Abstract
It is generally assumed that hypersaline environments with sodium chloride concentrations close to saturation are dominated by halophilic members of the domain Archaea, while Bacteria are not considered to be relevant in this kind of environment. Here, we report the high abundance and growth of a new group of hitherto-uncultured Bacteria in crystallizer ponds (salinity, from 30 to 37%) from multipond solar salterns. In the present study, these Bacteria constituted from 5 to 25% of the total prokaryotic community and were affiliated with the Cytophaga-Flavobacterium-Bacteroides phylum. Growth was demonstrated in saturated NaCl. A provisional classification of this new bacterial group as “Candidatus Salinibacter gen. nov.” is proposed. The perception that Archaea are the only ecologically relevant prokaryotes in hypersaline aquatic environments should be revised.
Hypersaline environments, such as the crystallizer ponds (i.e., ponds where sodium chloride precipitates) of multipond solar salterns, have been shown to harbor a very low prokaryotic diversity, to the point of having been described as “almost monospecific cultures of halophilic archaea” (9). The use of molecular techniques has also revealed a very low diversity, although the most abundant haloarchaeon (5) corresponding to the unique archaeal 16S ribosomal DNA (rDNA) sequences most frequently recovered from several crystallizer ponds by PCR-based methods did not correspond to any previously described microorganism (5, 6, 26). This apparent discrepancy between molecular and culture-based data has been found in other environments (4).
When fluorescence in situ hybridization (FISH) was used to analyze the prokaryotic community inhabiting crystallizer ponds (around 37% salinity) of a marine solar saltern located in Alicante, Spain (5), Bacteria in high numbers (around 3 × 106/ml) were unexpectedly found. The cells, which accounted for 18% of total cell counts, contained large numbers of ribosomes, as indicated by the intense and uniform FISH signals obtained with the 16S rRNA-targeted oligonucleotide probe EUB338 (3), which is specific for members of the Bacteria domain. The finding of abundant Bacteria with high cellular rRNA content in such a hypersaline environment was unexpected in light of previous reports (20–23) suggesting that almost all the active biomass was of archaeal origin.
Solar salterns consist of a series of shallow ponds connected in a sequence of increasingly saline brines. Crystallizers are the last ponds and have a salinity above 30% (6). Therefore, the presence in these ponds of Bacteria could be due either to their import from previous ponds with lower salinity or to their active growth in the crystallizers. The key point that would determine the relevance of finding Bacteria in the crystallizers was, then, whether they were in fact extreme halophiles forming part of the autochthonous microbiota of these ponds.
In order to characterize this bacterial community and assess this point, we used the rRNA approach (4, 19), which allows direct, cultivation-independent 16S rDNA sequence retrieval and the design of probes specific for these sequences. As shown in the present study, this approach has been successfully used for quantifying, monitoring growth, and studying the distribution of these newly discovered, extremely halophilic Bacteria.
MATERIALS AND METHODS
Sampling.
Water samples were collected from the multipond solar saltern “Braç del Port” located in Santa Pola (Alicante, Spain). Samples from crystallizer CR-30 (37% salinity) collected in June 1998 were used for FISH analysis, retrieval of the most abundant phylotype, and growth experiments. For analysis of the bacterial community along the salinity gradient, samples of six different ponds (salinities, 11, 15, 22.4, 25, 31.6, and 37%) were taken in May 1999. Analysis of the geographical distribution of extremely halophilic Bacteria (EHB) was carried out with crystallizer samples from solar salterns located in Grand Canary (Canary Islands) and Ibiza and Majorca (Balearic Islands). The salinity of each sample was determined with a hand refractometer (S-28; Atago, Tokyo, Japan).
Nucleic acid extraction, DGGE, cloning, and sequencing.
For DNA extraction, cells from 1 ml of water from CR-30 were collected by centrifugation for 5 min at 13,000 rpm in a bench-top microcentrifuge (Heraeus, Osterode, Germany), resuspended in 200 μl of sterilized deionized water (Millipore Corporation), and boiled for 10 min. After cell debris was eliminated by centrifugation, supernatant was used for PCR amplification. Amplification of 16S rDNA fragments between Escherichia coli positions 341 and 907 (7), denaturing gradient gel electrophoresis (DGGE), excision of bands, reamplification, and sequencing were performed as previously described (16). DNA crude extract was used as a template for PCR with primers that allowed the amplification of the complete bacterial 16S rRNA gene (14). PCR products were purified, ligated into the vector, and cloned using the pGEM-T easy vector (Promega Corporation, Madison, Wis.) following the manufacturer's recommendations. After the clones had been screened for redundancies by restriction analysis with the enzymes NotI and Sau3AI (29), two clones were selected for complete sequencing.
16S rDNA sequence analysis.
Partial 16S rDNA sequences corresponding to the two DGGE bands were analyzed by BLAST at the National Center for Biotechnology Information web page (2). The corresponding complete sequences were added to an alignment of about 13,000 homologous bacterial 16S rRNA primary structures (13) by using the aligning tool of the ARB program package (Department of Microbiology, Technische Universität München, Munich, Germany [http://www.mikro.biologie.tu-muenchen.de]). Distance matrix, maximum parsimony, and maximum likelihood methods were applied as implemented in the ARB software package. Phylogenetic trees were constructed using subsets of data that included outgroup reference sequences, as well as representative sequences of members of the Cytophaga-Flavobacterium-Bacteroides (CFB) phylum (13); topologies were evaluated by using the different approaches to elaborate a consensus tree (12).
Probe design.
Three probes were designed using the probe-designing tool of the ARB software package (see above): probe EHB412 (E. coli [7] positions 412 to 429) specifically targeted both EHB-1 and EHB-2 sequences, probe EHB586 (positions 586 to 603) was specifically designed for EHB-1, and probe EHB1451 (positions 1451 to 1468) was designed for EHB-2.
FISH.
Sample fixation was carried out as previously described (5) using the protocol optimized for fixation of extremely halophilic microorganisms. Hybridization, DAPI (4′,6′-diamidino-2-phenylindole) staining, and microscopy were carried out as described previously (30). Different formamide concentrations (from 0 to 80%) in the hybridization buffer were assayed for every probe in order to determine optimum hybridization conditions that give both sufficient specificity and good sensitivity. For every sample, one filter was analyzed and at least 700 cells were counted.
Enrichment studies.
Water from CR-30 was used to inoculate (at 1% [vol/vol]) seven different media containing a salt mixture (“sw” in reference 27) at different concentrations (10, 15, 20, 25, and 30% and 30% plus NaCl up to saturation) and 0.1% yeast extract (Difco). For every salt concentration, four different incubation temperatures were assayed (20, 28, 37, and 47°C). All the cultures were incubated without shaking. Growth was monitored by total cell counts (DAPI staining) and hybridization with probe EHB412, specific for EHB.
Nucleotide sequence accession numbers.
The sequences we call EHB-1 and EHB-2 (see below) have received EMBL accession numbers AJ133744 and AJ242998, respectively.
RESULTS
Composition of the bacterial community in crystallizer CR-30.
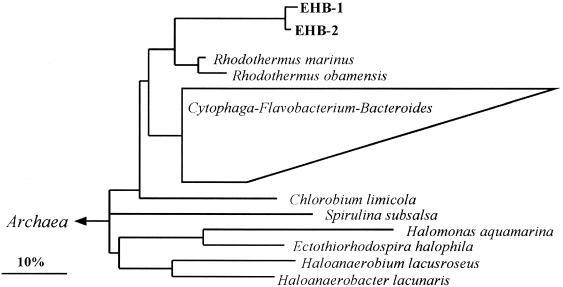
An overview of the bacterial diversity in crystallizer CR-30 was obtained by DGGE analysis (16) of PCR-amplified 16S rDNA fragments. When DNA from CR-30 was used as a PCR template for DGGE analysis, only two bands were detected (data not shown). The sequencing of these bands revealed that they were very similar to each other (around 98%) and were related (around 89% similarity in the analyzed stretch) to Rhodothermus marinus. Thus, a very low bacterial diversity was found in CR-30. In order to get these complete 16S rDNA sequences, total DNA was used as a template for PCR with primers that allowed the amplification of the complete 16S rRNA gene (14). Thus, we obtained an almost-complete sequence for the clones corresponding to the DGGE bands. We refer to these two sequences as EHB-1 and EHB-2, naming the bacterial community they represent as EHB. The two sequences had 97.6% similarity. Phylogenetic reconstructions loosely affiliated them with the CFB phylum (Fig. 1). Their closest relatives were both members of the genus Rhodothermus, with overall 16S rRNA sequence similarities of 86% between EHB-1/EHB-2 and R. marinus and 83% between EHB-1/EHB-2 and Rhodothermus obamensis. EHB-1 and EHB-2, together with both members of the genus Rhodothermus, represent the only available sequences of a deep branch within this phylum.
FIG. 1.
Phylogenetic tree based on 16S rDNA sequences from the complete EHB sequences and all almost-complete sequences of the CFB phylum available (over 370) and representatives of the domain Bacteria. The multifurcation indicates a topology that could not be unambiguously resolved. The phylogenetic position of EHB sequences did not differ in any of the treeing approaches. The bar indicates 10% of estimated sequence divergence. Sequence accession numbers: Chlorobium limicola, Y10640; Halomonas aquamarina, M93352; Haloanaerobacter lacunaris, X89075; Haloanaerobium lacusroseus, L39787; E. halophila, M26630; R. marinus, X77140; R. obamensis, X95071; and Spirulina subsalsa, AB003166.
Probe design and optimization of the FISH conditions.
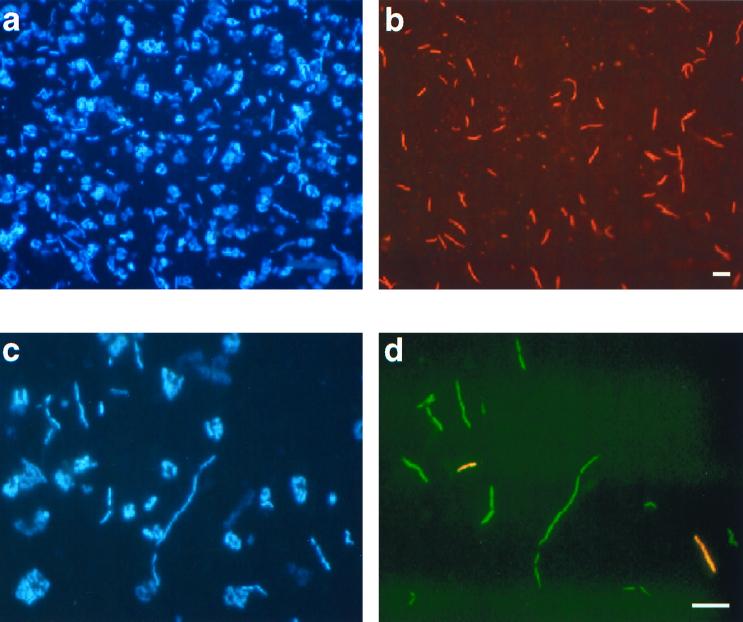
An 18-nucleotide sequence (Table 1) was chosen as the target site for a FISH probe for both EHB-1 and EHB-2 using the ARB software package (see above). This probe (labeled EHB412) targeted both DGGE bands and the complete 16S rDNA clone sequences obtained. For probe EHB412, optimum hybridization was found at 45% formamide. In all the samples analyzed, EHB412 hybridized with slender long rods (Fig. 2) endowed with a high ribosomal content, as shown by the intense hybridization signal.
TABLE 1.
Difference alignments of probe target sites indicating the discriminatory mismatchesa
| Probe (positionb) and organism | Target |
|---|---|
| EHB412c (412–429) | |
| EHBs | 5′-ACACCCCUAUGGGGCGUA-3′ |
| Hydrogenobacter thermophilus | 5′-.........C....G.C.-3′ |
| Sphingobacterium heparinum | 5′-...G....C....U....-3′ |
| Rhodothermus marinus | 5′-GA.G.U..UC....U...-3′ |
| Rhodothermus obamensis | 5′-GA.G....UC....U...-3′ |
| EHB586 (586–603) | |
| EHB-1 | 5′-GGGCAGCAAGUCGGAUGU-3′ |
| EHB-2 | 5′-.....A............-3′ |
| EHB1451 (1451–1468) | |
| EHB-2 | 5′-AGCCGGAGGGAGAGCGGC-3′ |
| EHB-1 | 5′-UUUG........C.GCCG-3′ |
Results are based on analysis of probe checks against at least 13,000 complete and partial sequences (ARB software; see text).
E. coli positions (7).
No sequences with one or two mismatches were found for probe EHB412.
FIG. 2.
Identification by FISH of Bacteria in samples from crystallizer ponds. Identical microscopic fields were visualized with an epifluorescence microscope using filter sets specific for DAPI (a and c) and the fluorochromes used for probe labeling (b and d). (a and b) Cells from the crystallizer in Majorca hybridized with Cy3-labeled probe EHB412. (c and d) Cells from CR-30 (Alicante) hybridized with fluorescein-labeled probe EHB586 and Cy3-labeled probe EHB1451. Bar, 5 μm.
EHB corresponded to the most abundant bacterial population in CR-30 since it hybridized with 14% of the prokaryotes in CR-30 detectable by DAPI staining, while probe EUB338, specific for members of the domain Bacteria, hybridized with 18% of the prokaryotic community. Thus, bacteria related to EHB sequences accounted for around 78% of the bacterial community.
In order to ascertain whether EHB-1 and EHB-2 sequences corresponded to two bacterial populations rather than to two different rRNA operons from a single bacterial population, two probes, EHB586 and EHB1451, that specifically targeted each of these sequences (Table 1) were designed. For both probes, optimum hybridization was found at 45% formamide. In no case did probes EHB586 and EHB1451 hybridize with the same cell; instead, they targeted different bacterial populations (Fig. 2).
Distribution of EHB throughout the salinity gradient.
The abundance of Bacteria hybridizing with probes EUB338 and EHB412 in several ponds of different salinities of the Alicante salterns was quantified. EHB could not be detected in ponds of 11 and 15% salinity. In the rest of the analyzed ponds (Table 2), the number of Bacteria detectable with EHB412 increased with salinity, while the total bacterial community (i.e., Bacteria detectable with probe EUB338) decreased. In fact, at the highest salinity (CR-30), the bacterial community was completely dominated by EHB. The distribution of the populations corresponding to EHB-1 and EHB-2 was also analyzed with probes EHB586 and EHB1451, respectively, and EHB-1 was found to be more abundant in all the ponds (Table 2).
TABLE 2.
Quantification of Bacteria, EHB, EHB-1, and EHB-2 in ponds of different salinity of the Alicante salterna
| Pond salinity (%)b | Total DAPI counts/ml | % of counts specific for:
|
|||
|---|---|---|---|---|---|
| Bacteria | EHB | EHB-1 | EHB-2 | ||
| 22.4 | 4.32 × 107 ± 0.65 × 107 | 35.6 ± 7.6 | Dc | D | D |
| 25 | 5.84 × 107 ± 0.91 × 107 | 17.1 ± 3.5 | 3.5 ± 0.7 | 2.2 ± 1.0 | D |
| 31.6 | 1.74 × 107 ± 0.22 × 107 | 11.4 ± 1.7 | 6.1 ± 2.4 | 5.1 ± 4.3 | 1.1 ± 0.9 |
| 37 | 2.78 × 107 ± 0.51 × 107 | 12.7 ± 4.6 | 12.0 ± 3.1 | 10.4 ± 4.0 | 2.4 ± 1.2 |
Values are expressed as DAPI counts detectable by FISH with probes EUB338 (specific for Bacteria), EHB412 (EHB), EHB586 (EHB-1), and EHB1451 (EHB-2). Values are means ± standard deviations.
EHB were not detectable in ponds of 11 and 15% salinity.
D, detectable—under 1%.
Growth at high salt concentrations.
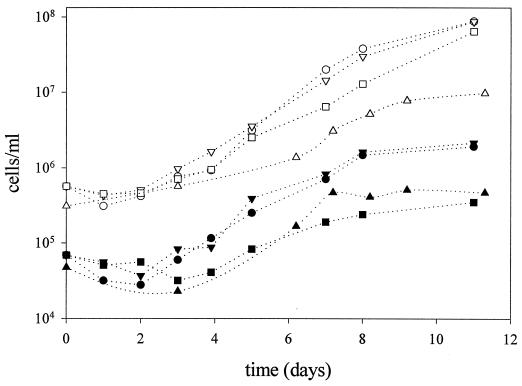
Once a specific FISH probe had been designed for EHB, their growth could be studied even in the absence of a pure culture. Culture media with different salt concentrations (10, 15, 20, 25, and 30% and NaCl saturation) were inoculated with water from CR-30 and incubated at different temperatures (20, 28, 37, and 47°C). Growth of EHB was monitored by FISH with probe EHB412, while total cell numbers were analyzed by DAPI staining. The optimum growth temperature of EHB was 37°C, while no growth was detected at 20°C after 2 weeks of incubation. As shown in Fig. 3, EHB were actually able to proliferate in saturated salt solution. Their optimum salinity for growth in the assayed conditions was between 20 and 25% total salts, which is typical of an extreme halophile (17) (growth rates of 0.66 ± 0.04 and 0.69 ± 0.02 day−1, respectively), while no growth was detected at 10 and 15% salinity.
FIG. 3.
Growth curve at 37°C of the halophilic bacteria detected with probe EHB412. Liquid media containing 0.1% (wt/vol) yeast extract and a salt mixture (“sw” in reference 27) at concentrations (vol/vol) of 20% (circles), 25% (inverted triangles), 30% (squares), and 30% plus NaCl up to saturation (triangles) were inoculated with crystallizer water (1% [vol/vol]) and incubated at 37°C. EHB were detected by hybridization with probe EHB412 (filled symbols). Empty symbols, total DAPI counts.
Geographical distribution of EHB.
In order to determine whether EHB were found only in the Alicante crystallizer pond, we looked for EHB in crystallizers from different solar salterns and found them in all the samples analyzed (Table 3). EHB accounted for 18 and 27% of the total counts in two crystallizer ponds (30 and 36% salinity) located in Ibiza and Majorca (Balearic Islands, west Mediterranean), respectively, and accounted for up to 5 and 8% in two crystallizer ponds (32% salinity) from solar salterns in the Canary Islands (east Atlantic Ocean). Furthermore, EHB could also be detected in samples from a natural hypersaline lagoon (28% salinity) in Sabkha (Gulf of Suez, Egypt). Thus, EHB were found in samples obtained from geographically isolated areas.
TABLE 3.
Quantification of EHB in crystallizers from different solar salterns
| Saltern | Salinity (%) | Counts/ml (mean ± SD)
|
% of counts specific for EHB (mean ± SD) | |
|---|---|---|---|---|
| DAPI | EHB412 | |||
| Grand Canary 1 | 32 | 2.4 × 106 ± 0.9 × 106 | 1.3 × 105 ± 1.0 × 105 | 5.1 ± 2.8 |
| Grand Canary 2 | 32 | 5.7 × 106 ± 2.0 × 107 | 4.7 × 105 ± 2.0 × 105 | 8.6 ± 3.9 |
| Majorca | 36 | 5.0 × 107 ± 0.6 × 107 | 1.3 × 107 ± 0.3 × 107 | 27.0 ± 6.8 |
| Ibiza | 30 | 2.8 × 107 ± 0.4 × 107 | 5.1 × 106 ± 1.6 × 106 | 18.7 ± 7.2 |
DISCUSSION
Recently, an unexpectedly high abundance of signals with the bacterial probe EUB338 in samples from a hypersaline crystallizer pond in Alicante (5) was reported. This result indicated that members of the domain Bacteria could form an important part of the autochthonous microbiota, in contrast with what had been reported previously (6, 20–23). DGGE results, and the subsequent retrieval of the most abundant bacterial 16S rRNA sequences of the hypersaline pond, suggested that most of the EUB338-positive signals in these samples corresponded to hitherto-uncultured members of the domain Bacteria. These samples had low bacterial diversity, as indicated by DGGE and subsequent 16S rDNA clone library analysis. Sequencing and phylogenetic reconstruction of the two analyzed clone sequences showed that both were affiliated with Rhodothermus species, forming a deep branch within the CFB phylum (33). R. marinus and R. obamensis are thermophilic (optimum growth temperatures, 65 and 80°C, respectively), moderately halophilic bacteria isolated from marine hydrothermal environments (1, 28). Thus, to date this deep CFB branch is composed only of extremophilic (e.g., thermophilic and halophilic) Bacteria. Given the low sequence similarity of EHB-1 and EHB-2 to their closest known relative, EHB can be considered a new genus within the phylum. The highest similarity (86%) between the two bacterial groups is far from the empirical limit of 94% of sequence identity that discriminates genera (12). Therefore, in accordance with the work of Murray and Schleifer (15), we propose provisional classification of EHB as “Candidatus Salinibacter gen. nov.,” with the following short description: phylogenetic position, Cytophaga-Flavobacterium-Bacteroides phylum; cultivation, noncultivated; gram reaction, negative; morphology, rod; basis of assignment, 16S rDNA sequences (EMBL accession numbers AJ133744 and AJ242998) and oligonucleotide probe EHB412, 5′-TACGCCCCATAGGGGTGT-3′; habitat, hypersaline environments; metabolism and unusual features, extremely halophilic; authors, Antón et al. (this study).
Since the two EHB sequences were closely related, a probe (EHB412) that specifically targeted the candidate genus was designed. This probe was used for quantifying EHB and studying their geographical distribution and growth. In addition, we also determined that these two sequences did not represent the same bacterium, since prokaryotes with different rRNA operons have been described for both the Archaea and the Bacteria domains (24, 32, 34). For this purpose, probes EHB586 and EHB1451, which specifically targeted EHB-1 and EHB-2, respectively, were designed. The results proved that the two 16S rDNA sequences retrieved by DGGE and cloning analysis corresponded to two different bacterial populations. However, these two populations were closely related (97.6% sequence similarity in the 16S rRNA gene), and therefore the candidate genus was monitored with probe EHB412.
The halophilic nature of these Bacteria has been shown without having isolated them in pure culture. The first evidence of the strict halophilicity of the EHB was given by their distribution and abundance along the salinity gradient (Table 2). EHB appeared only in ponds with at least 20% salinity, and their highest abundance was observed in the crystallizer pond, where they represented nearly the whole bacterial population. The second evidence was given by the enrichment experiments depicted in Fig. 3. The absence of growth at 15% total salts, together with optimum growth between 20 and 25% total salts, indicated that EHB were strict halophiles (17, 31). They grew with a generation time of about 0.7 day at these salinities.
To the best of our knowledge (17, 31), there are currently three known bacterial species which proliferate in saturated salts. One is Halorhodospira halophila (formerly Ectothiorhodospira halophila), a halophilic phototrophic bacterium isolated from soda lakes (10), which grows optimally in an environment around 25% NaCl. The second is Haloanaerobium lacusroseus (8), an anerobic bacterium isolated from sediments of a hypersaline lake. There is also one actinomycete, Actinopolyspora halophila, able to grow in saturated NaCl that was first isolated as a contaminant of culture medium containing 25% NaCl (11). The ecological relevance of the aforementioned bacteria is unknown, since no studies of their in situ abundance have been carried out.
Moreover, the occurrence of EHB as a significant part of the autochthonous microbiota in hypersaline environments in Alicante is not an isolated case. EHB412-positive bacteria with high rRNA content have been found in salterns of the Canary Islands and the Balearic Islands, and even in the east Mediterranean (Table 3). Abundances of these organisms varied with the salinity, but it is worth mentioning that in the salterns of Majorca, the population of a single phylotype represented one-fourth of the total prokaryotic population.
Thus, our results indicate that EHB are part of the autochthonous microbiota in hypersaline environments, which had been repeatedly described as dominated by halophilic Archaea (6, 20–23). However, it is not surprising that the presence of EHB was not detected before if we consider that the most abundant archaeon in these environments has never been cultured (5, 6). Even for an environment with such a low diversity, culture-dependent techniques have offered a very biased view of the prokaryotic community composition.
Conclusion.
Until recently (18), it had been commonly assumed that Archaea dwell in extreme habitats (e.g., high temperature, high salt, low oxygen), whereas Bacteria are restricted to moderate sites. Lately, molecular data have indicated the presence of Archaea in moderate environments, such as marine waters or soil (18), while Bacteria seem to be quite common, for example, at high temperatures that were once considered to be exclusive for Archaea (25). The present work is the first report indicating that Bacteria constitute a significant and important part of the microbiota that inhabit NaCl-saturated water, another classical habitat for Archaea. Both domains have obviously developed wide ecological competence. Bacteria seem to be as widespread as Archaea are.
ACKNOWLEDGMENTS
This work was supported by the Max Planck Society. J.A. was a recipient of an EMBO short-term fellowship.
We thank Miguel Cuervo-Arango, owner of the salterns, for his kind help, Enric Llobet-Brossa, Silvia G. Acinas, Jörg Wulf, and Hendrick Schäfer for technical assistance, and Yehuda Cohen, Marga Amat, Antonia Plovins, and Christian Knoblauch for providing some of the samples used for this work. We are grateful to K. O. Stetter and C. Pedrós-Alió for their critical reading of early versions of the manuscript and to H. G. Trüper for assistance in naming the candidate genus.
REFERENCES
- 1.Alfredsson G A, Kristjansson J K, Hjorleifsdottir S, Stetter K O. Rhodothermus marinus new-genus new-species, a thermophilic halophilic bacterium from submarine hot springs in Iceland. J Gen Microbiol. 1988;134:299–306. [Google Scholar]
- 2.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antón J, Llobet-Brossa E, Rodríguez-Valera F, Amann R. Fluorescence in situ hybridization analysis of the prokaryotic community inhabiting crystallizer ponds. Environ Microbiol. 1999;1:517–523. doi: 10.1046/j.1462-2920.1999.00065.x. [DOI] [PubMed] [Google Scholar]
- 6.Benlloch S, Martínez-Murcia A J, Rodríguez-Valera F. Sequencing and bacterial and archaeal 16S rRNA genes directly amplified from a hypersaline environment. Syst Appl Microbiol. 1995;18:574–581. [Google Scholar]
- 7.Brosius J, Dull T L, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 8.Cayol J-L, Ollivier B, Patel B K C, Ageron E, Grimont P A D, Prensier G, Garcia J-L. Haloanaerobium lacusroseus sp. nov., an extremely halophilic fermentative bacterium from the sediments of a hypersaline lake. Int J Syst Bacteriol. 1995;45:790–797. doi: 10.1099/00207713-45-4-790. [DOI] [PubMed] [Google Scholar]
- 9.Guixa-Boixereu N, Calderón-Paz J I, Heldal M, Bratbak G, Pedrós-Alió C. Viral lysis and bacterivory as prokaryotic loss factors along a salinity gradient. Aquat Microb Ecol. 1996;11:215–227. [Google Scholar]
- 10.Imhoff J F. The family Ectothiorhodospiraceae. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. New York, N.Y: Springer-Verlag; 1992. pp. 3222–3229. [Google Scholar]
- 11.Johnson K G, Lanthier P H, Gochnauer M B. Studies of two strains of Actinopolyspora halophila, an extremely halophilic actinomycete. Arch Microbiol. 1986;143:370–378. [Google Scholar]
- 12.Ludwig W, Strunk O, Klugbauer S, Klugbauer N, Weizenegger M, Neumaier J, Bachleitner M, Schleifer K-H. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis. 1998;19:554–568. doi: 10.1002/elps.1150190416. [DOI] [PubMed] [Google Scholar]
- 13.Maidak B L, Olsen G J, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martínez-Murcia A J, Acinas S G, Rodríguez-Valera F. Evaluation of prokaryotic diversity by restrictase digestion of 16S rDNA directly amplified from hypersaline environments. FEMS Microbiol Ecol. 1995;17:247–256. [Google Scholar]
- 15.Murray R G E, Schleifer K H. Taxonomic notes: a proposal for recording the properties of putative taxa of prokaryotes. Int J Syst Bacteriol. 1994;44:174–176. doi: 10.1099/00207713-44-1-174. [DOI] [PubMed] [Google Scholar]
- 16.Muyzer G, Hottenträger S, Teske A, Wawer C. Denaturing gradient gel electrophoresis of PCR-amplified 16S rDNA—a new molecular approach to analyse the genetic diversity of mixed microbial communities. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1996. pp. 3.4.4./1–3.4.4./23. [Google Scholar]
- 17.Ollivier B, Caumette P, Garcia J-L, Mah R A. Anaerobic bacteria from hypersaline environments. Microbiol Rev. 1994;58:27–38. doi: 10.1128/mr.58.1.27-38.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olsen G J. Archaea, archaea everywhere. Nature. 1994;731:657–658. doi: 10.1038/371657a0. [DOI] [PubMed] [Google Scholar]
- 19.Olsen G J, Lane D J, Giovannoni S J, Pace N R, Stahl D A. Microbial ecology and evolution: a ribosomal RNA approach. Annu Rev Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- 20.Oren A. Estimation of the contribution of halobacteria to the bacterial biomass and activity in solar salterns by the use of bile salts. FEMS Microbiol Ecol. 1990;73:41–48. [Google Scholar]
- 21.Oren A. The use of protein synthesis inhibitors in the estimation of the contribution of halophilic archaebacteria to bacterial activity in hypersaline environments. FEMS Microbiol Ecol. 1990;73:187–192. [Google Scholar]
- 22.Oren A. Thymidine incorporation in saltern ponds of different salinities: estimation of in situ growth rates of halophilic Archaeobacteria and Eubacteria. Microb Ecol. 1990;19:43–51. doi: 10.1007/BF02015052. [DOI] [PubMed] [Google Scholar]
- 23.Oren A. Ecology of extremely halophilic microorganisms. In: Vreeland R H, Hichstein L I, editors. The biology of halophilic bacteria. Boca Raton, Fla: CRC Press; 1994. pp. 25–53. [Google Scholar]
- 24.Oren A, Ventosa A, Gutiérrez M C, Kamekura M. Haloarcula quadrata sp. nov., a square, motile archaeon isolated from a brine pool in Sinai (Egypt) Int J Syst Bacteriol. 1999;49:1149–1155. doi: 10.1099/00207713-49-3-1149. [DOI] [PubMed] [Google Scholar]
- 25.Pace N R. Molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 26.Rodríguez-Valera F, Acinas S G, Antón J. Contribution of molecular techniques to the study of microbial diversity in hypersaline environments. In: Oren A, editor. Microbiology and biogeochemistry of hypersaline environments. Boca Raton, Fla: CRC Press; 1999. pp. 27–38. [Google Scholar]
- 27.Rodríguez-Valera F, Ventosa A, Juez G, Imhoff J F. Variation of environmental features and microbial populations with salt concentrations in a multi-pond saltern. Microb Ecol. 1985;11:107–115. doi: 10.1007/BF02010483. [DOI] [PubMed] [Google Scholar]
- 28.Sako Y, Takai K, Ishida Y, Uchida A, Katayama Y. Rhodothermus obamensis sp. nov., a modern lineage of extremely thermophilic marine bacteria. Int J Syst Bacteriol. 1996;46:1099–1104. doi: 10.1099/00207713-46-4-1099. [DOI] [PubMed] [Google Scholar]
- 29.Sambrock J, Fritsch E F, Maniatis T. Molecular cloning. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Snaidr J, Amann R, Huber I, Ludwig W, Schleifer K-H. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl Environ Microbiol. 1997;63:2884–2896. doi: 10.1128/aem.63.7.2884-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ventosa A, Nieto J J, Oren A. Biology of moderately halophilic aerobic bacteria. Microbiol Mol Biol Rev. 1998;62:504–544. doi: 10.1128/mmbr.62.2.504-544.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Zhang Z, Ramanan N. The actinomycete Thermobispora bispora contains two distinct types of transcriptionally active 16S rRNA genes. J Bacteriol. 1997;179:3270–3276. doi: 10.1128/jb.179.10.3270-3276.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yap W H, Zhang Z, Wang Y. Distinct types of rRNA operons exist in the genome of the actinomycete Thermomonospora chromogena and evidence for horizontal transfer of an entire rRNA operon. J Bacteriol. 1999;181:5201–5209. doi: 10.1128/jb.181.17.5201-5209.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]