Abstract
The culturability of abundant members of the domain Bacteria in North Sea bacterioplankton was investigated by a combination of various cultivation strategies and cultivation-independent 16S rRNA-based techniques. We retrieved 16S rRNA gene (rDNA) clones from environmental DNAs and determined the in situ abundance of different groups and genera by fluorescence in situ hybridization (FISH). A culture collection of 145 strains was established by plating on oligotrophic medium. Isolates were screened by FISH, amplified ribosomal DNA restriction analysis (ARDRA), and sequencing of representative 16S rDNAs. The majority of isolates were members of the genera Pseudoalteromonas, Alteromonas, and Vibrio. Despite being readily culturable, they constituted only a minor fraction of the bacterioplankton community. They were not detected in the 16S rDNA library, and FISH indicated rare (<1% of total cell counts) occurrence as large, rRNA-rich, particle-associated bacteria. Conversely, abundant members of the Cytophaga-Flavobacteria and gamma proteobacterial SAR86 clusters, identified by FISH as 17 to 30% and up to 10% of total cells in the North Sea bacterioplankton, respectively, were cultured rarely or not at all. Whereas SAR86-affiliated clones dominated the 16S rDNA library (44 of 53 clones), no clone affiliated to the Cytophaga-Flavobacterum cluster was retrieved. The only readily culturable abundant group of marine bacteria was related to the genus Roseobacter. The group made up 10% of the total cells in the summer, and the corresponding sequences were also present in our clone library. Rarefaction analysis of the ARDRA patterns of all of the isolates suggested that the total culturable diversity by our method was high and still not covered by the numbers of isolated strains but was almost saturated for the gamma proteobacteria. This predicts a limit to the isolation of unculturable marine bacteria, particularly the gamma-proteobacterial SAR86 cluster, as long as no new techniques for isolation are available and thus contrasts with more optimistic accounts of the culturability of marine bacterioplankton.
ZoBell's landmark paper on the taxonomy and abundance of marine bacteria (60) essentially defined this group for a long time. The species then most frequently cultured from marine water samples belonged to the genera Pseudomonas, Vibrio, Spirillum, Achromobacter, Flavobacterium, and Bacillus, and these were assumed to be dominant in marine waters.
The discrepancy between direct microscopic enumeration and plate counts of bacteria was first pointed out by Jannasch and Jones (27). They attributed it to the presence of bacteria in aggregates, to selective effects of the media used, and to the presence of inactive cells. In 1982, Colwell and coworkers developed the viable-but-nonculturable hypothesis (59). Ferguson et al. showed that >99.9% of the natural bacterioplankton community in seawater could not be cultured on Marine Agar 2216 (13). Cultivation failed to solve the discrepancy for a long time. It was left to the cultivation-independent rRNA approach, most notably to 16S rRNA gene (rDNA) clone libraries, to reveal the high additional diversity of marine bacterioplankton communities (7, 11, 15–17, 35, 42). In the Atlantic and Pacific Oceans, most of the sequences clustered within the alpha (e.g., SAR11 and SAR116) and gamma (e.g., SAR92 and SAR86) subclasses of Proteobacteria and two novel groups of Archaea were found. 16S rDNA sequences of previously isolated marine bacteria (e.g., Pseudomonas, Rhodobacter, and Arthrobacter spp.) were also occasionally retrieved, but their in situ abundances remained unknown (6, 55).
In view of the difficulty of isolating common marine bacteria, dilution culture methods were applied (10). This led to strategies for optimizing viability determinations and eventually to the pure culture of, so far, only one strain of a probably typical marine oligocarbophilic bacterium (48). In contrast, based on DNA-DNA hybridization of the genomic DNAs of isolates obtained with the traditional ZoBell medium against community DNA, it has been suggested that readily culturable bacteria are abundant in the marine water column (21, 22, 40, 44). The aim of this study was to address these discrepancies by evaluating which microorganisms in the North Sea bacterioplankton are readily culturable. For this, we combined cultivation on defined oligotrophic medium with cloning of PCR-amplified environmental 16S rDNAs and fluorescence in situ hybridization (FISH).
MATERIALS AND METHODS
Sampling and fixation.
In September and November 1997 and February and August 1998, surface water samples were collected at a 1-m depth in acid-washed and seawater-prerinsed 50-liter polyethylene containers. The sampling station Helgoland Roads (54°09′N, 7°52′E) is near the island of Helgoland, approximately 50 km offshore in the German Bay of the North Sea. Samples were stored at 4°C and further processed within approximately 5 h.
For DNA extraction, prefiltered picoplankton (cellulose nitrate filter; diameter, 47 mm; pore size, 5 μm; Sartorius AG, Göttingen, Germany) was collected in September 1997 and unfiltered picoplankton was collected in November 1997 by filtration of 1 to 3 liters of water on white polycarbonate filters (diameter, 47 mm; pore size, 0.2 μm; type GTTP2500; Millipore, Eschborn, Germany).
For FISH, 10- to 100-ml samples of unfiltered seawater were fixed with formaldehyde (final concentration, 2% [wt/vol]) for 30 min at room temperature, collected on white polycarbonate filters (diameter, 47 mm; pore size, 0.2 μm; type GTTP2500; Millipore), and rinsed with double-distilled water. Filters were stored at −20°C until further processing.
Enrichment and isolation of marine microorganisms.
For cultivation, synthetic seawater was prepared as described by Schut et al. (48). Trace elements and vitamins were added separately. A mixture of monomers (alanine, l-aspartate, dl-leucine, l-glutamate, l-ornithine, and dl-serine [all at 1 μM]; glucose, fructose, galactose, glycolate, succinate, and mannitol [all at 10 μM]; and acetate, lactate, ethanol, and glycerol [all at 15 μM]) was added as a substrate.
The cultivation conditions of this basic approach were modified, e.g., by varying the pH (5.7 and 8.3) or salinity (25 and 35 g of NaCl per liter), by the absence of vitamins and trace elements, and by replacing the monomers with a mixture of polymers (chitin, cellulose, xylan, and pectin [1 g of each per liter] and starch [5 g/liter]).
Aliquots (100 μl) of unfiltered and filtered (cellulose nitrate filter; diameter, 47 mm; pore sizes, 5.0, 1.2, 0.45, and 0.22 μm; Sartorius AG) seawater were either directly spread on plates containing 1% (wt/vol) agar (Difco) or preincubated in a dilution series of the corresponding medium. Colonies were selected randomly from agar plates and subcultured at least three times under the same conditions.
16S rDNA clone library construction.
Total nucleic acids were extracted by procedures described by Tsai and Olson (56) from the filters prepared in September and November 1997. Bacterial 16S rRNA primers 8f (5′-AGAGTTTGATCMTGGC-3′) and 1542r (5′-AAAGGAGGTGATCCA-3′) were used to amplify almost full-length 16S rDNAs from total community DNA (9) by PCR (46). The amplified rDNA was inserted into the pGEM-T vector (Promega Corp., Madison, Wis.) in accordance with the manufacturer's instructions. Competent Escherichia coli JM109 cells (Promega) were transformed and screened for plasmid insertions by following the manufacturer's instructions.
Sequencing and phylogenetic analysis.
Plasmid DNAs from selected 16S rDNA clones and amplified 16S rDNAs from isolates were sequenced by Taq Cycle Sequencing and universal 16S rRNA-specific primers using an ABI377 (Applied Biosystems, Inc.) sequencer. All sequences were checked for chimera formation with the CHECK_CHIMERA software of the Ribosomal Database Project (32), which compares the phylogenetic affiliations of the 5′ and 3′ ends. Sequence data were analyzed with the ARB software package (http://www.mikro.biologie.tu-muenchen.de). A phylogenetic tree was reconstructed using neighbor-joining, maximum-parsimony, and maximum-likelihood analyses. Only sequences at least 90% complete were used for tree construction. Alignment positions at which less than 50% of sequences of the entire set of data had the same residues were excluded from the calculations to prevent uncertain alignments within highly variable positions of the 16S rDNA, which cause mistakes in tree topology.
Amplified ribosomal DNA restriction analysis (ARDRA).
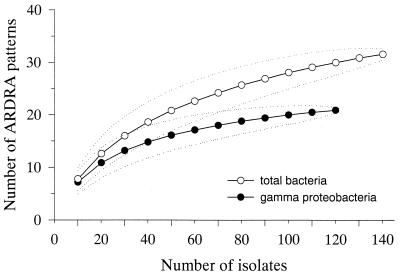
Purified (QIAquick Purification Kit; Qiagen, Hilden, Germany), amplified 16S rDNAs (approximately 1 μg) from all of the isolates were digested with 7.5 U of the restriction endonuclease HaeIII (Promega) for 3 h at 37°C. The fragments were analyzed by polyacrylamide gel electrophoresis, and restriction patterns were compared visually. The diversity of the isolates compared to total culturable diversity by our approach was analyzed by rarefaction analysis (51). This was performed for all isolates and for all isolated members of the gamma proteobacteria. Rarefaction curves were produced using the analytical approximation algorithm of Hurlbert (26), and 95% confidence intervals were estimated as described by Heck et al. (24). Calculations were performed using the freeware program “a RarefactWin” (http://www.uga.edu/∼strata/Software.html).
Cell counts, FISH, and oligonucleotide probe design.
Total bacterioplankton counts were determined by epifluorescence microscopy of acridine orange-stained cells (25). Screening of isolates and determination of North Sea bacterioplankton community structure were performed by FISH. Cells from a single colony of each isolate were transferred to Teflon-coated microscope slides and immobilized by air drying. After dehydration and fixation with 50, 80, and 96% (wt/vol) ethanol, cells on slides and on filter sections were hybridized with oligonucleotide probes EUB338 (3), ALF968 (36), GAM42a (34), and CF319a (33). Counterstaining with 4,6-diamidino-2-phenylindole (DAPI; 1 μg/ml) and mounting for microscopic evaluation were performed as described previously (3, 19).
Oligonucleotide probes ALT1413, PSA184, SAR86-1249, NOR1-56, NOR2-1453, and OCE232 (Table 1) were designed using the PROBE_FUNCTIONS tool of the ARB software package. Their specificity was evaluated with the PROBE_MATCH tool of the ARB package against the rRNA database of the Technical University of Munich (release 12/98). CY3-labeled probes were synthesized by Interactiva (Ulm, Germany). Hybridization conditions for the newly designed probes were optimized by varying the concentration of formamide (37).
TABLE 1.
Oligonucleotide probes used for FISH
| Probe | Specificity | Probe sequence (5′–3′) | Target sitea 16S rRNA positions | % FAb | Source or reference |
|---|---|---|---|---|---|
| ALT1413 | Alteromonas, Colwellia | TTTGCATCCCACTCCCAT | 1413–1430 | 40 | This study |
| G Rb | Rhodobacter, Roseobacter | GTCAGTATCGAGCCAGTGAG | 645–626 | 30 | 18 |
| G V | Vibrio | AGGCCACAACCTCCAAGTAG | 841–822 | 30 | 18 |
| NOR1-56 | NOR1 lineage | TTACCGCTCGGACTTGCA | 56–73 | 20 | This study |
| NOR2-1453 | NOR2 cluster group A | GGTCATCGCCATCCCC | 1453–1468 | 30 | This study |
| OCE232 | Oceanospirillum | AGCTAATCTCACGCAGGC | 232–249 | 40 | This study |
| PSA184 | Pseudoalteromonas, Colwellia | CCCCTTTGGTCCGTAGAC | 184–210 | 30 | This study |
| SAR86-1249 | SAR 86 clusterc | GGCTTAGCGTCCGTCTG | 1249–1265 | 50 | This study |
| SPH120 | Sphingomonas | GGGCAGATTCCCACGCGT | 120–137 | 30 | 36 |
Nucleotide sequence accession numbers.
The 16S rDNA sequences of the isolates and clones generated in this study were deposited in GenBank under accession numbers AF172840, AF173962 to AF173976, AF235107 to AF235131, AF239705 to AF239707, AF241653, and AF241654.
RESULTS
Diversity of isolated strains.
In September and November 1997 and February and August 1998, a total of 145 strains were isolated from North Sea surface water. Initial screening by FISH showed the hybridization of 9 with probe CF319a, 11 with ALF968, 110 with GAM42a, and 15 with none of the group-specific probe but only with EUB338. Sequencing and phylogenetic analysis of the latter 15 strains revealed that 1 was related to gram-positive bacteria with a high G+C DNA content (Arthrobacter spp.) and two strains were affiliated with epsilon proteobacteria (Arcobacter spp.). The remaining 12 strains were found to be gamma proteobacteria of the genera Pseudoalteromonas (10 strains) and Alteromonas (2 strains). Representatives from two out of three Pseudoalteromonas ARDRA patterns (10 isolates) and from one out of five Alteromonas ARDRA patterns (2 isolates) were not detected by probe GAM42a, although they are gamma proteobacteria. Sequencing of the 23S rDNAs of two representatives of each group revealed a single base change from C to T at position 1032 in helix 42 (31) (data not shown), which is the target site of probe GAM42a.
Subsequently, selected clones of each ARDRA pattern were sequenced. Altogether, 95 (35 nearly complete and 60 partial) 16S rRNA sequences of isolated strains were determined, including at least one full and several partial sequences for each ARDRA pattern. Identical HaeIII ARDRA patterns exhibit highly similar full-length or partial 16S rDNA sequences. This was experimentally verified for frequent isolates, i.e., the NOR2 cluster and the genera Vibrio and Oceanospirillum (data not shown). Comparative sequence analysis indicated that 142 of 145 strains were closely related to known marine bacteria (16S rRNA similarity, >93%; Table 2). Only three strains grouping within two gamma-proteobacterial clusters (referred to as NOR3 and NOR4 in Table 2) have no known close cultured relative. There was evidence of an effect of filtration and changes in cultivation conditions on the species composition of isolates. Prefiltration of water with a 1.2-μm filter favored Oceanospirillum spp. (five out of nine isolates) and Arcobacter spp. (two out of two). Inoculations with unfiltered water and water prefiltered with a 5-μm-cutoff filter predominantly resulted in isolation of strains related to Roseobacter spp. (6 out of 8), Sphingomonas spp. (2 out of 3), Vibrio spp. (15 out of 15), Pseudoalteromonas spp. (24 out of 29), Alteromonas spp. (18 out of 18), and the gamma-proteobacterial cluster NOR2 (23 out of 31). No preference concerning variations in cultivation conditions (pH, salinity, monomers or polymers, availability of vitamins and trace elements) was observed, except for Roseobacter, which was isolated only at pH 8.3 and 35‰ salinity. Attempts to increase the cultivation of strains affiliated with Cytophaga-Flavobacterium by using a mixture of polymers as the substrate were unsuccessful. The diversity of the isolates was further evaluated by ARDRA and rarefaction analysis (Fig. 1) as described by Ravenschlag et al. (43). From a total of 145 isolates, 32 HaeIII patterns were distinguished. The 122 gamma proteobacteria grouped into 21 different patterns (Fig. 1). Rarefaction indicated saturation of the number of ARDRA patterns within this group, but not for all of the isolates (Fig. 1).
TABLE 2.
Frequency and phylogenetic affiliations of North Sea isolatesa
| Phylogenetic group | No. (%) of isolates | Next relative to isolatesb | % 16S rRNA similarity |
|---|---|---|---|
| Bacteria | 145 (100) | ||
| Cyt/Flac | 9 (6.2) | ||
| Flexibacter | 2 | Cytophaga marinoflavaM58770 | 98.0 |
| 2 | Marine bacterium strain E110 AF052742 | 96.0 | |
| 2 | C. uliginosaM28238 | 93.6 | |
| 1 | Melosira-colonizing bacterium strain IC166 | 97.3 | |
| 1 | Flavobacterium salegenesM92279 | 92.7 | |
| Cytophaga johnsonae | 1 | Flavobacterium columnareM58781 | 96.2 |
| High GC, gram positive | 1 (0.7) | ||
| Arthrobacter | 1 | Micrococcus luteusM38242 | 99.3 |
| α-Proteobacteria | 11 (7.6) | ||
| Roseobacter | 8 | ||
| Group A | 7 | R. algicola | 94.0 |
| Group B | 1 | Unidentified alpha proteobacterium strain AF022392 | 95.7 |
| Sphingomonas | 3 | Sphingomonas sp. U85838 | 93.5 |
| ɛ-Proteobacteria | 2 (1.4) | ||
| Arcobacter | 2 | A. nitrofigilisL14627 | 98.6 |
| γ-Proteobacteria | 122 (84.1) | ||
| Pseudoalteromonas | 29 | P. atlanticaX82134 | 99.7 |
| Vibrio | 15 | V. splendidusZ31659 | 99.0 |
| Alteromonas | 18 | ||
| Group A | 10 | A. macleodiiX82145 | 97.8 |
| Group B | 8 | Colwellia psychroerythrusAB011364 | 94.5 |
| Oceanospirillum | 9 | O. commune ATCC 27118 | 95.7 |
| NOR1 | 2 | Unculturable Mariana eubacterium clone D87345 | 98.6 |
| NOR2 | 31 | ||
| Group A | 22 | Facultative barophile strain CNTP3 U91588 | 95.6 |
| Group B | 9 | Pseudoalteromonas haloplanktisD11172 | 92.9 |
| NOR3 | 2 | Unidentified gamma proteobacterium clone AB004573 | 94.0 |
| NOR4 | 1 | Clone of aggregate 47 L10949 | 87.0 |
| Marinobacter | 4 | Marinobacter sp. strain PCOB-2 AJ000647 | 98.1 |
| Halomonas | 9 | H. variabilisU85873 | 98.2 |
| Photobacterium | 1 | Photobacterium sp. strain SS9 U91586 | 95.2 |
| Shewanella | 1 | S. putrefaciensU91593 | 96.1 |
Obtained in September and November 1997 and February and August 1998.
In the ARB database.
Cytophaga-Flavobacterium cluster.
FIG. 1.
Rarefaction curves for the different ARDRA patterns of all of the isolates used in this study. The expected number of ARDRA patterns is plotted versus the number of isolates (○). Rarefaction curves were also calculated for the fraction of gamma Proteobacteria (●). The dotted lines represent 95% confidence intervals.
16S rDNA clones.
Fifty-four 16S rDNA clones were randomly selected for sequencing and phylogenetic analysis. Sequences belonged to several clusters: alpha-proteobacterial cluster SAR116, described by Mullins et al. (35), from Sargasso Sea samples; Roseobacter spp.; four gamma-proteobacterial lineages; and the epsilon-proteobacterial genus Arcobacter. The most frequent sequences in the clone library (44 of 53) were affiliated with the SAR86 cluster of gamma proteobacteria, first described by Mullins et al. (35) and Fuhrman et al. (16), from the Atlantic and Pacific Oceans, respectively (Table 3). We did not screen more clones, since the library was apparently biased toward the SAR86 cluster.
TABLE 3.
Frequencies and phylogenetic affiliations of North Sea 16S rRNA clonesa
| Phylogenetic group | No. (%) of clones | Next relative to cloneb | % 16S rRNA similarity |
|---|---|---|---|
| Bacteria | 54 (100) | ||
| α-Proteobacteria | 4 (7.5) | ||
| SAR116 clusterc | 3 | Unidentified alpha proteobacterium clone OM25d | 95.5 |
| Roseobacter | 1 | Marine bacterium strain SFR1e | 97.9 |
| γ-Proteobacteria | 49 (92.5) | ||
| SAR86 clusterc | 44 (83) | ||
| Group A | 17 | Unidentified gamma proteobacterium clone OM10d | 97.8–99.2 |
| Group B | 27 | Unidentified gamma proteobacterium clone OCS5 AF001651d | 96.9–98.1 |
| Thiomicrospira | 2 | Coxiella burnetiiD89795 | 88.4 |
| Unknown affiliation | 2 | Clone of macroaggregate clone 44f | 99.2 |
| Unknown affiliation | 1 | Unidentified gamma proteobacterium clone OM60d | 95.3 |
| ɛ-Proteobacteria | 1 (2) | ||
| Arcobacter | 1 | A. butzleri | 92.2 |
Oligonucleotide probe design.
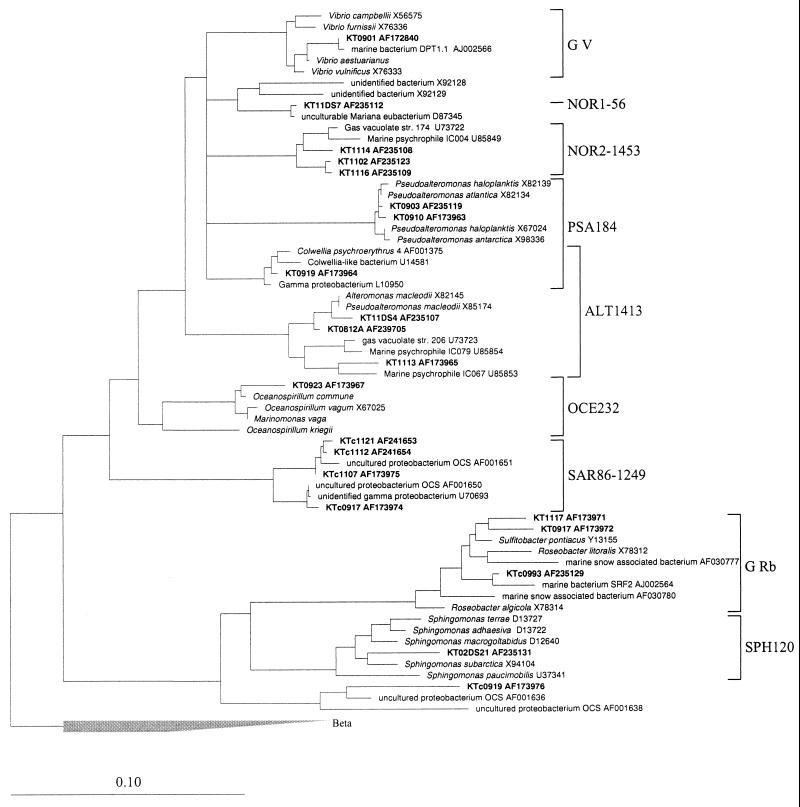
Probes were designed for some of the most abundant gamma-proteobacterial sequences obtained from isolates and direct 16S rDNA sequence retrieval. Probes ALT1413 and PSA184 target Alteromonas and Pseudoalteromonas spp., OCE232 targets Oceanospirillum spp., NOR1-56 targets the NOR1 lineage, SAR86-1249 targets the SAR86 cluster, and NOR2-1453 targets all members of the NOR2 clusters (Table 1). All of the probes have at least one strong central mismatch with a nontarget sequence (1.4 to 2.0 weighted mismatches) (37), with the exception of probe OCE232, which has only a weak mismatch (0.2 weighted mismatches) with Methylomicrobium album and M. agile. Optimized hybridization conditions are given in Table 1. In addition, we adapted for FISH the oligonucleotide probes G V and G Rb designed by Giuliano et al. (18), which are targeted to marine Vibrio spp. and to Roseobacter and Rhodobacter spp., respectively. These two probes encompass all 16 isolates affiliated with Vibrio spp. and all eight strains of Roseobacter spp.
The target groups of all of the probes are shown in the phylogenetic tree of full 16S rDNA sequences in Fig. 2. This tree was calculated only on the basis of nearly full-length sequences and was corrected by taking into consideration the different results of the various tree reconstruction algorithms. Bifurcations indicate branchings which appeared stable and well separated from neighboring branchings in all cases. Multifurcations indicate tree topologies which could not be significantly resolved based on the available data set.
FIG. 2.
Phylogenetic tree based on comparative analysis of 16S rDNA from selected clones and isolates of the alpha and gamma subclasses of Proteobacteria. Brackets indicate probe specificity. Selected sequences from the beta subclass of Proteobacteria were used to root the tree. The bar indicates 10% sequence divergence.
FISH of plankton samples.
For the samplings in September and November 1997, as well as February and August 1998, total bacterioplankton cell numbers and percentages of cells hybridizing with specific probes were determined (Table 4). The total cell numbers in the four samples were between 1.2 × 105 (February 1998) and 1.1 × 106 (September 1997) cells per ml. In a similar manner, the rate of detection by FISH varied during the year. Only 31% of DAPI-stained cells hybridized with the general bacterial probe EUB338 in November 1997. This rate increased to a maximum of 71% in August 1998.
TABLE 4.
Abundances of various phylogenetic groups in the North Sea as determined by FISH
| Datea collected | Total cell concn (105 ml−1) | % Detected by DAPI staining and probe:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EUB338 | ALF968 | GAM42a | CF319a | G Rb | ALT1413 | SAR86-1249 | G V | OCE232 | PSA184 | NOR1-56 | NOR2-1453 | SPH120 | ||
| 11.02.98 | 1.2 | 58.9 | 24.8 | 5.9 | 17.5 | 3.0 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| 20.08.98 | 8.0 | 71.4 | 15.1 | 9.2 | 30.3 | 9.2 | <1 | 10.2 | <1 | NDb | ND | <1 | <1 | <1 |
| 12.09.97 | 10.5 | 60.3 | 23.7 | 8.0 | 25.0 | 4.7 | <1 | 1.7 | 1.0 | <1 | <1 | <1 | <1 | <1 |
| 13.11.97 | 3.5 | 31.2 | 2.9 | 6.3 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
The day, month, and year are reported.
ND, not determined.
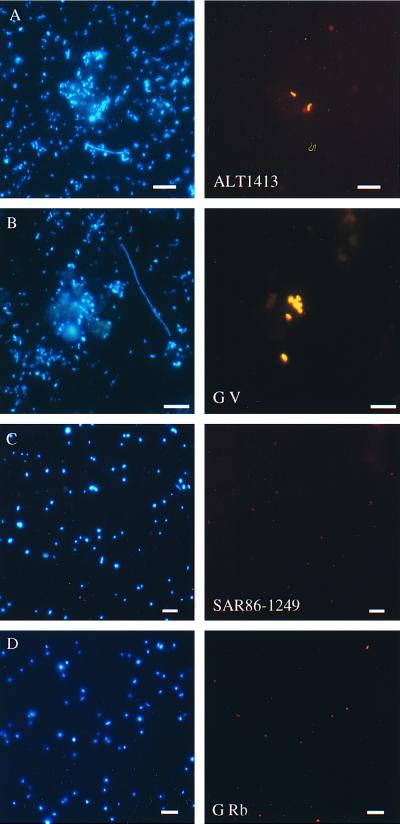
Bacteria hybridizing with group-specific probes CF319a, ALF968, and GAM42a were found to be abundant (Table 4). They made up more than half (54%) of all of the cells in August 1998. The most abundant bacteria detected in February 1998 were the alpha proteobacteria, with a maximum of 25%. In August 1998, members of the Cytophaga-Flavobacterium cluster constituted 30% of all of the cells collected and alpha proteobacteria constituted 15%. In September 1997, the percentages of alpha proteobacteria and members of the Cytophaga-Flavobacteria cluster were more or less equal at 24 and 25% of the DAPI counts, respectively. Probe G Rb for the alpha-proteobacterial genera Rhodobacter and Roseobacter hybridized with a significant fraction of the cells detected by probe ALF968. This group was most prominent during August 1998, when it constituted 9% of the total community or 60% of the ALF968 counts. Gamma proteobacteria were detected in a more or less constant fraction of 6 to 9% of the total cells in all of the samples examined. Using genus- and cluster-specific probes, we examined the abundances of readily culturable bacteria of the genera Vibrio, Oceanospirillum, Alteromonas, Pseudoalteromonas, and Sphingomonas and the NOR1 and NOR2 clusters. These never accounted for more than 1% of the total counts. Interestingly, cells detected by G V, ALT1413, PSA184, NOR1-56, and NOR2-1453 were generally large and frequently associated with small clusters of bacteria or particles. Their strong fluorescence with the probes indicated high rRNA contents of cells (Fig. 3). Members of the as yet uncultured SAR86 cluster were much smaller rods (1 to 2 by 0.5 μm) and were less fluorescent (Fig. 3). They showed a maximum in August 1998, when they represented 10% of the total cell numbers.
FIG. 3.
Epifluorescence micrographs of bacteria in bacterioplankton from the North Sea station Helgoland Roads. Hybridization with CY3-labeled probes (right) and the same microscopic field with UV excitation (DAPI staining, left). Panels: A, probe ALT1413 (18.08.98); B, probe G V (12.09.97); C, probe SAR86-1249 (20.08.98); D, probe G Rb (12.09.97). Scale bars, 5 μm.
DISCUSSION
The cultivation of microorganisms in combination with methods based on the 16S rRNA approach (4) gave insight into the culturable diversity and community structure of North Sea bacterioplankton at various seasons. Numerous isolates were screened and phylogenetically identified, thus assessing culturable diversity. Additional phylotypes were detected by cloning from environmental DNA, and the abundances of isolated bacteria and other phylogenetic groups were determined by FISH.
Culturable bacteria with high in situ abundance.
In our study, we found two groups of abundant marine bacteria to be culturable. Members of the genus Roseobacter, formerly Erythrobacter, have been frequently isolated from marine samples as aerobic, heterotrophic, often pigmented bacteria (50). Sequences related to this genus have also been routinely found in marine clone libraries (16, 18, 35, 42, 55). A study by González and Moran (21) suggested high abundance in coastal seawater of a large phylogenetic branch of marine alpha proteobacteria, including Roseobacter spp. In our study, we obtained isolates of Roseobacter spp. with oligotrophic medium, found clones related to this group in our North Sea 16S rDNA library, and identified up to 9% of the total counts with probe G Rb. This group was most abundant in the summer, when absolute numbers approach 105/ml.
High numbers of members of the Cytophaga-Flavobacterium cluster, from which we obtained nine isolates, can be identified by probe CF319a in North Sea bacterioplankton throughout the year. Our failure to retrieve them in the North Sea 16S rDNA library is likely due to a primer bias (20), or the number of clones examined may have been too low. Using other eubacterial primers (28), e.g., 8F and 1492R, Cytophaga and relatives have been cloned from macroaggregate samples in the Santa Barbara Channel (12) and from a Bermuda site (16).
Molecular techniques suggest low in situ abundance of frequently isolated bacterial groups.
Our extensive cultivation attempts, which included the use of defined artificial medium, enrichments in dilution series (48), and direct plating (60), proved to be highly selective for gamma proteobacteria. Most isolates were closely related to well-known gamma-proteobacterial genera such as Pseudoalteromonas, Alteromonas, Vibrio, and Oceanospirillum. We also obtained several gamma-proteobacterial clusters for which we had no close relatives in our 16S rRNA database (NOR1-4; Table 2). FISH with probes targeting the different genera and clusters of culturable gamma proteobacteria never detected more than 1% of the total counts. Except for Oceanospirillum spp., the cells detected were large, were attached to particles, and had high cellular rRNA contents. The predominant occurrence of Alteromonas and Pseudoalteromonas spp. as attached bacteria had been suggested before (1, 12). This was supported not only by our FISH data but also by the lack of isolation of these bacteria from the <1.2-μm fraction.
The genus Vibrio, one of the best-known marine taxa, was once claimed to be a major component of the bacterial flora of the sea and to account for nearly 80% of the bacterial community in surface waters of the western Pacific Ocean (52). Likewise, hybridization of community DNA with oligonucleotide probes targeting 16S rDNAs of culturable bacteria suggested the dominance of gamma proteobacteria, in particular, Vibrio and Photobacterium spp. (44). Yet, these bacteria could not be detected in situ in high numbers in our study and related sequences were not frequent in our 16S rDNA library. Our results thus suggest that these hybridizations to extracted community DNA overestimated the abundances of particular species.
The good growth on agar plates of some gamma proteobacteria is most likely the result of their specific life strategies, which have been studied in detail for Vibrio spp. (5, 38, 39). These marine bacteria, which survive carbon starvation for extended periods of time, can grow rapidly at high substrate concentrations with high cellular rRNA contents. It has been shown that upon the onset of carbon starvation a Vibrio strain maintains ribosomes for several days in large excess over the apparent demand for protein synthesis (14). The cells of Vibrio spp. we detected in situ were all particle attached. Particles are sites of higher nutrient availability, and the large size and high ribosome content of the cells detected could be the result of recent metabolic activity (45). In addition, colonies of Vibrio spp. were shiny, which is a characteristic of bacteria producing extracellular slime. Cells with the ability to produce a protective matrix seem to colonize surfaces at the solid-air interface more readily than bacteria that lack this feature (2).
We also isolated three strains of Sphingomonas spp. Abundances of 15 to 35% have been reported for this alpha-proteobacterial genus (48). In our North Sea samples, we obtained no FISH counts above the background with SPH120, a 16S rRNA-targeted probe for sphingomonads (36). As in the case of the culturable gamma proteobacteria, FISH data alone are insufficient to decide whether the probe target groups were absent or undetectable due to low rRNA contents (47).
Members of the abundant SAR86 cluster remain uncultured.
FISH identified up to 10% of the total cells as members of the SAR86 cluster. The small rods were usually not attached to particles, confirming earlier reports that they belong to the free-living fraction of bacterioplankton (1). Sequences related to the SAR86 cluster dominated our North Sea 16S rDNA clone library (Table 3). Nevertheless, no strains affiliated with the SAR86 cluster were among the culturable gamma proteobacteria. Rarefaction analysis of ARDRA patterns (Fig. 1) indicated a low probability of discovering new groups of pelagic gamma proteobacteria by analysis of additional North Sea isolates. After the screening of 70 isolates, 18 ± 3 (average ± 95% confidence interval) ARDRA groups were identified and after the screening of 52 additional isolates, only 3 ± 0.3 new patterns were found. Less than one new ARDRA pattern is predicted for the screening of an additional 20 gamma-proteobacterial isolates. We therefore stopped our efforts to cultivate SAR86.
With regard to the in situ abundance and culturability of heterotrophic marine bacteria, we have evidence for three groups: (i) abundant groups that are culturable, such as Roseobacter and members of the Cytophaga-Flavobacterium cluster; (ii) abundant bacteria that are still uncultured, such as members of the SAR86 cluster; and (iii) frequently isolated bacteria of the Vibrio sp. type with possibly low in situ abundance. Our findings are in obvious contrast to the more optimistic conclusions of Pinhassi et al. and Rehnstam et al. that the most abundant marine bacteria are readily culturable (40, 44).
Although our cultivation attempts failed to isolate new abundant marine pelagic bacteria, the rarefaction analysis of all of our ARDRA patterns (Fig. 1) (not just those from gamma proteobacteria) clearly indicated that further marine bacteria could have been isolated. With high-throughput molecular screening, and early rejection of laboratory weeds, future isolation efforts could be directed to groups such as the alpha proteobacteria and the Cytophaga-Flavobacterium cluster. This might provide additional good model organisms of marine aerobic heterotrophic bacteria.
Cultivation strategies.
Few new genera of marine bacteria have been cultured since ZoBell's experiments with substrate-amended seawater (23, 40, 55). Using a synthetic medium designed by Schut et al. (48), we may have extended the number of cultured marine species with the strains of our clusters NOR1 to NOR4. Sequences related to the NOR1 cluster have been found in samples from the deep Mariana trench and have been attributed to an unculturable bacterium (30). Applying an appropriate cultivation strategy, we were, however, able to obtain isolates from this phylogenetic lineage. We thus caution against the premature use of the term “unculturable” for bacteria that are only represented by their rDNA sequences in clone libraries.
We are unable to provide the chemical, nutritional, and physical prerequisites for the growth of all of the microorganisms present in natural seawater. Different marine bacteria react differently to confinement (13) and substrate quality and quantity (54). Active metabolism and multiplication might be terminated due to enrichment of toxic products, depletion of essential nutrients (53), and viral infection (58). The growth state of the bacteria at the time of sampling, whether they are active, starved, or dormant, may also strongly influence the success of cultivation.
In principle, for successful enrichments, the physiological requirements of the target microorganism should be known. Most marine bacteria face an oligotrophic environment, but the definitions of the needs of oligocarbophilic microorganisms are as diverse as they are difficult to justify (49). Not even the amount of organic carbon per liter sufficient for growth of oligocarbophilic bacteria is agreed upon, and the appropriate types of carbon sources are in dispute. It is not known if defined mixtures of monomers and polymers, undefined substrates like peptone and yeast extract, or naturally occurring substrates like DMSP (29) and algae lysate will be most suitable for the isolation of hitherto uncultured microorganisms. The quantity and quality of substrates may even play a subordinate role. Our oligotrophic medium, with 1 to 10 mg of C per liter, did not select against Vibrio spp. or Pseudoalteromonas spp., which also grow well on rich media (40, 60). These bacteria are known to resist nutrient deprivation for long periods of time and to regain active metabolism quite rapidly (5), which is a dilemma for cultivation. Strategies that attempt to prevent substrate-accelerated death (41) by initial incubation at very low substrate concentrations, followed by a gradual increase, will therefore be of little use for the isolation of slowly growing bacteria with potentially long lag phases. The role of phages in the control of CFU is also still unresolved. Bacteriophages of known microorganisms from the North Sea are very host specific and, in general, highly virulent (59). It has been suggested that many bacteria may be apparently unculturable because they are infected by lysogenic viruses (57).
A further problem could be that as yet uncultured bacteria do not form colonies at the air-solid interface. This should not be confused with the general ability to grow on surfaces or submerged particles. Future cultivation attempts could consider (i) filtration (pore size, <1.2 μm) of the inoculum to remove large, highly active, particle-associated bacteria, (ii) dilution to favor dominant bacteria (10), and (iii) colony isolation in semiliquid (soft-agar) medium and subsequent subculturing in liquid medium for bacteria unable to grow at the air-water interface.
ACKNOWLEDGMENTS
We acknowledge Steven M. Holland for providing the freeware program aRarefactWin. We thank Christian Schütt, Gunnar Gerdts, and Antje Wichels (Department of Microbiology, Biologische Anstalt Helgoland) for sampling and the use of the laboratory facility.
This work was supported by grants from the chemical industry and the Max Planck Society (Germany).
REFERENCES
- 1.Acinas S G, Antón J, Rodríguez-Valera F. Diversity of free-living and attached bacteria in offshore western Mediterranean waters as depicted by analysis of genes encoding 16S rRNA. Appl Environ Microbiol. 1999;65:514–522. doi: 10.1128/aem.65.2.514-522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison D G, Sutherland I W. The role of exopolysaccarides in adhesion of freshwater bacteria. J Gen Microbiol. 1987;133:1319–1327. [Google Scholar]
- 3.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amy P S, Morita R Y. Starvation-survival patterns of sixteen freshly isolated open-ocean bacteria. Appl Environ Microbiol. 1983;45:1109–1115. doi: 10.1128/aem.45.3.1109-1115.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowman J P, McCammon S A, Brown M V, Nichols D S, McMeekin T A. Diversity and association of psychrophilic bacteria in Antarctic Sea ice. Appl Environ Microbiol. 1997;63:3068–3078. doi: 10.1128/aem.63.8.3068-3078.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Britschgi T B, Giovannoni S J. Phylogenetic analysis of a natural marine bacterioplankton population by rRNA gene cloning and sequencing. Appl Environ Microbiol. 1991;57:1707–1713. doi: 10.1128/aem.57.6.1707-1713.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 9.Buchholz-Cleven B, Rattunde B, Straub K. Screening for genetic diversity of isolates of anaerobic Fe(II)-oxidizing bacteria using DGGE and whole-cell hybridization. Syst Appl Microbiol. 1997;20:301–309. [Google Scholar]
- 10.Button D K, Schut F, Quang P, Martin R, Robertson B. Viability and isolation of marine bacteria by dilution culture: theory, procedures, and initial results. Appl Environ Microbiol. 1993;59:881–891. doi: 10.1128/aem.59.3.881-891.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLong E F. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeLong E F, Franks D G, Alldredge A L. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol Oceanogr. 1993;38:924–934. [Google Scholar]
- 13.Ferguson R L, Buckley E N, Palumbo A V. Response of marine bacterioplankton to differential filtration and confinement. Appl Environ Microbiol. 1984;47:49–55. doi: 10.1128/aem.47.1.49-55.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flärdh K, Cohen P S, Kjelleberg S. Ribosomes exist in large excess over the apparent demand for protein synthesis during carbon starvation in marine Vibrio sp. strain CCUG 15956. J Bacteriol. 1992;174:6780–6788. doi: 10.1128/jb.174.21.6780-6788.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuhrman J A, McCallum K, Davis A A. Novel major archaebacterial group from marine plankton. Nature. 1992;356:148–149. doi: 10.1038/356148a0. [DOI] [PubMed] [Google Scholar]
- 16.Fuhrman J A, McCallum K, Davis A A. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific Oceans. Appl Environ Microbiol. 1993;59:1294–1302. doi: 10.1128/aem.59.5.1294-1302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giovannoni S J, Britschgi T B, Moyer C L, Field K G. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 18.Giuliano L, De Domenico E, Höfle M G, Yakimov M M. Identification of culturable oligotrophic bacteria within naturally occurring bacterioplankton communities of the Ligurian Sea by 16S rRNA sequencing and probing. Microb Ecol. 1999;37:77–85. doi: 10.1007/s002489900132. [DOI] [PubMed] [Google Scholar]
- 19.Glöckner F O, Amann R, Alfreider A, Pernthaler J, Psenner R, Trebesius K, Schleifer K-H. An in situ hybridization protocol for detection and identification of planktonic bacteria. Syst Appl Microbiol. 1996;19:403–406. [Google Scholar]
- 20.Glöckner F O, Fuchs B M, Amann R. Bacterioplankton composition of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl Environ Microbiol. 1999;65:3721–3726. doi: 10.1128/aem.65.8.3721-3726.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.González J M, Moran M A. Numerical dominance of a group of marine bacteria in the α-subclass of the class Proteobacteria in coastal seawater. Appl Environ Microbiol. 1997;63:4237–4242. doi: 10.1128/aem.63.11.4237-4242.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.González J M, Whitman W B, Hodson R E, Moran M A. Identifying numerically abundant culturable bacteria from complex communities: an example from a lignin enrichment culture. Appl Environ Microbiol. 1996;62:4433–4440. doi: 10.1128/aem.62.12.4433-4440.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hagström Å, Pinhassi J, Zweifel U L. Biogeographical diversity among marine bacterioplankton. In: Pinhassi J, editor. Population dynamics in marine bacterioplankton. PhD. Thesis. Umeå, Sweden: Umeå University; 1999. [Google Scholar]
- 24.Heck K L, van Belle G, Simberloff D. Expicit calculation of the rarefaction diversity measurement and the determination of sufficient sample size. Ecology. 1975;56:1459–1461. [Google Scholar]
- 25.Hobbie J E, Daley R J, Jasper S. Use of Nuclepore filters for counting bacteria by fluorescence microscopy. Appl Environ Microbiol. 1977;33:1225–1228. doi: 10.1128/aem.33.5.1225-1228.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hurlbert S H. The nonconcept of species diversity: a critique and alternative parameters. Ecology. 1971;52:577–586. doi: 10.2307/1934145. [DOI] [PubMed] [Google Scholar]
- 27.Jannasch H W, Jones G E. Bacterial populations in sea water as determined by different methods of enumeration. Limnol Oceanogr. 1959;4:128–139. [Google Scholar]
- 28.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, England: John Wiley & Sons; 1991. pp. 115–147. [Google Scholar]
- 29.Ledyard K M, DeLong E F, Dacey J W H. Characterization of a DMSP-degrading bacterial isolate from the Sargasso Sea. Arch Microbiol. 1993;160:312–318. [Google Scholar]
- 30.Li L N, Kato C, Horikoshi K. Bacterial diverity in deep-sea sediments from different depths. Biodivers Conserv. 1999;8:659–677. [Google Scholar]
- 31.Ludwig W, Schleifer K H. Bacterial phylogeny based on 16S and 23S rRNA sequence analysis. FEMS Microbiol Rev. 1994;15:155–173. doi: 10.1111/j.1574-6976.1994.tb00132.x. [DOI] [PubMed] [Google Scholar]
- 32.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer K-H. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology. 1996;142:1097–1106. doi: 10.1099/13500872-142-5-1097. [DOI] [PubMed] [Google Scholar]
- 34.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 35.Mullins T D, Britschgi T B, Krest R L, Giovannoni S J. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol Oceanogr. 1995;40:148–158. [Google Scholar]
- 36.Neef A. Ph.D. thesis. Munich, Germany: Technische Universität München; 1997. [Google Scholar]
- 37.Neef A, Zaglauer A, Meier H, Amann R, Lemmer H, Schleifer K-H. Population analysis in a denitrifying sand filter: conventional and in situ identification of Paracoccus spp. in methanol-fed biofilms. Appl Environ Microbiol. 1996;62:4329–4339. doi: 10.1128/aem.62.12.4329-4339.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nyström T, Albertson K, Flärdh K, Kjelleberg S. Physiological and molecular adaptation to starvation and recovery from starvation by the marine Vibrio sp. S14. FEMS Microbiol Ecol. 1990;74:129–140. [Google Scholar]
- 39.Nyström T, Flärdh K, Kjelleberg S. Responses to multiple-nutrient starvation in marine Vibrio sp. strain CCUG 15956. J Bacteriol. 1990;172:7085–7097. doi: 10.1128/jb.172.12.7085-7097.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pinhassi J, Zweifel U L, Hagström A. Dominant marine bacterioplankton species found among colony-forming bacteria. Appl Environ Microbiol. 1997;63:3359–3366. doi: 10.1128/aem.63.9.3359-3366.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Postgate J R, Hunter J R. Accelerated death of Aerobacter aerogenes starved in the presence of growth limiting substrates. J Gen Microbiol. 1964;34:459–473. doi: 10.1099/00221287-34-3-459. [DOI] [PubMed] [Google Scholar]
- 42.Rappé M S, Kemp P F, Giovannoni S J. Phylogenetic diversity of marine coastal picoplankton 16S rRNA genes cloned from the continental shelf off Cape Hatteras, North Carolina. Limnol Oceanogr. 1997;42:811–826. [Google Scholar]
- 43.Ravenschlag K, Sahm K, Pernthaler J, Amann R. High bacterial diversity in permanently cold marine sediments. Appl Environ Microbiol. 1999;65:3982–3989. doi: 10.1128/aem.65.9.3982-3989.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rehnstam A S, Backman S, Smith D C, Azam F, Hagström A. Blooms of sequence-specific culturable bacteria in the sea. FEMS Microbiol Ecol. 1993;102:161–166. [Google Scholar]
- 45.Rosselló-Mora R, Thamdrup B, Schäfer H, Weller R, Amann R. The response of the microbial community of marine sediments to organic input under anaerobic conditions. Syst Appl Microbiol. 1999;22:237–248. doi: 10.1016/S0723-2020(99)80071-X. [DOI] [PubMed] [Google Scholar]
- 46.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullins K B, Erlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 47.Schut F. Ph.D. thesis. Groningen, The Netherlands: University of Groningen; 1994. [Google Scholar]
- 48.Schut F, De Vries E J, Gottschal J C, Robertson B R, Harder W, Prins R A, Button D K. Isolation of typical marine bacteria by dilution culture growth maintenance and characteristics of isolates under laboratory conditions. Appl Environ Microbiol. 1993;59:2150–2160. doi: 10.1128/aem.59.7.2150-2160.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schut F, Prins R A, Gottschal J C. Oligotrophy and pelagic marine bacteria: facts and fiction. Aquat Microb Ecol. 1997;12:177–202. [Google Scholar]
- 50.Shiba T. The genus Roseobacter. In: Starr M P, Stolp H, Trüper H G, Balows A, Schlegel H G, editors. The prokaryotes. Berlin, Germany: Springer-Verlag; 1992. pp. 2156–2159. [Google Scholar]
- 51.Simberloff D. Use of rarefaction and related methods. In: Dickson K L, et al., editors. Biological data in water pollution assessment: quantitative and statistical analyses. Philadelphia, Pa: American Society for Testing and Materials; 1978. pp. 150–165. [Google Scholar]
- 52.Smidu R, Taga N, Colwell R R, Schwartz J R. Heterotrophic bacterial flora of the seawater from the Nansei Shoto (Ruyukyu Retto) area. Bull Jpn Soc Fish. 1980;46:505–510. [Google Scholar]
- 53.Stevenson L H. A case for bacterial dormancy in aquatic systems. Microb Ecol. 1978;4:127–133. doi: 10.1007/BF02014283. [DOI] [PubMed] [Google Scholar]
- 54.Straškrabová V. The effect of substrate shock on populations of starving aquatic bacteria. J Appl Bacteriol. 1983;54:217–224. [Google Scholar]
- 55.Suzuki M T, Rappé M S, Haimberger Z W, Winfield H, Adair N, Strobel J, Giovannoni S J. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl Environ Microbiol. 1997;63:983–989. doi: 10.1128/aem.63.3.983-989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsai Y-L, Olson B H. Rapid method for direct extraction of DNA from soil and sediments. Appl Environ Microbiol. 1991;57:1070–1074. doi: 10.1128/aem.57.4.1070-1074.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weinbauer M G, Suttle C A. Potential significance of lysogeny to bacteriophage production and bacterial mortality in coastal waters of the Gulf of Mexico. Appl Environ Microbiol. 1996;62:4374–4380. doi: 10.1128/aem.62.12.4374-4380.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wichels A, Biel S S, Gelderblom H R, Brinkhoff T, Muyzer G, Schütt C. Bacteriophage diversity in the North Sea. Appl Environ Microbiol. 1999;64:4128–4133. doi: 10.1128/aem.64.11.4128-4133.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu H S, Roberts N, Singleton F L, Attwell R W, Grimes D J, Colwell R R. Survival and viability of nonculturable Escherichia coli and Vibrio cholerae in the estuarine and marine environment. Microb Ecol. 1982;8:313–323. doi: 10.1007/BF02010671. [DOI] [PubMed] [Google Scholar]
- 60.ZoBell C E. Marine microbiology. A monograph on hydrobacteriology. Waltham, Mass: Chronica Botanica Company; 1946. [Google Scholar]