ABSTRACT
Cell wall peptidoglycan is a heteropolymeric mesh that protects the bacterium from internal turgor and external insults. In many rod-shaped bacteria, peptidoglycan synthesis for normal growth is achieved by two distinct pathways: the Rod complex, comprised of MreB, RodA, and a cognate class B penicillin-binding protein (PBP), and the class A PBPs (aPBPs). In contrast to laterally growing bacteria, pole-growing mycobacteria do not encode an MreB homolog and do not require SEDS protein RodA for in vitro growth. However, RodA contributes to the survival of Mycobacterium tuberculosis in some infection models, suggesting that the protein could have a stress-dependent role in maintaining cell wall integrity. Under basal conditions, we find here that the subcellular distribution of RodA largely overlaps that of the aPBP PonA1 and that both RodA and the aPBPs promote polar peptidoglycan assembly. Upon cell wall damage, RodA fortifies Mycobacterium smegmatis against lysis and, unlike aPBPs, contributes to a shift in peptidoglycan assembly from the poles to the sidewall. Neither RodA nor PonA1 relocalize; instead, the redistribution of nascent cell wall parallels that of peptidoglycan precursor synthase MurG. Our results support a model in which mycobacteria balance polar growth and cell-wide repair via spatial flexibility in precursor synthesis and extracellular insertion.
IMPORTANCE Peptidoglycan synthesis is a highly successful target for antibiotics. The pathway has been extensively studied in model organisms under laboratory-optimized conditions. In natural environments, bacteria are frequently under attack. Moreover, the vast majority of bacterial species are unlikely to fit a single paradigm of cell wall assembly because of differences in growth mode and/or envelope structure. Studying cell wall synthesis under nonoptimal conditions and in nonstandard species may improve our understanding of pathway function and suggest new inhibition strategies. Mycobacterium smegmatis, a relative of several notorious human and animal pathogens, has an unusual polar growth mode and multilayered envelope. In this work, we challenged M. smegmatis with cell wall-damaging enzymes to characterize the roles of cell wall-building enzymes when the bacterium is under attack.
KEYWORDS: cell envelope, cell wall, mycobacteria, peptidoglycan, stress response, tuberculosis
INTRODUCTION
Bacterial cell wall peptidoglycan is required for viability in most species under most conditions (1). Although peptidoglycan synthesis has been extensively studied, much of this work has been done under idealized growth conditions that do not reflect the variety of stressors found in the natural environment. Outside of the laboratory, the bacterial cell wall is under constant attack. In virtually all environments, competitors, predators, and unwilling hosts challenge bacteria with peptidoglycan-hydrolyzing enzymes (1–5). However, mechanisms to counteract cell wall damage are poorly defined. Studying peptidoglycan synthesis and remodeling under nonoptimal stress conditions may lead to a better understanding of pathogenesis and ecologically relevant pathways and interactions.
In laterally growing, rod-shaped organisms like Escherichia coli and Bacillus subtilis, the combined activity of two distinct peptidoglycan polymerization pathways ensures cell wall integrity during normal growth as well as hostile conditions. The final, lipid-linked peptidoglycan precursor lipid II is synthesized in the inner leaflet of the plasma membrane by MurG and then flipped to the outer leaflet by MurJ (6) and inserted into the existing cell wall by the action of transglycosylases and transpeptidases. In one pathway, two dedicated enzymes work as a cognate pair, SEDS family transglycosylase RodA (7, 8) and a monofunctional, class B penicillin-binding protein (bPBP) transpeptidase (9, 10). Along with cytoskeletal protein MreB, these proteins make up the Rod complex. This essential pathway contributes to elongation and rod-shape homeostasis by directed motion around the cell (10–13). A second pathway, nonessential in some organisms, utilizes bifunctional, class A PBPs (aPBPs) that perform both transglycosylation and transpeptidation (14, 15), move diffusively, and are thought to maintain and repair the cell wall (16–21). Despite a growing body of evidence suggesting that aPBPs are important for stress response while the Rod complex contributes to normal growth, there are also reports that Rod complex components can sense and respond to stress (22–24).
While RodA and its cognate bPBP are more conserved than the aPBPs (25–27), they are not found in all bacterial species (28). Even when they are encoded in the genome, RodA and its bPBP are not always essential for viability nor are they always associated with MreB. For example, mycobacteria and related organisms lack MreB and do not rely on RodA for in vitro growth (29–33). Individual aPBPs are also largely dispensable for in vitro growth in this genus, with Mycobacterium smegmatis PonA1 being a notable exception (9, 34–36). Why have these organisms retained enzymatically redundant systems for peptidoglycan synthesis? One clue may arise from work with the human pathogen Mycobacterium tuberculosis, where RodA and the aPBPs individually contribute to survival in immune cells, some mouse backgrounds, and in a guinea pig model (29, 37–41). These observations suggest that RodA and the aPBPs play unique roles in protecting mycobacteria from stress.
Another way that the mycobacterial cell wall differs from those of model organisms is its polar mode of elongation. Cell wall damage from external sources poses a spatial challenge to pole-growing bacteria, as it presumably is not confined to the normal sites of active peptidoglycan metabolism. We previously found that treatment with the peptidoglycan-digesting enzymes lysozyme and mutanolysin causes nascent cell wall in M. smegmatis to shift from the poles to the sidewall (42). Here, we show that M. smegmatis RodA and PonA1 largely overlap in localization and activity. Upon cell wall damage, peptidoglycan synthesis is redistributed from the pole to the sidewall. Neither RodA nor PonA1 relocalize in a manner that corresponds to this shift; instead, the redistribution of nascent cell wall correlates with that of peptidoglycan precursor synthase MurG. Although not essential for growth under normal laboratory conditions, RodA has a nonredundant role in damage-induced relocalization of cell wall synthesis and protects M. smegmatis from lysis under this condition. Our data support a model in which the location of precursor synthesis and use of specific transglycosylases can be tuned for growth or repair.
RESULTS
Substantial overlap in PonA1 and RodA localization under basal conditions.
In other organisms, aPBPs are hypothesized to contribute to cell wall integrity and the Rod complex is hypothesized to contribute to cell wall elongation (9, 10, 16, 17, 22, 43, 44). If this division of labor is true in pole-growing mycobacteria, we hypothesized that RodA may be more polar than aPBPs like PonA1. To test this hypothesis, we sought to compare the subcellular localization of fluorescent protein fusions to PonA1 and RodA. We previously confirmed the functionality of an monomeric Red Fluorescent Protein (mRFP) fusion to PonA1, an essential protein in M. smegmatis, by allele swapping (45). Here, we used the reduced cell length phenotype associated with rodA deletion (29) to test and confirm the functionality of our RodA-mRFP construct (see Fig. S1 in the supplemental material). We also showed that the fusion protein is membrane bound, as expected, and primarily detected as full length (Fig. S2).
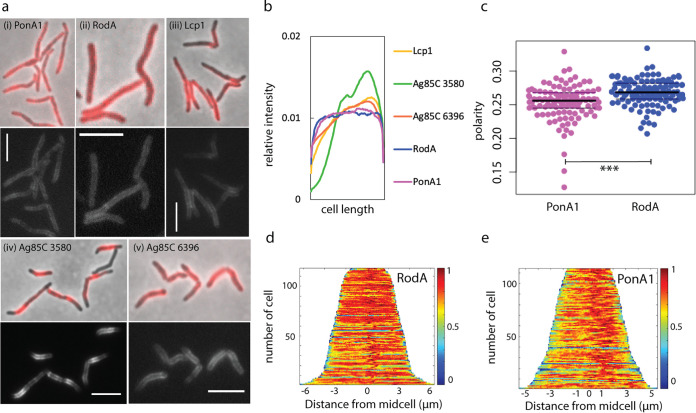
Under basal conditions, we found that RodA-mRFP and, as we and others previously reported, PonA1-mRFP are distributed along the perimeter of the mycobacterial cell (45, 46) (Fig. 1a and b). Neither enzyme showed clear polar enrichment, but RodA-mRFP localization extended further toward the poles than did that of PonA1-mRFP (Fig. 1b and c). mRFP fusions to extracellular synthetic enzymes for other layers of the mycobacterial cell envelope, including the arabinogalactan phosphotransferase Lcp1 (47) and two mycolyltransferases, MSMEG_3580 and MSMEG_6396 (48), showed very different patterns of localization from the peptidoglycan synthetic enzymes, indicating that the subcellular localization patterns of RodA-mRFP and PonA1-mRFP are specific (Fig. 1a and b). These data suggest that the cell-wide distributions of the proteins largely overlap, with RodA-mRFP slightly more polar than PonA1-mRFP.
FIG 1.
RodA and PonA1 localize around the cell periphery under basal conditions. (a) Enzymes involved in peptidoglycan (i and ii), arabinogalactan (iii), and mycomembrane (iv and v) synthesis were fused to mRFP and imaged at an exposure of 5 s (i) or 4 s (ii to v). Scale bars, 5 μm. (b) Normalized mean fluorescence intensity profiles for mRFP-tagged enzymes. Fluorescence signals of each nonseptating cell were detected using Oufti and analyzed using MATLAB. Each cell was divided into 100 fragments, with each assigned an intensity value to normalize for cell length. Fragments for all cells were aligned to generate a mean intensity value for each fragment. These values were divided by the mean total cell fluorescence. 93 < n < 121. (c) Localization of RodA-mRFP and PonA1-mRFP was compared by calculation of the polarity ratio as a signal from both poles divided by the total cell fluorescence. t test, P < 0.001. (d and e) Normalized demographs. 112 < n < 118 (number of cells analyzed).
aPBPs and RodA both promote polar cell wall synthesis.
Using a variety of metabolic labeling probes, we and others have found that active cell wall metabolism in mycobacteria occurs in asymmetric polar gradients (42, 46, 49–54). To test whether the modest difference in PonA1-mRFP and RodA-mRFP localization (Fig. 1) reflected their sites of activity, we labeled nascent cell wall using the dipeptide probe alkyne-d-alanine-d-alanine (55, 56). We previously showed that this probe incorporates into lipid-linked peptidoglycan precursors in M. smegmatis and therefore is a readout for cell wall biosynthesis in this species (42). To visualize aPBP activity, we labeled a rodA knockout mutant. To enrich for RodA activity, we reduced aPBP activity by moenomycin, which specifically inhibits transglycosylation by aPBPs but not by RodA (8, 57–60).
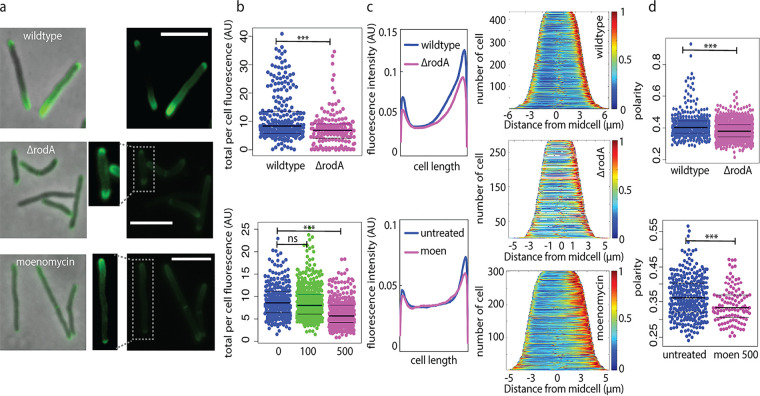
The overall amount of cell wall labeling from ΔrodA or moenomycin-treated wild-type cells was reduced compared to that of the untreated wild type (Fig. 2a and b). In the absence of RodA, labeling was reduced along the sidewall and, to an even greater extent, at the poles, such that there was a net decrease in the polarity of cell wall synthesis (Fig. 2c and d). We observed a similar phenotype when alkyne-d-alanine-d-alanine was detected by click chemistry ligation to a different fluorophore (Fig. S3). Inhibition of transglycosylation by aPBPs also resulted in a labeling decrease along the sidewall and, to a greater extent, at the poles (Fig. 2c and d). These data suggest that under basal conditions, RodA and aPBPs both contribute to polar cell wall synthesis.
FIG 2.
RodA and aPBPs promote polar peptidoglycan synthesis under basal conditions. (a) Wild-type M. smegmatis, ΔrodA M. smegmatis, and M. smegmatis cells treated with 500 μg/mL moenomycin for 30 min were incubated with alkyne-d-alanine-d-alanine for 10 min (~5 to 6% generation time). Probe was detected by click chemistry ligation to picolyl azide AF488. All images were acquired at 1 s of exposure. Dim signal from boxed cells was enhanced for visibility. Scale bars, 5 μm. (b) Total fluorescence per cell. Moenomycin was added at 100 or 500 μg/mL. Significance was determined using t test (top) or analysis of variance (ANOVA), followed by a Tukey post hoc test to conduct pairwise comparisons (bottom). ***, P < 0.001. AU, arbitrary units. (c) Fluorescence signal localization represented by mean raw fluorescence intensity profile from nonseptating cells detected using Oufti and analyzed using MATLAB (each cell was divided into 100 fragments, with each assigned an intensity value to normalize for cell length) (left) and demographs (right). (d) Polarity ratios. Moen 500, 500 μg/mL moenomycin treatment. t test, P < 0.001 for 168 < n < 433 (number of cells analyzed).
Mutanolysin/lysozyme-mediated cell wall damage occurs along the cell periphery.
We previously showed that when M. smegmatis cells are treated with a combination of lysozyme and mutanolysin, nascent peptidoglycan redistributes from the poles to the sidewall (42). These enzymes are glycoside hydrolases and break the linkages that connect neighboring glycans N-acetylglucosamine and N-acetylmuramic acid in the peptidoglycan backbone (Fig. S4) (3–5). We hypothesized that cell wall assembly can shift to places where the cell wall is damaged, presumably for repair. Implicit in this hypothesis is the assumption that enzymes added to the bacterial growth medium attack the cell wall indiscriminately, i.e., without preference for poles versus sidewall. In support, a previous scanning electron microscopy study showed that lysozyme-associated surface irregularities occur along the entire M. smegmatis cell periphery (61). We also observed that wild-type M. smegmatis treated with lysozyme and mutanolysin often has bumps around the cell, which we interpret as areas of weakened peptidoglycan (Fig. S4), and that loss of fluorescently labeled cell wall occurs along the sidewall (Fig. S4).
MurG relocalizes to the sidewall in response to cell wall damage, but PonA1, RodA, and DivIVA do not.
We next considered what element(s) of cell wall assembly machinery might be responsible for redistributing peptidoglycan synthesis from sites of polar growth to sites of sidewall damage. After treatment with mutanolysin/lysozyme, we initially found that the localization of RodA-mRFP, and, to a lesser extent, PonA1-mRFP, became more polar (Fig. S5). This was unexpected, since nascent cell wall localization shifts in the opposite direction, i.e., becomes less polar (42). However, when we stained enzyme-treated cells with SYTOX green, a dye that preferentially labels dead bacteria, we did not observe any viable cells with relocalized RodA-mRFP (Fig. 3a; Fig. S6). While we do not yet understand this phenotype, quantification of RodA-mRFP fluorescence from SYTOX green-negative cells suggests that cell wall damage likely does not change the localization of extracellular synthesis proteins (Fig. 3b).
FIG 3.
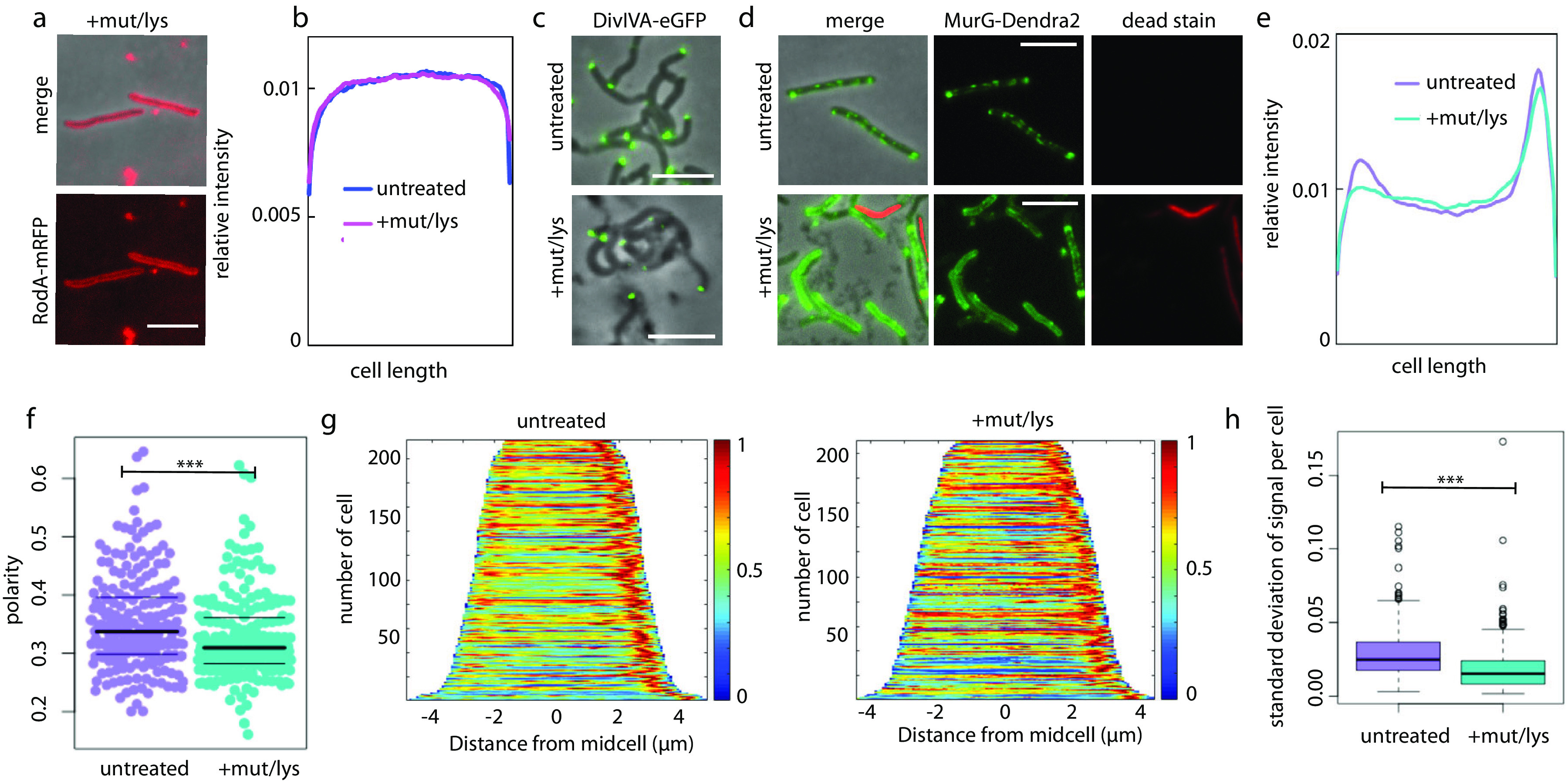
MurG relocalizes upon cell wall damage. (a) RodA-mRFP remains cell-wide in viable cells treated with mutanolysin/lysozyme. Scale bars, 5 μm. (b) Mean fluorescence intensity profiles of RodA-mRFP were generated as described in the legend to Fig. 1. Cells expressing RodA-mRFP were treated with or without mutanolysin/lysozyme and then stained with SYTOX green. Only cells that did not stain green, i.e., were deemed viable, were included in Oufti followed by MATLAB analysis. 58 < n < 116. (c) Cells expressing the DivIVA-eGFP (52) were treated with or without mutanolysin/lysozyme. Scale bars, 5 μm. (d) Cells expressing the functional fusion MurG-Dendra2 (45) were treated with or without mutanolysin/lysozyme and then stained with propidium iodide for detection of dead cells. Three-second exposure for the green channel; 500-ms exposure for the red channel. Scale bars, 5 μm. (e) Normalized MurG-Dendra2 mean fluorescence intensity profiles. (f) Polarity ratio of MurG-Dendra2 signal. t test, P < 0.001. (g) MurG-Dendra2 demographs. (h) Standard deviation calculated for 100 fluorescence values per cell in untreated and treated cells. t test, P < 0.001. 210 < n < 215 (number of cells analyzed).
In contrast to well-studied, rod-shaped species, mycobacteria coordinate cell wall synthesis via the cytoskeletal protein DivIVA (Wag31) rather than MreB (52, 62–64). We wondered whether DivIVA might also organize cell wall synthesis in response to sidewall damage. However, the location of the functional fluorescent protein fusion DivIVA-eGFP (42, 52, 64), like those of PonA1 and RodA, did not change upon mutanolysin/lysozyme treatment (Fig. 3c).
We next turned our attention to the source of peptidoglycan precursor substrates for the aPBPs and RodA. Synthesis of the final precursor lipid II is carried out by MurG. Accordingly, we treated cells expressing the functional fluorescent protein fusion MurG-Dendra2 (45) with mutanolysin/lysozyme. Imaging of cells before and after treatment revealed that MurG-Dendra2 signal transitions from a predominantly subpolar and patchy signal to a more uniform signal that often surrounds the entire periphery of the cell (Fig. 3d). Because relocalization of RodA-mRFP was observed only in dead cells, we stained both untreated and treated cells with propidium iodide, another dye that preferentially labels dead bacteria. After omission of propidium iodide-stained cells from our analysis, fluorescence quantitation supported our observation that MurG-Dendra2 relocalizes away from the polar region upon cell wall damage (Fig. 3e to g) and that it becomes less patchy (Fig. 3h).
Taken together, our data indicate that MurG, but not PonA1, RodA, or DivIVA, relocalizes upon cell wall damage.
RodA, but not aPBPs, contributes to redistribution of peptidoglycan synthesis upon cell wall damage.
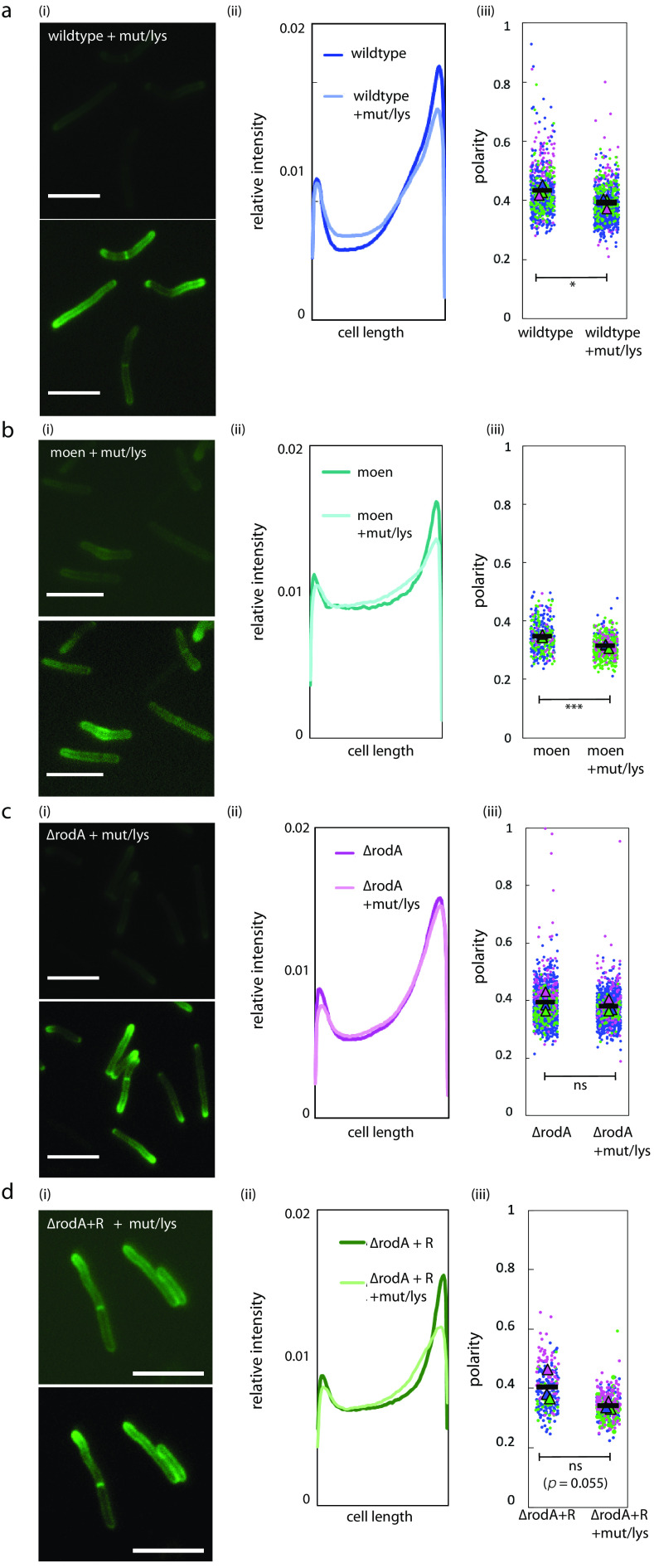
The distribution of MurG, and therefore lipid II, likely contributes to spatial flexibility in peptidoglycan assembly. The location of RodA (and, likely, PonA1) did not obviously change upon cell wall damage (Fig. S5 and S6). However, given the basal, cell-wide distribution of both transglycosylases, we asked if they might contribute to damage-induced pole-to-sidewall redistribution of nascent peptidoglycan. We first reproduced the cell wall labeling phenotype in mutanolysin/lysozyme-treated wild-type M. smegmatis (42) and showed that there was a decrease in nascent peptidoglycan polarity (Fig. 4a). Pretreating wild-type cells with the aPBP-transglycosylation inhibitor moenomycin did not alter this pole-to-sidewall damage response (Fig. 4b). In contrast, mutanolysin/lysozyme treatment of M. smegmatis lacking RodA prevented the shift in nascent peptidoglycan localization (Fig. 4c; Fig. S3), a phenotype that we could partially complement by expression of rodA-mRFP (Fig. 4d). These data suggest that RodA, but not aPBPs, contributes to stress-induced spatial flexibility in peptidoglycan synthesis.
FIG 4.
RodA contributes to damage-induced relocalization of peptidoglycan assembly. (a to d) At the end of lysozyme/mutanolysin treatment, the nascent cell wall in wild-type cells (a), moenomycin-treated cells (b), ΔrodA cells (c), or ΔrodA cells complemented with rodA-mRFP (d) was labeled with alkyne-d-alanine-d-alanine, as described in the legend to Fig. 2a, imaged at 1 s of exposure (the bottom panel is the same image with enhanced signal for visualization), and compared to that of untreated cells, as described in the legend to Fig. 2a (i). (ii) Normalized mean fluorescence intensity profiles comparing relative signals from untreated and treated cells. (iii) Super plots of cell wall labeling polarity ratio (signal from both poles divided by total cell fluorescence) from 3 independent experiments, with each color representing a biological replicate. t test, P < 0.05 (a); P > 0.1 (b); P < 0.01 (c). 102 < n < 826 (number of cells analyzed). Scale bars, 5 μm.
A nonredundant role for RodA in protection against cell wall damage.
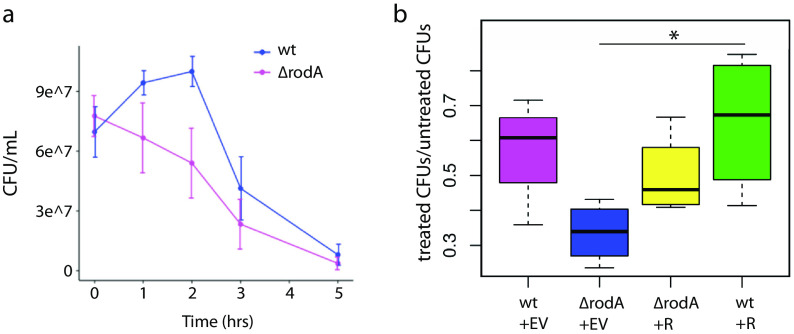
The contribution of RodAMtb to M. tuberculosis survival in different in vivo models (29, 37–39, 41) and the requirement for RodAMsm in the damage-induced sidewall shift in M. smegmatis peptidoglycan synthesis (Fig. 4) suggested that the enzyme could play a nonredundant role in protecting against cell wall stress. To test, we challenged wild-type and ΔrodA M. smegmatis cultures with mutanolysin/lysozyme and performed CFU assays and microscopy at several time points. Upon addition of mutanolysin/lysozyme to the growth medium, wild-type M. smegmatis grew for 2 h prior to lysing (Fig. 5a) albeit more slowly than untreated cells (Fig. 5b). In contrast, ΔrodA M. smegmatis immediately began to lyse, a phenotype evident both by CFU and by microscopy (Fig. 5a; Fig. S4). ΔrodA M. smegmatis lysis was partially complemented by expression of rodA-mRFP (Fig. 5b). Expression of rodA-mRFP in a wild-type background, moreover, enhanced survival compared to that of the wild type alone (Fig. 5b). Thus, while RodA is dispensable for normal growth (29) (Fig. S7), it plays a nonredundant role in protection from cell wall damage.
FIG 5.
RodA protects against cell wall damage. (a) CFU/ml over time for wild-type (wt) and ΔrodA cells treated with mutanolysin/lysozyme. n = 3. Compare these findings to Fig. S7 in the supplemental material. (b) Ratios of treated/untreated CFU after 2 h of mutanolysin/lysozyme treatment, comparing wild-type cells plus empty vector (EV), ΔrodA cells plus empty vector, ΔrodA cells plus rodA-mRFP (R), and wild-type cells plus rodA-mRFP. n = 4 independent experiments, two of which were done in triplicate. Significance was determined using analysis of variance (ANOVA), followed by a Tukey post hoc test to conduct pairwise comparisons. *, P < 0.05, the only significant relationship portrayed.
DISCUSSION
We have previously shown that upon cell wall insult, peptidoglycan assembly in M. smegmatis relocalizes from the growing poles to the nongrowing sidewall (42). Given that bacteria are likely to encounter such insults frequently in their natural habitats, we sought to better understand the factors that drive relocalization. We focused on the roles of two peptidoglycan transglycosylases, the aPBP PonA1 and SEDS family transglycosylase RodA.
In laterally growing, rod-shaped bacteria, the emerging narrative is that RodA lays the template for elongation and aPBPs fill in the gaps for maintenance and repair (10, 16–19, 21). Unlike the organisms in which this model has been tested, pole-growing bacteria like members of the Mycobacteriales and Hyphomicrobiales lack the cytoskeletal protein MreB and either lack or do not require RodA for viability or shape (28, 29). If the division of labor that has been reported in laterally growing bacilli were employed by mycobacteria, we would predict that localization and activity of RodA would be enriched at the poles, while localization and activity of aPBPs like PonA1 would be distributed around the cell periphery. This is reminiscent of the model for transpeptidation in mycobacterial peptidoglycan, where the d,d-transpeptidases that catalyze 4,3-cross-links associated with lipid II insertion are likely to be enriched at the poles and the l,d-transpeptidases that catalyze 3,3-cross-links associated with cell wall maturation localize along the cell periphery (42, 46, 65, 66). Instead, we observed that the distributions of functional fluorescent protein fusions to RodA and PonA1 are similar, as are their enzymatic activities (Fig. 1 and 2). As recent findings in pole-growing Corynebacterium glutamicum (31) also suggest, the division of labor for M. smegmatis peptidoglycan polymerases under basal conditions is subtle.
While RodA and aPBPs make similar contributions to polar growth, our data suggest that their roles diverge upon cell wall damage (Fig. 5). Specifically, RodA plays a nonredundant role in damage-associated pole-to-sidewall redistribution of peptidoglycan synthesis, which is concomitant with a similar redistribution of the lipid II synthase MurG (Fig. 3). It is not yet clear if the change in substrate availability is the cause or consequence or simply occurs in parallel with the change in transglycosylase usage. In the future, localization of lipid II flippase MurJ—which bridges lipid II synthesis in the inner leaflet and lipid II insertion in the outer leaflet—may help us to distinguish between these models. In Staphylococcus aureus, MurJ recruitment redirects peptidoglycan synthesis from the cell periphery, for expansion, to midcell, for division (67).
Our data suggest that RodA promotes a pole-to-sidewall shift in peptidoglycan synthesis (Fig. 4) and survival during cell wall damage (Fig. 5). The nonredundant role(s) for RodA in resistance to lysis (Fig. 5) is consistent with at least two models. In the first, RodA provides on-demand repair of cell wall damage. A similar stress-specific, peptidoglycan-building role for RodA has been suggested in Listeria monocytogenes (24), where the absence of a RodA homolog also sensitizes bacteria to cell wall damage (68). Loss of damage-induced sidewall shift supports this type of active role for RodA in mycobacteria. However, if true, this would be in contrast to the repair function of aPBPs, rather than RodA, in other organisms (16–19, 21). Thus, a second model to explain the lysis phenotype of the ΔrodA mutant is that RodA builds a cell wall with an architecture that is inherently more resistant to damage or that is more amenable to repair.
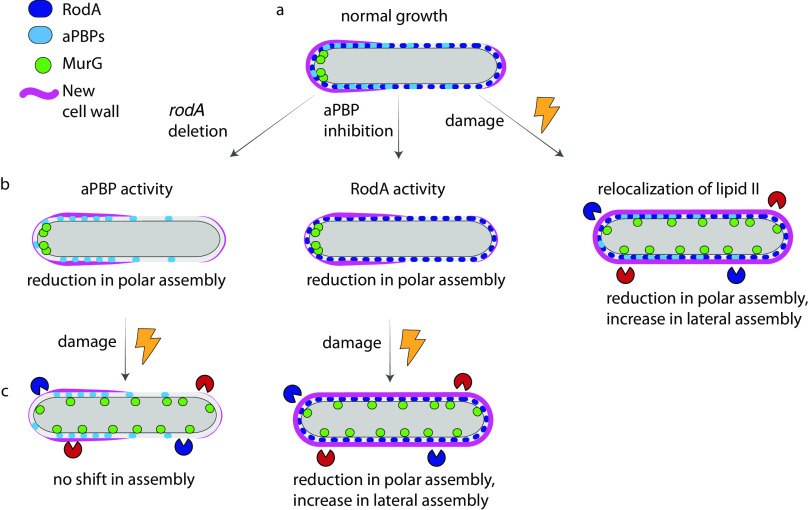
The utility of two pathways for the same enzymatic reactions is not always apparent under laboratory-optimized growth conditions. By studying the requirements for peptidoglycan synthesis during cell wall damage, we have uncovered spatial flexibility in precursor synthesis and extracellular insertion and a nonredundant role for RodA in protection (Fig. 6). These factors may enable mycobacteria to balance polar growth with cell-wide repair in the host and soil environments.
FIG 6.
Spatial flexibility model for peptidoglycan assembly. (a) Under basal conditions, PonA1 and RodA overlap substantially but not completely; new cell wall is assembled asymmetrically and enriched at the poles. (b, left and middle) Upon rodA deletion and aPBP inhibition, new cell wall assembly is disproportionally reduced at the poles. (b, right) Upon cell wall damage, MurG redistributes from the poles to the cell periphery, as does new cell wall. (c, left) In the absence of RodA, cell wall assembly is not shifted to the lateral cell under stress. (c, right) When aPBPs are inhibited, cell wall assembly redistributes upon damage.
MATERIALS AND METHODS
Strain construction.
Genes of interest were amplified from M. smegmatis mc2155 genomic DNA. mRFP was amplified from a pL5pTetO plasmid, with primers given in Table S1 in the supplemental material. Backbone plasmid pL5pTetO was linearized by PCR. Fifteen nanograms of plasmid backbone, 20 ng of the gene of interest, and 20 ng of mRFP PCR products were incubated with Gibson master mix (New England Biolabs; catalog no. E2611S) at 50°C for 1 h. Five microliters of Gibson product was transformed into 50 μL of XL1-Blue E. coli competent cells by heat shock. Transformants on 50-μg/mL kanamycin plates were confirmed by colony PCR and sequencing and then electroporated into M. smegmatis mc2155 or into the ΔrodA mutant.
Culture conditions.
Unless described otherwise, all experiments performed in liquid culture were done in 7H9 medium (Middlebrook), and all plating was done on lysogeny broth (LB) agar (Thermo) plates.
Cell wall damage.
Wild-type or ΔrodA cells were grown to stationary phase and then back-diluted and allowed to grow overnight to log phase (optical density at 600 nm [OD600], 0.5 to 0.8). Lysozyme (Sigma-Aldrich; catalog no. L6876) was freshly resuspended in phosphate-buffered saline (PBS), filter sterilized, and added to cultures at a final concentration of 500 μg/mL. Mutanolysin (Sigma-Aldrich catalog no. M9901) was added simultaneously at a final concentration of 500 U/mL. The hydrolases were added to bacteria in 7H9 growth medium. Cultures were incubated at 37°C with shaking at 300 rpm for 1 h.
Peptidoglycan labeling.
Peptidoglycan precursor probe alkyne-d-alanine-d-alanine (2 mM; custom synthesized by WuXi AppTech) was added to cultures for the final 10 min of incubation. Cells were washed three times in cold PBS and fixed in 2% formaldehyde for 10 min. Cells were washed in PBS and then subjected to Copper-catalysed azide-alkyne cycloaddition (CuAAC) with picolyl azide‐AF488 or picolyl azide‐TAMRA (Click Chemistry Tools) as described previously (69, 70).
CFU and growth curves.
Wild-type plus pL5pTetO, wild-type plus pL5pTetO-rodA-mRFP, ΔrodA plus pL5pTetO, and ΔrodA plus pL5pTetO-rodA-mRFP cells were grown to stationary phase and then back-diluted and allowed to grow overnight to log phase (OD600, 0.5 to 0.8). Cultures were back-diluted once more to an OD600 of 0.05. Lysozyme and mutanolysin were added as described above. Triplicate cultures were incubated at 37°C with shaking at 150 rpm for 5 h. Aliquots were periodically plated for determination of CFU.
Moenomycin treatment.
Wild-type cells were grown to stationary phase, back-diluted, and allowed to grow overnight to log phase (OD600, 0.5 to 0.8). Moenomycin (Cayman Chemicals; catalog no. 15506) was added at concentrations described in the text. Cultures were incubated at 37°C with shaking at 400 rpm for 30 min in a Benchmark Scientific MultiTherm H5000-H shaker.
Viability staining.
Staining was calibrated using untreated cells as a live control and 70% isopropanol-treated cells as a dead control. Following 90 min of treatment with lysozyme and mutanolysin, cells expressing RodA-mRFP were washed with HEPES-buffered saline (HBS) twice and resuspended in HBS plus SYTOX green (Fisher Scientific; catalog no. S7020) at a final concentration of 2.5 μM. For cells expressing MurG-Dendra2, propidium iodide was added to a final concentration of 4 μM. Cells were then incubated in the dark for an additional 30 min and imaged immediately.
Imaging.
Cells were placed on pads made of 1% agarose in water. Images were acquired on a Nikon Eclipse E600 fluorescence microscope at exposure times detailed in the text.
Image analysis.
Cell outlines were traced using Oufti (71). Demographs were generated using tools built into the program. Intensity profiles of nonseptating labeled cells only were generated using MATLAB code described previously (42). Polarity ratios were calculated by combining signal values for 15% of the cell length on either pole and dividing the sum by the total cell fluorescence. Beeswarm plots and boxplots were generated on R studio. Super plots were generated as described previously (72).
Membrane fractionation and Western blotting.
Wild-type M. smegmatis and cells expressing RodA-mRFP were grown to an OD600 of ~0.6 and lysed by nitrogen cavitation. Lysates were separated into cytoplasm and membrane fractions by ultracentrifugation at 35,000 rpm for 2 h. Protein concentration was adjusted to 560 μg/mL. Cell lysate or fractionated samples were separated by SDS-PAGE on a 4-to-20% polyacrylamide gel and transferred to a polyvinylidene difluoride (PVDF) membrane. The membrane was blocked with 3% skim milk in PBS plus 0.05% Tween 80 (PBST) and then incubated overnight with primary monoclonal mouse anti-RFP. Antibodies were detected with appropriate secondary antibodies conjugated to horseradish peroxidase (GE Healthcare, Chicago, IL). Membranes were rinsed in PBS plus 0.05% Tween 20 before visualization.
ACKNOWLEDGMENTS
Research was supported by funds from the National Institutes of Health (NIH) under awards funding R21 AI144748, R01 AI148255, and DP2 AI138238. E.S.M. was supported by NIH T32 GM008515 administered to the Chemistry Biology Interface Program at the University of Massachusetts Amherst.
We are grateful to Emily Bechtold for technical assistance.
Footnotes
For a commentary on this article, see https://doi.org/10.1128/JB.00125-22.
Supplemental material is available online only.
Contributor Information
M. Sloan Siegrist, Email: siegrist@umass.edu.
Michael J. Federle, University of Illinois at Chicago
REFERENCES
- 1.Fernandes S, Sao-Jose C. 2018. Enzymes and mechanisms employed by tailed bacteriophages to breach the bacterial cell barriers. Viruses 10:396. 10.3390/v10080396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamada S, Torii M, Kotani S, Masuda N, Ooshima T, Yokogawa K, Kawata S. 1978. Lysis of Streptococcus mutans cells with mutanolysin, a lytic enzyme prepared from a culture liquor of Streptomyces globisporus 1829. Arch Oral Biol 23:543–549. 10.1016/0003-9969(78)90268-6. [DOI] [PubMed] [Google Scholar]
- 3.Ragland SA, Criss AK. 2017. From bacterial killing to immune modulation: recent insights into the functions of lysozyme. PLoS Pathog 13:e1006512. 10.1371/journal.ppat.1006512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stamp GW, Poulsom R, Chung LP, Keshav S, Jeffery RE, Longcroft JA, Pignatelli M, Wright NA. 1992. Lysozyme gene expression in inflammatory bowel disease. Gastroenterology 103:532–538. 10.1016/0016-5085(92)90843-N. [DOI] [PubMed] [Google Scholar]
- 5.Nash JA, Ballard TN, Weaver TE, Akinbi HT. 2006. The peptidoglycan-degrading property of lysozyme is not required for bactericidal activity in vivo. J Immunol 177:519–526. 10.4049/jimmunol.177.1.519. [DOI] [PubMed] [Google Scholar]
- 6.Barreteau H, Kovac A, Boniface A, Sova M, Gobec S, Blanot D. 2008. Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol Rev 32:168–207. 10.1111/j.1574-6976.2008.00104.x. [DOI] [PubMed] [Google Scholar]
- 7.Emami K, Guyet A, Kawai Y, Devi J, Wu LJ, Allenby N, Daniel RA, Errington J. 2017. RodA as the missing glycosyltransferase in Bacillus subtilis and antibiotic discovery for the peptidoglycan polymerase pathway. Nat Microbiol 2:16253. 10.1038/nmicrobiol.2016.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meeske AJ, Riley EP, Robins WP, Uehara T, Mekalanos JJ, Kahne D, Walker S, Kruse AC, Bernhardt TG, Rudner DZ. 2016. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 537:634–638. 10.1038/nature19331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao H, Patel V, Helmann JD, Dorr T. 2017. Don't let sleeping dogmas lie: new views of peptidoglycan synthesis and its regulation. Mol Microbiol 106:847–860. 10.1111/mmi.13853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho H, Wivagg CN, Kapoor M, Barry Z, Rohs PDA, Suh H, Marto JA, Garner EC, Bernhardt TG. 2016. Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nat Microbiol 1:16172. 10.1038/nmicrobiol.2016.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dominguez-Escobar J, Chastanet A, Crevenna AH, Fromion V, Wedlich-Soldner R, Carballido-Lopez R. 2011. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science 333:225–228. 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- 12.Garner EC, Bernard R, Wang W, Zhuang X, Rudner DZ, Mitchison T. 2011. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science 333:222–225. 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Teeffelen S, Wang S, Furchtgott L, Huang KC, Wingreen NS, Shaevitz JW, Gitai Z. 2011. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc Natl Acad Sci USA 108:15822–15827. 10.1073/pnas.1108999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pazos M, Vollmer W. 2021. Regulation and function of class A penicillin-binding proteins. Curr Opin Microbiol 60:80–87. 10.1016/j.mib.2021.01.008. [DOI] [PubMed] [Google Scholar]
- 15.McPherson DC, Popham DL. 2003. Peptidoglycan synthesis in the absence of class A penicillin-binding proteins in Bacillus subtilis. J Bacteriol 185:1423–1431. 10.1128/JB.185.4.1423-1431.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vigouroux A, Cordier B, Aristov A, Alvarez L, Ozbaykal G, Chaze T, Oldewurtel ER, Matondo M, Cava F, Bikard D, van Teeffelen S. 2020. Class-A penicillin binding proteins do not contribute to cell shape but repair cell-wall defects. Elife 9:e51998. 10.7554/eLife.51998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mueller EA, Egan AJ, Breukink E, Vollmer W, Levin PA. 2019. Plasticity of Escherichia coli cell wall metabolism promotes fitness and antibiotic resistance across environmental conditions. Elife 8:e45704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paradis-Bleau C, Markovski M, Uehara T, Lupoli TJ, Walker S, Kahne DE, Bernhardt TG. 2010. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell 143:1110–1120. 10.1016/j.cell.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Typas A, Banzhaf M, van den Berg van Saparoea B, Verheul J, Biboy J, Nichols RJ, Zietek M, Beilharz K, Kannenberg K, von Rechenberg M, Breukink E, den Blaauwen T, Gross CA, Vollmer W. 2010. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell 143:1097–1109. 10.1016/j.cell.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greene NG, Fumeaux C, Bernhardt TG. 2018. Conserved mechanism of cell-wall synthase regulation revealed by the identification of a new PBP activator in Pseudomonas aeruginosa. Proc Natl Acad Sci USA 115:3150–3155. 10.1073/pnas.1717925115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy SG, Murtha AN, Zhao Z, Alvarez L, Diebold P, Shin JH, VanNieuwenhze MS, Cava F, Dorr T. 2021. Class A penicillin-binding protein-mediated cell wall synthesis promotes structural integrity during peptidoglycan endopeptidase insufficiency in Vibrio cholerae. mBio 12:e03596-20. 10.1128/mBio.03596-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dersch S, Mehl J, Stuckenschneider L, Mayer B, Roth J, Rohrbach A, Graumann PL. 2020. Super-resolution microscopy and single-molecule tracking reveal distinct adaptive dynamics of MreB and of cell wall-synthesis enzymes. Front Microbiol 11:1946. 10.3389/fmicb.2020.01946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Billaudeau C, Chastanet A, Yao Z, Cornilleau C, Mirouze N, Fromion V, Carballido-Lopez R. 2017. Contrasting mechanisms of growth in two model rod-shaped bacteria. Nat Commun 8:15370. 10.1038/ncomms15370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wamp S, Rothe P, Holland G, Halbedel S. 2021. MurA escape mutations uncouple peptidoglycan biosynthsis from PrkA singaling. bioRxiv. 10.1101/2021.09.09.459578. [DOI] [PMC free article] [PubMed]
- 25.Henrichfreise B, Brunke M, Viollier PH. 2016. Bacterial surfaces: the wall that SEDS built. Curr Biol 26:R1158–R1160. 10.1016/j.cub.2016.09.028. [DOI] [PubMed] [Google Scholar]
- 26.Otten C, Brilli M, Vollmer W, Viollier PH, Salje J. 2018. Peptidoglycan in obligate intracellular bacteria. Mol Microbiol 107:142–163. 10.1111/mmi.13880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Atwal S, Chuenklin S, Bonder EM, Flores J, Gillespie JJ, Driscoll TP, Salje J. 2021. Discovery of a diverse set of bacteria that build their cell walls without the canonical peptidoglycan polymerase aPBP. mBio 12:e0134221. 10.1128/mBio.01342-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams MA, Aliashkevich A, Krol E, Kuru E, Bouchier JM, Rittichier J, Brun YV, VanNieuwenhze MS, Becker A, Cava F, Brown PJB. 2021. Unipolar peptidoglycan synthesis in the Rhizobiales requires an essential class A penicillin-binding protein. mBio 12:e02346-21. 10.1128/mBio.02346-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arora D, Chawla Y, Malakar B, Singh A, Nandicoori VK. 2018. The transpeptidase PbpA and noncanonical transglycosylase RodA of Mycobacterium tuberculosis play important roles in regulating bacterial cell lengths. J Biol Chem 293:6497–6516. 10.1074/jbc.M117.811190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cole ST. 1998. Comparative mycobacterial genomics. Curr Opin Microbiol 1:567–571. 10.1016/S1369-5274(98)80090-8. [DOI] [PubMed] [Google Scholar]
- 31.Sher JW, Lim HC, Bernhardt TG. 2021. Polar growth in Corynebacterium glutamicum has a flexible cell wall synthase requirement. mBio 12:e00682-21. 10.1128/mBio.00682-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sassetti CM, Rubin EJ. 2003. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci USA 100:12989–12994. 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dragset MS, Ioerger TR, Zhang YJ, Mærk M, Ginbot Z, Sacchettini JC, Flo TH, Rubin EJ, Steigedal M. 2019. Genome-wide phenotypic profiling identifies and categorizes genes required for mycobacterial low iron fitness. Sci Rep 9:11394. 10.1038/s41598-019-47905-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hett EC, Chao MC, Rubin EJ. 2010. Interaction and modulation of two antagonistic cell wall enzymes of mycobacteria. PLoS Pathog 6:e1001020. 10.1371/journal.ppat.1001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goffin C, Ghuysen JM. 1998. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol Mol Biol Rev 62:1079–1093. 10.1128/MMBR.62.4.1079-1093.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. 2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev 32:234–258. 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- 37.Vandal OH, Roberts JA, Odaira T, Schnappinger D, Nathan CF, Ehrt S. 2009. Acid-susceptible mutants of Mycobacterium tuberculosis share hypersusceptibility to cell wall and oxidative stress and to the host environment. J Bacteriol 191:625–631. 10.1128/JB.00932-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith CM, Baker RE, Proulx MK, Mishra BB, Long JE, Park SW, Lee H-N, Kiritsy MC, Bellerose MM, Olive AJ, Murphy KC, Papavinasasundaram K, Boehm FJ, Reames CJ, Meade RK, Hampton BK, Linnertz CL, Shaw GD, Hock P, Bell TA, Ehrt S, Schnappinger D, Pardo-Manuel de Villena F, Ferris MT, Ioerger TR, Sassetti CM. 2021. Host-pathogen genetic interactions underlie tuberculosis susceptibility. bioRxiv. 10.1101/2020.12.01.405514. [DOI] [PMC free article] [PubMed]
- 39.Zhang YJ, Reddy MC, Ioerger TR, Rothchild AC, Dartois V, Schuster BM, Trauner A, Wallis D, Galaviz S, Huttenhower C, Sacchettini JC, Behar SM, Rubin EJ. 2013. Tryptophan biosynthesis protects mycobacteria from CD4 T-cell-mediated killing. Cell 155:1296–1308. 10.1016/j.cell.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kieser KJ, Boutte CC, Kester JC, Baer CE, Barczak AK, Meniche X, Chao MC, Rego EH, Sassetti CM, Fortune SM, Rubin EJ. 2015. Phosphorylation of the peptidoglycan synthase PonA1 governs the rate of polar elongation in mycobacteria. PLoS Pathog 11:e1005010. 10.1371/journal.ppat.1005010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rengarajan J, Bloom BR, Rubin EJ. 2005. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc Natl Acad Sci USA 102:8327–8332. 10.1073/pnas.0503272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garcia-Heredia A, Pohane AA, Melzer ES, Carr CR, Fiolek TJ, Rundell SR, Lim HC, Wagner JC, Morita YS, Swarts BM, Siegrist MS. 2018. Peptidoglycan precursor synthesis along the sidewall of pole-growing mycobacteria. Elife 7:e37243. 10.7554/eLife.37243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daitch AK, Goley ED. 2020. Uncovering unappreciated activities and niche functions of bacterial cell wall enzymes. Curr Biol 30:R1170–R1175. 10.1016/j.cub.2020.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dion MF, Kapoor M, Sun Y, Wilson S, Ryan J, Vigouroux A, van Teeffelen S, Oldenbourg R, Garner EC. 2019. Bacillus subtilis cell diameter is determined by the opposing actions of two distinct cell wall synthetic systems. Nat Microbiol 4:1294–1305. 10.1038/s41564-019-0439-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garcia-Heredia A, Kado T, Sein CE, Puffal J, Osman SH, Judd J, Gray TA, Morita YS, Siegrist MS. 2021. Membrane-partitioned cell wall synthesis in mycobacteria. Elife 10:e60263. 10.7554/eLife.60263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baranowski C, Welsh MA, Sham LT, Eskandarian HA, Lim HC, Kieser KJ, Wagner JC, McKinney JD, Fantner GE, Ioerger TR, Walker S, Bernhardt TG, Rubin EJ, Rego EH. 2018. Maturing Mycobacterium smegmatis peptidoglycan requires non-canonical crosslinks to maintain shape. Elife 7:e37516. 10.7554/eLife.37516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harrison J, Lloyd G, Joe M, Lowary TL, Reynolds E, Walters-Morgan H, Bhatt A, Lovering A, Besra GS, Alderwick LJ. 2016. Lcp1 is a phosphotransferase responsible for ligating arabinogalactan to peptidoglycan in Mycobacterium tuberculosis. mBio 7:e00972-16. 10.1128/mBio.00972-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belisle JT, Vissa VD, Sievert T, Takayama K, Brennan PJ, Besra GS. 1997. Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science 276:1420–1422. 10.1126/science.276.5317.1420. [DOI] [PubMed] [Google Scholar]
- 49.Aldridge BB, Fernandez-Suarez M, Heller D, Ambravaneswaran V, Irimia D, Toner M, Fortune SM. 2012. Asymmetry and aging of mycobacterial cells lead to variable growth and antibiotic susceptibility. Science 335:100–104. 10.1126/science.1216166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Botella H, Yang G, Ouerfelli O, Ehrt S, Nathan CF, Vaubourgeix J. 2017. Distinct spatiotemporal dynamics of peptidoglycan synthesis between Mycobacterium smegmatis and Mycobacterium tuberculosis. mBio 8:e01183-17. 10.1128/mBio.01183-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Joyce G, Williams KJ, Robb M, Noens E, Tizzano B, Shahrezaei V, Robertson BD. 2012. Cell division site placement and asymmetric growth in mycobacteria. PLoS One 7:e44582. 10.1371/journal.pone.0044582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meniche X, Otten R, Siegrist MS, Baer CE, Murphy KC, Bertozzi CR, Sassetti CM. 2014. Subpolar addition of new cell wall is directed by DivIVA in mycobacteria. Proc Natl Acad Sci USA 111:E3243–E3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rego EH, Audette RE, Rubin EJ. 2017. Deletion of a mycobacterial divisome factor collapses single-cell phenotypic heterogeneity. Nature 546:153–157. 10.1038/nature22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh B, Nitharwal RG, Ramesh M, Pettersson BM, Kirsebom LA, Dasgupta S. 2013. Asymmetric growth and division in Mycobacterium spp.: compensatory mechanisms for non-medial septa. Mol Microbiol 88:64–76. 10.1111/mmi.12169. [DOI] [PubMed] [Google Scholar]
- 55.Liechti GW, Kuru E, Hall E, Kalinda A, Brun YV, VanNieuwenhze M, Maurelli AT. 2014. A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature 506:507–510. 10.1038/nature12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sarkar S, Libby EA, Pidgeon SE, Dworkin J, Pires MM. 2016. In vivo probe of lipid II-interacting proteins. Angew Chem Int Ed Engl 55:8401–8404. 10.1002/anie.201603441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Welzel P. 2005. Syntheses around the transglycosylation step in peptidoglycan biosynthesis. Chem Rev 105:4610–4660. 10.1021/cr040634e. [DOI] [PubMed] [Google Scholar]
- 58.Gampe CM, Tsukamoto H, Doud EH, Walker S, Kahne D. 2013. Tuning the moenomycin pharmacophore to enable discovery of bacterial cell wall synthesis inhibitors. J Am Chem Soc 135:3776–3779. 10.1021/ja4000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ostash B, Walker S. 2010. Moenomycin family antibiotics: chemical synthesis, biosynthesis, and biological activity. Nat Prod Rep 27:1594–1617. 10.1039/c001461n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rebets Y, Lupoli T, Qiao Y, Schirner K, Villet R, Hooper D, Kahne D, Walker S. 2014. Moenomycin resistance mutations in Staphylococcus aureus reduce peptidoglycan chain length and cause aberrant cell division. ACS Chem Biol 9:459–467. 10.1021/cb4006744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang S, Zhang F, Kang J, Zhang W, Deng G, Xin Y, Ma Y. 2014. Mycobacterium tuberculosis Rv1096 protein: gene cloning, protein expression, and peptidoglycan deacetylase activity. BMC Microbiol 14:174. 10.1186/1471-2180-14-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kang CM, Nyayapathy S, Lee JY, Suh JW, Husson RN. 2008. Wag31, a homologue of the cell division protein DivIVA, regulates growth, morphology and polar cell wall synthesis in mycobacteria. Microbiology (Reading) 154:725–735. 10.1099/mic.0.2007/014076-0. [DOI] [PubMed] [Google Scholar]
- 63.Nguyen L, Scherr N, Gatfield J, Walburger A, Pieters J, Thompson CJ. 2007. Antigen 84, an effector of pleiomorphism in Mycobacterium smegmatis. J Bacteriol 189:7896–7910. 10.1128/JB.00726-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Melzer ES, Sein CE, Chambers JJ, Sloan Siegrist M. 2018. DivIVA concentrates mycobacterial cell envelope assembly for initiation and stabilization of polar growth. Cytoskeleton (Hoboken) 75:498–507. 10.1002/cm.21490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thanky NR, Young DB, Robertson BD. 2007. Unusual features of the cell cycle in mycobacteria: polar-restricted growth and the snapping-model of cell division. Tuberculosis (Edinb) 87:231–236. 10.1016/j.tube.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 66.Pidgeon SE, Apostolos AJ, Nelson JM, Shaku M, Rimal B, Islam MN, Crick DC, Kim SJ, Pavelka MS, Kana BD, Pires MM. 2019. l,d-Transpeptidase specific probe reveals spatial activity of peptidoglycan cross-linking. ACS Chem Biol 14:2185–2196. 10.1021/acschembio.9b00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Monteiro JM, Pereira AR, Reichmann NT, Saraiva BM, Fernandes PB, Veiga H, Tavares AC, Santos M, Ferreira MT, Macario V, VanNieuwenhze MS, Filipe SR, Pinho MG. 2018. Peptidoglycan synthesis drives an FtsZ-treadmilling-independent step of cytokinesis. Nature 554:528–532. 10.1038/nature25506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rismondo J, Halbedel S, Grundling A. 2019. Cell shape and antibiotic resistance are maintained by the activity of multiple FtsW and RodA enzymes in Listeria monocytogenes. mBio 10:e01448-19. 10.1128/mBio.01448-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Siegrist MS, Swarts BM, Fox DM, Lim SA, Bertozzi CR. 2015. Illumination of growth, division and secretion by metabolic labeling of the bacterial cell surface. FEMS Microbiol Rev 39:184–202. 10.1093/femsre/fuu012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Siegrist MS, Whiteside S, Jewett JC, Aditham A, Cava F, Bertozzi CR. 2013. (D)-Amino acid chemical reporters reveal peptidoglycan dynamics of an intracellular pathogen. ACS Chem Biol 8:500–505. 10.1021/cb3004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Paintdakhi A, Parry B, Campos M, Irnov I, Elf J, Surovtsev I, Jacobs-Wagner C. 2016. Oufti: an integrated software package for high-accuracy, high-throughput quantitative microscopy analysis. Mol Microbiol 99:767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lord SJ, Velle KB, Mullins RD, Fritz-Laylin LK. 2020. SuperPlots: communicating reproducibility and variability in cell biology. J Cell Biol 219:e202001064. 10.1083/jcb.202001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1-S7 and Table S1. Download jb.00540-21-s0001.pdf, PDF file, 3.3 MB (3.4MB, pdf)