23S rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1990;56:2927–2929. doi: 10.1128/aem.56.9.2927-2929.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boivin-Jahns V, Ruimy D, Bianchi A, Daumas S, Christen R. Bacterial diversity in a deep-subsurface clay environment. Appl Environ Microbiol. 1996;63:3405–3412. doi: 10.1128/aem.62.9.3405-3412.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borneman J, Skroch P W, O'Sullivan K M, Palus J A, Rumjanek N G, Janse J L, Nienhuis J, Triplett E W. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl Environ Microbiol. 1996;62:1935–1943. doi: 10.1128/aem.62.6.1935-1943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun-Howland E B, Vescio P A, Nierzwicki-Bauer S A. Use of a simplified cell blot technique and 16S rRNA-directed probes for identification of common environmental isolates. Appl Environ Microbiol. 1993;59:3219–3224. doi: 10.1128/aem.59.10.3219-3224.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeLong E F, Wickham G S, Pace N R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 1989;243:1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- 7.Devereux R, Delaney M, Widdel F, Stahl D A. Natural relationships among sulfate-reducing bacteria. J Bacteriol. 1989;171:6689–6695. doi: 10.1128/jb.171.12.6689-6695.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devereux R, Kane M D, Winfrey J, Stahl D A. Genus- and group-specific hybridization probes for determinative and environmental studies of sulfate-reducing bacteria. Syst Appl Microbiol. 1992;15:601–609. [Google Scholar]
- 9.Devereux R, Mundfrom G W. A phylogenetic tree of 16S rRNA sequences from sulfate-reducing bacteria in a sandy marine sediment. Appl Environ Microbiol. 1994;60:3437–3439. doi: 10.1128/aem.60.9.3437-3439.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devereux R, Hines M E, Stahl D A. S cycling: characterization of natural communities of sulfate-reducing bacteria by 16S rRNA sequence comparisons. Microb Ecol. 1996;32:283–292. doi: 10.1007/BF00183063. [DOI] [PubMed] [Google Scholar]
- 11.Dye A H. A method for the quantitative estimation of bacteria from mangrove sediments. Estuar Coast Shelf Sci. 1983;17:207–212. [Google Scholar]
- 12.Edgcomb V P, McDonald J H, Devereux R, Smith D W. Estimation of bacterial cell numbers in humic acid-rich salt marsh sediments with probes directed to 16S ribosomal DNA. Appl Environ Microbiol. 1999;65:1516–1523. doi: 10.1128/aem.65.4.1516-1523.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellery W N, Schleyer M H. Comparison of homogenization and ultrasonication as techniques in extracting attached sedimentary bacteria. Mar Ecol Prog Ser. 1984;15:247–250. [Google Scholar]
- 14.Epstein S S, Rossel J. Enumeration of sandy sediment bacteria: search for optimal protocol. Mar Ecol Prog Ser. 1995;117:289–298. [Google Scholar]
- 15.Frischer M E, Floriani P J, Nierzwicki-Bauer S A. Differential sensitivity of 16S rRNA targeted oligonucleotide probes used for fluorescence in situ hybridization is a result of ribosomal higher order structure. Can J Microbiol. 1996;42:1061–1071. doi: 10.1139/m96-136. [DOI] [PubMed] [Google Scholar]
- 16.Giovannoni S J, DeLong E F, Olsen G J, Pace N R. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J Bacteriol. 1988;170:720–726. doi: 10.1128/jb.170.2.720-726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hines M E, Evans R S, Sharak Genthner B R, Willis S G, Friedman S, Rooney-Varga J N, Devereux R. Molecular phylogenetic and biogeochemical studies of sulfate-reducing bacteria in the rhizosphere of Spartina alterniflora. Appl Environ Microbiol. 1999;65:2209–2216. doi: 10.1128/aem.65.5.2209-2216.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kannan K, Maruya K A, Tanabe S. Distribution and characterization of polychlorinated biphenyl congeners in soil and sediment from a Superfund site contaminated with Aroclor 1268. Environ Sci Technol. 1997;31:1483–1488. [Google Scholar]
- 19.King J K. Quantitative assessment of mercury methylation by phylogenetically diverse consortia of sulfate-reducing bacteria in salt marsh systems. Ph.D. thesis. Atlanta: Georgia Institute of Technology; 1999. [Google Scholar]
- 20.King J K, Kostka J E, Frischer M E, Saunders F M. Sulfate-reducing bacteria methylate mercury at variable rates in pure culture and in marine sediments. Appl Environ Microbiol. 2000;66:2430–2437. doi: 10.1128/aem.66.6.2430-2437.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee R F. Bioremediation studies at the Skidaway Institute of Oceanography. Skidaway Scenes: Newsl. Skidaway Mar Sci Found. 1997;13:2–4. [Google Scholar]
- 22.Lonergan D J, Jenter H L, Coates J D, Phillips E J, Schmidt T M, Lovely D R. Phylogenetic analysis of dissimilatory Fe(III)-reducing bacteria. J Bacteriol. 1996;178:2402–2408. doi: 10.1128/jb.178.8.2402-2408.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacGregor B J, Moser D P, Alm E W, Nealson K H, Stahl D A. Crenarchaeota in Lake Michigan sediment. Appl Environ Microbiol. 1997;63:1178–1181. doi: 10.1128/aem.63.3.1178-1181.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manz W, Eisenbrecher M, Neu T R, Szewzyk U. Abundance and spatial organization of Gram-negative sulfate-reducing bacteria in activated sludge investigated by in situ probing with specific 16S rRNA targeted oligonucleotides. FEMS Microbiol Ecol. 1998;25:43–61. [Google Scholar]
- 25.Moran M A, Torsvic V L, Torsvick T, Hodson R E. Direct extraction and purification of rRNA for ecological studies. Appl Environ Microbiol. 1993;59:915–918. doi: 10.1128/aem.59.3.915-918.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mostajir B, Dolan J R, Rassoulzadegan F. A simple method for the quantification of a class of labile marine pico-sized and nano-sized detritus DAPI yellow particles (DYP) Aquat Microbiol Ecol. 1995;9:259–266. [Google Scholar]
- 27.Paul J H. The use of Hoechst dyes 33258 and 33342 for enumeration of attached and planktonic bacteria. Appl Environ Microbiol. 1982;43:939–944. doi: 10.1128/aem.43.4.939-944.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramsing N G, Kuhl M, Jorgensen B B. Distribution of sulfate-reducing bacteria, O2, and H2S in photosynthetic biofilms determined by oligonucleotide probes and microelectrodes. Appl Environ Microbiol. 1993;59:3840–3849. doi: 10.1128/aem.59.11.3840-3849.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramsing N B, Fossing H, Ferdelman T G, Andersen F, Thamdrup B. Distribution of bacterial populations in a stratified fjord (Mariager fjord, Denmark) quantified by in situ hybridization and related to chemical gradients in the water column. Appl Environ Microbiol. 1996;62:1391–1404. doi: 10.1128/aem.62.4.1391-1404.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rooney-Varga J N, Devereux R, Evans R S, Hines M E. Seasonal changes in the relative abundance of uncultivated sulfate-reducing bacteria in a salt marsh sediment and in the rhizosphere of Spartina alterniflora. Appl Environ Microbiol. 1997;63:3895–3901. doi: 10.1128/aem.63.10.3895-3901.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rublee P, Dornseif B E. Direct counts of bacteria in the sediments of North Carolina saltmarsh. Estuaries. 1978;1:188–191. [Google Scholar]
- 32.Rublee P A. Bacteria and microbial distribution in estuarine sediments. In: Dennedy V S, editor. Estuarine comparisons. New York, N.Y: Academic Press, Inc.; 1982. pp. 159–182. [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Schallenberg M, Kalff J, Rasmussen J B. Solutions to problems in enumerating sediment bacteria by direct counts. Appl Environ Microbiol. 1989;55:1214–1219. doi: 10.1128/aem.55.5.1214-1219.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stahl D A. Application of phylogenetically based hybridization probes to microbial ecology. Mol Ecol. 1995;4:535–542. [Google Scholar]
- 36.Tao S F, Taghon G L. Enumeration of protozoa and bacteria in muddy sediment. Microbial Ecol. 1997;33:144–148. doi: 10.1007/s002489900016. [DOI] [PubMed] [Google Scholar]
- 37.U.S. Environmental Protection Agency. Phase I site characterization sampling map and chemical analysis supplement, LCP Chemicals. Washington, D.C.: U.S. Environmental Protection Agency; 1995. [Google Scholar]
- 38.Velji M I, Albright L J. Microscopic enumeration of attached marine bacteria of seawater, marine sediment, fecal matter, and kelp blade samples following pyrophosphate and ultrasound treatments. Can J Microbiol. 1986;32:121–126. [Google Scholar]
- 39.Vescio P A, Nierzwicki-Bauer S A. Extraction and purification of CR amplifiable DNA from lacustrine subsurface sediments. J Microbiol Methods. 1995;21:225–233. [Google Scholar]
- 40.Williams S C, Hong Y, Danavall D C A, Howard-Jones M H, Gibson D, Frischer M E, Verity P G. Distinguishing between living and nonliving bacteria: evaluation of the vital stain propidium iodide and the combined use with molecular probes in aquatic samples. J Microbiol Methods. 1998;32:225–236. [Google Scholar]
- 41.Zepp Falz, K., C. Holliger, R. Großkopt, W. Liesack, A. N. Nozhevnikova, B. Müller, B. Wehrli, and D. Hahn. Vertical distribution of methanogens in the anoxic sediment of Rotsee (Switzerland). Appl. Environ. Microbiol. 65:2402–2408. [DOI] [PMC free article] [PubMed]